Abstract
Background:
The number of neurodegenerative diseases associated with pathological aggregates of TAR DNA-binding protein 43 (TDP-43) has increased, leading to the new designation “TDP-43 proteinopathy.” Biochemically, TDP-43 proteinopathies are characterized by decreased solubility, hyperphosphorylation, and cleavage of TDP-43 into 25- and 35-kD fragments, as well as altered cellular localization.
Objective:
This review summarizes the extensive research characterizing the distribution of TDP-43 pathology in human postmortem brain tissue and discusses possible therapeutic strategies based on genetic and in vitro studies.
Methods:
We reviewed recently published studies of TDP-43 proteinopathy in the following manuscript.
Results/conclusion:
Given that several different genetic mutations can lead to TDP-43 proteinopathies, including mutations in progranulin and valosin-containing protein, research is needed to decipher and potentially exploit the link between these mutations and TDP-43 pathology.
Keywords: amyotrophic lateral sclerosis, frontotemporal dementia, progranulin, TAR DNA binding protein-43 (TDP-43), valosin-containing protein
1. Background
Neumann and colleagues discovered TAR DNA binding protein-43 (TDP-43) in the inclusions of frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U) and amyotrophic lateral sclerosis (ALS) [2]. Subsequently, numerous retrospective studies have evaluated the frequency of TDP-43 in other neurodegenerative diseases. From the following collective review of these studies, including new insights into TDP-43 function and regulation, the mystery of TDP-43 proteinopathies is beginning to unravel, but this rapid progression of scientific understanding has generated additional questions that will be discussed within this review.
2. Genetics, protein structures, and biological functions of TDP-43
TDP-43 was identified from a genomic screen for novel transcriptional inactivators that bind to the TAR-DNA element of the HIV-1 virus, where it functions as a transcriptional repressor [3]. The human TDP-43 gene, which is located on chromosome 1 and contains 6 exons, is alternatively spliced to generate at least four isoforms that have been identified to date [4]. Given that the TDP-43 gene is highly conserved throughout evolution, it is likely that TDP-43 fulfills a crucial and vital role in maintaining cellular function and survival.
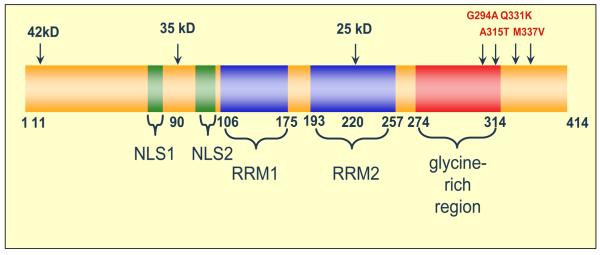
TDP-43, a 414-amino acid protein, has a theoretical molecular weight of 44.74 kDa (Figure 1). In addition to full-length TDP-43 (observed at 43 kDa), three isoforms of TDP-43 have also been detected in human brain and spinal cord tissue [5], but the functional significance of these isoforms is currently unknown. The expressed protein contains two highly conserved RNA recognition motifs (RRM1 and RRM2), as well as a glycine-rich C-terminal sequence. Little is known about the function of the N-terminal region, only that the RRM1 domain of TDP-43 binds single-stranded UG or TG repeat motifs with very high affinity [6, 7]. Although not completely characterized, the RRM2 domain is believed to participate in RNA binding, and also contains a leucine-rich nuclear export signal located between the amino acid residues 239-250 of the human TDP-43 sequence [5, 7, 8]. The glycine-rich C-terminus, a region that mediates protein-protein interactions, is required for TDP-43 to participate in alternative splicing, and also enables TDP-43 to bind to several proteins of the heterogeneous nuclear ribonucleoprotein (hnRNP) family, a class of proteins involved in the biogenesis of mRNA [4, 9]. Thus, given the role of TDP-43 in alternative splicing, it is not surprising that in cell culture models and human postmortem brain tissue, full-length TDP-43 has been localized predominately to the nuclear compartment. However, first reported by Buratti and Baralle as unpublished observations [10], a recent report has verified the presence of small amounts of cytosolic TDP-43 under normal, physiologic conditions [8].
Figure 1.
Schematic of TDP-43 Molecule
Although the functions of TDP-43 are not completely characterized, we do know that TDP-43 possesses the capacity to regulate biological systems through multiple mechanisms due to its ability to bind to both single-stranded DNA and RNA, as well as various proteins. Currently, TDP-43 has been implicated in regulating gene transcription and alternative splicing, in addition to maintaining mRNA stability. As a transcriptional repressor, TDP-43 binds to a polypyrimidine-rich motif in TAR-DNA [3]. In mice, TDP-43 binds and regulates the expression of the proximal promoter of the SP-10 gene, which is involved in spermatogenesis [11]. TDP-43 also forms part of a complex involved in the alternative splicing of the cystic fibrosis transmembrane conductance regulator gene [12, 13], as well as the apolipoprotein A2 gene [14]. In addition, Ayala and colleagues recently identified TDP-43 as a negative regulator of cdk6 expression, such that loss of TDP-43 led to a concomitant increase in cdk6 expression and phosphorylation of retinoblastoma protein (pRb), a substrate for cdk6 and apoptosis [15]. Further expanding the role of TDP-43 in cellular function, a recent study demonstrated that TDP-43 stabilizes the human low-molecular-weight neurofilament (hNFL) mRNA transcript through direct interactions with the 3′ untranslated region [5]. It has also been suggested to play a role as a neuronal activity-responsive factor by binding to mRNA species in P bodies near synaptic structures [16]. However, we still need to investigate the effects of posttranslational modifications of TDP-43, including phosphorylation, ubiquitination, and cleavage, on its regulation of various cellular processes. Thus given the diversity of physiological functions already ascribed to TDP-43, future research into the continued characterization of this protein is essential to fully appreciate the cellular consequences of TDP-43 pathology.
