Abstract
Class switch recombination (CSR) diversifies antibodies by joining highly repetitive DNA elements, which are separated by 60–200 kbp. CSR is initiated by activation-induced cytidine deaminase, an enzyme that produces multiple DNA double-strand breaks (DSBs) in switch regions. Switch regions are joined by a mechanism that requires an intact DNA damage response and classical or alternative nonhomologous end joining (A-NHEJ). Among the DNA damage response factors, 53BP1 has the most profound effect on CSR. We explore the role of 53BP1 in intrachromosomal DNA repair using I-SceI to introduce paired DSBs in the IgH locus. We find that the absence of 53BP1 results in an ataxia telangiectasia mutated–dependent increase in DNA end resection and that resected DNA is preferentially repaired by microhomology-mediated A-NHEJ. We propose that 53BP1 favors long-range CSR in part by protecting DNA ends against resection, which prevents A-NHEJ–dependent short-range rejoining of intra–switch region DSBs.
Class switch recombination (CSR) is an Ig gene diversification reaction that alters the Ig effector function while retaining antibody specificity. During CSR, one constant region gene is exchanged for another by a deletional recombination reaction between switch regions, which are highly repetitive DNA sequences that precede constant region genes (Chaudhuri et al., 2007; Stavnezer et al., 2008). CSR occurs in the G1 phase of the cell cycle and proceeds through obligate double-strand break (DSB) intermediates (Petersen et al., 2001; Rada et al., 2002). The reaction is initiated by activation-induced cytidine deaminase (AID), an enzyme that deaminates cytidine residues in single-stranded switch region DNA that is exposed during Ig transcription (Petersen-Mahrt et al., 2002; Bransteitter et al., 2003; Chaudhuri et al., 2003; Dickerson et al., 2003; Pham et al., 2003; Ramiro et al., 2003). AID produces multiple lesions in switch DNA, and the resulting U/G mismatches are processed to DSBs via the base excision and mismatch repair pathways (Catalan et al., 2003; Schrader et al., 2005; Stavnezer et al., 2008).
There are several ways to resolve switch region DSBs. For example, a single DSB can be repaired by religation, or two paired DSBs within a single switch region can be ligated together to produce an intra–switch region deletion (ISD; Dudley et al., 2002). Alternatively, synapsis and ligation of paired DSBs in two different switch regions leads to productive CSR (Stavnezer et al., 2008). Finally, in rare instances, an Ig DSB can be joined to a DSB on a heterologous chromosome to produce a translocation (Ramiro et al., 2004, 2006; Franco et al., 2006).
Switch region DSBs occur during G1, and they are ligated by either classical nonhomologous end joining (C-NHEJ; Manis et al., 2002; Lieber, 2008; Stavnezer et al., 2008) or alternative nonhomologous end joining (A-NHEJ; Yan et al., 2007; Haber, 2008; Boboila et al., 2010a,b). C-NHEJ is an evolutionarily conserved pathway that utilizes Ku70/80 and DNA-PKcs for DSB recognition, and ligase IV–XRCC4 for ligation (Lieber, 2008). Deletion of any of these factors impairs CSR (Callén et al., 2007; Soulas-Sprauel et al., 2007; Yan et al., 2007; Boboila et al., 2010a,b). C-NHEJ ligates DSBs with little or no microhomology and appears to be the dominant pathway involved in CSR, based on the physiological predominance of blunt or small microhomology switch joins (Yan et al., 2007; Boboila et al., 2010a,b). In contrast, little is known about the factors that mediate A-NHEJ (Haber, 2008). However, this is a robust pathway that makes extensive use of junctional microhomologies and can reconstitute up to 20–50% of CSR in the absence of C-NHEJ (Yan et al., 2007; Boboila et al., 2010a,b). A-NHEJ appears to be kinetically slower than C-NHEJ and mediates many of the translocations that are rare byproducts of V(D)J recombination and CSR (Zhu et al., 2002; Han and Yu, 2008; Wang et al., 2008, 2009; Xie et al., 2009; Boboila et al., 2010b).
DSBs incurred during CSR activate the DNA damage response, as indicated by the accumulation of foci of Mre11/Rad50/Nbs1 (MRN), H2AX, and 53BP1 on the IgH locus during CSR (Petersen et al., 2001; Reina-San-Martin et al., 2003). DNA damage response factors are also required for efficient CSR. Deficiency in any of these or ataxia telangiectasia mutated (ATM), a key mediator of the DNA damage response, leads to inefficient switching and concomitant accumulation of DNA damage on chromosome 12 (Reina-San-Martin et al., 2004; Franco et al., 2006; Ramiro et al., 2006; Jankovic et al., 2007). Among DNA damage response factors, the most pronounced defect in CSR occurs upon loss of 53BP1, a chromatin binding protein that is also an ATM and DNA-PKcs substrate (DiTullio et al., 2002; Manis et al., 2004; Ward et al., 2004; Callén et al., 2007). Similarly, the absence of 53BP1 leads to a joining defect between distal DSBs during V(D)J recombination and a reduced rate of transchromosomal fusions of deprotected telomeres (Difilippantonio et al., 2008; Dimitrova et al., 2008). In contrast, ISDs and chromosome translocations, both of which appear to be mediated primarily by A-NHEJ, are either increased or not affected by the loss of 53BP1 (Ramiro et al., 2006; Reina-San-Martin et al., 2007). In addition, overexpression of a dominant-negative fragment of 53BP1 enhances homologous recombination (HR), as determined using a reporter substrate (Xie et al., 2007).
Several nonmutually exclusive models have been proposed to account for the effects of 53BP1 on CSR, V(D)J recombination, HR, telomere fusion, and chromosome translocation. 53BP1 may enhance synapsis and long-range interactions between two distal DSBs (Manis et al., 2004; Ward et al., 2004; Reina-San-Martin et al., 2007), possibly by altering local chromatin structure (Difilippantonio et al., 2008) or increasing chromatin mobility (Dimitrova et al., 2008). Alternatively, it may regulate repair by favoring one end-joining pathway or another, and thereby control the relative importance of microhomology in the joining process. However, there is little direct experimental data to support these ideas.
