Abstract
Although CD103-expressing dendritic cells (DCs) are widely present in nonlymphoid tissues, the transcription factors controlling their development and their relationship to other DC subsets remain unclear. Mice lacking the transcription factor Batf3 have a defect in the development of CD8α+ conventional DCs (cDCs) within lymphoid tissues. We demonstrate that Batf3−/− mice also lack CD103+CD11b− DCs in the lung, intestine, mesenteric lymph nodes (MLNs), dermis, and skin-draining lymph nodes. Notably, Batf3−/− mice displayed reduced priming of CD8 T cells after pulmonary Sendai virus infection, with increased pulmonary inflammation. In the MLNs and intestine, Batf3 deficiency resulted in the specific lack of CD103+CD11b− DCs, with the population of CD103+CD11b+ DCs remaining intact. Batf3−/− mice showed no evidence of spontaneous gastrointestinal inflammation and had a normal contact hypersensitivity (CHS) response, despite previous suggestions that CD103+ DCs were required for immune homeostasis in the gut and CHS. The relationship between CD8α+ cDCs and nonlymphoid CD103+ DCs implied by their shared dependence on Batf3 was further supported by similar patterns of gene expression and their shared developmental dependence on the transcription factor Irf8. These data provide evidence for a developmental relationship between lymphoid organ–resident CD8α+ cDCs and nonlymphoid CD103+ DCs.
DCs comprise a heterogeneous group of APCs found throughout the body that include plasmacytoid DCs (pDCs) and CD11chigh conventional DCs (cDCs). Mouse cDCs in lymphoid organs include CD8α+ and CD8α− subsets that have distinct functional properties (Steinman, 2007; Naik, 2008). DC populations present in nonlymphoid organs also include several unique DC subsets. Although there has been some progress in defining the phenotypic properties of DCs in the lung, gastrointestinal tract, and skin, the relationship between peripheral DCs and lymphoid-resident DCs is unclear (GeurtsvanKessel and Lambrecht, 2008; Kelsall, 2008; Udey and Nagao, 2008). In these tissues, CD103+ DCs have been identified that generally share the phenotype CD103+CD11blow/−DEC205+, and in the lung and skin also express the lectin Langerin. These CD103+ subsets appear to represent migratory cells of similar phenotype and are identified in lymph nodes draining these tissues. In some studies, peripheral CD103+ DCs have exhibited the specialized features of TLR3 responsiveness and the capacity for cross-presentation, suggesting a relationship with classical CD8α+ DCs in lymphoid tissues, which themselves are CD103+CD11blow/negDEC205+Langerinlow (Sung et al., 2006; del Rio et al., 2007; Bedoui et al., 2009).
We recently identified the transcription factor Batf3 as being highly expressed in cDCs and selectively required for the development of CD8α+ DCs of lymphoid tissues. Batf3−/− mice had selective defects in generating CD8+ T cell responses to a virus, cross-presentation, and rejection of syngeneic fibrosarcomas (Hildner et al., 2008). Batf3−/− mice also had reduced numbers of CD103+CD8α− cDCs in skin-draining lymph nodes (SDLNs; Hildner et al., 2008), a recently identified subset of peripheral cDCs (Bursch et al., 2007; Ginhoux et al., 2007; Poulin et al., 2007). Because the developmental stages of DCs are still incompletely defined, the relationships between peripheral and lymphoid subsets of cDCs are unclear. Other transcription factors have been identified as important in the development of DCs (Merad and Manz, 2009). In particular, Irf8 mutant and Irf8-deficient mice also lack CD8α+ cDCs but have additional immune defects (Holtschke et al., 1996; Aliberti et al., 2003; Tsujimura et al., 2003; Turcotte et al., 2005; Gabriele and Ozato, 2007; Tailor et al., 2008).
In this study, we extend our analysis of the roles of the transcription factors Batf3 and Irf8 in the development of lymphoid and nonlymphoid cDCs in mice. We determine that classical CD8α+ cDCs and CD103-expressing DCs present in diverse nonlymphoid tissues appear developmentally related based on similar requirements for both Batf3 and Irf8 and similar patterns of gene expression. Further, we use Batf3−/− mice to examine the role of CD103+ DCs in Sendai virus (SeV) infection of the lung, intestinal homeostasis, and skin-based contact hypersensitivity (CHS).