3. TDP-43 pathology in neurodegenerative disease
3.1 FTLD-U/MND
Frontotemporal lobar degeneration (FTLD) is the third most common cause of dementia in some series and one of the major causes of dementia in young adults [17, 18]. FTLD encompasses a class of heterogeneous neurodegenerative disorders that are occasionally associated with motor neuron disease (MND) [19, 20]. Neuropathologically, FTLD can be subdivided into two major categories: tauopathies and non-tauopathies. Tauopathies include Pick's disease, corticobasal degeneration, progressive supranuclear palsy, and neurofibrillary tangle-only dementia, as well as frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17). The most common non-tauopathy is FTLD-U, a form of FTLD in which neuronal and sometimes glial inclusions are positive for ubiquitin and negative for tau and α-synuclein [21, 22]. Cases of FTLD-U associated with motor neuron disease (MND), sometimes resembling ALS, are referred to as FTLDMND. Although TDP-43 appears to be the major constituent of inclusions in motor and non-motor neurons in ALS and FTLD-MND [2, 23-26], some inclusions in ALS, particularly familial forms of ALS, show no TDP-43 immunoreactivity [27, 28]. Other non-tauopathies include rare disorders associated with inclusions composed of neuronal intermediate filaments [29], disorders in which there are inclusions composed of yet-tobe-identified proteins [30], and disorders with no inclusions and only nonspecific neuronal loss and gliosis [31].
Two groups independently identified TDP-43 as the major constituent in FTLD-U by using similar yet distinctly different approaches. Sampathu and colleagues isolated the highly insoluble, high-molecular-weight material from brain extracts of FTLD-U patients and generated a series of antibodies that was subsequently used in histological studies [1]. A subset of these antibodies colocalized with ubiquitin inclusions solely in FTLD-U, with no immunoreactivity detected in other neurodegenerative diseases, demonstrating the specificity of these novel antibodies. In a subsequent study, two of those antibodies were used for immunoblotting to determine which protein species was being recognized; both antibodies detected 24-26-kDa bands only in FTLD-U patients [2]. When these 24-26-kDa bands were extracted and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS), the C-terminal fragments of TDP-43 were identified. Simultaneously, Arai and colleagues isolated insoluble material from FTLD-U patients and performed LC-MS/MS on all bands, also identifying TDP-43 as the major protein component of ubiquitin inclusions in FTLD-U patients [23].
Specific TDP-43 antibodies were used to histologically determine the localization and distribution of TDP-43 and ubiquitin pathology in FTLD-U patients, which has led to further subclassification of FTLD-U cases into 4 types (see Table 1) [1, 24, 32, 33]. Although Mackenzie and colleagues and Sampathu and colleagues assigned different characteristics to subtypes 1-3, as demonstrated in Table 1 and Figure 2, the FTLD-U subtypes identified by each group are presumably the same. Given that most researchers utilized the Sampathu classification system, the remainder of this review will refer to the subtypes as defined by that group [1, 32]. Briefly, type 1 is characterized by long, dystrophic neurites in superficial and middle cortical layers, with few to no neuronal cytosolic inclusions (NCI) or neuronal intranuclear inclusions (NII). In type 2, abundant NCIs are observed in both superficial and deep cortical layers, with few to no NII and fewer dystrophic neurites. In contrast, type 3 is characterized by numerous NCIs and short dystrophic neurites in superficial layers of the cortex. Rod-shaped (lentiform) NIIs are also present in affected regions, particularly in patients with a positive family history [33]. In all 3 subtypes NCIs are observed in the dentate gyrus of the hippocampus. Although NCIs are uncommon in the cortex in type 1, they are present in the dentate gyrus. Other consistent observations across subtypes are the nuclear localization of TDP-43 in unaffected cells and the absence of TDP-43 in the nuclear compartment of cells with cytosolic TDP-43 inclusions. Those observations indicate that in TDP-43 pathologies, TDP-43 is redistributed from the nuclear compartment to the cytosolic compartment. A potential mechanism by which this occurs is just beginning to be made clear by recent in vitro studies, as discussed below [34].
Table 1.
FTLD-U Subtypes according to Cairns et al [32].
| Type I | Type II | Type III | Type IV | |
|---|---|---|---|---|
|
Characteristic histology |
Long dystrophic neurites |
NCI, including granular “pre- inclusions” |
NCI and short neurites; NII in some |
Many NII |
|
Cortical layers most affected |
Superficial and middle | Superficial and deep | Superficial | Superficial and middle |
| Dentate gyrus | Round NCI | Variable granular, crescent or round NCI |
Variable crescent or round NCI |
No NCI |
| Clinical feature | Many patients have semantic dementia |
Some patients have motor neuron disease |
Most patients have frontotemporal dementia |
Most patients have frontotemporal dementia |
| Genetic linkage | None known | 9p chromosome linkage |
PGRN | VCP |
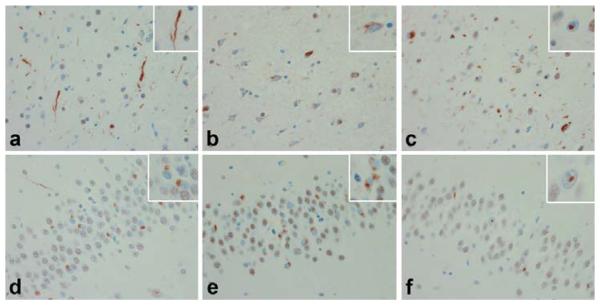
Figure 2.
Cortex (a, b & c) and dentate gyrus (d, e & f) of Type 1 (a & d), Type 2 (b & e), and Type 3 (c & f) FTLD-U according to Sampathu & Cairns [1]. Type 1. Predominance of dystrophic neurites (DN) (a, inset) in cortex with round neuronal cytoplasmic inclusions (NCI) (d, inset) in the dentate gyrus. Type 2. Predominance of NCI, including granular cytoplasmic TDP-43 immunoreactivity, with sparse DN in cortex (b, inset) and dentate gyrus (e, inset). Type 3. NCI, DN & neuronal intranuclear inclusions (NII) (c, inset) and NCI and NII in dentate gyrus (f, inset).