In this paper, we report experiments that investigate the function of 53BP1 using the I-SceI meganuclease to introduce site-specific DSBs in IgH in B lymphocytes undergoing CSR. We show that 53BP1 regulates the choice of end-joining pathways by interfering with resection, thereby favoring C-NHEJ.
RESULTS
IgH locus modification
To examine DSB repair in the context of IgH during CSR, we used sequential gene targeting in embryonic stem cells to introduce paired loxP and I-SceI sites in mouse chromosome 12. One set of loxP and I-SceI sites was placed 3′ of the intronic enhancer Eμ (Robbiani et al., 2008), and the second set of sites was introduced 3′ of the IgG1 switch region, 95.9 kb downstream of the first. (IgHI-96k; Fig. S1, A and B). IgHI-96k mice showed normal B cell development and CSR to IgG1 upon stimulation with LPS and IL-4 (Fig. S1, C and D).
In contrast to the DNA lesions introduced by AID, which can involve almost any cytidine residue in the switch region and are processed by a variety of different pathways (Stavnezer et al., 2008), the introduced loxP and I-SceI sites are in specific locations and are targeted by enzymes with well-defined mechanisms of action. Cre is a bacteriophage enzyme that mediates recombination between loxP sites via Holliday junction intermediates (Gopaul et al., 1998). Synapsis between two molecules of Cre bound to separate loxP sites precedes and is required for catalysis (Van Duyne, 2001). Because synapsis is limiting in this reaction, the rate of recombination between loxP sites in mammalian cells expressing Cre is inversely proportional to the distance between the sites and can therefore serve as a measure of chromosome topology (Yu and Bradley, 2001; Egli et al., 2004). In contrast, I-SceI, a yeast-derived restriction meganuclease not normally present in the mouse genome, produces a single DSB in an 18-bp consensus sequence (Stoddard, 2005). When this enzyme is expressed in B lymphocytes containing an I-SceI site integrated into the IgH locus, DNA breaks are detectable at a high frequency at IgH, and translocations can form between I-SceI– and AID-generated breaks (Zarrin et al., 2007; Robbiani et al., 2008).
Recombination between loxP or I-SceI sites in IgHI-96k B lymphocytes should result in class switching from IgM to IgG1. However, IgH genes are allelically excluded (Chowdhury and Sen, 2004), with only 50% of the alleles encoding a functional variable region. Therefore, the maximum rate of Cre- or I-SceI–mediated CSR measured by IgG1 surface expression in heterozygous mice is 50% of the true recombination frequency.
Switching by Cre-mediated recombination
Although the mechanism that mediates synapsis between switch regions is poorly understood, chromosome conformation capture PCR experiments suggest that AID may contribute to long-range interactions between switch regions (Wuerffel et al., 2007). To determine whether AID expression alters IgH topology sufficiently to change the rate of Cre-mediated recombination between switch regions, we compared Cre-induced CSR in AID-deficient IgHI-96k B cells and control IgHI-96k B cells (Fig. 1).
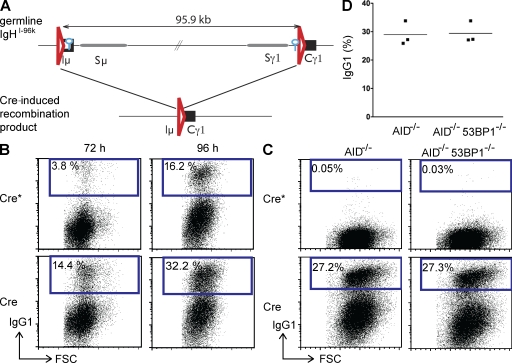
Figure 1.
Cre recombinase induces efficient CSR to IgG1 independently of AID and 53BP1. (A) Schematic representation of the IgHI-96k allele (top) and the Cre-induced recombinant that encodes IgG1 (bottom). LoxP sites are indicated as red triangles, and I-SceI sites are indicated as blue circles. (B) Representative flow cytometry experiments showing CSR to IgG1 of IgHI-96k B cells infected with retroviruses encoding Cre or catalytically inactive Cre*. IgG1 expression was analyzed at 72 and 96 h after LPS and IL-4 stimulation. (C) Experiment as in B but for IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells analyzed at 96 h after LPS and IL-4 stimulation. (D) Graph shows the results of three independent flow cytometry experiments measuring CSR to IgG1 after Cre infection of IgHI-96AID−/− and IgHI-96kAID−/−53BP1−/− B cells. The means are shown as horizontal lines. FSC, forward scatter.
B cells were stimulated with LPS and IL-4, and infected with a Cre-encoding retrovirus or an inactive retrovirus Cre*. Infected cells were identified by GFP expression and CSR was measured by flow cytometry. Cre expression in AID-sufficient IgHI-96k B lymphocytes increased CSR to IgG1 from 3.8 to 14.4% and from 16.2 to 32.2% at 72 and 96 h after stimulation, respectively (Fig. 1 B). However, the rate of CSR in IgHI-96k B cells was similar in the presence or absence of AID 96 h after stimulation (Fig. 1, B and C).
To determine whether the loss of 53BP1 alters the efficiency of Cre-mediated CSR, we compared the frequency of recombination between IgHI-96kAID−/−53BP1−/− and IgHI-96kAID−/− B cells. We found that CSR in Cre-infected IgHI-96kAID−/−53BP1−/− B cells was indistinguishable from 53BP1-proficient cells (Fig. 1, C and D). We conclude that neither AID nor 53BP1 alters the overall structure of the IgH locus sufficiently to change the rate of synapsis between the IgHI-96k loxP sites.
I-SceI endonuclease–induced CSR
To further investigate the role of 53BP1 in CSR, we made use of I-SceI, which produces DSBs independently of synapsis. IgHI-96kAID−/− B cells were stimulated with LPS and IL-4, and infected with retroviruses encoding I-SceI or an inactive form of the enzyme I-SceI* (Fig. S2). Switching to IgG1 by retrovirally infected B cells was initially measured by flow cytometry (Fig. 2 B).
