Abstract
Currently, there are more than 30 million people infected with HIV-1 and thousands more are infected each day. Vaccination is the single most effective mechanism for prevention of viral disease, and after more than 25 years of research, one vaccine has shown somewhat encouraging results in an advanced clinical efficacy trial. A modified intent-to-treat analysis of trial results showed that infection was approximately 30% lower in the vaccine group compared to the placebo group. The vaccine was administered using a heterologous prime-boost regimen in which both target antigens and delivery vehicles were changed during the course of inoculations. Here we examine the complexity of heterologous prime-boost immunizations. We show that the use of different delivery vehicles in prime and boost inoculations can help to avert the inhibitory effects caused by vector-specific immune responses. We also show that the introduction of new antigens into boost inoculations can be advantageous, demonstrating that the effect of ‘original antigenic sin’ is not absolute. Pre-clinical and clinical studies are reviewed, including our own work with a three-vector vaccination regimen using recombinant DNA, virus (Sendai virus or vaccinia virus) and protein. Promising preliminary results suggest that the heterologous prime-boost strategy may possibly provide a foundation for the future prevention of HIV-1 infections in humans.
Keywords: HIV-1, prime-boost, heterologous, Sendai virus, clinical trials
1. Introduction
Tens of millions of individuals are currently living with HIV-1. In the United States alone, despite plentiful resources, there are an estimated one million individuals infected with HIV-1, and approximately 55,000 new infections each year [1,2]. Vaccination remains among the highest health priorities, and despite many past disappointments in HIV-1 vaccine clinical trials, recent results from a phase III study suggest that progress is being made toward the development of a protective product [3–7]. The trial was named RV 144 and was conducted in Thailand with more than 16,000 study participants. The vaccine regimen employed a heterologous prime-boost strategy. One vaccine component was a recombinant canarypox vector, ALVAC-HIV (vCP1521), produced by Aventis Pasteur. This vector expressed HIV-1 envelope (from CRF01_AE 92TH023 and LAI viruses), gag (LAI) and protease (LAI) sequences. The second vaccine component was a mixture of two CHO-derived HIV-1 envelope proteins formulated in alum, named AIDSVAX B/E, produced by Vaxgen. These protein sequences derived from CRF01_AE A244 and MN [8]. Trial participants were first inoculated with vCP1521 on weeks 0 and 4. They were then inoculated with vCP1521 mixed with AIDSVAX B/E on weeks 12 and 24. When the outcome of vaccinations was evaluated by a modified intent-to-treat analysis, results showed that there were approximately 30% fewer infections in the vaccinated group compared to the placebo control group. These somewhat encouraging results provide a stepping stone for the production of improved vaccines. Here we will examine the obstacles presented by HIV-1 and the means by which a heterologous prime-boost vaccine might overcome them.
2. The obstacles presented by HIV-1
2.1. Antigen diversity
One of the biggest challenges in HIV-1 vaccine design is posed by the striking diversity of HIV-1 proteins. This virus mutates at an impressive rate due to its error-prone reverse transcriptase and lack of proof-reading capacity [9–11]. The envelope protein is most diverse [10–12], but none of the viral proteins, either internal or external, are entirely conserved. Animal studies have shown repeatedly that HIV-1 readily escapes a focused immune response, whether B-cell or T-cell mediated [13,14].
One might be inclined to describe the diversity of HIV-1 proteins as limitless, but requirements for protein function impose some limits. For example, the envelope protein must bind the highly conserved CD4 and co-receptor molecules to mediate the fusion of virus and host cell target membranes [15]. These requirements levy constraints on the number of mutually exclusive three-dimensional structures that a functional protein can assume.
2.1.1. Conserved determinants within HIV-1 as targets for vaccine design
A popular strategy in the HIV-1 field is to identify a B-cell or T-cell determinant that is conserved or semi-conserved across all HIV-1 isolates and to mark this as a central target for vaccine design. If a conserved determinant can be associated with a protective response and then rendered immunogenic in all humans, this could be the basis of a successful vaccine. In the 1990s, for example, vaccine developers began to target semi-conserved determinants in the V3 loop [16,17], once termed the principal neutralizing determinant (PND) of HIV-1. Numerous additional neutralizing determinants have since been targeted [18–21]. Once a conserved determinant is recognized, researchers seek methods to present that determinant to the immune system, in some cases by unmasking the determinant via HIV-1 gene manipulation (e.g., by removal of a variable region on the envelope protein [22,23] or by stabilization of an envelope-CD4 fusion intermediate [24,25]).
Attempts to render a conserved determinant immunogenic for all humans have not yet been successful, a situation which may reflect the many years of co-evolution between HIV-1 and its human host. In essence, evolution has selected HIV-1 proteins that accept variation among important B-cell and T-cell target epitopes and can thus escape a focused immune response. Conserved determinants, by definition, are those that resist immune pressure. Either their recognition by the immune system is inconsequential to virus growth, or they evade immune responses (e.g., neutralizing antibodies) by (i) concealment beneath variable protein fragments or sugar groups, or (ii) mimicry of ‘self’ proteins toward which humans are tolerant [26]. The unfortunate outcome is that protective immune responses toward conserved HIV-1 determinants are rare in healthy humans and difficult to induce.
2.1.2. Harnessing diverse lymphocytes to target a diverse pathogen
An alternative strategy, based on impressive successes in other vaccine fields, is to formulate vaccine cocktails representative of pathogen diversity. This strategy is aimed at harnessing the full capacity of the immune system rather than a few rare antibodies or T-cell receptors (TCR). Indeed, the strength of the immune system lies in its capacity to respond to an enormous repertoire of antigens. A sophisticated mechanism of rearrangement among immunoglobulin or TCR variable, diversity, joining and constant region genes makes this possible, as each combination yields a different receptor with capacity to recognize a different pathogen or foreign particle via a lock-and-key type interaction. The number of distinct receptors is further increased by imprecise gene joining mechanisms as well as n-and p-region additions [27]. Because of the impressive diversity of receptors in the immune system, virtually every pathogen in nature can be countered by a ‘specific’ set of B-cell and T-cell populations. It may be particularly useful to harness this army of immune effectors when tackling a diverse pathogen like HIV-1.
To create vaccine cocktails, researchers in other fields generally group pathogens by antigenic structure, most often defined by antibody recognition patterns. Once antigens are grouped, representative members from each group are assembled into vaccines to activate respective B-cell and T-cell populations. It is noteworthy that in past years, antigenicity was generally defined by functional assays rather than by evaluation of amino acid similarities/differences between two proteins. This was in part due to technological limitations, but provided some advantage given that (i) two proteins may differ at only one amino acid, but may assume different 3-dimensional antigenic structures, and (ii) conversely, two proteins with multiple amino acid changes may be structurally and antigenically alike [28].
The individual antibodies induced by cocktail vaccines need not be ‘broadly-neutralizing’ provided that they function together to target the breadth of pathogen diversity [29]. As examples, cocktail approaches have been successfully employed to tackle polio virus (trivalent), seasonal influenza virus (trivalent), rotavirus (pentavalent [30]), papillomavirus (quadravalent[31]) and pneumococcus (7,9,10,11 or 23-valent [32,33]). In the HIV-1 vaccine field, cocktail approaches are gaining favor. The vaccine in the recent RV 144 clinical trial included proteins from several different viruses and conferred a hint of protection to vaccinees. Perhaps larger cocktail vaccines, currently under development, will enhance protective efficacy in future clinical trials.
2.2. The chronic nature of HIV-1 infection
The chronic nature of HIV-1 infection is another obstacle often perceived as a major hindrance to HIV-1 vaccine development. However, this feature is perhaps more problematic for the development of viral therapies than for the development of vaccines. In recent years, successful vaccines have been developed and licensed against viruses that are associated with chronic infection, even though the development of therapies or cures has been difficult (e.g., Varicella zoster virus and papilloma virus [31,34]). Like HIV-1, these viruses can persist for long periods without disease, co-existing with the immune response. However, an insult to the immune system (whether related or unrelated to the virus infection) can associate with virus flare, sometimes with a fatal outcome [35,36].
The complexity of chronic virus disease highlights the importance of preventive measures, as it is far easier to block virus at the moment of first exposure than after the establishment of chronic infection. The history of effective vaccines against herpes and papilloma viruses demonstrates that even when certain viruses cannot be cleared from infected humans, successful vaccines can be designed.
3. HIV-1 vaccine development: non recombinant vaccine strategies
3.1. Killed vaccines
One of the first vaccines to be tested in the HIV-1 field was the killed virus vaccine. At first glance, this strategy appeared successful when tested in an SIV macaque model [37], but excitement turned to disappointment when it was discovered that the protective response was a consequence of experimental design. As it turned out, the vaccine and challenge virus stocks had been propagated on human cells and the protective immune responses in macaques were generated toward human MHC proteins that were incorporated into viral membranes [38,39]. An additional disappointment came when certain viral purification methods were found to release envelope proteins from the virus surface. Finally, there was the lingering concern that virus inactivation might not be 100% complete and that ‘killed’ HIV-1 vaccines might infect healthy volunteers. Despite the original disappointments associated with the killed vaccine approach, research continues in this area. Inactivation and purification procedures have been refined so that there is convincing kill with retention of immunogenic envelope proteins [40]. Perhaps these results will one day renew enthusiasm for advanced testing of the killed HIV-1 vaccine approach.
3.2. Attenuated Virus Vaccines
The attenuation of virus was a second method used in early research in the HIV-1 field. As with the killed vaccine approach, first results were promising. A nef-deletion mutant was produced that protected macaques from SIV infection, provided that there was a several month delay between vaccination and challenge [41]. However, success was again transient. Disappointment came when a fraction of animals infected with the attenuated virus vaccines experienced morbidity and mortality [42]. These incidents validated previous concerns that attenuated vaccines might not be safe [43,44]. The virus was capable of repairing the nef deletion mutation and reverting to wildtype phenotype [43,45]. Investigators next sought to strike a balance between virus attenuation and immunogenicity by introducing additional mutations into vaccines. Enhanced safety was achieved when double or triple mutations were introduced into the virus genome, but this additional crippling of virus replication also resulted in weaker immune responses that failed to confer complete protective immunity [46].
Numerous laboratories have now demonstrated that infection with one immunodeficiency virus (SIV or SHIV) can protect against subsequent infection with another [47–50]. How can this be explained? Perhaps the protection induced by live virus vaccines is dependent on a complex interplay between virus and the immune response. Once a virus establishes itself in a safe sanctuary (e.g. in the brain) it may mutate repeatedly. Over the course of months, the immune system is therefore exposed to a cocktail of new antigens created by escape mutation [14]. Possibly, cocktails induce a variety of heterogeneous B cells and T cells, each with a different antigenic specificity and each able to tackle a different virus subset. Together the lymphocytes work as an army [29] to survey the full population of variant viruses. Viral infections from an exogenous source may then be blocked, because each challenge virus matches at least one of the escape variants in the natural vaccine by antigenicity.
This explanation for virus-induced protective immunity might explain the failure of the double- or triple-deletion viral vaccines. When virus growth is severely attenuated by the introduction of multiple mutations, the generation of escape mutants is curtailed and diverse antigens do not evolve. The vaccine is no longer a cocktail, and responding lymphocytes are not diverse. Vaccine efficacy is lost when a focused response cannot recognize the numerous viral variants of nature.
As with the killed vaccine, the attenuated virus vaccine approach remains a topic of active research. Scientists will continue to seek a balance between vaccine safety and the induction of robust, heterogeneous immune cells.
4. Recombinant vaccine strategies
4.1. Introduction of recombinant vectors
The discovery of HIV-1 as the cause of AIDS coincided temporally with an explosive use of recombinant nucleic acid technology [51–53]. It is not surprising that vaccine developers harnessed this technology to create recombinant vaccines with dozens of different vector systems. To date, recombinant SIV or HIV-1 vaccine products have been based on vectors including leishmania [54], baculovirus [55], Semlicki Forest virus [56,57], Venezuelan equine encephalitis virus [58], adenovirus [59], adeno-associated virus [60,61], vaccinia virus [62], modified vaccinia ankara (MVA [63,64]), canarypox, fowlpox [65–67], yeast [68], vesicular stomatitis virus [69], Listeria monocytogenes [70], phage [71], ovine atadenovirus [72], Mycobacterium tuberculosis [73], foamy virus [74], influenza virus [75], coxsackievirus [76], lentivirus [77], Salmonella [78], BCG [79], herpes simplex virus [80], Australian Flavivirus Kunjin [81], measles virus [82], mumps virus [83], rabies virus [84] and plant plastids [85]. Naked DNA was also proven to be an effective vaccine by Dr. Webster of St. Jude Children’s Research Hospital and Dr. Robinson of the University of Massachusetts when DNA-primed chickens and ferrets were protected from challenge with influenza virus, and has since been used as a vector in the HIV-1 vaccine field [86–89].
Some of the first recombinant vector systems were used to produce soluble viral antigens in culture, often formulated with adjuvants for pre-clinical and clinical research [90]. Vaccinations with soluble recombinant protein products typically elicit B-cell and CD4+ T-cell activities. With these products, CD8+ T-cell function is generally weak, because the classical mode of antigen processing for CD8+ T-cell responses requires endogenous protein expression [27]. Certain adjuvants have been shown to enhance CD8+ T-cell activity by promoting antigen cross-presentation, thus heightening the attraction of the soluble protein vaccine approach [91]. The DNA and live viral vector vaccines provide added benefit to soluble protein vaccines in that they instruct endogenous protein expression by the vaccinated host cell, and in the case of virus, can induce remarkably durable CD8+ T-cell, CD4+ T-cell and B-cell activities [27,92–94].