The genetic causes of FTLD have been studied extensively. Although mutations in tau are known to FTDP-17 [35, 36], the most common familial tauopathy, an additional linkage to chromosome 17 was observed for familial FTLD-U. Recently, mutations in progranulin (PGRN) were identified in FTLD-U patients [37-39]. The PGRN gene encodes a secreted growth factor involved in cell cycle regulation. Additional genetic mutations that have been linked to FTLD-U include valosin-containing protein (VCP) [40-42], charged multivesicular body protein 2B (CHMP2B) [43], as well as a newly identified genetic locus on chromosome 9p, which might result from mutations in intraflagellar transport protein 74 [44]. In addition, although a number of groups have evaluated the TDP-43 gene in FTLD patients, currently no pathogenic mutations in TDP-43 have been linked to FTLD-U [45-47].
Analysis of TDP-43 immunoreactivity in familial FTLD-U cases revealed that PGRN mutations are associated with type 3 pathology, whereas VCP mutations produce an atypical pattern of TDP-43 pathology that led to the designation of a fourth subtype (see Table 1) [32, 40]. In type 4 FTLD-U, the characteristic findings are scarcity of NCIs in proportion to NIIs, and lack of inclusions in the dentate gyrus of the hippocampus. Although TDP-43 pathology was not detected in FTLD-U cases with CHMP2B mutations, chromosome 9p-linked FTLD-U cases were classified as type 2, with most of those cases also exhibiting ubiquitin-positive and TDP-43-positive inclusions in both upper and lower motor neurons [24]. This finding is consistent with the high incidence of MND in patients with FTLD-U type 2 pathology [24, 33, 48]. Cairns and colleagues also observed TDP-43 cytoplasmic immunoreactivity that was not immunoreactive for ubiquitin, which they hypothesized might represent an early stage of TDP-43 inclusion formation [24]. These ubiquitin-negative TDP-43 deposits also appear to be particularly common in motor neurons. Although further analysis of these deposits is needed, if they are in fact pre-inclusions, this finding could be exploited to characterize the different stages of TDP-43 inclusion formation. However, it is still unclear how three different genetic mutations result in a similar frontotemporal dementia yet have distinct pathological phenotypes.
3.2 Amyotrophic lateral sclerosis (ALS)
ALS is a rare neurodegenerative disorder that affects both upper and lower motor neurons, leading to progressive weakness and spasticity, with eventual paralysis and death within 3-5 years [49]. Although most ALS cases are sporadic, approximately 10% of patients have a positive family history [49]. The most common genetic cause of familial ALS, accounting for at least 20% of identified familial ALS cases, is missense mutations in the Cu/Zn superoxide dismutase gene (SOD1) [50, 51]. Interestingly, 20% of ALS patients also develop clinical features suggestive of FTLD [52, 53], and 50% of ALS patients have coincident deterioration of both motor and cognitive function [54, 55]. Pathologically, ALS patients have tau-negative ubiquitin inclusions identical to those of FTLD-U patients [21]. Given the considerable overlap between these two very heterogeneous disorders, TDP-43 pathology was evaluated in ALS. Both reports that initially identified TDP-43 as a major protein component of ubiquitin inclusions in FTLD-U also found abnormal TDP-43 inclusions in the motor neurons of ALS patients [2, 23]. Intriguingly, although no genetic mutations in TDP-43 have yet been linked to FTLD, two groups have independently identified several mutations in TDP-43 that segregate with disease in MND/ALS [56, 57]. Specifically, Gitcho and colleagues detected a mutation at A315T that was present only in affected family members [56], and Sreedharan and colleagues identified the mutations Q331K and M337V that also segregated with disease [57]. Although this group also identified the additional mutations G294A and A90V, the G294A mutation has been detected in a single ALS patient to date, while the A90V mutation has only been observed in a young, healthy individual [57], thus future studies are needed to evaluate the pathogenicity of these additional mutations. However, expression of TDP-43 mutants Q331K and M337V in developing chick embryos led to impaired maturation and an increase in neuronal apoptosis compared to wild-type TDP-43 expression, implying a toxic gain of function of these TDP-43 mutations [57].
Since the initial reports detecting the presence of TDP-43 in ubiquitin inclusions in motor neurons of ALS patients, TDP-43 pathology has been confirmed in both sporadic and non-SOD1-linked familial ALS cases. However, TDP-43 is not present in ubiquitin inclusions of familial ALS caused by SOD1 mutations [25-27]. From these findings, it has been hypothesized that different mechanisms control degeneration of motor neurons in SOD1-linked ALS compared with sporadic and non-SOD1-linked ALS [58]; however, it is also possible that TDP-43 pathology and mutant SOD1 produce or contribute to the same downstream consequences. In one mechanism, motor neuropathy with neurofilament (NF) aggregate formation results from disruption of the stoichiometry between the three NF isoforms, including the low-molecular-weight NF (NFL), middle-molecular-weight NF (NFM), and the high-molecular-weight NF (NFH) [59, 60]. In addition, the MND induced in transgenic mice overexpressing NFH was dose-dependently rescued by overexpressing NFL [61]. Thus, since TDP-43 binds and stabilizes NFL mRNA [5] whereas mutant SOD1 binds but destabilizes NFL mRNA [62], it might explain how loss of normal TDP-43 function or mutations in SOD1 could produce a similar pathological phenotype.