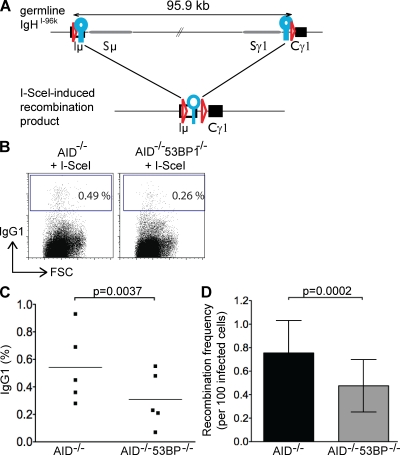
Figure 2.
Loss of 53BP1 decreases the joining efficiency of two distal I-SceI–induced DSBs. (A) Schematic representation of the IgHI-96k allele (top) and the I-SceI–induced recombinant that encodes IgG1 (bottom). LoxP sites are indicated as red triangles, and I-SceI sites are indicated as blue circles. (B) Representative flow cytometry experiments showing CSR to IgG1 of IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells 72 h after the first infection with an I-SceI–encoding retrovirus. (C) Graph shows the results of five independent flow cytometry experiments, with each dot representing an individual experiment. The p-value was calculated using a two-tailed paired Student’s t test. The means are shown as horizontal lines. (D) Bar graph showing I-SceI to I-SceI recombination frequency in the presence and absence of 53BP1, determined by nine independent PCR experiments. Error bars indicate standard deviation. The p-value was calculated using a two-tailed paired Student’s t test. FSC, forward scatter.
In contrast to Cre, I-SceI mediated CSR was inefficient in IgHI-96kAID−/− B cells, with a mean recombination frequency of 0.54% 3 d after retroviral infection (five independent experiments; Fig. 2 C and Table S1). The observed rate of I-SceI–induced CSR was somewhat lower than the rate obtained in B cells derived by RAG-blastocyst complementation (Zarrin et al., 2007). However, our experiments differ from previous experiments in several respects: AID was deleted and switch regions were present in our experiments, also, we used only a single I-SceI site in each location as opposed to two I-SceI sites, which doubles the number of breaks.
To determine whether loss of 53BP1 affects the joining efficiency of I-SceI breaks, we assayed IgHI-96kAID−/−53BP1−/− B cells. In contrast to Cre, where we observed no effects of 53BP1 deficiency, the I-SceI recombination frequency was significantly reduced in IgHI-96kAID−/−53BP1−/− B cells to 0.31% (five independent experiments; P = 0.0037; Fig. 2 C and Table S1). To obtain an independent measure of recombination frequency, we examined joining between I-SceI sites by sample dilution PCR. In agreement with the flow cytometry analysis, we found that the joining efficiency in stimulated IgHI-96kAID−/− B lymphocytes was significantly reduced from 0.76 to 0.48% in the absence of 53BP1 (P = 0.0002; Fig. 2 D and Table S2). We conclude that loss of 53BP1 decreases the efficiency of recombination between I-SceI sites in IgHI-96kAID−/− B cells; however, the effect is far less penetrant than it is for CSR.
53BP1 prevents processing of broken ends
53BP1 has been implicated in regulating the choice between HR and NHEJ DNA repair pathways in the S or G2 phase of the cell cycle. CSR does not appear to involve HR and occurs in the G1 phase of the cell cycle (Petersen et al., 2001). Nevertheless, CSR-induced DSBs can be joined by two different pathways: C-NHEJ or A-NHEJ (Yan et al., 2007; Boboila et al., 2010a,b).
To determine whether 53BP1 affects the choice between C-NHEJ and A-NHEJ, we characterized the joins between I-SceI sites from IgHI-96kAID−/−53BP1−/− B cells and IgHI-96kAID−/− controls. Precise joining between I-SceI sites yields a 336-nt PCR product (Fig. 3 A), which was the predominant species in control IgHI-96kAID−/− B cells (Fig. 3 B). End processing results in lower molecular weight species, which can be scored directly by counting the number of products that run ≤300 nt (i.e., more than ∼30 nt total end resection). The loss of 53BP1 resulted in an overall increase in the number of PCR products, with lower than expected molecular weight to 51.6% of all products. (P = 0.0001; Fig. 3 C and Table S3).
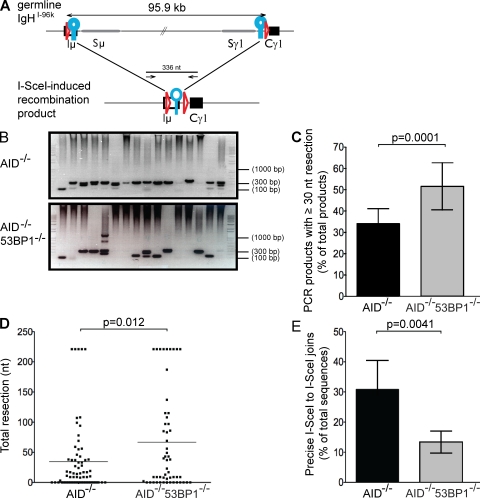
Figure 3.
Loss of 53BP1 leads to increased end resection. (A) Schematic representation of IgHI-96k allele (top) with the PCR primers used to amplify a 336-nt recombination product, indicated as arrows (bottom). LoxP sites are indicated as red triangles, and I-SceI sites are indicated as blue circles. (B) Representative ethidium bromide–stained argarose gels showing PCR products obtained after I-SceI–induced recombination in IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells. (C) Bar graphs showing frequency of I-SceI–induced recombination products running ≤300 nt for IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells, determined by 12 independent PCR experiments. Error bars indicate standard deviation. The p-value was calculated using a two-tailed Student’s t test. (D) Dot plot showing total resection of I-SceI–infected IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells. Each dot represents one sequence. The p-value was calculated using a two-tailed Student’s t test. The means are shown as horizontal lines. (E) Bar graph shows the frequency of perfect I-SceI joins in I-SceI–infected IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells in five independent experiments. Error bars indicate standard deviation. The p-value was calculated using a two-tailed Student’s t test.