4.2. HIV-1 gene modifications
Molecular biology enables construction of multiple delivery vehicles and also assists the modification of vaccine antigens. Therefore, researchers have tested a great number of HIV or SIV antigen modifications. Genetic manipulations include the deletion of selected protein fragments [95], the creation of computer-designed sequences (e.g., ancestral, consensus, mosaic sequences [96–99]) and/or the alteration of post-translational modifications (e.g., removal of potential di-sulfide bonds or glycosylation sites [23,100]). In fact, proteins can be entirely scrambled to yield products with little similarity to their original template [101].
The study of modified proteins has been highly instructive, and has demonstrated that protein modifications may significantly impact antigenicity, both positively and negatively. Even when mutations are introduced into positions distant from a targeted epitope, they may alter epitope presentation. For example, the three dimensional structure upon which many B-cell epitopes are dependent, may be altered when a mutation is introduced at a distant site [102]. T-cell determinants can also be affected by mutations outside the target peptide, because the release of peptide for association with MHC depends on protein fragmentation, which in turn depends on disulfide bonds, protease cut-sites and glycosylation [103–105].
Debates continue as to how extensively epitopes should be modified from their natural context in candidate vaccines. It has been thought preferable to mimic the post-translational modifications typical of HIV-1-infected mammalian cells, discouraging the early use of recombinant baculovirus or yeast vectors [90,106]. Nevertheless, HIV-1 epitopes have since been expressed by numerous vectors that do not instruct post-translational modifications typical of the mammalian cell [70,71]. Discussions are ongoing as researchers strive to balance the flexibility of recombinant technology with their charge to mimic the natural epitopes of HIV-1.
5. Prime-boost vaccine regimens
5.1. Prime-boost with one or two delivery vehicles
The prime-boost strategy is routinely used in vaccination regimens to increase the magnitude of an immune response. Classical immune tests show that when the immune system is activated upon delivery of a foreign antigen, allowed to rest, and then reactivated, there can be an enormous improvement in both B-cell and T-cell responsiveness [27]. Boosting is relatively simple when a preformed antigen is used, but becomes more difficult when the same live virus vector is used repeatedly to direct antigen expression by host cells. In the latter instance, an anti-vector immune response (induced by the priming vaccine) can block efficacy of the boost. For example, when vaccinia virus is used to prime an immune response, vaccinia virus-specific immune responses can be so robust that a boost inoculation with vaccinia virus will not ‘take’. Essentially, the boost vaccine is quelled before it can instruct host cells to express foreign protein, and therefore provides little improvement to the vaccination regimen.
To overcome the vector-specific immunity induced by an initial prime, one can use a different vector for the boost. This strategy protects the boost vaccine from vector-specific antibodies and also protects infected host cells from vector-specific cytotoxic attack. One further advantage of the heterologous prime-boost strategy is that two different vectors may be selected to target two different lymphocyte subsets [107].
In the early 1990s, the value of the heterologous prime boost in the context of HIV-1 vaccinations was demonstrated by Hu et al. Briefly, investigators used a recombinant vaccinia virus that expressed an HIV-1 (BRU) envelope for the priming of small animals, and followed this with a purified HIV-1 envelope protein boost [108]. They next tested the heterologous prime-boost strategy in non-human primates. In this experiment, they demonstrated that an envelope vaccine administered by heterologous prime-boost was fully protective against SIV infection provided that the SIV antigens were identical between vaccine and challenge virus [109].
In the mid-1990s, our own group collaborated with researchers from the University of Massachusetts, to test a prime-boost regimen using DNA and vaccinia viruses each expressing HIV-1 envelope proteins. We demonstrated that although repeated inoculations with the recombinant DNA vaccine induced relatively weak responses, a single boost with recombinant vaccinia viruses improved immune responses by 1–2 orders of magnitude. This response also exceeded that which could be induced with vaccinia virus alone [110]. At the same time, Ramsay et al. tested DNA and fowlpox virus in a prime-boost regimen, with similar results [111–113].
The prime boost strategy has now been adopted by many research groups in HIV-1 and other fields [71,83,99,114,115]. There are many variations on the theme such as the use of a DNA prime and NYVAC boost [116], a DNA prime and adenovirus boost [117], a DNA prime and vesicular stomatitis virus boost [118], a mumps virus prime and a vesicular stomatitis virus boost [83], a DNA prime and protein boost [115,119], a DNA prime and recombinant phage boost [71], an MVA prime and a fowlpox boost [120] or an adenovirus prime and a protein, peptide or chimeric alphavirus replicon particle boost [121–124]. As new vectors are developed, these are often tested in prime-boost strategies. For example, adenoviruses from chimpanzees have been tested as a means to overcome the ad5-specific pre-existing immunity in humans that is known to hamper vaccine efficacy (as was the case in the disappointing Merck-sponsored STEP trial [7]). If used in a prime-boost protocol, the vector specific response induced by one serotype of adenovirus (e.g., adC6) need not inhibit the ‘take’ of a boost vaccine (e.g., adC7, [125,126]).
The prime-boost strategy with heterologous vectors is also showing promise in clinical trials, as indicated by the moderately successful RV 144 trial [3]. Another example of a prime-boost protocol in the clinic is the PAVE 100 study, redesigned as HVTN 505. This DNA-adenovirus prime-boost vaccine includes 3 HIV-1 envelopes (clades A, B, and C), as well as gag, pol and nef [127,128]. Still another prime-boost strategy uses DNA and MVA vectors. An early test of DNA-MVA was with recombinants expressing T-cell peptides. Results were discouraging in that immune responses were induced in only a small fraction of vaccinated individuals [4]. More recently, the DNA-MVA prime-boost protocol has been employed to deliver proteins rather than peptides [129–131], yielding better outcomes. A GeoVax DNA-MVA clade B vaccine is now in a Phase IIa trial, HVTN 205. The DNA component expresses gag, protease, reverse transcriptase, envelope, vpu, tat and rev sequences, while the MVA component expresses gag, protease, reverse transcriptase and envelope sequences [132]. Another group is using the DNA-MVA prime-boost strategy to present 4 envelopes plus rev, gag, pol and reverse transcriptase (described in more detail below [130,131,133–135]). An additional vaccine strategy forwarded by investigators at the University of Massachusetts Medical School and Advanced Bioscience Laboratories involves the use of a DNA-protein prime-boost regimen to deliver 5 envelope proteins from 4 different clades and gag [107,119,136,137].
As time progresses, researchers are becoming increasingly aware of the need to represent more than one virus isolate in HIV-1 vaccines [138,139], explaining the inclusion of 3, 4 or more envelopes in several current clinical vaccine formulations. The advancement of this strategy to clinical trials was prompted by a number of small and large animal pre-clinical studies comparing individual envelopes with envelope cocktails. Results demonstrated that the cocktails elicited qualitatively improved immune responses compared to single-envelope vaccines [130,133,134,140–143]. The strategy was also prompted by successes in other vaccine fields with the cocktail approach (e.g., influenza virus, rotavirus, papilloma virus, varicella virus and polio virus fields). Clinical trial data show that envelope cocktail vaccines can elicit responses against heterologous and diverse panels of HIV-1 [136,144,145]. While the heterologous target viruses are not precisely represented by sequences in the vaccines, there is sufficient similarity between at least one of the vaccine components and each target to render immune responses cross-reactive.
5.2. Three or more vectors in prime-boost protocols
In the late 1990s, we first tested a prime-boost-boost strategy, using three different vector systems to express HIV-1 envelope proteins [145–149]. Our vectors included recombinant DNA (D), recombinant virus (V, in this case vaccinia virus) and recombinant protein (P). Results demonstrated that three vaccinations with different vectors elicited durable B-cell and T-cell responses [147,148]. Research with three-vector systems have since been conducted by other groups in the context of HIV-1 and non-HIV-1 vaccines [150].
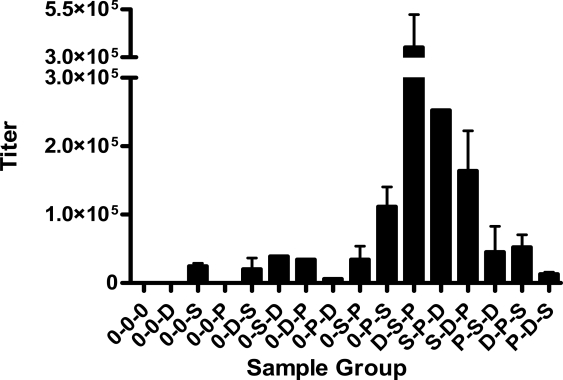
What is the best order of vaccinations when three vectors are used? When using recombinant vaccinia virus as the recombinant virus, we found that the order D-V-P was best [146]. We then asked if the optimal order of vector delivery would change if Sendai virus (SeV) was used as the viral component. To answer this question, we grouped mice to receive one, two or three of the D, SeV and P vaccines in different orders. Each vector was a UG92005 envelope recombinant. After vaccinations, mice were rested for eight months and then anti-envelope antibodies were tested in a UG92005-based envelope ELISA. The results of ELISAs are shown in Figure 1.
Figure 1.
Durable antibody responses elicited by successive HIV-1 envelope immunizations with three different recombinant delivery systems. Female C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed under specific pathogen-free conditions in a BL1 or BL2+ containment area at St. Jude Children’s Research Hospital animal facilities, with adherence to AAALAC Guidelines. Mice were grouped (n = 3) for immunizations with UG92005 (UG) envelope using recombinant vectors: (i) DNA (‘D’, 100 μg, carrying a gp140 envelope sequence) administered i.m. in the gastrocnemius muscles, (ii) CHO-derived recombinant UG envelope protein (‘P’, 2 μg, 100 μl gp140 envelope in PBS mixed with 100 μl CFA) administered i.p., and (iii) SeV (‘S’, 104 plaque forming units, carrying a gp120 envelope sequence for expression in infected cells) administered i.n.. Groups of mice received one, two or three of the recombinant vectors (or no vaccine, ‘0’), administered with one month intervals. Eight months after the completion of inoculations, ELISAs were performed with serially diluted serum samples. Antibody binding titers were determined using Prism Software (Non-Linear Regression, GraphPad Prism®, San Diego, CA). Mean titers and standard errors are shown. Some data points represent fewer than 3 animals due to mouse death or non-conformance of data to the non-linear regression curve (O-S-D, O-D-P, O-P-D, O-S-P, S-P-D, P-S-D). To perform the ELISA, 96-well plates (BD Biosciences, Franklin Lakes, NJ) were coated overnight at 4 °C with 2 μg/ml of purified CHO-derived UG gp140 envelope protein in PBS. The plates were washed three times with 0.05% Tween 20 in PBS, blocked with 1% BSA/PBS at room temperature for 1 h, and washed an additional three times. Samples were serially diluted in PBS to a final volume of 50 μl and were incubated in wells for 2 h at room temperature. After three washes, alkaline phosphatase-conjugated anti-mouse IgG1 (50 μl/well; Southern Biotechnology Associates, Birmingham, AL) diluted 1/1000 in 1% BSA/0.05% Tween 20/PBS was added for 1 h at room temperature. Following five washes, the assay was developed with 75 μl/well of p-nitrophenyl phosphate (Sigma-Aldrich) substrate (1 mg/ml in diethanolamine buffer) and was read at OD450nm after 1 hour at 37 °C.
We noted that virtually all of the animals that received recombinant SeV generated long-term immunity, a result which highlighted the well known association of live virus vaccines with durable immune responses [92–94,151]. The preliminary study results in Figure 1 further showed that the more durable antibody immune responses were elicited when the SeV immunization was administered as the first or second inoculation (see D-S-P or S-P-D) rather than as the third immunication (see D-P-S or P-D-S). The result was confirmed in a repeat experiment. This phenomenon might be explained by our previous finding that a robust pre-existing T-cell response toward HIV-1 envelope can significantly inhibit the growth or ‘take’ of recombinant SeV [152]. The situation of vaccine inhibition is similar to that described previously in that a vector-specific response can quell the ‘take’ of a live virus boost. The difference in this case is that the pre-existing immune response targets the HIV-1 envelope passenger gene product rather than the delivery vehicle. If the immune response to the passenger gene product is too strong (in this case following D or P vaccines), the SeV vaccine may not ‘take’, yielding an inferior overall outcome. Results highlight the complexities of the prime-boost strategy and demonstrate how each inoculation can impact the efficacy of the next.
Yu et al. have also examined a three-vectored prime-boost-boost regimen, in this case using DNA, adenovirus and SeV recombinants expressing gag antigens. They found that the vaccines delivered in the order DNA-SeV-adenovirus yielded slightly higher gag-specific CD8+ T-cell activities than vaccines delivered in the order DNA-adenovirus-SeV [150,153].
5.3. Complex factors impact prime-boost vaccine outcomes
A great number of factors will impact the final outcome of a prime-boost strategy, due to the important interplay between responses induced by each inoculation. A subtle balance must therefore be struck. On the one hand, the activation of naïve cells by a priming vaccine is desired. On the other hand, an overly robust primary immune response may be counter-productive if it inhibits the efficacy of a boost. DNA and virus vaccines are perhaps most sensitive to pre-existing immune responses, because they must transfect or infect host cells prior to production of the desired antigen. If/when host cells are susceptible to immune attack due to pre-existing immunity, the boost function may be reduced or eliminated. It is perhaps the case that a protein boost is most resilient to the effects of pre-existing immunity, because the antigen is preformed and its expression is not dependent on infection or transfection of the host cell.