Intriguingly, a recent study illustrated that phosphorylated Smad2/3, transcription factors phosphorylated in response to TGF-β signaling, colocalize with TDP-43 in round hyaline inclusions in sporadic ALS cases; while like TDP-43, there was no detection of Smad2/3 in inclusions from SOD-1 linked familial ALS cases [63]. Phosphorylation of Smad2/3 promotes association with Smad4, leading to the subsequent translocation of these factors into the nucleus [64]. Although Smad4 immunoreactivity was not assessed within this study, either the presence or absence of Smad4 in these inclusions could be enlightening and help to reveal the level of impairment in TGF-β-Smad signal transduction. Although the colocalization of Smad2/3 with TDP-43 in sporadic ALS is hypothesized to be indicative of a general impairment in the regulation of nuclear-cytosolic transport, it is unclear why the abnormal cytosolic colocalization of these proteins would only occur in sporadic ALS patients, highlighting the need for further research to more extensively evaluate this relationship.
In-depth analyses of the distribution of TDP-43 pathology in the brainstem of FTLDMND and ALS patients found that NCIs are present in nuclei of the facial, trigeminal, and hypoglossal cranial nerves [65, 66], but not in nuclei of the oculomotor, trochlear, abducens, or vestibular nerves [65]. Because the nuclei of the cranial nerves are innervated directly (facial, trigeminal, and hypoglossal nuclei) or indirectly by the corticobulbar tract (a pathway that originates from the motor cortex and other areas of the frontal and parietal lobes), it is intriguing that TDP-43 pathology has only been observed in nuclei that receive direct projections from the cortex, and not in cranial nerve nuclei that are indirectly innervated by the cortex through interneurons of the reticular formation [67]. In addition, given that FTLD-MND is most commonly associated with type 2 pathology [24, 48], which is characterized by TDP-43 inclusions in deep cortical layers, one might speculate that cortical output pathways, including the corticobulbar and corticospinal tracts originating from deep layers of the cortex, may be particularly affected in this subtype. This hypothesis awaits confirmation but might provide a link between selective vulnerability of distinct neuronal populations and clinical phenotype.
3.3 TDP-43 pathology in other neurodegenerative diseases
Since the discovery of TDP-43 aggregate formation in FTLD-U and ALS, several other neurodegenerative diseases have been evaluated for the presence of TDP-43-positive lesions. Attesting to the specificity of the involvement of TDP-43 pathology in neurodegenerative disease, Lee and associates recently demonstrated the lack of TDP-43 inclusion formation in both anoxic and ischemic lesions [68]. Although investigators have not detected TDP-43 pathology in classic tauopathies, including corticobasal degeneration and progressive supranuclear palsy, as well as familial FTLD cases with tau mutations [65], tau pathology associated with Alzheimer's disease (AD) is sometimes associated with TDP-43 pathology [69]. Amador-Ortiz and colleagues screened a large number of AD cases with and without hippocampal sclerosis (HpScl), which is characterized by neuronal loss and gliosis in the subiculum and CA1 region of the hippocampus. They detected TDP-43 pathology in approximately 75% of AD cases with HpScl and 30% of AD cases without HpScl [69]. Although Cairns and colleagues detected TDP-43 pathology in just 15% of AD patients, only 20 AD patients were included in the analysis, which due to small sample size may not be an accurate representation of the patient population [24]. In a subsequent study that evaluated the incidence of TDP-43 pathology in Lewy body disorders, TDP-43-positive inclusions were observed in 30% of AD cases with concomitant dementia with Lewy bodies (DLB), 7% of Parkinson disease (PD) cases, and 19% of PD cases with dementia [70]. Although a number of studies have failed to observe colocalization between TDP-43 and tau pathology, both Arai and colleagues as well as the aforementioned studies detected TDP-43 immunoreactivity in a small number of neurofibrillary tangles [23, 69, 70]. In addition, Nakashima-Yasuda and colleagues did not detect TDP-43 in Lewy bodies, but α-synuclein and TDP-43 were occasionally colocalized within dystrophic neurites [70]. These findings, along with the discovery of a PGRN mutation in individuals with heterogeneous pathology, including tau-related neurofibrillary pathology, α-synuclein-positive Lewy bodies, and TDP-43-positive NCIs and NIIs [71], show a common pathogenic link between a variety of neurodegenerative conditions. This link is further supported by the demonstration of TDP-43 pathology in neurodegenerative disorders of the Chamorro population of Guam, particularly Guamanian parkinsonism-dementia complex (G-PDC) and Guamanian ALS (G-ALS) cases [72, 73]. Although the etiology of G-PDC and G-ALS remains unknown, we believe that a greater understanding of disease pathogenesis in these disorders could provide insight into the mechanisms of neurodegeneration in ALS, AD, and PD. Thus, the detection of TDP-43 pathology in GPDC and G-ALS indicates that this protein will have far-reaching pathophysiologic ramifications on the field of neurodegeneration.
4. TDP-43 in disease
Biochemical analyses of human postmortem brain and spinal cord from patients with various neurodegenerative diseases showed a similar pattern of TDP-43 immunoreactivity. Specifically, in cases with TDP-43 inclusions, biochemical evaluation revealed decreased solubility of TDP-43, as well as ubiquitination, phosphorylation, and cleavage into 35-kDa and 25-kDa fragments in affected regions [2, 22, 23, 26, 34, 40, 69, 72, 73]. Although the effects of ubiquitination or phosphorylation on TDP-43 function and cellular localization have not been determined, a recent report demonstrates that caspase-dependent cleavage of TDP-43 mediates abnormal redistribution of TDP-43 to the cytosol and thus loss of nuclear TDP-43 [34]. In addition, siRNA-mediated knockdown of PGRN led to a decrease in the solubility of TDP-43, as well as an increase in cleavage that paralleled caspase-3 activation [34]. Because PGRN mutations associated with FTLD-U are hypothesized to lead to a loss of functional PGRN [37-39], the demonstration that decreased PGRN expression in vitro increases TDP-43 cleavage via caspase activation provides the first mechanistic link between PGRN haploinsufficiency and TDP-43 pathology; however a subsequent study reported an inability to replicate these findings [74], A parsimonious explanation to account for these differences could be the influence of variations in methods (e.g. extraction buffers, knockdown efficiency between various targeted PGRN siRNA, and treatment duration). Regardless, strategies to promote the overexpression or inhibit degradation of PGRN could potentially yield a very promising therapeutic approach to either prevent or alleviate TDP-43 pathology.