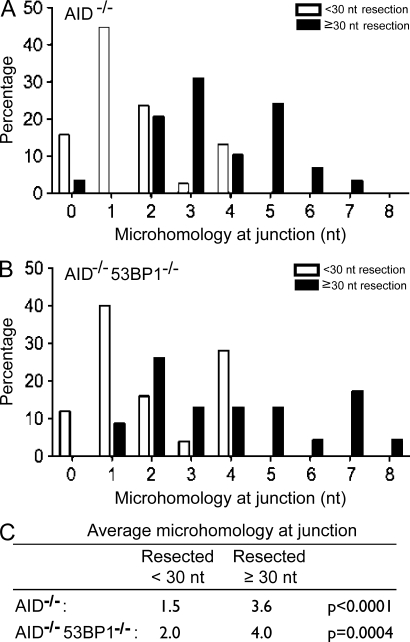
The precise molecular structure of the joins was determined by sequencing. Although the mean extent of end processing was 34.7 nt in IgHI-96kAID−/− B cells, it was 66.8 nt in the absence of 53BP1 (P = 0.012; Fig. 3 D and Table S4). Consistent with the idea that ends are more frequently processed in the absence of 53BP1, the number of precise joins that reconstituted the I-SceI site also decreased from 30.9 to 13.3% in 53BP1−/− B cells (P = 0.0041; Fig. 3 E and Table S5). Interestingly, all of the minimally processed joins showed 0–4 nt of junctional microhomology, with a mean of 1.5 and 2 nt for AID−/− and AID−/−53BP1−/− B cells, respectively, which is characteristic of C-NHEJ (Fig. 4 and Fig. S3). In contrast, the majority of joining events that involved extensive resection (≥30 nt) used 3 nt or more of junctional microhomology, with a mean of 3.6 and 4 nt for AID−/− and AID−/−53BP1−/− B cells, respectively, indicative of A-NHEJ (P < 0.0001 for AID−/−; P = 0.0004 for AID−/−53BP1−/−; Fig. 4 C and Fig. S3). These data show that loss of 53BP1 leads to increased resection of DNA ends, and that resection is associated with microhomology-mediated end joining irrespective of 53BP1.
Figure 4.
Resected DSBs are joined using microhomology-mediated end joining independently of 53BP1. (A) Bar graph shows microhomology at junctions of PCR products cloned from I-SceI–infected IgHI-96kAID−/− B cells. White bars indicate sequences with little resection (<30 nt), and black bars indicate sequences with resection ≥30 nt. A total of 67 individual sequences were analyzed from three different mice. (B) Bar graph shows microhomology at junctions of PCR products cloned from I-SceI–infected IgHI-96kAID−/−53BP1−/− B cells. White bars indicate sequences with little resection (<30 nt), and black bars indicate sequences with resection ≥30 nt. A total of 48 individual sequences were analyzed from three different mice. (C) Table indicates the mean number of nucleotides of microhomology at junctions in I-SceI–infected IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells.
Inhibition of ATM kinase decreases resection
DNA end resection by combined action of MRN, CtIP, Bloom’s helicase, and Exo1 is essential for HR (Mimitou and Symington, 2009). Optimal resection and formation of the single-stranded DNA (ssDNA) substrate for HR requires ATM, because this kinase facilitates the recruitment and activation of the nucleases that attack DNA ends (Jazayeri et al., 2006; You et al., 2009). Although HR does not appear to be involved in CSR, we investigated whether a related mechanism is involved in processing DNA ends for microhomology-based A-NHEJ.
To determine how the loss of ATM activity might affect the processing of I-SceI breaks, we treated IgHI-96kAID−/− B cells with a small molecule ATM inhibitor (ATMi). In contrast to loss of 53BP1, ATMi did not significantly affect the joining rate of paired I-SceI breaks on the IgH locus, as measured by flow cytometry (Fig. 5, A and B; and Table S6). However, ATMi-treated IgHI-96kAID−/− B cells showed significantly reduced DNA end resection (17.8% ATMi vs. 34.1% control; P = 0.0024; Fig. 5, C and D; and Tables S3 and S7) and a concomitant increase in the number of precise I-SceI joins (47.6% ATMi treated vs. 30.9% control; P = 0.026; Fig. 5 E; and Tables S5 and S8).
Figure 5.
Increased DNA end resection in the absence of 53BP1 is dependent on ATM. (A) Representative flow cytometry experiment showing CSR to IgG1 by IgHI-96kAID−/− B cells infected with an I-SceI–encoding retrovirus in the presence or absence of ATMi. (B) Graph shows the results of three independent flow cytometry experiments, with each dot representing an individual experiment. The means are shown as horizontal lines. (C) Representative ethidium bromide–stained argarose gels showing the PCR amplification products after I-SceI–induced recombination of IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells in the presence or absence of ATMi. (D) Bar graph showing the frequency of I-SceI–induced recombination products running ≤300 nt for IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells in the presence or absence of ATMi. Error bars indicate standard deviation. p-values were calculated using a two-tailed Student’s t test (three independent experiments). (E) Frequency of direct I-SceI joins, reconstituting an I-SceI site, determined by sequencing of individual molecular products of I-SceI–infected IgHI-96kAID−/− and IgHI-96kAID−/− 53BP1−/− B cells in the presence or absence of ATMi. Error bars indicate standard deviation. p-values were calculated using a two-tailed Student’s t test (three independent sequencing experiments). FSC, forward scatter.
To determine whether the increased end resection in the absence of 53BP1 is ATM dependent, we examined end resection in ATMi-treated IgHI-96kAID−/−53BP1−/− B cells. ATMi induced a pronounced decrease in end resection in IgHI-96kAID−/−53BP1−/− B cells (18.1% ATMi treated vs. 51.6% untreated IgHI-96kAID−/−53BP1−/− control; P = 0.0003; Fig. 5, C and D; and Tables S3 and S7) and a concomitant increase in precise I-SceI joins (34.2% ATMi vs. 13.3% untreated control; P = 0.0021; Fig. 5 E; and Tables S5 and S8). Thus, the increase in I-SceI–induced DNA end processing observed in the absence of 53BP1 is dependent on ATM.