Not only will the immune response toward prime-boost vaccination depend on the selection of recombinant vectors and the order in which they are administered, but vaccine dose will also affect outcome. Again, a balance must be reached to successfully ‘prime’ the immune response without inhibiting the boost. The selection of a priming dose can sometimes be counter-intuitive in that an increase in dose does not always improve immunogenicity. This is because high-dose priming antigens can in some cases hinder a response to the boost inoculation and can also hinder the establishment of long-term immune memory [154,155]. In a hepatitis vaccine clinical study, researchers found that a dose as small as 2 μg DNA could be effective as part of a prime-boost regimen [156].
To complicate vaccine formulation further, it is noteworthy that a dose study in one species need not predict vaccine outcome in a second. As an example, a DNA dose escalation study in mice suggested that increases in DNA dose would improve immune activities, yet in macaques a 10-fold DNA dose escalation was not advantageous [157,158]. The situation becomes more complicated with each experimental change. Another example is provided by the opposing outcomes in studies of DNA-adenovirus prime-boost regimens in macaques. Whereas research in one system demonstrated complementary effects of a DNA prime and adenovirus boost [117], a more recent protection study showed no advantage conferred by a 400 mg DNA prime administered prior to an adenovirus boost [159].
Among the many factors influencing outcome in a prime-boost study are: (i) the vaccine vectors, (ii) vaccine doses, (iii) the interval between prime and boost, (iv) virus tropism for the host species, (v) non-vaccine sources of pre-existing immunity, (vi) the type of desired immune activity (e.g., B-cell versus T-cell, Th1 versus Th2), and (vii) the timing of desired immune function (acute versus long-term). When prime-boost regimens are newly designed, each factor deserves attention to ensure vaccine efficacy.
6. Changing antigens during a prime-boost regimen
6.1. Introducing variant antigens in prime-boost vaccine protocols: lessons from the influenza virus vaccine field and original antigenic sin
The influenza virus, like HIV-1, encompasses a large diversity of viral proteins. The review of nucleic acid or amino acid sequences from influenza virus variants may be mind-boggling to the vaccine developer, but researchers have grouped viruses by antigenicity rather than by sequence to produce effective vaccines.
Influenza virus vaccine developers must accommodate antigenic drift (moderate changes in viral proteins) as well as antigenic shift (major changes in viral proteins incurred when the virus jumps species) [160]. To deal with these temporal changes, vaccine developers produce new vaccines annually. They also produce novel vaccines in response to shifting viral variants, as for the recent H1N1 swine flu pandemic [161]. Humans who receive vaccines each year are exposed to ‘new’ heterologous influenza virus hemagglutinin and neuraminidase proteins with each boost inoculation.
Original antigenic sin (OAS) was demonstrated decades ago in the context of immune responses toward influenza antigens. When humans or experimental animals experienced infections with related influenza viruses, their sera repeatedly showed higher neutralizing titers against the earlier encountered (‘original’) virus strain [162,163]. The common theme of pre-existing immunity may be central to the explanation of OAS. In this case, the immune system has memory for a portion of (but not all) epitopes on the challenge virus and may therefore hamper boost function. The concept is similar to that described above in the context of vector-specific or HIV-1-envelope-specific activities which can clear and weaken the boost inoculum. As before, clearance can be mediated by both humoral (B-cell) and cellular (T-cell) responses. In the case of humoral immunity, pre-existing antibodies may recognize and clear virus particles. In the case of cellular immunity, effectors can kill virus-infected targets that display viral peptides on membrane surfaces in the context of MHC. Together, immune effectors can reduce antigen load and durability of the boost. In this instance, whereas the transient appearance of boost antigen may suffice to drive re-activation and replication of memory cells responsive to the ‘old’ epitopes, the antigen load and persistence may be insufficient to activate naïve lymphocytes responsive to ‘new’ epitopes. The outcome is an improved response toward ‘old’ or ‘original’ epitopes, but a relatively weak response toward epitopes that are newly presented.
Is OAS absolute? In a recent influenza virus study in mice, researchers confirmed the phenomenon of profound OAS [164]. In contrast, a recent study in humans revealed the opposite result [165]. In the latter case, humans who were vaccinated with the seasonal influenza virus vaccine responded better to ‘new’ versus ‘old’ epitopes. Results supported the theory upon which influenza virus vaccines are based, that the quality of immune responses can be improved by successive immunizations with related, but dissimilar influenza viral antigens. Contrasting results in OAS studies again illustrate that the rules driving immune responses to a prime-boost vaccine are not simple. As stated above, outcomes will depend on complex variables including (i) vaccine dose, (ii) interval between first and second antigen exposure, and (iii) relatedness of epitopes between old and new antigens.
6.2. T-cells responsive to an epitope shared by prime and boost vaccines can assist B-cells responsive to a unique epitope in the boost
Even though a vigorous pre-existing immune response may limit the load and persistence of boost antigens, memory cells can also play a positive role. Specifically, the CD4+ T cells that are primed by a first dose of vaccine may ‘help’ naïve lymphocytes to respond to novel epitopes in the boost inoculum.
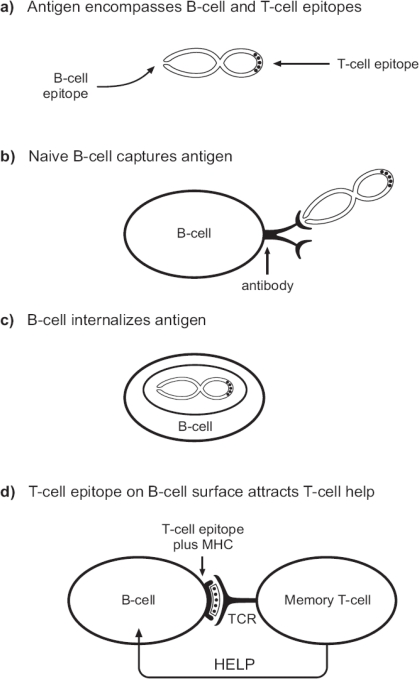
T-cell help is conferred by a variety of mechanisms including secretion of interleukins and activation of antigen presenting cells [27]. In Figure 2 is illustrated the cognate B-cell:T-cell interactions that can enhance naive B-cell stimulation. Briefly, when a naïve B cell encounters antigen for the first time (Figure 2A), its antibody will bind and internalize the antigenic protein or particle (Figure 2A–C). Antigens are then fragmented for ultimate display on the cell surface in association with MHC class II. The peptide-MHC complex can then be bound by the TCR of a memory T cell (Figure 2D). Cognate B-cell:T-cell interactions then trigger the relay of ‘help’ (e.g., interleukin 4) for support of B-cell activation. It is noteworthy that the epitopes recognized by the B cells and T cells need not be the same. The T-cell epitope may be shared by the prime and boost vaccines, whereas the B-cell eptiope may be unique to the boost. It is only necessary that the two epitopes are linked on the antigen or particle to which antibody is bound [27]. The phenomenon can be observed when mice are primed with a vaccine in vivo, after which lymphocytes are exposed to a new determinant(s) linked to that vaccine in vitro. In this scenario, naïve B cells with specificity for the new determinant(s) are activated when they receive ‘help’ from the vaccine-induced memory T cells [166,167].
Figure 2.
Cognate B-cell:T-cell interactions support T-cell help. This cartoon illustrates a situation in which a B cell and T cell recognize different determinants on the same particle (panel A). In this circumstance, the B-cell antibody may capture antigen via a lock-and-key type interaction (panel B). The antigen is then fragmented (panel C), after which the T-cell epitope is presented on the B-cell membrane as a complex with MHC. The peptide-MHC complex is bound by the T-cell receptor, after which ‘help’ may be relayed from the T cell to its B cell partner (panel D).
The lesson from the above experiments is that a pre-existing immune response elicited by a priming vaccine can have both positive and negative effects on naïve responses to novel epitopes in a boost. The balance yields striking OAS in some circumstances, and relatively little OAS in others.
6.3. HIV and OAS
In 1991, Klinman et al. [168] described a study in which mice received sequential inoculations with gp120 proteins from independent isolates of HIV-1. The specificities of vaccine-induced plasma cells were then tested by positioning cells between two antigen coated surfaces. Secreted antibodies were examined by spot analyses for simultaneous recognition of two different antigens. Researchers found that OAS was a factor, just as had been the case in the influenza virus studies, but was not absolute; immune responses toward shared determinants were dominant, but responses to new antigens, presented in the boost inocula, were also observed.
Zhan et al. later evaluated T-cell activities toward a heterologous prime-boost regimen in mice after successive immunizations with two different HIV-1 envelope gp140 constructs [169]. In this case, the two gp140 antigens 1007 and UG92005 carried very different immunodominant peptides [170,171]. Despite the inoculation of mice with one immunogen (1007) before the other (UG92005), T-cell responses toward the unique epitopes on each of the antigens were generated [169].
Wang et al. [172] also tested a heterologous prime-boost approach, in which peptides were used as a prime and a recombinant vaccinia virus was used to deliver the boost. They found that the first immunization had insignificant effect on the CD8+ T-cell dominance hierarchies induced by the second. Taken together, these results suggest that novel proteins and epitopes can be introduced in the boost phase of a prime-boost regimen to improve the quality of the HIV-1-specific response in both B-cell and T-cell populations.
As stated above, the vaccine evaluated in the recent RV 144 clinical trial in Thailand presented heterologous proteins upon prime and boost. This prime-boost vaccine demonstrated a hint of protection whereas individual vaccines were not protective when administered alone [173]. While the precise mechanism of protection is not yet known, it is likely that the combination of ‘old’ and ‘new’ responses toward antigens in prime and boost vaccines lent to the moderate success.
Perhaps the best example of incomplete OAS is demonstrated by infections with SIV, SHIV or HIV. As described above, an infected subject is exposed to a variety of viral variants over the course of time and responds with an ever-increasing quality of immune activity. That immune activity is often sufficient to confer protection against virus from an exogenous source [14,41,47,174,175]. It is worth contemplating that if a safe vaccine could recapitulate the immune responses induced by the successive antigens presented by natural infection, uninfected individuals might be fully protected from HIV-1.
7. Changing both vectors and antigens during heterologous prime-boost immunizations in HIV-1 vaccine clinical trials
Several clinical studies have now tested strategies that change both vectors and antigens during prime-boost regimens, as was the case in the RV 144 trial. For example, Gudmundsdotter et al. are currently testing a DNA vaccine that expresses envelopes from Subtypes A, B and C (A92UG031, LAI and 92BR025), rev from subtype B, gag from subtypes A and B, reverse transcriptase from subtype B, and a boost vaccine that is an MVA-Chiang Mai double recombinant (MVA-CMDR)(HIV MVA) expressing envelope and gag/pol sequences from Thai isolates CM235 and CM240 [131].
Our own research has involved clinical testing of a multi-envelope heterologous prime-boost approach using DNA, virus (vaccinia virus) and purified protein recombinants. The selection of envelopes for inclusion in our vaccine was based on (i) differing patterns in antibody binding studies [176], (ii) longitudinal capture from infected persons [138], and (iii) differing clades. This strategy was first tested in macaques and found to confer protection against a heterologous SHIV challenge [177] which was not represented by sequence in the vaccine. To test the concept in humans, we first demonstrated the safety of each of the three vectors D, V, and P in FDA- and IRB-approved phase I clinical studies [144,145,178]. We next designed a protocol, approved by the FDA and our local IRB, to vaccinate individuals with dozens of envelopes. Six successive immunizations (D-D-V-P-D-P) separated by 1 month intervals, were planned for each individual. The D vaccine included 51 recombinant plasmids each expressing a different envelope protein, administered as an i.m. inoculation of 100 micrograms; the vaccinia virus vaccine included 23 recombinant viruses, each expressing a different envelope protein (a subset of which were shared with the D vaccine), administered by s.q. inoculation with 107 total plaque forming units; the recombinant CHO-derived protein vaccine was 100 microgram purified envelope from HIV-1 isolate UG92005 (shared with D and V vaccines), administered i.m. as a formulation with 500 micrograms alum. The single protein immunization was administered to enhance both B-cell and T-cell responses. We emphasize that based on the mechanism described in Figure 2, a T cell which has responded to one envelope immunogen can ‘help’ a B cell which has responded to another, provided that the B cell displays the T-cell epitope on its surface.
Based on a local administrative decision, only three individuals were ultimately enrolled in the study. The first participant received all 6 inoculations (D-D-V-P-D-P); the second participant received four inoculations (D-D-V-P), and the third recipient received three inoculations (D-D-V). All inoculations were well tolerated. Adverse events that were possibly, probably or definitely vaccine related were grade 1 only.
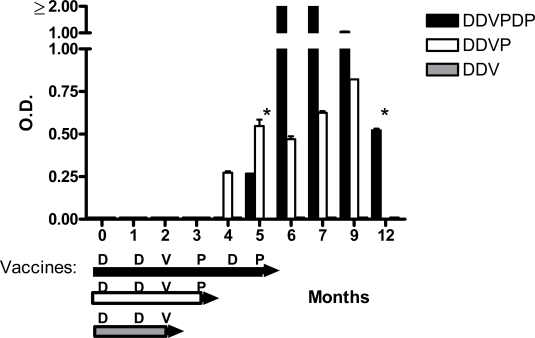
Preliminary tests of envelope-specific immune responses were conducted with the Abbott enzyme-linked immunosorbent assay (HIVAB HIV-1/HIV-2 (recombinant DNA) Abbott Laboratories, Abbott Park, IL, USA). This standardized assay is used worldwide as a diagnostic for HIV-1 infection and was conducted in the Clinical Pathology Department of St. Jude Children’s Research Hospital. There is a cut-off score for each test (ranging between 0.1 and 0.2 OD450nm). Absolute responses for HIV-1-infected persons in this test can be variable and often reach the assay peak value of ≥2. In Figure 3, results are shown for the three vaccinees.