Another approach to treat TDP-43 proteinopathies might be to accelerate the clearance of TDP-43 inclusions. Recent data have demonstrated that TDP-43 pathology increased when either proteasomal or autophagic pathways were inhibited in vitro [34, 75]. Because the precise cellular mechanism(s) of TDP-43 degradation are unknown, these findings suggest that TDP-43 aggregates might be cleared by stimulating either the ubiquitin-proteasome system (UPS) or autophagy. Stimulation of autophagy by overexpressing histone deacetylase 6 (HDAC6) in Drosophila suppressed degenerative phenotypes caused by UPS impairment resulting from the overexpression of either Aβ fragments (unpublished observations reported in [76]) or polyQ-expanded proteins [76]. Other inducers of autophagy, including treatment with trehalose and rapamycin, alleviated toxicity and enhanced clearance of mutant huntingtin and α-synuclein in both in vitro and in vivo models [77, 78]. Thus the UPS and lysosomal/autophagy pathways might regulate the clearance of pathological aggregates. Stimulation of either pathway could increase degradation of TDP-43 pathology. To evaluate the utility of this approach in TDP-43 proteinopathies, future studies to determine how TDP-43 expression is regulated, in particular the mechanism(s) of TDP-43 degradation, will be imperative.
5. Expert opinion
As the list of TDP-43 proteinopathies expands, it is becoming increasingly necessary to study normal and pathological functions of TDP-43. Although several studies have investigated the physiological functions of TDP-43, the role of TDP-43 in cellular function is still not completely characterized. The effects of posttranslational modifications, such as ubiquitination, phosphorylation, and cleavage, on TDP-43 function are also unclear. Given that the cleaved 25-kDa and 35-kDa fragments of TDP-43 do not contain the N-terminus, which has an RNA-binding domain and a nuclear localization signal, cleavage most likely results in altered function of TDP-43 through loss of the N-terminus and nuclear localization, although future studies will need to investigate this hypothesis. In addition, the identification of a putative nuclear export signal in the RRM2 domain [8], which upon cleavage of TDP-43 would remain intact in C-terminal fragments, may explain how caspase-mediated cleavage of TDP-43 leads to a cytosolic localization. The recent demonstration that siRNA-mediated knockdown of TDP-43 leads to an increase in both cdk6 expression and phosphorylation of the cdk6 substrate pRb, as well as an increase in apoptosis and disruption of the nuclear membrane, further emphasizes the dramatic effects of loss of normal TDP-43 function in disease [15].
Evidence from histological studies suggests that there is heterogeneity in the morphology, distribution, and biochemical correlates of TDP-43 immunoreactive inclusions. Specific monoclonal antibodies recognize the 25-kDa fragment of TDP-43 [2]. Although these antibodies detected ubiquitinated inclusions in cases of FTLD-U types 1 and 2, they failed to recognize ubiquitinated inclusions from type 3 cases [1]. The full implications of this are not yet completely understood, but it is possible that in FTLD-U type 3 cases, the TDP-43 present in inclusions has been modified to block a critical epitope that is otherwise exposed in inclusions from type 1 and 2 cases. Thus additional research is needed to characterize the spectrum of TDP-43 inclusions, in order to fully understand the mechanism(s) by which TDP-43 inclusions form, and to identify specific kinases, phosphatases, and ubiquitin ligases/hydrolases that regulate the phosphorylation and ubiquitination state of TDP-43. It will also be important to determine if and how these modifications are linked and if phosphorylation alters the biological function of TDP-43. Whether these post-translational modifications are preferential to full-length or cleaved fragments of TDP-43 also remains to be determined.
Despite the failure to identify mutations in TDP-43 linked to FTLD-U, the recent detection of genetic mutations in TDP-43 associated with ALS and MND have already yielded extremely enlightening findings regarding disease pathogenesis [56, 57]. Given the precedent from other neurodegenerative disorders characterized by the detection of genetic mutations in genes encoding proteins that accumulate aberrantly, such continued efforts are considerably important, especially in familial cases. Furthermore, genetic engineering of TDP-43 will illuminate TDP-43 biology. For example, mutations in residues critical for phosphorylation or ubiquitination of TDP-43 might alter its function and subcellular localization, as would mutations or deletions in the nuclear localization signal and putative caspase cleavage sites Deletion constructs composed of the 25-kDa and 35-kDa fragments can be used to characterize and differentiate the properties of these fragments and to determine whether full-length TDP-43 or these fragments might form insoluble aggregates similar to fibrillar aggregates that are characteristic of tauopathies, synucleinopathies, and amyloidoses. To conduct such research, we will need to generate animal models that will help advance the current understanding of TDP-43 pathobiology. In addition, given the link between TDP-43 proteinopathies and mutations in PGRN and VCP, evaluating TDP-43 expression, function, and localization in various mutant and knockout models of PGRN and VCP might explain the link between these proteins. The effects of both a loss and overexpression of PGRN and VCP on TDP-43 should also be investigated to determine whether manipulation of either PGRN or VCP levels may be a suitable therapeutic strategy to treat TDP-43 proteinopathies.
Acknowledgements
This work was supported by the Mayo Clinic Foundation and funding by NIA R01-AG-026251-01.
Bibliography
Papers of special note have been highlighted as either of interest (·) or of considerable interest (··) to readers.