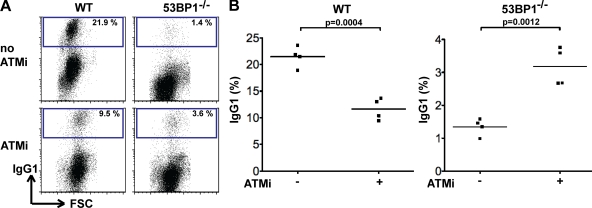
Loss of 53BP1 interferes with CSR but enhances recombination between repeat DNA within switch regions (Manis et al., 2004; Ward et al., 2004; Ramiro et al., 2006; Reina-San-Martin et al., 2007). Because the latter requires DNA end processing, we examined whether inhibition of DNA end processing by ATMi might enhance CSR in 53BP1−/− B cells. In agreement with published studies (Lumsden et al., 2004; Reina-San-Martin et al., 2004; Callén et al., 2007), ATMi reduces CSR to IgG1 in WT B cells (11.6% ATMi vs. 21.5% untreated control; P = 0.0004; Fig. 6, A and B; and Table S9). In striking contrast, ATMi enhanced CSR in 53BP1−/− B cells (3.2% ATMi vs. 1.3% untreated controls; P = 0.0012; Fig. 6, A and B; and Table S9). We conclude that ATMi ameliorates the severe defect in CSR observed in the absence of 53BP1.
Figure 6.
Inhibition of ATM leads to partial rescue of the CSR defect in 53BP1−/− B cells. (A) Representative flow cytometry experiment showing CSR to IgG1 by WT and 53BP1−/− B cells 96 h after LPS, IL-4, and RP105 stimulation in the presence or absence of ATMi. (B) Graph summarizes CSR to IgG1 by WT and 53BP1−/− B cells in four independent experiments in the presence or absence of ATMi. The means are shown as horizontal lines. p-values were calculated using a two-tailed Student’s t test. FSC, forward scatter.
DISCUSSION
DSBs are dangerous lesions, which, if left unrepaired, can lead to genomic instability and genome rearrangements. These potentially cytotoxic lesions are repaired either by HR, which is conservative and error free, or by NHEJ, which frequently leads to deletions and insertions. NHEJ is mediated by at least two separate pathways: C-NHEJ, which does not require DNA end resection and produces direct joins with little or no microhomology (Lieber, 2008), and A-NHEJ, which is a robust alternative pathway that makes extensive use of microhomology and therefore requires resection of ends (Haber, 2008). Of the two NHEJ pathways, C-NHEJ is the more conservative because there is no obligatory loss of genetic material.
DSBs can be repaired with a high degree of fidelity during the S and G2 phases of the cell cycle by HR, which uses the undamaged sister chromatid as a template for repair. HR requires 5′ to 3′ resection of DNA ends to produce ssDNA, which recruits replication protein A, leading to deposition of Rad51, a factor that is essential in homology search (Sung and Klein, 2006). End resection is believed to occur during two stages: in the first phase, relatively short stretches of ssDNA are produced by the combined action of MRN and CtIP; in the second phase, longer stretches of ssDNA are produced by the combined action of Bloom’s helicase and Exo1 (Sartori et al., 2007; Gravel et al., 2008; Mimitou and Symington, 2008; Nimonkar et al., 2008; Zhu et al., 2008; Rass et al., 2009; Xie et al., 2009). ATM is implicated as a regulator of the resection process because it phosphorylates all of the enzymes known to be involved in resection (Cortez et al., 2001), and is required for optimal ATR activation and for CtIP recruitment to DNA ends (Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006; Sung and Klein, 2006). Recent work from several laboratories indicates that the critical choice between HR and NHEJ in the S/G2/M phases of the cell cycle is regulated at the level of DNA end resection (Huertas et al., 2008; Huertas and Jackson, 2009; Yun and Hiom, 2009).
In the absence of a template sister chromatid during CSR in G1, DSBs are repaired by error-prone C-NHEJ or A-NHEJ. However, little is known about the choice between C-NHEJ and A-NHEJ. Under physiological circumstances, the majority of switch joins are blunt or show minimal microhomology, suggesting that C-NHEJ is the dominant pathway (Stavnezer et al., 2008). Nevertheless, A-NHEJ is a robust pathway that can reconstitute up to 50% of normal levels of CSR in the absence of core C-NHEJ factors such as ligase IV, XRCC4, or Ku70/80, or even the combination of Ku70 and ligase IV (Corneo et al., 2007; Yan et al., 2007; Boboila et al., 2010a,b).
A-NHEJ can also mediate plasmid recircularization in transfected cells (Kabotyanski et al., 1998), joining of I-SceI breaks (Guirouilh-Barbat et al., 2007), oncogenic translocations (Zhu et al., 2002; Ramiro et al., 2006; Wang et al., 2008, 2009), and, finally, V(D)J recombination when the end protection function of the RAG recombinase is disabled (Lee et al., 2004). However, other than the preponderance of microhomologies found at the junctions (Haber, 2008; Zha et al., 2009) and the suggestion that MRN is required for A-NHEJ (Deng et al., 2009; Dinkelmann et al., 2009; Rass et al., 2009; Xie et al., 2009), this pathway remains poorly defined.
In addition to NHEJ, the DNA damage response is also essential for physiological CSR (Jankovic et al., 2007). Among the factors that mediate this response, 53BP1 has the most profound effect on CSR and specifically affects long-range joining between different switch regions (Manis et al., 2004; Reina-San-Martin et al., 2007). In contrast, 53BP1-deficient B cells show increased short-range intra-switch joining, which involves ligation of highly repetitive DNA (Reina-San-Martin et al., 2007). These observations and the related findings that loss of 53BP1 results in decreased V(D)J recombination between distal gene segments (Difilippantonio et al., 2008) and transchromosomal fusions of deprotected telomeres (Dimitrova et al., 2008) support the hypothesis that 53BP1 plays a role in synapsis during CSR. An alternative, nonexclusive explanation of the dominant effect of 53BP1 on long-range joining, which is strongly supported by our data, is that this protein disfavors short-range joining between repetitive sequences in the switch region by blocking the processing of DNA ends. Specifically, end processing is essential for the production of the ssDNA required for microhomology-based A-NHEJ. Therefore, by interfering with end processing, 53BP1 normally impairs A-NHEJ, which leads to enhanced C-NHEJ. Conversely, loss of 53BP1 favors ISDs by enhancing resection-dependent A-NHEJ and disfavors C-NHEJ–dependent long-range recombination between two nonhomologous switch regions. Consistent with this idea, inhibition of end resection by interfering with ATM activity enhances CSR in 53BP1-deficient B cells.