Figure 3.
Antibody responses elicited by heterologous prime-boost vaccination of humans. Three study participants received three different vaccination regimens. Study Participant 1 (black bars) received D (month 0), D (month 1), V (month 2), P (month 3), D (month 4) and P (month 5). Participant 2 (clear bars) received the first four inoculations and Participant 3 (grey bars) received the first three inoculations. Sera were collected and tests were conducted with the Abbott enzyme-linked immunosorbent assay (HIVAB HIV-1/HIV-2 (recombinant DNA) Abbott Laboratories, Abbott Park, IL, USA). Asterisks indicate an absence of sample.
Participant 1 who received all six inoculations, generated the best response. An Abbott-positive response was first identified in this volunteer one month after the fifth immunization (D-D-V-P-D). One month after the sixth immunization (P), the participant exhibited peak absorbance levels in the Abbott assay (OD450nm ≥ 2). Significant responses persisted for at least seven months following the last inoculation.
Participant 2, who received four immunizations (D-D-V-P) also generated significant envelope-specific activity. This response continued to improve throughout the observation period, but did not achieve peak values (within the observation period which ended 6 months after the final immunization).
Participant 3, who received only D-D-V did not generate a positive response in the Abbott assay. Apparently, the combination of at least 4 immunizations and 3 vector systems was advantageous, distinguishing the positive responses in Participants 1 and 2 from the negative response in Participant 3.
The antibodies with the strongest immune activity (from Participant 1 taken 1 month following the completion of all vaccinations) were also tested for neutralizing activity in a preliminary GHOST cell-based assay [145]. To conduct this study, antibodies were first purified over a protein G column to remove non-specific factors. Although immunoglobulin purification is not standard practice in the field, it may be essential to avoid the non-specific inhibitory or enhancing effects of serum components in the HIV-1 neutralization assay [179–182]. Such factors are particularly problematic when sera are tested at high concentrations and/or when results from parallel analyses with panels of unselected negative control sera are not available. In our study, of eight tested heterologous viruses, four viruses were neutralized at the 50% level by samples taken post-vaccination (diluted 1:5 relative to the original serum value), but not pre-vaccination. Positive tests were with viruses HIV-1IIIB (CXCR4), HIV-130e (CXCR4), HIV-1SF2 (CCR5) and HIV-192HT593 (CCR5), but not viruses HIV-196ZM651 (CCR5), HIV-1ZM53M (CCR5), HIV-192UG029 (CCR5) and HIV-193UG082 (CCR5). The neutralization of four heterologous viruses was promising and indicated the sharing of antigenic structures between at least one envelope in the vaccine and envelopes on test viruses.
The relevance of neutralizing responses to protection in vivo is not known. Currently, there are no clear correlates of protection in the HIV-1 field. In fact, antibodies can demonstrate a much improved protective capacity in vivo versus in vitro. For example, in a macaque study, passive transfer of hyperimmune sera to naïve animals was fully protective against SIV, even when the same antibodies showed no neutralizing activity against the same challenge virus in vitro [183]. The neutralization assay is clearly able to measure some vaccine-induced antibody potentials, but misses critical antibody functions such as antibody-dependent cell-mediated cytotoxicity (ADCC) and antibody-dependent cell-mediated virus inhibition (ADCVI, [184–186]). Taken together, the preliminary clinical results with D-V-P vaccines demonstrate a degree of immune breadth and encourage continued testing of the vaccine concept with larger participant groups and larger virus panels. Preliminary results also reveal safety and immunogenicity of the vaccine in humans and show that responses are durable.
What might be the mechanism responsible for antibody (and T-cell) durability, particularly in the context of a live virus infection (a single vaccinia virus inoculation elicits antibody responses that can persist for more than 70 years in humans [92–94,187–190])? Immune response durability has long been a topic of much debate. Some researchers argue that antigens (particularly viral antigens) are maintained in vivo for the long term and constitutively drive B cells to end-differentiate into antibody forming cells [191]. Other researchers, however, have shown that when B cells have lost their capacity to recognize an antigen, they still persist. The latter feature was best illustrated using a cre/lox recombination system with which antibody genes were intentionally altered among memory B cells in vivo. B-cells that had lost their capacity to recognize the priming antigen were nonetheless maintained [192]. These results suggest that immune responses are maintained by a combination of factors, including antigen, memory B cell, and antibody-forming cell persistence [191].
The proof-of-concept findings described by ourselves and others in pre-clinical and phase I/II clinical trials cannot predict protective efficacy, but in combination with the recent hint of success in the RV 144 trial, encourage further advancement of vaccine strategies that combine heterologous vectors and heterologous antigens in a prime-boost approach.
8. Conclusion
The heterologous prime-boost vaccination strategy can be used to harness robust and durable immune activity, and is particularly useful as a means to circumvent vector-specific inhibitory effects. In the context of a heterologous prime-boost, there are also inhibitory effects directed toward the passenger gene which can lend to ‘original antigenic sin’. Fortunately, this phenomenon is not absolute. The final outcome of prime-boost vaccinations is complex and will be influenced by multiple factors including: (i) the vaccine vectors, (ii) the vaccine doses, (iii) the interval between prime and boost, (iv) virus tropism for the host species, (v) non-vaccine sources of pre-existing immunity, (vi) the type of desired immune activity, and (vii) the timing of desired immune function. Despite the complexities posed by heterologous prime-boost vaccine protocols, the strategy holds enormous promise and might ultimately prevent the morbidity and mortality caused by HIV-1.
Acknowledgments
We thank Harold Stamey from the Tennessee Blood Services for providing samples for this study, and R.V. Srinivas, the NIH AIDS Reference Reagent Program (NARRP, Rockville, MD) and the World Health Organization Network for HIV Isolation and Characterization, for providing viruses. We thank Brita Brown, Amy Zirkel, Samantha D. Rencher, Suzette Wingo and Queen E. Rodgers for excellent technical assistance. We thank Randy Hayden (Pathology Department, St. Jude) for the conduct of Abbott ELISAs. We thank the volunteers who participated in the clinical studies. We thank C. Coleclough and S. Naron for critical reading of the manuscript and editorial suggestions. We thank the Animal Resource Center and Hartwell Center of St. Jude Children’s Research Hospital for support. This work was supported in part by NIH NIAID grants P01-AI45142, R21-AI056974 and R01-AI078819, NCI Cancer Center Support core grant P30-CA21765, the Federated Department Stores, the Mitchell Fund, the Carl C. Anderson Sr. and Marie Joe Anderson Charitable Foundation, the James B. Pendleton Charitable Trust, the Pioneer Fund and the American Lebanese Syrian-Associated Charities (ALSAC).
References and Notes
- 1.Hall HI, Song R, Rhodes P, Prejean J, An Q, Lee LM, Karon J, Brookmeyer R, Kaplan EH, McKenna MT, Janssen RS. Estimation of HIV incidence in the United States. JAMA. 2008;300:520–529. doi: 10.1001/jama.300.5.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.HIV prevalence estimates--United States, 2006. MMWR Morb Mortal Wkly Rep. 2008;57:1073–1076. [PubMed] [Google Scholar]
- 3.McNeil DG., Jr For first time, AIDS Vaccine shows some success. The New York Times. 2009 [Google Scholar]
- 4.Cohen J. AIDS vaccines. HIV dodges one-two punch. Science. 2004;305:1545–1547. doi: 10.1126/science.305.5690.1545. [DOI] [PubMed] [Google Scholar]
- 5.Burton DR, Desrosiers RC, Doms RW, Feinberg MB, Gallo RC, Hahn B, Hoxie JA, Hunter E, Korber B, Landay A, Lederman MM, Lieberman J, McCune JM, Moore JP, Nathanson N, Picker L, Richman D, Rinaldo C, Stevenson M, Watkins DI, Wolinsky SM, Zack JA. Public health. A sound rationale needed for phase III HIV-1 vaccine trials. Science. 2004;303:316. doi: 10.1126/science.1094620. [DOI] [PubMed] [Google Scholar]
- 6.Belshe R, Franchini G, Girard MP, Gotch F, Kaleebu P, Marthas ML, McChesney MB, McCullough R, Mhalu F, Salmon-Ceron D, Sekaly RP, Van RK, Verrier B, Wahren B, Weissenbacher M. Support for the RV144 HIV vaccine trial. Science. 2004;305:177–180. doi: 10.1126/science.305.5681.177b. [DOI] [PubMed] [Google Scholar]
- 7.Robb ML. Failure of the Merck HIV vaccine: an uncertain step forward. Lancet. 2008;372:1857–1858. doi: 10.1016/S0140-6736(08)61593-7. [DOI] [PubMed] [Google Scholar]
- 8.Nitayaphan S, Pitisuttithum P, Karnasuta C, Eamsila C, de SM, Morgan P, Polonis V, Benenson M, vanCott T, Ratto-Kim S, Kim J, Thapinta D, Garner R, Bussaratid V, Singharaj P, el-Habib R, Gurunathan S, Heyward W, Birx D, McNeil J, Brown AE. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J Infect Dis. 2004;190:702–706. doi: 10.1086/422258. [DOI] [PubMed] [Google Scholar]
- 9.Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 10.Kuiken C, Korber B, Shafer RW. HIV sequence databases. AIDS Rev. 2003;5:52–61. [PMC free article] [PubMed] [Google Scholar]
- 11.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 12.Lockey TD, Hurwitz JL. Size-heterogeneous sequences mark hot spots for asparagine, serine, and threonine insertions in HIV type 1 envelope. Aids Res Hum Retroviruses. 1998;14:717–719. doi: 10.1089/aid.1998.14.717. [DOI] [PubMed] [Google Scholar]
- 13.Barouch DH, Kunstman J, Glowczwskie J, Kunstman KJ, Egan MA, Peyerl FW, Santra S, Kuroda MJ, Schmitz JE, Beaudry K, Krivulka GR, Lifton MA, Gorgone DA, Wolinsky SM, Letvin NL. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J Virol. 2003;77:7367–7375. doi: 10.1128/JVI.77.13.7367-7375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrin T, Crawford L, Sawyer L, Weber P, Sheppard HW, Hanson CV. Neutralizing antibody responses to autologous and heterologous isolates of human immunodeficiency virus. J Acquir Immune Defic Syndr. 1994;7:211–219. [PubMed] [Google Scholar]
- 15.Siciliano SJ, Kuhmann SE, Weng Y, Madani N, Springer MS, Lineberger JE, Danzeisen R, Miller MD, Kavanaugh MP, DeMartino JA, Kabat D. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274:1905–1913. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- 16.LaRosa GJ, Davide JP, Weinhold K, Waterbury JA, Profy AT, LEwis JA, Lanlois AJ, Dreesman GR, Boswell RN, Shadduck P, Holley LH, Karplus M, Bolognesi DP, Matthews TJ, Emini EA, Putney SD. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science. 1990;249:932–935. doi: 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- 17.White-Scharf ME, Potts BJ, Smith LM, Sokolowski KA, Rusche JR, Silver S. Broadly neutralizing monoclonal antibodies to the V3 region of HIV-1 can be elicited by peptide immunization. Virology. 1993;192:197–206. doi: 10.1006/viro.1993.1022. [DOI] [PubMed] [Google Scholar]
- 18.Pinter A, Honnen WJ, Kayman SC, Trochev O, Wu Z. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine. 1998;16:1803–1811. doi: 10.1016/s0264-410x(98)00182-0. [DOI] [PubMed] [Google Scholar]
- 19.Ferrari G, Kostyu DD, Cox J, Dawson DV, Flores J, Weinhold KJ, Osmanov S. Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines. Aids Res Hum Retroviruses. 2000;16:1433–1443. doi: 10.1089/08892220050140982. [DOI] [PubMed] [Google Scholar]