- 1.Sampathu DM, Neumann M, Kwong LK, Chou TT, Micsenyi M, Truax A, Bruce J, Grossman M, Trojanowski JQ, Lee VM. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am J Pathol. 2006;169:1343–1352. doi: 10.2353/ajpath.2006.060438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 3.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang HY, Wang IF, Bose J, Shen CK. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 5.Strong MJ, Volkening K, Hammond R, Yang W, Strong W, LeystraLantz C, Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Ayala YM, Pantano S, D'Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE. Human, Drosophila, and C.elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 7.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 8.Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM. Disturbance of nuclear and cytoplasmic Tar DNA binding protein (TDP-43) induces disease-like redistribution, sequestration and aggregate formation. J Biol Chem. 2008 doi: 10.1074/jbc.M800342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 10.Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 11.Acharya KK, Govind CK, Shore AN, Stoler MH, Reddi PP. cis-requirement for the maintenance of round spermatid-specific transcription. Dev Biol. 2006;295:781–790. doi: 10.1016/j.ydbio.2006.04.443. [DOI] [PubMed] [Google Scholar]
- 12.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. Embo J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am J Hum Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayala YM, Misteli T, Baralle FE. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci U S A. 2008;105:3785–3789. doi: 10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang IF, Wu LS, Chang HY, James Shen CK. TDP -43, the Signature Protein of FTLD-U, Is A Neuronal Activity-Responsive Factor. J Neurochem. 2007 doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 17.Ratnavalli E, Brayne C, Dawson K, Hodges JR. The prevalence of frontotemporal dementia. Neurology. 2002;58:1615–1621. doi: 10.1212/wnl.58.11.1615. [DOI] [PubMed] [Google Scholar]
- 18.Snowden JS, Neary D, Mann DM. Frontotemporal dementia. Br J Psychiatry. 2002;180:140–143. doi: 10.1192/bjp.180.2.140. [DOI] [PubMed] [Google Scholar]
- 19.Neary D, Snowden JS, Mann DM. Classification and description of frontotemporal dementias. Ann N Y Acad Sci. 2000;920:46–51. doi: 10.1111/j.1749-6632.2000.tb06904.x. [DOI] [PubMed] [Google Scholar]
- 20.Neary D, Snowden JS, Mann DM, Northen B, Goulding PJ, Macdermott N. Frontal lobe dementia and motor neuron disease. J Neurol Neurosurg Psychiatry. 1990;53:23–32. doi: 10.1136/jnnp.53.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forman MS, Farmer J, Johnson JK, Clark CM, Arnold SE, Coslett HB, Chatterjee A, Hurtig HI, Karlawish JH, Rosen HJ, Van Deerlin V, Lee VM, Miller BL, Trojanowski JQ, Grossman M. Frontotemporal dementia: clinicopathological correlations. Ann Neurol. 2006;59:952–962. doi: 10.1002/ana.20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann M, Kwong LK, Sampathu DM, Trojanowski JQ, Lee VM. TDP-43 proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis: protein misfolding diseases without amyloidosis. Arch Neurol. 2007;64:1388–1394. doi: 10.1001/archneur.64.10.1388. [DOI] [PubMed] [Google Scholar]
- 23.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 24.Cairns NJ, Neumann M, Bigio EH, Holm IE, Troost D, Hatanpaa KJ, Foong C, White CL, 3rd, Schneider JA, Kretzschmar HA, Carter D, Taylor-Reinwald L, Paulsmeyer K, Strider J, Gitcho M, Goate AM, Morris JC, Mishra M, Kwong LK, Stieber A, Xu Y, Forman MS, Trojanowski JQ, Lee VM, Mackenzie IR. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007;171:227–240. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan CF, Eguchi H, Tagawa A, Onodera O, Iwasaki T, Tsujino A, Nishizawa M, Kakita A, Takahashi H. TDP-43 immunoreactivity in neuronal inclusions in familial amyotrophic lateral sclerosis with or without SOD1 gene mutation. Acta Neuropathol. 2007;113:535–542. doi: 10.1007/s00401-007-0206-9. [DOI] [PubMed] [Google Scholar]
- 26.Mackenzie IR, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, Kwong LK, Forman MS, Ravits J, Stewart H, Eisen A, McClusky L, Kretzschmar HA, Monoranu CM, Highley JR, Kirby J, Siddique T, Shaw PJ, Lee VM, Trojanowski JQ. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 27.Robertson J, Sanelli T, Xiao S, Yang W, Horne P, Hammond R, Pioro EP, Strong MJ. Lack of TDP-43 abnormalities in mutant SOD1 transgenic mice shows disparity with ALS. Neurosci Lett. 2007;420:128–132. doi: 10.1016/j.neulet.2007.03.066. [DOI] [PubMed] [Google Scholar]
- 28.Sanelli T, Xiao S, Horne P, Bilbao J, Zinman L, Robertson J. Evidence that TDP-43 is not the major ubiquitinated target within the pathological inclusions of amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2007;66:1147–1153. doi: 10.1097/nen.0b013e31815c5edd. [DOI] [PubMed] [Google Scholar]
- 29.Josephs KA, Holton JL, Rossor MN, Braendgaard H, Ozawa T, Fox NC, Petersen RC, Pearl GS, Ganguly M, Rosa P, Laursen H, Parisi JE, Waldemar G, Quinn NP, Dickson DW, Revesz T. Neurofilament inclusion body disease: a new proteinopathy? Brain. 2003;126:2291–2303. doi: 10.1093/brain/awg231. [DOI] [PubMed] [Google Scholar]