However, a switch from C-NHEJ to A-NHEJ is not likely to account for the profound defect in CSR in 53BP1 mutant B cells, as there is persistent CSR at up to 50% of normal levels despite increased ISDs in the complete absence of C-NHEJ (Yan et al., 2007; Han and Yu, 2008; Boboila et al., 2010a,b). We therefore propose a model in which 53BP1 has two roles in CSR: it favors synapsis between distal DSBs and also interferes with end resection and A-NHEJ. Alternatively, the rapid formation of a synapse in the presence of 53BP1 could prevent nuclease access and extensive DNA end resection. Conversely, absence of 53BP1 would lead to prolonged access of nucleases, leading to excessive DNA end resection and A-NHEJ, and decreased CSR. Mechanistically, 53BP1 could block DNA end resection either directly by binding to H4K20me2 in the vicinity of the DSB or by acting as an adaptor that mediates the recruitment of additional factors, which in turn would prevent nuclease access. The combined effect of 53BP1 on synapsis and DNA end resection may explain why the absence of 53BP1 leads to a more pronounced defect in CSR than other DNA damage response factors.
Like the choice between HR and NHEJ during the S and G2 phases of the cell cycle (Huertas et al., 2008; Huertas and Jackson, 2009; Yun and Hiom, 2009), DNA end resection appears to be a key event in the choice between C-NHEJ and A-NHEJ. Furthermore, as is the case for HR, where ATM is required for the efficient recruitment and activation of the factors mediating DNA end resection (Cuadrado et al., 2006; Jazayeri et al., 2006; Myers and Cortez, 2006; Sung and Klein, 2006; You et al., 2009), ATM also regulates the resection that precedes A-NHEJ. However, in contrast to S and G2, where end resection stimulates HR and preservation of genetic material, resection in G1 leads to its obligate loss. Our data suggest a model wherein 53BP1 blocks access of nucleases to DNA ends, which favors C-NHEJ. We speculate that the requirement for obligate deletion of genetic information during A-NHEJ may explain why C-NHEJ, which is more conservative, has evolved as the preferential end-joining pathway in the G1 phase of the cell cycle.
MATERIALS AND METHODS
Mice.
IgHI-96k mice were produced by sequential gene targeting of C57BL/6 embryonic stem cells (Fig. S1). AID−/− mice (Muramatsu et al., 2000) were backcrossed to C57BL/6 for at least 10 generations, and 53BP1−/− mice (Ward et al., 2003) were backcrossed to C57BL/6 for at least 5 generations. All experiments were in agreement with protocols approved by the Rockefeller University and the National Institutes of Health Institutional Animal Care and Use Committee.
B cell cultures.
B lymphocytes were isolated from mouse spleens using anti-CD43 MicroBeads (Miltenyi Biotec) and cultured at 0.5 × 106 cells/ml in R10 medium (RPMI 1640 supplemented with l-glutamine, sodium pyruvate, Hepes, 50 µM 2-mercaptoethanol, antibiotic/antimytotic, and 10% fetal calf serum; Hyclone). For B cell stimulation 25 µg/ml LPS (Sigma-Aldrich) and 5 ng/ml of mouse recombinant IL-4 (Sigma-Aldrich) were added to the culture. Where indicated in the figures, 0.5 µg/ml RP105 (BD) or 2.5 µM ATMi Ku55933 was added to the culture.
Retroviral infection.
Retroviral supernatants for B cell infections were produced in BOSC23 by transfection with Fugene 6 with pCL-Eco and pMX-IRES-GFP plasmids carrying either Cre, Cre*, I-SceI, or I-SceI* 72 h before the first B cell infection (Robbiani et al., 2008). Retroviral supernatants were filtered and added to B cell cultures at 20 and 44 h after start of stimulation, and spinoculated at 1,150 g for 90 min in the presence of 2.5 µg/ml polybrene. After 6 h, retroviral supernatants were replaced with LPS- and IL-4–supplemented R10.
FACS analysis.
For FACS analysis, spleen cell suspensions were stained with fluorochrome-conjugated anti-CD19, -CD3, -IgM, -IgD, and -IgG1 (BD). Labeling for cell division was at 37°C for 10 min in 5 mM CFSE.
PCR assays.
For the intrachromosomal recombination PCR assay, nested PCR reactions were performed with the Expand Long Template PCR System (Roche). The first round of PCR was performed with the following primers and conditions: 5′-CCAATACCCGAAGCATTTACAGT-3′ and 5′-AGACAGGACAGGACAGGACCAAAC-3′ for 10 cycles at 92°C for 10 s, 56°C for 30 s, and 68°C for 40 s, followed by 25 cycles at 92°C for 15 s, 56°C for 30 s, and 68°C for 40 s, with 2 s of additional extension time per cycle. The second round of PCR was performed with the following primers and conditions: 5′-ATGTATGGTTGTGGCTTCTGGG-3′ and 5′-CCTGCTTTCCAGTATGGGTATCTG-3′ for 10 cycles at 92°C for 10 s, 55°C for 30 s, and 68°C for 30 s, followed by 15 cycles at 92°C for 15 s, 55°C for 30 s, and 68°C for 30 s, with 2 s of additional extension time per cycle. PCR products were isolated from gels and sequenced.
Online supplemental material.
Fig. S1 shows the gene targeting strategy and phenotype analysis of IgHI-96k mice. Fig. S2 shows that catalytically inactive I-SceI* does not induce recombination between I-SceI sites. Fig. S3 is a summary of all junctional analysis data for IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells. Tables S1 and S2 are summaries of I-SceI–induced recombination frequency by FACS and PCR. Tables S3–S5 show the resection data of I-SceI–infected IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells. Tables S6–S8 are summaries of joining frequencies and resection data of I-SceI–infected IgHI-96kAID−/− and IgHI-96kAID−/−53BP1−/− B cells in the presence of ATMi. Table S9 is a summary of IgG1 expression in WT and 53BP1−/− B cells in the presence or absence of ATMi. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100244/DC1.
Acknowledgments
We thank all members of the Nussenzweig laboratory for discussion, and D. Bosque and T. Eisenreich for help in managing mouse colonies.