- 20.Chow YH, Wei OL, Phogat S, Sidorov IA, Fouts TR, Broder CC, Dimitrov DS. Conserved structures exposed in HIV-1 envelope glycoproteins stabilized by flexible linkers as potent entry inhibitors and potential immunogens. Biochemistry. 2002;41:7176–7182. doi: 10.1021/bi025646d. [DOI] [PubMed] [Google Scholar]
- 21.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Miiro G, Serwanga J, Pozniak A, McPhee D, Manigart O, Mwananyanda L, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Allen S, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New HIV-1 Vaccine Target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava IK, Stamatatos L, Kan E, Vajdy M, Lian Y, Hilt S, Martin L, Vita C, Zhu P, Roux KH, Vojtech L, Montefiori C, Donnelly J, Ulmer JB, Barnett SW. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J Virol. 2003;77:11244–11259. doi: 10.1128/JVI.77.20.11244-11259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 2003;313:387–400. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 24.Chakrabarti BK, Kong WP, Wu BY, Yang ZY, Friborg J, Ling X, King SR, Montefiori DC, Nabel GJ. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76:5357–5368. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaCasse RA, Follis KE, Trahey M, Scarborough JD, Littman DR, Nunberg JH. Fusion-competent vaccines: broad neutralization of primary isolates of HIV (Retraction: Science 2002, 296, 1025) Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]
- 26.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 27.Murphy K, Travers P, Walport M. Janeway’s Immunobiology. 7th ed. Garland Science; New York, NY, USA: 2008. [Google Scholar]
- 28.Gerhard W, Yewdell J, Frankel ME, Webster R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature. 1981;290:713–717. doi: 10.1038/290713a0. [DOI] [PubMed] [Google Scholar]
- 29.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 30.Clark HF, Offit PA, Plotkin SA, Heaton PM. The new pentavalent rotavirus vaccine composed of bovine (strain WC3) -human rotavirus reassortants. Pediatr Infect Dis J. 2006;25:577–583. doi: 10.1097/01.inf.0000220283.58039.b6. [DOI] [PubMed] [Google Scholar]
- 31.Krogstad P, Cherry JD. Quadrivalent human vaccine - a call to action and for additional research. Pediatr Res. 2007;62:527. doi: 10.1203/PDR.0b013e31815b445b. [DOI] [PubMed] [Google Scholar]
- 32.Biagini RE, Schlottmann SA, Sammons DL, Smith JP, Snawder JC, Striley CA, MacKenzie BA, Weissman DN. Method for simultaneous measurement of antibodies to 23 pneumococcal capsular polysaccharides. Clin Diagn Lab Immunol. 2003;10:744–750. doi: 10.1128/CDLI.10.5.744-750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vesikari T, Wysocki J, Chevallier B, Karvonen A, Czajka H, Arsene JP, Lommel P, Dieussaert I, Schuerman L. Immunogenicity of the 10-valent pneumococcal non-typeable Haemophilus influenzae protein D conjugate vaccine (PHiD-CV) compared to the licensed 7vCRM vaccine. Pediatr Infect Dis J. 2009;28:S66–S76. doi: 10.1097/INF.0b013e318199f8ef. [DOI] [PubMed] [Google Scholar]
- 34.Hambleton S. Prevention of varicella and zoster by live attenuated VZV vaccine. Front Biosci. 2008;13:2696–2704. doi: 10.2741/2876. [DOI] [PubMed] [Google Scholar]
- 35.Varicella-related deaths--United States, January 2003– June 2004. MMWR Morb Mortal Wkly Rep. 2005;54:272–274. [PubMed] [Google Scholar]
- 36.Bjorklund A, Aschan J, Labopin M, Remberger M, Ringden O, Winiarski J, Ljungman P. Risk factors for fatal infectious complications developing late after allogeneic stem cell transplantation. Bone Marrow Transpl. 2007;40:1055–1062. doi: 10.1038/sj.bmt.1705856. [DOI] [PubMed] [Google Scholar]
- 37.Murphey-Corb M, Martin LN, vison-Fairburn B, Montelaro RC, Miller M, West M, Ohkawa S, Baskin GB, Zhang JY, Putney SD. A formalin-inactivated whole SIV vaccine confers protection in macaques. Science. 1989;246:1293–1297. doi: 10.1126/science.2555923. [DOI] [PubMed] [Google Scholar]
- 38.Carmichael AJ, Sissons JG. Vaccines against HIV. QJM. 1995;88:77–79. [PubMed] [Google Scholar]
- 39.Stott EJ. Anti-cell antibody in macaques. Nature. 1991;353:393. doi: 10.1038/353393a0. [DOI] [PubMed] [Google Scholar]
- 40.Sheppard HW. Inactivated- or Killed-Virus HIV/AIDS Vaccines. Curr Drug Targets Infect Disord. 2005;5:131–141. doi: 10.2174/1568005054201599. [DOI] [PubMed] [Google Scholar]
- 41.Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 42.Hofmann-Lehmann R, Vlasak J, Williams AL, Chenine AL, McClure HM, Anderson DC, O’Neil S, Ruprecht RM. Live attenuated, nef-deleted SIV is pathogenic in most adult macaques after prolonged observation. AIDS. 2003;17:157–166. doi: 10.1097/00002030-200301240-00004. [DOI] [PubMed] [Google Scholar]
- 43.Stahl-Hennig C, Dittmer U, Nisslein T, Pekrun K, Petry H, Jurkiewicz E, Fuchs D, Wachter H, Rud EW, Hunsmann G. Attenuated SIV imparts immunity to challenge with pathogenic spleen-derived SIV but cannot prevent repair of the nef deletion. Immunol Lett. 1996;51:129–135. doi: 10.1016/0165-2478(96)02567-9. [DOI] [PubMed] [Google Scholar]
- 44.Whatmore AM, Cook N, Hall GA, Sharpe S, Rud EW, Cranage MP. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J Virol. 1995;69:5117–5123. doi: 10.1128/jvi.69.8.5117-5123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baba TW, Jeong YS, Pennick D, Bronson R, Greene MF, Ruprecht RM. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 46.Wyand MS, Manson KH, Garcia-Moll M, Montefiori D, Desrosiers RC. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sealy R, Zhan X, Lockey TD, Martin L, Blanchard J, Traina-Dorge V, Hurwitz JL. SHIV infection protects against heterologous pathogenic SHIV challenge in macaques: a gold-standard for HIV-1 vaccine development. Curr HIV Res. 2009;7:497–503. doi: 10.2174/157016209789346255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cranage MP, Whatmore AM, Sharpe SA, Cook N, Polyanskaya N, Leech S, Smith JD, Rud EW, Dennis MJ, Hall GA. Macaques infected with live attenuated SIVmac are protected against superinfection via the rectal mucosa. Virology. 1997;229:143–154. doi: 10.1006/viro.1996.8419. [DOI] [PubMed] [Google Scholar]
- 49.Stahl-Hennig C, Dittmer U, Nisslein T, Petry H, Jurkiewicz E, Fuchs D, Wachter H, Matz-Rensing K, Kuhn EM, Kaup FJ, Rud EW, Hunsmann G. Rapid development of vaccine protection in macaques by live-attenuated simian immunodeficiency virus. J Gen Virol. 1996;77:2969–2981. doi: 10.1099/0022-1317-77-12-2969. [DOI] [PubMed] [Google Scholar]
- 50.Stephens EB, Joag SV, Atkinson B, Sahni M, Li Z, Foresman L, Adany I, Narayan O. Infected macaques that controlled replication of SIVmac or nonpathogenic SHIV developed sterilizing resistance against pathogenic SHIV(KU-1) Virology. 1997;234:328–339. doi: 10.1006/viro.1997.8662. [DOI] [PubMed] [Google Scholar]
- 51.Maniatis T, Hardison RC, Lacy E, Lauer J, O’Connell C, Quon D, Sim GK, Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978;15:687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- 52.Grunstein M, Hogness DS. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci USA. 1975;72:3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shubeita HE, Sambrook JF, McCormick AM. Molecular cloning and analysis of functional cDNA and genomic clones encoding bovine cellular retinoic acid-binding protein. Proc Natl Acad Sci USA. 1987;84:5645–5649. doi: 10.1073/pnas.84.16.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breton M, Zhao C, Ouellette M, Tremblay MJ, Papadopoulou B. A recombinant non-pathogenic Leishmania vaccine expressing human immunodeficiency virus 1 (HIV-1) Gag elicits cell-mediated immunity in mice and decreases HIV-1 replication in human tonsillar tissue following exposure to HIV-1 infection. J Gen Virol. 2007;88:217–225. doi: 10.1099/vir.0.81995-0. [DOI] [PubMed] [Google Scholar]
- 55.Farmer JL, Hampton RG, Boots E. Flow cytometric assays for monitoring production of recombinant HIV-1 gp160 in insect cells infected with a baculovirus expression vector. J Virol Meth. 1989;26:279–290. doi: 10.1016/0166-0934(89)90110-9. [DOI] [PubMed] [Google Scholar]
- 56.Hanke T, Barnfield C, Wee EG, Agren L, Samuel RV, Larke N, Liljestrom P. Construction and immunogenicity in a prime-boost regimen of a Semliki Forest virus-vectored experimental HIV clade A vaccine. J Gen Virol. 2003;84:361–368. doi: 10.1099/vir.0.18738-0. [DOI] [PubMed] [Google Scholar]
- 57.Berglund P, Quesada-Rolander M, Putkonen P, Biberfeld G, Thorstensson R, Liljestrom P. Outcome of immunization of cynomolgus monkeys with recombinant Semliki Forest virus encoding human immunodeficiency virus type 1 envelope protein and challenge with a high dose of SHIV-4 virus. Aids Res Hum Retroviruses. 1997;13:1487–1495. doi: 10.1089/aid.1997.13.1487. [DOI] [PubMed] [Google Scholar]
- 58.Davis NL, West A, Reap E, MacDonald G, Collier M, Dryga S, Maughan M, Connell M, Walker C, McGrath K, Cecil C, Ping LH, Frelinger J, Olmsted R. Alphavirus replicon particles as candidate HIV vaccines. IUBMB Life. 2002;53:209–211. doi: 10.1080/15216540212657. [DOI] [PubMed] [Google Scholar]
- 59.Lubeck MD, Natuk RJ, Chengalvala M, Chanda PK, Murthy KK, Murthy S, Mizutani S, Lee SG, Wade MS, Bhat BM. Immunogenicity of recombinant adenovirus-human immunodeficiency virus vaccines in chimpanzees following intranasal administration. Aids Res Hum Retroviruses. 1994;10:1443–1449. doi: 10.1089/aid.1994.10.1443. [DOI] [PubMed] [Google Scholar]
- 60.Xin KQ, Urabe M, Yang J, Nomiyama K, Mizukami H, Hamajima K, Nomiyama H, Saito T, Imai M, Monahan J, Okuda K, Ozawa K, Okuda K. A novel recombinant adeno-associated virus vaccine induces a long-term humoral immune response to human immunodeficiency virus. Hum Gene Ther. 2001;12:1047–1061. doi: 10.1089/104303401750214276. [DOI] [PubMed] [Google Scholar]
- 61.Lin J, Calcedo R, Vandenberghe LH, Bell P, Somanathan S, Wilson JM. A new genetic vaccine platform based on an adeno-associated virus isolated from a rhesus macaque. J Virol. 2009;83:12738–12750. doi: 10.1128/JVI.01441-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rencher SD, Lockey TD, Srinivas RV, Owens RJ, Hurwitz JL. Eliciting HIV-1 envelope-specific antibodies with mixed vaccinia virus recombinants. Vaccine. 1997;15:265–272. doi: 10.1016/s0264-410x(96)00185-5. [DOI] [PubMed] [Google Scholar]
- 63.Amara RR, Villinger F, Altman JD, Lydy SL, O’Neil SP, Staprans SI, Montefiori DC, Xu Y, Herndon JG, Wyatt LS, Candido MA, Kozyr NL, Earl PL, Smith JM, Ma HL, Grimm BD, Hulsey ML, McClure HM, McNicholl JM, Moss B, Robinson HL. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Vaccine. 2002;20:1949–1955. doi: 10.1016/s0264-410x(02)00076-2. [DOI] [PubMed] [Google Scholar]
- 64.Amara RR, Smith JM, Staprans SI, Montefiori DC, Villinger F, Altman JD, O’Neil SP, Kozyr NL, Xu Y, Wyatt LS, Earl PL, Herndon JG, McNicholl JM, McClure HM, Moss B, Robinson HL. Critical role for Env as well as Gag-Pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J Virol. 2002;76:6138–6146. doi: 10.1128/JVI.76.12.6138-6146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clements-Mann ML, Weinhold K, Matthews TJ, Graham BS, Gorse GJ, Keefer MC, McElrath MJ, Hsieh R-H, Mestecky J, Zolla-Pazner S, Mascola J, Schwartz D, Siliciano R, Corey L, Wright PF, Belshe R, Dolin R, Jackson S, Xu S, Fast P, Walker MC, Stablein D, Excler J-L, Tartaglia J, Duliege A-M, Sinangil F, Paoletti E. NIAID AIDS VACCINE EVALUATION GROUP, Immune responses to human immunodeficiency virus (HIV) type 1 induced by canarypox expressing HIV-1-MN gp120, HIV-1-SF2 recombinant gp120, or both vaccines in seronegative adults. J Infect Dis. 1998;177:1246. doi: 10.1086/515288. [DOI] [PubMed] [Google Scholar]
- 66.Pialoux G, Excler J-L, Rivière Y, Gonzalez-Canali G, Feuillie V, Coulaud P, Gluckman J-C, Matthews TJ, Meignier B, Kieny M-P, Gonnet P, Diaz I, Méric C, Paoletti E, Tartaglia J, Salomon H, Plotkin S, The Agis Group L’Agence Natl de Recherche sur le Sida, A prime-boost approach to HIV preventive vaccine using a recombinant canarypox virus expressing glycoprotein 160 (MN) followed by a recombinant glycoprotein 160 (MN/LAI) Aids Res Hum Retroviruses. 1995;11:373–381. doi: 10.1089/aid.1995.11.373. [DOI] [PubMed] [Google Scholar]