- 30.Yokota O, Tsuchiya K, Terada S, Ishizu H, Uchikado H, Ikeda M, Oyanagi K, Nakano I, Murayama S, Kuroda S, Akiyama H. Basophilic inclusion body disease and neuronal intermediate filament inclusion disease: a comparative clinicopathological study. Acta Neuropathol. 2007 doi: 10.1007/s00401-007-0329-z. [DOI] [PubMed] [Google Scholar]
- 31.Mackenzie IR, Shi J, Shaw CL, Duplessis D, Neary D, Snowden JS, Mann DM. Dementia lacking distinctive histology (DLDH) revisited. Acta Neuropathol. 2006;112:551–559. doi: 10.1007/s00401-006-0123-3. [DOI] [PubMed] [Google Scholar]
- 32.Cairns NJ, Bigio EH, Mackenzie IR, Neumann M, Lee VM, Hatanpaa KJ, White CL, 3rd, Schneider JA, Grinberg LT, Halliday G, Duyckaerts C, Lowe JS, Holm IE, Tolnay M, Okamoto K, Yokoo H, Murayama S, Woulfe J, Munoz DG, Dickson DW, Ince PG, Trojanowski JQ, Mann DM. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 2007;114:5–22. doi: 10.1007/s00401-007-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackenzie IR, Baborie A, Pickering-Brown S, Du Plessis D, Jaros E, Perry RH, Neary D, Snowden JS, Mann DM. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 2006;112:539–549. doi: 10.1007/s00401-006-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang YJ, Xu YF, Dickey CA, Buratti E, Baralle F, Bailey R, Pickering-Brown S, Dickson D, Petrucelli L. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 36.Poorkaj P, Bird TD, Wijsman E, Nemens E, Garruto RM, Anderson L, Andreadis A, Wiederholt WC, Raskind M, Schellenberg GD. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol. 1998;43:815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- 37.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 38.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, van Duijn C, Peeters K, Sciot R, Santens P, De Pooter T, Mattheijssens M, Van den Broeck M, Cuijt I, Vennekens K, De Deyn PP, Kumar-Singh S, Van Broeckhoven C. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 39.Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, Crook R, Melquist S, Kuntz K, Petersen R, Josephs K, Pickering-Brown SM, Graff-Radford N, Uitti R, Dickson D, Wszolek Z, Gonzalez J, Beach TG, Bigio E, Johnson N, Weintraub S, Mesulam M, White CL, 3rd, Woodruff B, Caselli R, Hsiung GY, Feldman H, Knopman D, Hutton M, Rademakers R. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- 40.Neumann M, Mackenzie IR, Cairns NJ, Boyer PJ, Markesbery WR, Smith CD, Taylor JP, Kretzschmar HA, Kimonis VE, Forman MS. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J Neuropathol Exp Neurol. 2007;66:152–157. doi: 10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- 41.Forman MS, Mackenzie IR, Cairns NJ, Swanson E, Boyer PJ, Drachman DA, Jhaveri BS, Karlawish JH, Pestronk A, Smith TW, Tu PH, Watts GD, Markesbery WR, Smith CD, Kimonis VE. Novel ubiquitin neuropathology in frontotemporal dementia with valosin-containing protein gene mutations. J Neuropathol Exp Neurol. 2006;65:571–581. doi: 10.1097/00005072-200606000-00005. [DOI] [PubMed] [Google Scholar]
- 42.Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 43.Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, Hodges JR, Spillantini MG, Thusgaard T, Brandner S, Brun A, Rossor MN, Gade A, Johannsen P, Sorensen SA, Gydesen S, Fisher EM, Collinge J. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 44.Momeni P, Schymick J, Jain S, Cookson MR, Cairns NJ, Greggio E, Greenway MJ, Berger S, Pickering-Brown S, Chio A, Fung HC, Holtzman DM, Huey ED, Wassermann EM, Adamson J, Hutton ML, Rogaeva E, St George-Hyslop P, Rothstein JD, Hardiman O, Grafman J, Singleton A, Hardy J, Traynor BJ. Analysis of IFT74 as a candidate gene for chromosome 9p-linked ALS-FTD. BMC Neurol. 2006;6:44. doi: 10.1186/1471-2377-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gijselinck I, Sleegers K, Engelborghs S, Robberecht W, Martin JJ, Vandenberghe R, Sciot R, Dermaut B, Goossens D, van der Zee J, De Pooter T, Del-Favero J, Santens P, De Jonghe P, De Deyn PP, Van Broeckhoven C, Cruts M. Neuronal inclusion protein TDP-43 has no primary genetic role in FTD and ALS. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Schumacher A, Friedrich P, Diehl-Schmid J, Ibach B, Perneczky R, Eisele T, Vukovich R, Foerstl H, Riemenschneider M. No association of TDP-43 with sporadic frontotemporal dementia. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Rollinson S, Snowden JS, Neary D, Morrison KE, Mann DM, Pickering-Brown SM. TDP-43 gene analysis in frontotemporal lobar degeneration. Neurosci Lett. 2007;419:1–4. doi: 10.1016/j.neulet.2007.03.044. [DOI] [PubMed] [Google Scholar]
- 48.Forman MS, Trojanowski JQ, Lee VM. TDP-43: a novel neurodegenerative proteinopathy. Curr Opin Neurobiol. 2007 doi: 10.1016/j.conb.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 50.Gros-Louis F, Gaspar C, Rouleau GA. Genetics of familial and sporadic amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:956–972. doi: 10.1016/j.bbadis.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 52.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 53.Lomen-Hoerth C, Murphy J, Langmore S, Kramer JH, Olney RK, Miller B. Are amyotrophic lateral sclerosis patients cognitively normal? Neurology. 2003;60:1094–1097. doi: 10.1212/01.wnl.0000055861.95202.8d. [DOI] [PubMed] [Google Scholar]