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH; A. Nussenzweig), NIH grants 1ROAI072529-01 and 5RO1AI037526 (to M.C. Nussenzweig), and the Howard Hughes Medical Institute (M.C. Nussenzweig). A. Bothmer is a predoctoral fellow of the Cancer Research Institute. D.F. Robbiani was and N. Feldhahn is a fellow of the Leukemia and Lymphoma Society.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced cytidine deaminase
- A-NHEJ
- alternative nonhomologous end joining
- ATM
- ataxia telangiectasia mutated
- ATMi
- ATM inhibitor
- C-NHEJ
- classical nonhomologous end joining
- CSR
- class switch recombination
- DSB
- double-strand break
- HR
- homologous recombination
- ISD
- intra–switch region deletion
- MRN
- Mre11/Rad50/Nbs1
- ssDNA
- single-stranded DNA
References
- Boboila C., Jankovic M., Yan C.T., Wang J.H., Wesemann D.R., Zhang T., Fazeli A., Feldman L., Nussenzweig A., Nussenzweig M., Alt F.W. 2010a. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl. Acad. Sci. USA. 107:3034–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila C., Yan C.T., Wesemann D.R., Jankovic M., Wang J.H., Manis M.J., Nussenzweig A., Nussenzweig M., Alt F.W. 2010b. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J. Exp. Med. 207:417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Scharff M.D., Goodman M.F. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA. 100:4102–4107 10.1073/pnas.0730835100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callén E., Jankovic M., Difilippantonio S., Daniel J.A., Chen H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L., et al. 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 130:63–75 10.1016/j.cell.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Catalan N., Selz F., Imai K., Revy P., Fischer A., Durandy A. 2003. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J. Immunol. 171:2504–2509 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F.W. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730 10.1038/nature01574 [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Basu U., Zarrin A., Yan C., Franco S., Perlot T., Vuong B., Wang J., Phan R.T., Datta A., et al. 2007. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94:157–214 10.1016/S0065-2776(06)94006-1 [DOI] [PubMed] [Google Scholar]
- Chowdhury D., Sen R. 2004. Mechanisms for feedback inhibition of the immunoglobulin heavy chain locus. Curr. Opin. Immunol. 16:235–240 10.1016/j.coi.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Corneo B., Wendland R.L., Deriano L., Cui X., Klein I.A., Wong S.Y., Arnal S., Holub A.J., Weller G.R., Pancake B.A., et al. 2007. Rag mutations reveal robust alternative end joining. Nature. 449:483–486 10.1038/nature06168 [DOI] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S.J. 2001. ATR and ATRIP: partners in checkpoint signaling. Science. 294:1713–1716 10.1126/science.1065521 [DOI] [PubMed] [Google Scholar]
- Cuadrado M., Martinez-Pastor B., Murga M., Toledo L.I., Gutierrez-Martinez P., Lopez E., Fernandez-Capetillo O. 2006. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 203:297–303 10.1084/jem.20051923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Guo X., Ferguson D.O., Chang S. 2009. Multiple roles for MRE11 at uncapped telomeres. Nature. 460:914–918 10.1038/nature08196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson S.K., Market E., Besmer E., Papavasiliou F.N. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291–1296 10.1084/jem.20030481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S., Gapud E., Wong N., Huang C.Y., Mahowald G., Chen H.T., Kruhlak M.J., Callen E., Livak F., Nussenzweig M.C., et al. 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 456:529–533 10.1038/nature07476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N., Chen Y.C., Spector D.L., de Lange T. 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 456:524–528 10.1038/nature07433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelmann M., Spehalski E., Stoneham T., Buis J., Wu Y., Sekiguchi J.M., Ferguson D.O. 2009. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat. Struct. Mol. Biol. 16:808–813 10.1038/nsmb.1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTullio R.A., Jr., Mochan T.A., Venere M., Bartkova J., Sehested M., Bartek J., Halazonetis T.D. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4:998–1002 10.1038/ncb892 [DOI] [PubMed] [Google Scholar]
- Dudley D.D., Manis J.P., Zarrin A.A., Kaylor L., Tian M., Alt F.W. 2002. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc. Natl. Acad. Sci. USA. 99:9984–9989 10.1073/pnas.152333499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D., Hafen E., Schaffner W. 2004. An efficient method to generate chromosomal rearrangements by targeted DNA double-strand breaks in Drosophila melanogaster. Genome Res. 14:1382–1393 10.1101/gr.2279804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S., Alt F.W., Manis J.P. 2006. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst.). 5:1030–1041 10.1016/j.dnarep.2006.05.024 [DOI] [PubMed] [Google Scholar]
- Gopaul D.N., Guo F., Van Duyne G.D. 1998. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 17:4175–4187 10.1093/emboj/17.14.4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., Jackson S.P. 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22:2767–2772 10.1101/gad.503108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat J., Rass E., Plo I., Bertrand P., Lopez B.S. 2007. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl. Acad. Sci. USA. 104:20902–20907 10.1073/pnas.0708541104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. 2008. Alternative endings. Proc. Natl. Acad. Sci. USA. 105:405–406 10.1073/pnas.0711334105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Yu K. 2008. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV–deficient B cells. J. Exp. Med. 205:2745–2753 10.1084/jem.20081623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Jackson S.P. 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284:9558–9565 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Cortés-Ledesma F., Sartori A.A., Aguilera A., Jackson S.P. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 455:689–692 10.1038/nature07215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic M., Nussenzweig A., Nussenzweig M.C. 2007. Antigen receptor diversification and chromosome translocations. Nat. Immunol. 8:801–808 10.1038/ni1498 [DOI] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C., Lukas J., Jackson S.P. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8:37–45 10.1038/ncb1337 [DOI] [PubMed] [Google Scholar]
- Kabotyanski E.B., Gomelsky L., Han J.O., Stamato T.D., Roth D.B. 1998. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 26:5333–5342 10.1093/nar/26.23.5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.S., Neiditch M.B., Salus S.S., Roth D.B. 2004. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 117:171–184 10.1016/S0092-8674(04)00301-0 [DOI] [PubMed] [Google Scholar]
- Lieber M.R. 2008. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283:1–5 10.1074/jbc.R700039200 [DOI] [PubMed] [Google Scholar]