RESULTS
Batf3−/− mice lack lung CD103+ DCs
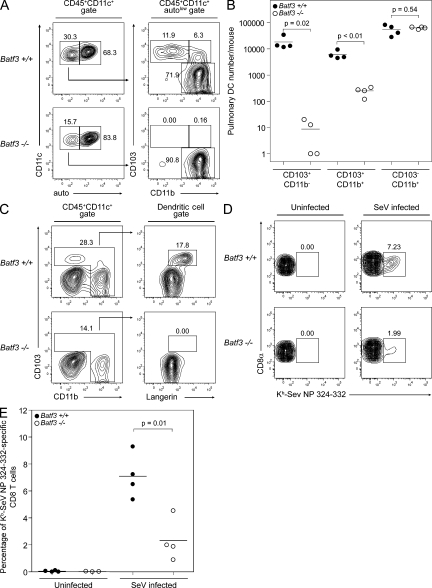
The mouse lung and associated airways are known to contain two distinct CD11c+ DC subsets (CD103+CD11b− and CD103−CD11b+) in addition to highly autofluorescent CD11c+CD103−CD11b− macrophages (GeurtsvanKessel and Lambrecht, 2008). We performed flow cytometry on cell suspensions of perfused, digested lungs from Batf3+/+ (WT) and Batf3−/− mice (Fig. 1 A). After gating on CD45+CD11c+ cells (a mixture of DCs and macrophages), DCs were identified in the autofluorescentlow fraction and subdivided by CD103 and CD11b expression. Lungs from WT 129S6/SvEv mice contain the two typical DC subsets in addition to a third CD103+CD11b+ (double-positive [DP]) population. This DP population has not been described previously but was consistently present in the control 129S6/SvEv mice used in analyzing Batf3−/− mice. Strikingly, 129S6/SvEv background Batf3−/− mice lacked both CD103+CD11b− and DP lung DCs but had normal numbers of CD103−CD11b+ lung DCs (Fig. 1, A and B). CD103+ lung DCs from WT mice expressed Langerin by intracellular staining. In contrast, DCs from Batf3−/− lungs showed no Langerin expression in any population. These results indicate that Batf3−/− mice lack the CD103+ Langerin+ population of lung DCs rather than simply lacking expression of a single marker (Fig. 1 C). Finally, the loss of this CD103+ population was caused by a cell-intrinsic defect, because in mixed BM chimeras, donor-derived Batf3−/− lung DCs selectively lacked this population despite the normal development of CD103+ lung DCs from cotransferred WT BM (Fig. S1 A).
Figure 1.
Batf3−/− mice lack lung CD103+ DCs. (A) Cell suspensions of lung digests from Batf3+/+ and Batf3−/− mice were stained for CD45, CD11c, autofluorescence (FITC channel; auto), CD11b, and CD103. Live CD45+CD11c+ cells, representing a mixture of lung macrophages and DCs, were initially gated (left), with the low autofluorescent population (DCs) further analyzed for the discrimination of three pulmonary DC subsets (right). Numbers represent the percentage of cells within the indicated gates. (B) Numbers of pulmonary DCs per mouse for each of the three pulmonary DC subsets as gated in A. Data represent the combination of two separate experiments, with each point representing the pooled lung cells from an individual mouse. Horizontal lines represent the mean cell number. (C) Cell suspensions of lung digests from Batf3+/+ and Batf3−/− mice were stained for CD45, CD11c, CD11b, CD103, and intracellular Langerin. Live CD45+CD11c+ cells, representing a mixture of lung macrophages and DCs, were initially gated (left). A further gate was drawn to encompass all pulmonary DCs and exclude CD11b−CD103− lung macrophages. These DCs were analyzed for the expression of CD103 and Langerin (right). Numbers represent the percentage of cells within the indicated gates. Staining was performed on two individual mice per group in one independent trial. (D) Batf3+/+ and Batf3−/− mice were infected with 100 PFU SeV intranasally or left uninfected (three or four uninfected mice per genotype; four infected mice per genotype). At day 8 after infection, SeV-specific CD8+ T cells were identified in cell suspensions of lung digests stained for CD19, Mac3, CD8α, and Kb-SeV NP 324–332 MHC pentamers. CD8α+CD19−Mac3− cells were gated. Numbers represent the percentage of SeV-specific CD8+ T cells. Data are representative of two separate experiments (one in which staining was performed at day 9 after infection). (E) Percentage of SeV-specific CD8+ T cells in the lungs of uninfected or infected mice, as stained in D. Horizontal lines represent the mean percentage.
To analyze the role for lung CD103+ DCs in a viral pathogen model, we infected WT and Batf3−/− mice intranasally with SeV (Fig. 1, D and E; and Fig. S2). On day 8 after infection, Batf3−/− mice showed markedly decreased numbers of SeV-specific T cells, as measured by Kb-SeV NP 324–332 MHC pentamer staining (Cole et al., 1994). In agreement with reduced priming of CD8 T cells, Batf3−/− mice showed greater weight loss after infection and had increased pulmonary inflammatory infiltrates on days 9 and 15 after infection.
Microarray expression analysis of lung DCs
Based on shared functional properties between classical CD8α+ cDCs and lung CD103+ DCs (Sung et al., 2006; del Rio et al., 2007; Kim and Braciale, 2009), and their shared developmental requirement for Batf3, we next examined the transcriptional profiles of lung and splenic cDC subsets by expression microarray analysis (Sung et al., 2006; Dudziak et al., 2007). Specifically, we compared purified CD103+CD11b− and CD103−CD11b+ lung DCs with splenic DEC205+ (surrogates for CD8α+) or 33D1+ (surrogates for CD8α−) cDCs.