- 54.Morita M, Al-Chalabi A, Andersen PM, Hosler B, Sapp P, Englund E, Mitchell JE, Habgood JJ, de Belleroche J, Xi J, Jongjaroenprasert W, Horvitz HR, Gunnarsson LG, Brown RH., Jr A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66:839–844. doi: 10.1212/01.wnl.0000200048.53766.b4. [DOI] [PubMed] [Google Scholar]
- 55.Talbot K, Ansorge O. Recent advances in the genetics of amyotrophic lateral sclerosis and frontotemporal dementia: common pathways in neurodegenerative disease. Hum Mol Genet. 2006;15(Spec No 2):R182–187. doi: 10.1093/hmg/ddl202. [DOI] [PubMed] [Google Scholar]
- 56.Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, 3rd, Bigio EH, Caselli R, Baker M, AlLozi MT, Morris JC, Pestronk A, Rademakers R, Goate AM, Cairns NJ. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008 doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Science. 2008 doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwong LK, Neumann M, Sampathu DM, Lee VM, Trojanowski JQ. TDP-43 proteinopathy: the neuropathology underlying major forms of sporadic and familial frontotemporal lobar degeneration and motor neuron disease. Acta Neuropathol. 2007;114:63–70. doi: 10.1007/s00401-007-0226-5. [DOI] [PubMed] [Google Scholar]
- 59.Ge WW, Leystra-Lantz C, Wen W, Strong MJ. Selective loss of trans-acting instability determinants of neurofilament mRNA in amyotrophic lateral sclerosis spinal cord. J Biol Chem. 2003;278:26558–26563. doi: 10.1074/jbc.M302886200. [DOI] [PubMed] [Google Scholar]
- 60.Mendonca DM, Chimelli L, Martinez AM. Quantitative evidence for neurofilament heavy subunit aggregation in motor neurons of spinal cords of patients with amyotrophic lateral sclerosis. Braz J Med Biol Res. 2005;38:925–933. doi: 10.1590/s0100-879x2005000600015. [DOI] [PubMed] [Google Scholar]
- 61.Meier J, Couillard-Despres S, Jacomy H, Gravel C, Julien JP. Extra neurofilament NF-L subunits rescue motor neuron disease caused by overexpression of the human NF-H gene in mice. J Neuropathol Exp Neurol. 1999;58:1099–1110. [PubMed] [Google Scholar]
- 62.Ge WW, Wen W, Strong W, Leystra-Lantz C, Strong MJ. Mutant copper-zinc superoxide dismutase binds to and destabilizes human low molecular weight neurofilament mRNA. J Biol Chem. 2005;280:118–124. doi: 10.1074/jbc.M405065200. [DOI] [PubMed] [Google Scholar]
- 63.Nakamura M, Ito H, Wate R, Nakano S, Hirano A, Kusaka H. Phosphorylated Smad2/3 immunoreactivity in sporadic and familial amyotrophic lateral sclerosis and its mouse model. Acta Neuropathol. 2008;115:327–334. doi: 10.1007/s00401-007-0337-z. [DOI] [PubMed] [Google Scholar]
- 64.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 65.Davidson Y, Kelley T, Mackenzie IR, Pickering-Brown S, Du Plessis D, Neary D, Snowden JS, Mann DM. Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol. 2007;113:521–533. doi: 10.1007/s00401-006-0189-y. [DOI] [PubMed] [Google Scholar]
- 66.Dickson DW, Josephs KA, Amador-Ortiz C. TDP-43 in differential diagnosis of motor neuron disorders. Acta Neuropathol. 2007;114:71–79. doi: 10.1007/s00401-007-0234-5. [DOI] [PubMed] [Google Scholar]
- 67.Nolte J. The human brain: an introduction to its functional anatomy. Mosby, Inc; St Louis, MO: 2002. [Google Scholar]
- 68.Lee EB, Lee VM, Trojanowski JQ, Neumann M. TDP-43 immunoreactivity in anoxic, ischemic and neoplastic lesions of the central nervous system. Acta Neuropathol. 2008;115:305–311. doi: 10.1007/s00401-007-0331-5. [DOI] [PubMed] [Google Scholar]
- 69.Amador-Ortiz C, Lin WL, Ahmed Z, Personett D, Davies P, Duara R, Graff-Radford NR, Hutton ML, Dickson DW. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer's disease. Ann Neurol. 2007;61:435–445. doi: 10.1002/ana.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakashima-Yasuda H, Uryu K, Robinson J, Xie SX, Hurtig H, Duda JE, Arnold SE, Siderowf A, Grossman M, Leverenz JB, Woltjer R, Lopez OL, Hamilton R, Tsuang DW, Galasko D, Masliah E, Kaye J, Clark CM, Montine TJ, Lee VM, Trojanowski JQ. Comorbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007;114:221–229. doi: 10.1007/s00401-007-0261-2. [DOI] [PubMed] [Google Scholar]
- 71.Leverenz JB, Yu CE, Montine TJ, Steinbart E, Bekris LM, Zabetian C, Kwong LK, Lee VM, Schellenberg GD, Bird TD. A novel progranulin mutation associated with variable clinical presentation and tau, TDP43 and alpha-synuclein pathology. Brain. 2007;130:1360–1374. doi: 10.1093/brain/awm069. [DOI] [PubMed] [Google Scholar]
- 72.Geser F, Winton MJ, Kwong LK, Xu Y, Xie SX, Igaz LM, Garruto RM, Perl DP, Galasko D, Lee VM, Trojanowski JQ. Pathological TDP-43 in parkinsonism-dementia complex and amyotrophic lateral sclerosis of Guam. Acta Neuropathol. 2008;115:133–145. doi: 10.1007/s00401-007-0257-y. [DOI] [PubMed] [Google Scholar]
- 73.Hasegawa M, Arai T, Akiyama H, Nonaka T, Mori H, Hashimoto T, Yamazaki M, Oyanagi K. TDP-43 is deposited in the Guam parkinsonism-dementia complex brains. Brain. 2007;130:1386–1394. doi: 10.1093/brain/awm065. [DOI] [PubMed] [Google Scholar]
- 74.Shankaran SS, Capell A, Hruscha AT, Fellerer K, Neumann M, Schmid B, Haass C. FTLD-U linked missense mutations in the progranulin gene reduce progranulin production and secretion. J Biol Chem. 2007 doi: 10.1074/jbc.M705115200. [DOI] [PubMed] [Google Scholar]
- 75.Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 77.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 78.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]