- Lumsden J.M., McCarty T., Petiniot L.K., Shen R., Barlow C., Wynn T.A., Morse H.C., III, Gearhart P.J., Wynshaw-Boris A., Max E.E., Hodes R.J. 2004. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J. Exp. Med. 200:1111–1121 10.1084/jem.20041074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J.P., Tian M., Alt F.W. 2002. Mechanism and control of class-switch recombination. Trends Immunol. 23:31–39 10.1016/S1471-4906(01)02111-1 [DOI] [PubMed] [Google Scholar]
- Manis J.P., Morales J.C., Xia Z., Kutok J.L., Alt F.W., Carpenter P.B. 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5:481–487 10.1038/ni1067 [DOI] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 455:770–774 10.1038/nature07312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst.). 8:983–995 10.1016/j.dnarep.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Myers J.S., Cortez D. 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 281:9346–9350 10.1074/jbc.M513265200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A.V., Ozsoy A.Z., Genschel J., Modrich P., Kowalczykowski S.C. 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA. 105:16906–16911 10.1073/pnas.0809380105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S., Casellas R., Reina-San-Martin B., Chen H.T., Difilippantonio M.J., Wilson P.C., Hanitsch L., Celeste A., Muramatsu M., Pilch D.R., et al. 2001. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 414:660–665 10.1038/414660a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103 10.1038/nature00862 [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107 10.1038/nature01760 [DOI] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755 10.1016/S0960-9822(02)01215-0 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Stavropoulos P., Jankovic M., Nussenzweig M.C. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456 10.1038/ni920 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Eisenreich T., Difilippantonio S., Chen-Kiang S., Muramatsu M., Honjo T., Nussenzweig A., Nussenzweig M.C. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438 10.1016/j.cell.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Callen E., Difilippantonio S., Chen H.T., McBride K.M., Eisenreich T.R., Chen J., Dickins R.A., Lowe S.W., et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109 10.1038/nature04495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., Lopez B.S. 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16:819–824 10.1038/nsmb.1641 [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767–1778 10.1084/jem.20030569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen H.T., Nussenzweig A., Nussenzweig M.C. 2004. ATM is required for efficient recombination between immunoglobulin switch regions. J. Exp. Med. 200:1103–1110 10.1084/jem.20041162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen J., Nussenzweig A., Nussenzweig M.C. 2007. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. Eur. J. Immunol. 37:235–239 10.1002/eji.200636789 [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Bothmer A., Callen E., Reina-San-Martin B., Dorsett Y., Difilippantonio S., Bolland D.J., Chen H.T., Corcoran A.E., Nussenzweig A., Nussenzweig M.C. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 135:1028–1038 10.1016/j.cell.2008.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C.E., Linehan E.K., Mochegova S.N., Woodland R.T., Stavnezer J. 2005. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J. Exp. Med. 202:561–568 10.1084/jem.20050872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulas-Sprauel P., Le Guyader G., Rivera-Munoz P., Abramowski V., Olivier-Martin C., Goujet-Zalc C., Charneau P., de Villartay J.P. 2007. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 204:1717–1727 10.1084/jem.20070255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E., Schrader C.E. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard B.L. 2005. Homing endonuclease structure and function. Q. Rev. Biophys. 38:49–95 10.1017/S0033583505004063 [DOI] [PubMed] [Google Scholar]
- Sung P., Klein H. 2006. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7:739–750 10.1038/nrm2008 [DOI] [PubMed] [Google Scholar]
- Van Duyne G.D. 2001. A structural view of cre-loxp site-specific recombination. Annu. Rev. Biophys. Biomol. Struct. 30:87–104 10.1146/annurev.biophys.30.1.87 [DOI] [PubMed] [Google Scholar]
- Wang J.H., Alt F.W., Gostissa M., Datta A., Murphy M., Alimzhanov M.B., Coakley K.M., Rajewsky K., Manis J.P., Yan C.T. 2008. Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J. Exp. Med. 205:3079–3090 10.1084/jem.20082271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H., Gostissa M., Yan C.T., Goff P., Hickernell T., Hansen E., Difilippantonio S., Wesemann D.R., Zarrin A.A., Rajewsky K., et al. 2009. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 460:231–236 10.1038/nature08159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Minn K., van Deursen J., Chen J. 2003. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556–2563 10.1128/MCB.23.7.2556-2563.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J.S., Cascalho M., Chen L., Nussenzweig A., Livak F., et al. 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459–464 10.1083/jcb.200403021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerffel R., Wang L., Grigera F., Manis J., Selsing E., Perlot T., Alt F.W., Cogne M., Pinaud E., Kenter A.L. 2007. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 27:711–722 10.1016/j.immuni.2007.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F.W., Chen J., et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 28:1045–1057 10.1016/j.molcel.2007.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A., Kwok A., Scully R. 2009. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16:814–818 10.1038/nsmb.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.T., Boboila C., Souza E.K., Franco S., Hickernell T.R., Murphy M., Gumaste S., Geyer M., Zarrin A.A., Manis J.P., et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482 10.1038/nature06020 [DOI] [PubMed] [Google Scholar]
- You Z., Shi L.Z., Zhu Q., Wu P., Zhang Y.W., Basilio A., Tonnu N., Verma I.M., Berns M.W., Hunter T. 2009. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 36:954–969 10.1016/j.molcel.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Bradley A. 2001. Engineering chromosomal rearrangements in mice. Nat. Rev. Genet. 2:780–790 10.1038/35093564 [DOI] [PubMed] [Google Scholar]
- Yun M.H., Hiom K. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 459:460–463 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin A.A., Del Vecchio C., Tseng E., Gleason M., Zarin P., Tian M., Alt F.W. 2007. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 315:377–381 10.1126/science.1136386 [DOI] [PubMed] [Google Scholar]
- Zha S., Boboila C., Alt F.W. 2009. Mre11: roles in DNA repair beyond homologous recombination. Nat. Struct. Mol. Biol. 16:798–800 10.1038/nsmb0809-798 [DOI] [PubMed] [Google Scholar]
- Zhu C., Mills K.D., Ferguson D.O., Lee C., Manis J., Fleming J., Gao Y., Morton C.C., Alt F.W. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 109:811–821 10.1016/S0092-8674(02)00770-5 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 134:981–994 10.1016/j.cell.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]