- 67.Kent SJ, Zhao A, Best SJ, Chandler JD, Boyle DB, Ramshaw IA. Enhanced T-cell immunogenicity and protective efficacy of a human immunodeficiency virus type 1 vaccine regimen consisting of consecutive priming with DNA and boosting with recombinant fowlpox virus. J Virol. 1998;72:10180–10188. doi: 10.1128/jvi.72.12.10180-10188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stubbs AC, Martin KS, Coeshott C, Skaates SV, Kuritzkes DR, Bellgrau D, Franzusoff A, Duke RC, Wilson CC. Whole recombinant yeast vaccine activates dendritic cells and elicits protective cell-mediated immunity. Nat Med. 2001;7:625–629. doi: 10.1038/87974. [DOI] [PubMed] [Google Scholar]
- 69.Haglund K, Leiner I, Kerksiek K, Buonocore L, Pamer E, Rose JK. Robust recall and long-term memory T-cell responses induced by prime-boost regimens with heterologous live viral vectors expressing human immunodeficiency virus type 1 Gag and Env proteins. J Virol. 2002;76:7506–7517. doi: 10.1128/JVI.76.15.7506-7517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z, Zhang M, Zhou C, Zhao X, Iijima N, Frankel FR. Novel vaccination protocol with two live mucosal vectors elicits strong cell-mediated immunity in the vagina and protects against vaginal virus challenge. J Immunol. 2008;180:2504–2513. doi: 10.4049/jimmunol.180.4.2504. [DOI] [PubMed] [Google Scholar]
- 71.Humbert M, Rasmussen RA, Ong H, Kaiser FM, Hu SL, Ruprecht RM. Inducing cross-clade neutralizing antibodies against HIV-1 by immunofocusing. PLoS ONE. 2008;3:e3937. doi: 10.1371/journal.pone.0003937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bridgeman A, Roshorm Y, Lockett LJ, Xu ZZ, Hopkins R, Shaw J, Both GW, Hanke T. Ovine atadenovirus, a novel and highly immunogenic vector in prime-boost studies of a candidate HIV-1 vaccine. Vaccine. 2009;28:474–483. doi: 10.1016/j.vaccine.2009.09.136. [DOI] [PubMed] [Google Scholar]
- 73.Ranganathan UD, Larsen MH, Kim J, Porcelli SA, Jacobs WR, Jr, Fennelly GJ. Recombinant pro-apoptotic Mycobacterium tuberculosis generates CD8+ T cell responses against human immunodeficiency virus type 1 Env and M. tuberculosis in neonatal mice. Vaccine. 2009;28:152–161. doi: 10.1016/j.vaccine.2009.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kiem HP, Wu RA, Sun G, von LD, Rossi JJ, Trobridge GD. Foamy combinatorial anti-HIV vectors with MGMTP140K potently inhibit HIV-1 and SHIV replication and mediate selection in vivo. Gene Ther. 2010;17:37–49. doi: 10.1038/gt.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Goede AL, Boers PH, Dekker LJ, Osterhaus AD, Gruters RA, Rimmelzwaan GF. Characterization of recombinant influenza A virus as a vector for HIV-1 p17Gag. Vaccine. 2009;27:5735–5739. doi: 10.1016/j.vaccine.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 76.Miller JP, Geng Y, Ng HL, Yang OO, Krogstad P. Packaging limits and stability of HIV-1 sequences in a coxsackievirus B vector. Vaccine. 2009;27:3992–4000. doi: 10.1016/j.vaccine.2009.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lemiale F, Korokhov N. Lentiviral vectors for HIV disease prevention and treatment. Vaccine. 2009;27:3443–3449. doi: 10.1016/j.vaccine.2009.01.055. [DOI] [PubMed] [Google Scholar]
- 78.Chin’ombe N, Bourn WR, Williamson AL, Shephard EG. Oral vaccination with a recombinant Salmonella vaccine vector provokes systemic HIV-1 subtype C Gag-specific CD4+ Th1 and Th2 cell immune responses in mice. Virol J. 2009;6:87. doi: 10.1186/1743-422X-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chege GK, Thomas R, Shephard EG, Meyers A, Bourn W, Williamson C, Maclean J, Gray CM, Rybicki EP, Williamson AL. A prime-boost immunisation regimen using recombinant BCG and Pr55(gag) virus-like particle vaccines based on HIV type 1 subtype C successfully elicits Gag-specific responses in baboons. Vaccine. 2009;27:4857–4866. doi: 10.1016/j.vaccine.2009.05.064. [DOI] [PubMed] [Google Scholar]
- 80.Kaur A, Sanford HB, Garry D, Lang S, Klumpp SA, Watanabe D, Bronson RT, Lifson JD, Rosati M, Pavlakis GN, Felber BK, Knipe DM, Desrosiers RC. Ability of herpes simplex virus vectors to boost immune responses to DNA vectors and to protect against challenge by simian immunodeficiency virus. Virology. 2007;357:199–214. doi: 10.1016/j.virol.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kent SJ, De RR, Mokhonov VV, Mokhonova EI, Fernandez CS, Alcantara S, Rollman E, Mason RD, Loh L, Peut V, Reece JC, Wang XJ, Wilson KM, Suhrbier A, Khromykh A. Evaluation of recombinant Kunjin replicon SIV vaccines for protective efficacy in macaques. Virology. 2008;374:528–534. doi: 10.1016/j.virol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 82.Liniger M, Zuniga A, Morin TN, Combardiere B, Marty R, Wiegand M, Ilter O, Knuchel M, Naim HY. Recombinant measles viruses expressing single or multiple antigens of human immunodeficiency virus (HIV-1) induce cellular and humoral immune responses. Vaccine. 2009;27:3299–3305. doi: 10.1016/j.vaccine.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu R, Nasar F, Megati S, Luckay A, Lee M, Udem SA, Eldridge JH, Egan MA, Emini E, Clarke DK. Prime-Boost Vaccination with Recombinant Mumps Virus and Recombinant Vesicular Stomatitis Virus Vectors Elicits an Enhanced HIV-1 Gag-Specific Cellular Immune Response in Rhesus Macaques. J Virol. 2009 doi: 10.1128/JVI.00550-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Faul EJ, Wanjalla CN, McGettigan JP, Schnell MJ. Interferon-beta expressed by a rabies virus-based HIV-1 vaccine vector serves as a molecular adjuvant and decreases pathogenicity. Virology. 2008;382:226–238. doi: 10.1016/j.virol.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCabe MS, Klaas M, Gonzalez-Rabade N, Poage M, Badillo-Corona JA, Zhou F, Karcher D, Bock R, Gray JC, Dix PJ. Plastid transformation of high-biomass tobacco variety Maryland Mammoth for production of human immunodeficiency virus type 1 (HIV-1) p24 antigen. Plant Biotechnol J. 2008 doi: 10.1111/j.1467-7652.2008.00365.x. [DOI] [PubMed] [Google Scholar]
- 86.Robinson HL, Hunt LA, Webster RG. Protection against a lethal influenza virus challenge by immunization with a haemagglutinin-expressing plasmid DNA. Vaccine. 1993;11:957–960. doi: 10.1016/0264-410x(93)90385-b. [DOI] [PubMed] [Google Scholar]
- 87.Webster RG, Fynan EF, Santoro JC, Robinson H. Protection of ferrets against influenza challenge with a DNA vaccine to the haemagglutinin. Vaccine. 1994;12:1495–1498. doi: 10.1016/0264-410x(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 88.Peet N, Delves PJ, de Souza B, Cambridge G, Wilkinson L, Klonisch T, Matear P, Vyakarnam A, Lund T. The immune response to HIV gp120 induced by nucleic acid immunization (NAI) Ann NY Acad Sci. 1995;772:257–260. doi: 10.1111/j.1749-6632.1995.tb44753.x. [DOI] [PubMed] [Google Scholar]
- 89.Yao Q, Bu Z, Vzorov A, Yang C, Compans RW. Virus-like particle and DNA-based candidate AIDS vaccines. Vaccine. 2003;21:638–643. doi: 10.1016/s0264-410x(02)00572-8. [DOI] [PubMed] [Google Scholar]
- 90.Bristow RG, Douglas AR, Skehel JJ, Daniels RS. Analysis of murine antibody responses to baculovirus-expressed human immunodeficiency virus type 1 envelope glycoproteins. J Gen Virol. 1994;75:2089–2095. doi: 10.1099/0022-1317-75-8-2089. [DOI] [PubMed] [Google Scholar]
- 91.Raychaudhuri S, Tonks M, Carbone F, Ryskamp T, Morrow WJ, Hanna N. Induction of antigen-specific class I-restricted cytotoxic T cells by soluble proteins in vivo. Proc Natl Acad Sci USA. 1992;89:8308–8312. doi: 10.1073/pnas.89.17.8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crotty S, Felgner P, Davies H, Glidewell J, Villarreal L, Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 93.Amanna IJ, Slifka MK, Crotty S. Immunity and immunological memory following smallpox vaccination. Immunol Rev. 2006;211:320–337. doi: 10.1111/j.0105-2896.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- 94.Hyland L, Sangster M, Sealy R, Coleclough C. Respiratory virus infection of mice provokes a permanent humoral immune response. J Virol. 1994;68:6083–6086. doi: 10.1128/jvi.68.9.6083-6086.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barnett SW, Srivastava IK, Ulmer JB, Donnelly JJ, Rappuoli R. Development of V2-deleted trimeric envelope vaccine candidates from human immunodeficiency virus type 1 (HIV-1) subtypes B and C. Microbes Infect. 2005;7:1386–1391. doi: 10.1016/j.micinf.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 96.Novitsky V, Smith UR, Gilbert P, McLane MF, Chigwedere P, Williamson C, Ndung’u T, Klein I, Chang SY, Peter T, Thior I, Foley BT, Gaolekwe S, Rybak N, Gaseitsiwe S, Vannberg F, Marlink R, Lee TH, Essex M. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design. J Virol. 2002;76:5435–5451. doi: 10.1128/JVI.76.11.5435-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 98.Thurmond J, Yoon H, Kuiken C, Yusim K, Perkins S, Theiler J, Bhattacharya T, Korber B, Fischer W. Web-based design and evaluation of T-cell vaccine candidates. Bioinformatics. 2008;24:1639–1640. doi: 10.1093/bioinformatics/btn251. [DOI] [PubMed] [Google Scholar]
- 99.Weaver EA, Lu Z, Camacho ZT, Moukdar F, Liao HX, Ma BJ, Muldoon M, Theiler J, Nabel GJ, Letvin NL, Korber BT, Hahn BH, Haynes BF, Gao F. Cross-subtype T-cell immune responses induced by a human immunodeficiency virus type 1 group m consensus env immunogen. J Virol. 2006;80:6745–6756. doi: 10.1128/JVI.02484-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Earl PL, Koenig S, Moss B. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J Virol. 1991;65:31–41. doi: 10.1128/jvi.65.1.31-41.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thomson SA, Jaramillo AB, Shoobridge M, Dunstan KJ, Everett B, Ranasinghe C, Kent SJ, Gao K, Medveckzy J, Ffrench RA, Ramshaw IA. Development of a synthetic consensus sequence scrambled antigen HIV-1 vaccine designed for global use. Vaccine. 2005;23:4647–4657. doi: 10.1016/j.vaccine.2005.04.045. [DOI] [PubMed] [Google Scholar]
- 102.D’Costa S, Slobod KS, Webster RG, White SW, Hurwitz JL. Structural features of HIV envelope defined by antibody escape mutant analysis. Aids Res Hum Retroviruses. 2001;17:1205–1209. doi: 10.1089/088922201316912808. [DOI] [PubMed] [Google Scholar]
- 103.Sealy R, Chaka W, Surman S, Brown SA, Cresswell P, Hurwitz JL. Target peptide sequence within infectious human immunodeficiency virus type 1 does not ensure envelope-specific T-helper cell reactivation: influences of cysteine protease and gamma interferon-induced thiol reductase activities. Clin Vaccine Immunol. 2008;15:713–719. doi: 10.1128/CVI.00412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maric M, Arunachalam B, Phan UT, Dong C, Garrett WS, Cannon KS, Alfonso C, Karlsson L, Flavell RA, Cresswell P. Defective antigen processing in GILT-free mice. Science. 2001;294:1361–1365. doi: 10.1126/science.1065500. [DOI] [PubMed] [Google Scholar]
- 105.Sjolander S, Bolmstedt A, Akerblom L, Horal P, Olofsson S, Morein B, Sjolander A. N-linked glycans in the CD4-binding domain of human immunodeficiency virus type 1 envelope glycoprotein gp160 are essential for the in vivo priming of T cells recognizing an epitope located in their vicinity. Virology. 1996;215:124–133. doi: 10.1006/viro.1996.0015. [DOI] [PubMed] [Google Scholar]
- 106.Esparza J, Heyward WL, Osmanov S. HIV vaccine development: from basic research to human trials. AIDS. 1996;10(Suppl. A):S123–S132. [PubMed] [Google Scholar]
- 107.Lu S. Heterologous prime-boost vaccination. Curr Opin Immunol. 2009;21:346–351. doi: 10.1016/j.coi.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hu SL, Klaniecki J, Dykers T, Sridhar P, Travis BM. Neutralizing antibodies against HIV-1 BRU and SF2 isolates generated in mice immunized with recombinant vaccinia virus expressing HIV-1 (BRU) envelope glycoproteins and boosted with homologous gp160. Aids Res Hum Retroviruses. 1991;7:615–620. doi: 10.1089/aid.1991.7.615. [DOI] [PubMed] [Google Scholar]
- 109.Hu S-L, Abrams K, Barber GN, Moran P, Zarling JM, Langlois AJ, Kuller L, Morton WR, Benveniste RE. Protection of macaques against SIV infection by subunit vaccines of SIV envelope glycoprotein gp160. Science. 1992;255:456–459. doi: 10.1126/science.1531159. [DOI] [PubMed] [Google Scholar]
- 110.Richmond JFL, Mustafa F, Lu S, Santoro JC, Weng J, O’Connell M, Fenyo EM, Hurwitz JL, Montefiori DC, Robinson HL. Screening of HIV-1 Env glycoproteins for the ability to raise neutralizing antibody using DNA immunization and recombinant vaccinia virus boosting. Virology. 1997;230:265–274. doi: 10.1006/viro.1997.8478. [DOI] [PubMed] [Google Scholar]
- 111.Ramsay AJ, Leong KH, Ramshaw IA. DNA vaccination against virus infection and enhancement of antiviral immunity following consecutive immunization with DNA and viral vectors. Immunol Cell Biol. 1997;75:382–388. doi: 10.1038/icb.1997.60. [DOI] [PubMed] [Google Scholar]
- 112.Ramshaw IA, Ramsay AJ. The prime-boost strategy: exciting prospects for improved vaccination. Immunol Today JID - 8008346. 2000;21:163–165. doi: 10.1016/s0167-5699(00)01612-1. [DOI] [PubMed] [Google Scholar]
- 113.Estcourt MJ, Ramsay AJ, Brooks A, Thomson SA, Medveckzy CJ, Ramshaw IA. Prime-boost immunization generates a high frequency, high-avidity CD8(+) cytotoxic T lymphocyte population. Int Immunol. 2002;14:31–37. doi: 10.1093/intimm/14.1.31. [DOI] [PubMed] [Google Scholar]
- 114.McConkey SJ, Reece WH, Moorthy VS, Webster D, Dunachie S, Butcher G, Vuola JM, Blanchard TJ, Gothard P, Watkins K, Hannan CM, Everaere S, Brown K, Kester KE, Cummings J, Williams J, Heppner DG, Pathan A, Flanagan K, Arulanantham N, Roberts MT, Roy M, Smith GL, Schneider J, Peto T, Sinden RE, Gilbert SC, Hill AV. Enhanced T-cell immunogenicity of plasmid DNA vaccines boosted by recombinant modified vaccinia virus Ankara in humans. Nat Med. 2003;9:729–735. doi: 10.1038/nm881. [DOI] [PubMed] [Google Scholar]
- 115.Shu Y, Winfrey S, Yang ZY, Xu L, Rao SS, Srivastava I, Barnett SW, Nabel GJ, Mascola JR. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine. 2007;25:1398–1408. doi: 10.1016/j.vaccine.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harari A, Bart PA, Stohr W, Tapia G, Garcia M, Medjitna-Rais E, Burnet S, Cellerai C, Erlwein O, Barber T, Moog C, Liljestrom P, Wagner R, Wolf H, Kraehenbuhl JP, Esteban M, Heeney J, Frachette MJ, Tartaglia J, McCormack S, Babiker A, Weber J, Pantaleo G. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J Exp Med. 2008;205:63–77. doi: 10.1084/jem.20071331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seaman MS, Xu L, Beaudry K, Martin KL, Beddall MH, Miura A, Sambor A, Chakrabarti BK, Huang Y, Bailer R, Koup RA, Mascola JR, Nabel GJ, Letvin NL. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J Virol. 2005;79:2956–2963. doi: 10.1128/JVI.79.5.2956-2963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Egan MA, Chong SY, Megati S, Montefiori DC, Rose NF, Boyer JD, Sidhu MK, Quiroz J, Rosati M, Schadeck EB, Pavlakis GN, Weiner DB, Rose JK, Israel ZR, Udem SA, Eldridge JH. Priming with plasmid DNAs expressing interleukin-12 and simian immunodeficiency virus gag enhances the immunogenicity and efficacy of an experimental AIDS vaccine based on recombinant vesicular stomatitis virus. Aids Res Hum Retroviruses. 2005;21:629–643. doi: 10.1089/aid.2005.21.629. [DOI] [PubMed] [Google Scholar]
- 119.Pal R, Kalyanaraman VS, Nair BC, Whitney S, Keen T, Hocker L, Hudacik L, Rose N, Mboudjeka I, Shen S, Wu-Chou TH, Montefiori D, Mascola J, Markham P, Lu S. Immunization of rhesus macaques with a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine elicits protective antibody response against simian human immunodeficiency virus of R5 phenotype. Virology. 2006;348:341–353. doi: 10.1016/j.virol.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 120.Santra S, Sun Y, Parvani JG, Philippon V, Wyand MS, Manson K, Gomez-Yafal A, Mazzara G, Panicali D, Markham PD, Montefiori DC, Letvin NL. Heterologous prime/boost immunization of rhesus monkeys by using diverse poxvirus vectors. J Virol. 2007;81:8563–8570. doi: 10.1128/JVI.00744-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lubeck MD, Natuk R, Myagkikh M, Kalyan N, Aldrich K, Sinangil F, Alipanah S, Murthy SC, Chanda PK, Nigida SM, Jr, Markham PD, Zolla-Pazner S, Steimer K, Wade M, Reitz MS, Jr, Arthur LO, Mizutani S, Davis A, Hung PP, Gallo RC, Eichberg J, Robert-Guroff M. Long-term protection of chimpanzees against high-dose HIV-1 challenge induced by immunization. Nat Med. 1997;3:651–658. doi: 10.1038/nm0697-651. [DOI] [PubMed] [Google Scholar]
- 122.Zhao J, Pinczewski J, Gomez-Roman VR, Venzon D, Kalyanaraman VS, Markham PD, Aldrich K, Moake M, Montefiori DC, Lou Y, Pavlakis GN, Robert-Guroff M. Improved protection of rhesus macaques against intrarectal simian immunodeficiency virus SIV(mac251) challenge by a replication-competent Ad5hr-SIVenv/rev and Ad5hr-SIVgag recombinant priming/gp120 boosting regimen. J Virol. 2003;77:8354–8365. doi: 10.1128/JVI.77.15.8354-8365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peng B, Wang LR, Gomez-Roman VR, vis-Warren A, Montefiori DC, Kalyanaraman VS, Venzon D, Zhao J, Kan E, Rowell TJ, Murthy KK, Srivastava I, Barnett SW, Robert-Guroff M. Replicating rather than nonreplicating adenovirus-human immunodeficiency virus recombinant vaccines are better at eliciting potent cellular immunity and priming high-titer antibodies. J Virol. 2005;79:10200–10209. doi: 10.1128/JVI.79.16.10200-10209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Patterson LJ, Robert-Guroff M. Replicating adenovirus vector prime/protein boost strategies for HIV vaccine development. Expert Opin Biol Ther. 2008;8:1347–1363. doi: 10.1517/14712598.8.9.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tatsis N, Lasaro MO, Lin SW, Xiang ZQ, Zhou D, Dimenna L, Li H, Bian A, Abdulla S, Li Y, Giles-Davis W, Engram J, Ratcliffe SJ, Silvestri G, Ertl HC, Betts MR. Adenovirus vector-induced immune responses in nonhuman primates: responses to prime boost regimens. J Immunol. 2009;182:6587–6599. doi: 10.4049/jimmunol.0900317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Santra S, Sun Y, Korioth-Schmitz B, Fitzgerald J, Charbonneau C, Santos G, Seaman MS, Ratcliffe SJ, Montefiori DC, Nabel GJ, Ertl HC, Letvin NL. Heterologous prime/boost immunizations of rhesus monkeys using chimpanzee adenovirus vectors. Vaccine. 2009;27:5837–5845. doi: 10.1016/j.vaccine.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fix A. Vaccine Briefs: Phase II prime-boost trial begins in the US. IAVI Report, The publication on AIDS Vaccine Research July– August 2009.
- 128.Catanzaro AT, Roederer M, Koup RA, Bailer RT, Enama ME, Nason MC, Martin JE, Rucker S, Andrews CA, Gomez PL, Mascola JR, Nabel GJ, Graham BS. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine. 2007;25:4085–4092. doi: 10.1016/j.vaccine.2007.02.050. [DOI] [PubMed] [Google Scholar]
- 129.Robinson HL. Working towards an HIV/AIDS vaccine. Hum Vaccin. 2009;5:436–438. doi: 10.4161/hv.9324. [DOI] [PubMed] [Google Scholar]
- 130.Brave A, Boberg A, Gudmundsdotter L, Rollman E, Hallermalm K, Ljungberg K, Blomberg P, Stout R, Paulie S, Sandstrom E, Biberfeld G, Earl P, Moss B, Cox JH, Wahren B. A new multi-clade DNA prime/recombinant MVA boost vaccine induces broad and high levels of HIV-1-specific CD8(+) T-cell and humoral responses in mice. Mol Ther. 2007;15:1724–1733. doi: 10.1038/sj.mt.6300235. [DOI] [PubMed] [Google Scholar]
- 131.Gudmundsdotter L, Nilsson C, Brave A, Hejdeman B, Earl P, Moss B, Robb M, Cox J, Michael N, Marovich M, Biberfeld G, Sandstrom E, Wahren B. Recombinant Modified Vaccinia Ankara (MVA) effectively boosts DNA-primed HIV-specific immune responses in humans despite pre-existing vaccinia immunity. Vaccine. 2009;27:4468–4474. doi: 10.1016/j.vaccine.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Robinson HL, Sharma S, Zhao J, Kannanganat S, Lai L, Chennareddi L, Yu T, Montefiori DC, Amara RR, Wyatt LS, Moss B. Immunogenicity in macaques of the clinical product for a clade B DNA/MVA HIV vaccine: elicitation of IFN-gamma, IL-2, and TNF-alpha coproducing CD4 and CD8 T cells. Aids Res Hum Retroviruses. 2007;23:1555–1562. doi: 10.1089/aid.2007.0165. [DOI] [PubMed] [Google Scholar]
- 133.Rollman E, Hinkula J, Arteaga J, Zuber B, Kjerrstrom A, Liu M, Wahren B, Ljungberg K. Multi-subtype gp160 DNA immunization induces broadly neutralizing anti-HIV antibodies. Gene Ther. 2004;11:1146–1154. doi: 10.1038/sj.gt.3302275. [DOI] [PubMed] [Google Scholar]
- 134.Rollman E, Brave A, Boberg A, Gudmundsdotter L, Engstrom G, Isaguliants M, Ljungberg K, Lundgren B, Blomberg P, Hinkula J, Hejdeman B, Sandstrom E, Liu M, Wahren B. The rationale behind a vaccine based on multiple HIV antigens. Microbes Infect. 2005;7:1414–1423. doi: 10.1016/j.micinf.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 135.Brave A, Ljungberg K, Boberg A, Rollman E, Isaguliants M, Lundgren B, Blomberg P, Hinkula J, Wahren B. Multigene/multisubtype HIV-1 vaccine induces potent cellular and humoral immune responses by needle-free intradermal delivery. Mol Ther. 2005;12:1197–1205. doi: 10.1016/j.ymthe.2005.06.473. [DOI] [PubMed] [Google Scholar]
- 136.Wang S, Kennedy JS, West K, Montefiori DC, Coley S, Lawrence J, Shen S, Green S, Rothman AL, Ennis FA, Arthos J, Pal R, Markham P, Lu S. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine. 2008;26:1098–1110. doi: 10.1016/j.vaccine.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bansal A, Jackson B, West K, Wang S, Lu S, Kennedy JS, Goepfert PA. Multifunctional T-cell characteristics induced by a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine regimen given to healthy adults are dependent on the route and dose of administration. J Virol. 2008;82:6458–6469. doi: 10.1128/JVI.00068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rencher SD, Slobod KS, Dawson D, Lockey TD, Hurwitz JL. Does the key to a successful HIV vaccine lie among the envelope sequences of infected individuals. Aids Res Hum Retroviruses. 1995;11:1131–1133. doi: 10.1089/aid.1995.11.1131. [DOI] [PubMed] [Google Scholar]
- 139.Slobod KS, Hurwitz JL. Does envelope V3 peptide comprise the principal neutralizing determinant for HIV. Vaccine. 1995;13:129. doi: 10.1016/0264-410x(95)80027-b. [DOI] [PubMed] [Google Scholar]
- 140.Ljungberg K, Rollman E, Eriksson L, Hinkula J, Wahren B. Enhanced immune responses after DNA vaccination with combined envelope genes from different HIV-1 subtypes. Virology. 2002;302:44–57. doi: 10.1006/viro.2002.1547. [DOI] [PubMed] [Google Scholar]
- 141.Hurwitz JL, Slobod KS, Lockey TD, Wang S, Chou TH, Lu S. Application of the Polyvalent Approach to HIV-1 Vaccine Development. Curr Drug Targets Infect Disord. 2005;5:143–156. doi: 10.2174/1568005054201517. [DOI] [PubMed] [Google Scholar]
- 142.Chakrabarti BK, Ling X, Yang ZY, Montefiori DC, Panet A, Kong WP, Welcher B, Louder MK, Mascola JR, Nabel GJ. Expanded breadth of virus neutralization after immunization with a multiclade envelope HIV vaccine candidate. Vaccine. 2005;23:3434–3445. doi: 10.1016/j.vaccine.2005.01.099. [DOI] [PubMed] [Google Scholar]
- 143.Kong W-P, Huang Y, Yang Z-Y, Chakrabarti BK, Moodie Z, Nabel GJ. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J Virol. 2003;77:12764–12772. doi: 10.1128/JVI.77.23.12764-12772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Slobod KS, Lockey TD, Howlett N, Srinivas RV, Rencher SD, Freiden PJ, Doherty PC, Hurwitz JL. Subcutaneous administration of a recombinant vaccinia virus vaccine expressing multiple envelopes of HIV-1. Eur J Clin Microbiol Infect Dis. 2004;23:106–110. doi: 10.1007/s10096-003-1075-3. [DOI] [PubMed] [Google Scholar]
- 145.Sealy R, Slobod KS, Flynn P, Branum K, Surman S, Jones B, Freiden P, Lockey T, Howlett N, Hurwitz JL. Preclinical and clinical development of a multi-envelope, DNA-virus-protein (D-V-P) HIV-1 vaccine. Int Rev Immunol. 2009;28:49–68. doi: 10.1080/08830180802495605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Caver TE, Lockey TD, Srinivas RV, Webster RG, Hurwitz JL. A novel vaccine regimen utilizing DNA, vaccinia virus and protein immunizations for HIV-1 envelope presentation. Vaccine. 1999;17:1567–1572. doi: 10.1016/s0264-410x(98)00355-7. [DOI] [PubMed] [Google Scholar]
- 147.Stambas J, Brown SA, Gutierrez A, Sealy R, Yue W, Jones B, Lockey TD, Zirkel A, Freiden P, Brown B, Surman S, Coleclough C, Slobod KS, Doherty PC, Hurwitz JL. Long lived multi-isotype anti-HIV antibody responses following a prime-double boost immunization strategy. Vaccine. 2005;23:2454–2464. doi: 10.1016/j.vaccine.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 148.Hurwitz JL, Zhan X, Brown SA, Bonsignori M, Stambas J, Lockey TD, Sealy R, Surman S, Freiden P, Jones B, Martin L, Blanchard J, Slobod KS. HIV-1 vaccine development: Tackling virus diversity with a multi-envelope cocktail. Frontiers Bioscience. 2008;13:609–620. doi: 10.2741/2706. [DOI] [PubMed] [Google Scholar]
- 149.Slobod KS, Brown SA, Surman S, Zirkel A, Jones B, Zhan X, Sealy B, Stambas J, Brown B, Bonsignori M, Lockey TD, Freiden P, Hurwitz JL, Doherty PC, Coleclough C. Overcoming diversity with a multi-envelope HIV vaccine. Current Topics in Virology. 2004;4:159–168. [Google Scholar]
- 150.Yu S, Feng X, Shu T, Matano T, Hasegawa M, Wang X, Ma H, Li H, Li Z, Zeng Y. Potent specific immune responses induced by prime-boost-boost strategies based on DNA, adenovirus, and Sendai virus vectors expressing gag gene of Chinese HIV-1 subtype B. Vaccine. 2008;26:6124–6131. doi: 10.1016/j.vaccine.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 151.Slifka MK, Antia R, Whitmire JK, Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity. 1998;8:363–372. doi: 10.1016/s1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- 152.Brown SA, Hurwitz JL, Zirkel A, Surman S, Takimoto T, Alymova I, Coleclough C, Portner A, Doherty PC, Slobod KS. A recombinant Sendai virus is controlled by CD4+ effector T cells responding to a secreted human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 2007;81:12535–12542. doi: 10.1128/JVI.00197-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Matano T, Kano M, Nakamura H, Takeda A, Nagai Y. Rapid appearance of secondary immune responses and protection from acute CD4 depletion after a highly pathogenic immunodeficiency virus challenge in macaques vaccinated with a DNA prime/Sendai virus vector boost regimen. J Virol. 2001;75:11891–11896. doi: 10.1128/JVI.75.23.11891-11896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wherry EJ, McElhaugh MJ, Eisenlohr LC. Generation of CD8(+) T cell memory in response to low, high, and excessive levels of epitope. J Immunol. 2002;168:4455–4461. doi: 10.4049/jimmunol.168.9.4455. [DOI] [PubMed] [Google Scholar]
- 155.Park SO, Han YW, Aleyas AG, George JA, Yoon HA, Lee JH, Kang HY, Kang SH, Eo SK. Low-dose antigen-experienced CD4(+) T cells display reduced clonal expansion but facilitate an effective memory pool in response to secondary exposure. Immunology. 2008;123:426–437. doi: 10.1111/j.1365-2567.2007.02707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Moyes CD, Milne A, Waldon J. Very low dose hepatitis B vaccination in the newborn: anamnestic response to booster at four years. J Med Virol. 1990;30:216–218. doi: 10.1002/jmv.1890300314. [DOI] [PubMed] [Google Scholar]
- 157.Liu J, Hellerstein M, McDonnel M, Amara RR, Wyatt LS, Moss B, Robinson HL. Dose-response studies for the elicitation of CD8 T cells by a DNA vaccine, used alone or as the prime for a modified vaccinia Ankara boost. Vaccine. 2007;25:2951–2958. doi: 10.1016/j.vaccine.2006.05.081. [DOI] [PubMed] [Google Scholar]
- 158.Smith JM, Amara RR, McClure HM, Patel M, Sharma S, Yi H, Chennareddi L, Herndon JG, Butera ST, Heneine W, Ellenberger DL, Parekh B, Earl PL, Wyatt LS, Moss B, Robinson HL. Multiprotein HIV type 1 clade B DNA/MVA vaccine: construction, safety, and immunogenicity in Macaques. Aids Res Hum Retroviruses. 2004;20:654–665. doi: 10.1089/0889222041217419. [DOI] [PubMed] [Google Scholar]
- 159.Barouch DH, Liu J, Lynch DM, O’Brien KL, La PA, Simmons NL, Riggs AM, Clark S, Abbink P, Montefiori DC, Landucci G, Forthal DN, Self SG, Carville A, Mansfield K, Goudsmit J. Protective efficacy of a single immunization of a chimeric adenovirus vector-based vaccine against simian immunodeficiency virus challenge in rhesus monkeys. J Virol. 2009;83:9584–9590. doi: 10.1128/JVI.00821-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. [DOI] [PubMed] [Google Scholar]
- 161.Stone R. Swine flu outbreak. China first to vaccinate against novel H1N1 virus. Science. 2009;325:1482–1483. doi: 10.1126/science.325_1482. [DOI] [PubMed] [Google Scholar]
- 162.Fazekas de St.Groth S, Webster RG. Disquisitions on original antigenic sin. I. Evidence in man. J Exp Med. 1966;124:331–345. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fazekas de St.Groth S, Webster RG. Disquisitions on original antigenic sin. II. Proof in lower creatures. J Exp Med. 1966;124:347–361. doi: 10.1084/jem.124.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim JH, Skountzou I, Compans R, Jacob J. Original antigenic sin responses to influenza viruses. J Immunol. 2009;183:3294–3301. doi: 10.4049/jimmunol.0900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gearhart PJ, Hurwitz JL, Cebra JJ. Successive switching of antibody isotype expressed within the lines of a B-cell clone. Proc Natl Acad Sci USA. 1980;77:5424–5428. doi: 10.1073/pnas.77.9.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gearhart PJ, Sigal NH, Klinman NR. Heterogeneity of the BALB/c antiphosphorylcholine antibody response at the precursor cell level. J Exp Med. 1975;141:56–71. doi: 10.1084/jem.141.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Klinman DM, Higgins KW, Conover J. Sequential immunizations with rgp120s from independent isolates of human immunodeficiency virus type 1 induce the preferential expansion of broadly crossreactive B cells. JExpMed. 1991;173:881–887. doi: 10.1084/jem.173.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhan X, Slobod KS, Surman S, Brown SA, Coleclough C, Hurwitz JL. Minor components of a multi-envelope HIV vaccine are recognized by type-specific T-helper cells. Vaccine. 2004;22:1206–1213. doi: 10.1016/j.vaccine.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 170.Surman S, Lockey TD, Slobod KS, Jones B, Riberdy JM, White SW, Doherty PC, Hurwitz JL. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc Natl Acad Sci USA. 2001;98:4587–4592. doi: 10.1073/pnas.071063898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Brown SA, Stambas J, Zhan X, Slobod KS, Coleclough C, Zirkel A, Surman S, White SW, Doherty PC, Hurwitz JL. Clustering of Th cell epitopes on exposed regions of HIV envelope despite defects in antibody activity. J Immunol. 2003;171:4140–4148. doi: 10.4049/jimmunol.171.8.4140. [DOI] [PubMed] [Google Scholar]
- 172.Wang Y, Flesch IE, Tscharke DC. Vaccinia virus CD8+ T-cell dominance hierarchies cannot be altered by prior immunization with individual peptides. J Virol. 2009;83:9008–9012. doi: 10.1128/JVI.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Berman PW, Gray AM, Wrin T, Vennari JC, Eastman DJ, Nakamura GR, Francis DP, Gorse G, Schwartz DH. Genetic and immunologic characterization of viruses infecting MN-rgp120-vaccinated volunteers. J Infect Dis. 1997;176:384–397. doi: 10.1086/514055. [DOI] [PubMed] [Google Scholar]
- 174.Gonzales MJ, Delwart E, Rhee SY, Tsui R, Zolopa AR, Taylor J, Shafer RW. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J Infect Dis. 2003;188:397–405. doi: 10.1086/376534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tsui R, Herring BL, Barbour JD, Grant RM, Bacchetti P, Kral A, Edli BR, Delwart EL. Human immunodeficiency virus type 1 superinfection was not detected following 215 years of injection drug user exposure. J Virol. 2004;78:94–103. doi: 10.1128/JVI.78.1.94-103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Slobod KS, Coleclough C, Bonsignori M, Brown SA, Zhan X, Surman S, Zirkel A, Jones BG, Sealy RE, Stambas J, Brown B, Lockey TD, Freiden PJ, Doherty PC, Blanchard JL, Martin LN, Hurwitz JL. HIV Vaccine Rationale, Design and Testing. Curr HIV Res. 2005;3:107–112. doi: 10.2174/1570162053506928. [DOI] [PubMed] [Google Scholar]
- 177.Zhan X, Martin LN, Slobod KS, Coleclough C, Lockey TD, Brown SA, Stambas J, Bonsignori M, Sealy RE, Blanchard JL, Hurwitz JL. Multi-envelope HIV-1 vaccine devoid of SIV components controls disease in macaques challenged with heterologous pathogenic SHIV. Vaccine. 2005;23:5306–5320. doi: 10.1016/j.vaccine.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 178.Hurwitz JL, Lockey TD, Jones B, Freiden P, Sealy R, Coleman J, Howlett N, Branum K, Slobod KS. First phase I clinical trial of an HIV-1 subtype D gp140 envelope protein vaccine: immune activity induced in all study participants. AIDS. 2008;22:149–151. doi: 10.1097/QAD.0b013e3282f174ed. [DOI] [PubMed] [Google Scholar]
- 179.Nara PL, Lin G. HIV-1: The Confounding Variables of Virus Neutralization. Curr Drug Targets Infect Disord. 2005;5:157–170. doi: 10.2174/1568005054201535. [DOI] [PubMed] [Google Scholar]
- 180.Lian Y, Srivastava I, Gomez-Roman VR, zur MJ, Sun Y, Kan E, Hilt S, Engelbrecht S, Himathongkham S, Luciw PA, Otten G, Ulmer JB, Donnelly JJ, Rabussay D, Montefiori D, van Rensburg EJ, Barnett SW. Evaluation of envelope vaccines derived from the South African subtype C human immunodeficiency virus type 1 TV1 strain. J Virol. 2005;79:13338–13349. doi: 10.1128/JVI.79.21.13338-13349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bures R, Gaitan A, Zhu T, Graziosi C, McGrath KM, Tartaglia J, Caudrelier P, El Habib R, Klein M, Lazzarin A, Stablein DM, Deers M, Corey L, Greenberg ML, Schwartz DH, Montefiori DC. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. Aids Res Hum Retroviruses. 2000;16:2019–2035. doi: 10.1089/088922200750054756. [DOI] [PubMed] [Google Scholar]
- 182.Hosoi S, Borsos T, Dunlop N, Nara PL. Heat-labile, complement-like factor(s) of animal sera prevent(s) HIV-1 infectivity in vitro. JAIDS. 1990;3:366–371. [PubMed] [Google Scholar]
- 183.Van Rompay KK, Berardi CJ, Dillard-Telm S, Tarara RP, Canfield DR, Valverde CR, Montefiori DC, Cole KS, Montelaro RC, Miller CJ, Marthas ML. Passive immunization of newborn rhesus macaques prevents oral simian immunodeficiency virus infection. J Infect Dis. 1998;177:1247–1259. doi: 10.1086/515270. [DOI] [PubMed] [Google Scholar]
- 184.Forthal DN, Landucci G, Cole KS, Marthas M, Becerra JC, Van RK. Rhesus macaque polyclonal and monoclonal antibodies inhibit simian immunodeficiency virus in the presence of human or autologous rhesus effector cells. J Virol. 2006;80:9217–9225. doi: 10.1128/JVI.02746-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Holl V, Hemmerter S, Burrer R, Schmidt S, Bohbot A, Aubertin AM, Moog C. Involvement of Fc gamma RI (CD64) in the mechanism of HIV-1 inhibition by polyclonal IgG purified from infected patients in cultured monocyte-derived macrophages. J Immunol. 2004;173:6274–6283. doi: 10.4049/jimmunol.173.10.6274. [DOI] [PubMed] [Google Scholar]
- 186.Gomez-Roman VR, Florese RH, Patterson LJ, Peng B, Venzon D, Aldrich K, Robert-Guroff M. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J Immunol Methods. 2006;308:53–67. doi: 10.1016/j.jim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 187.Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, Hanifin JM, Slifka MK. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 188.Jones PD, Ada GL. Persistence of influenza virus-specific antibody-secreting cells and B-cell memory after primary murine influenza virus infection. Cell Immunol. 1987;109:53–64. doi: 10.1016/0008-8749(87)90291-7. [DOI] [PubMed] [Google Scholar]
- 189.Schwarze J, O’Donnell DR, Rohwedder A, Openshaw PJ. Latency and persistence of respiratory syncytial virus despite T cell immunity. Am J Respir Crit Care Med. 2004;169:801–805. doi: 10.1164/rccm.200308-1203OC. [DOI] [PubMed] [Google Scholar]
- 190.Turner SJ, Diaz G, Cross R, Doherty PC. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8+ T cell response. Immunity. 2003;18:549–559. doi: 10.1016/s1074-7613(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 191.Gray D. A role for antigen in the maintenance of immunological memory. Nat Rev Immunol. 2002;2:60–65. doi: 10.1038/nri706. [DOI] [PubMed] [Google Scholar]
- 192.Maruyama M, Lam KP, Rajewsky K. Memory B-cell persistence is independent of persisting immunizing antigen. Nature. 2000;407:636–642. doi: 10.1038/35036600. [DOI] [PubMed] [Google Scholar]