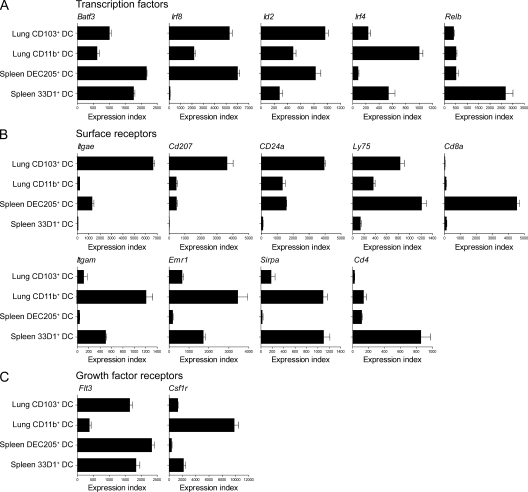
We compared the expression of transcription factors and receptors that are selectively expressed by subsets of splenic cDCs with their expression in subsets of lung DCs (Fig. 2, A and B). Batf3, which is highly expressed in both CD8α+ and CD8α− cDCs (Hildner et al., 2008), was also highly expressed in both lung DC populations (Fig. 2 A). Irf8 and Id2 (Hacker et al., 2003; Kusunoki et al., 2003), which are required for CD8α+ cDC development, are selectively expressed in the CD103+ subset of lung DCs. In a corresponding manner, Irf4, which is required for development of CD8α− splenic cDCs, is selectively expressed in the CD11b+ subset of lung DCs. Relb, reported to be required for development of CD8α− cDC, (Wu et al., 1998), in contrast, was not differentially expressed between lung DC subsets.
Figure 2.
Comparative microarray expression analysis of lung and splenic DC subsets. Microarray data for two lung DC and two splenic cDC subsets were normalized and modeled (three replicate arrays for each DC subset). Model-based expression indices were calculated using dChip software for selected (A) transcription factors, (B) surface receptors, and (C) growth factor receptors. Values represent the mean index, with error bars representing the standard deviation.
Several surface markers also showed a correspondence between splenic and lung DC subsets. Itgae (CD103) and Cd207 (Langerin) were highly expressed in lung CD103+ DCs and also showed significant expression in CD8α+ splenic cDCs. Likewise, CD24a and Ly75 (DEC205) also show a similar correspondence (Fig. 2 B). Several other receptors were selectively expressed more highly in CD11b+ lung DCs and splenic CD8α− cDCs than in CD103+ lung DCs or CD8α+ cDCs, including Itgam (CD11b), Emr1 (F4/80), and Sirpa (CD172a). In contrast, Cd8a and Cd4, which are useful for the discrimination of splenic cDCs, were not significantly expressed in lung DCs. Lastly, we examined the expression of the growth factor receptors Flt3 (the receptor for the DC growth factor Flt3L) and Csf1r (the receptor for M-CSF). Flt3 transcript was most abundant in both splenic subsets and in lung CD103+ cells, whereas Csf1r showed highest expression in lung CD11b+ cells (Fig. 2 C).
Batf3−/− mice lack intestinal CD103+CD11b− DCs
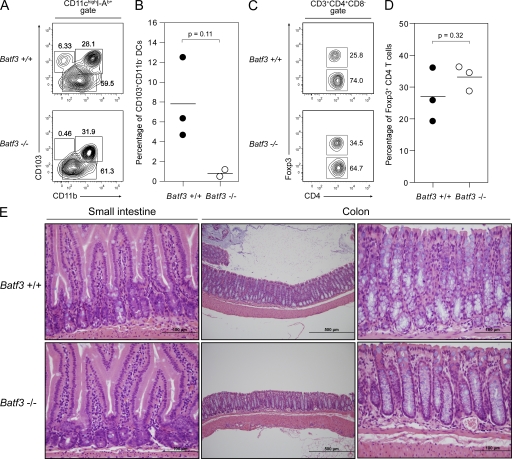
Several intestinal DC subsets have been described in mice (Kelsall, 2008; Bogunovic et al., 2009; Varol et al., 2009). CD103+ DCs of the lamina propria (LP) and mesenteric lymph nodes (MLNs) have been shown in vitro to promote the expression of specific integrins and chemokine receptors on T cells, and to direct the differentiation of CD4+Foxp3+ T reg cells, and are thought to be important components of intestinal homeostasis (Coombes and Powrie, 2008). Analysis of the LP and MLNs showed the absence of CD103+CD11b− DCs in Batf3−/− mice (Fig. 3, A and B; and Fig. S3, A–C), whereas CD103+CD11b+ DCs remained present in these mice. MLNs of Batf3−/− mice were found to lack the classical lymphoid organ–resident CD8α+CD103low cDCs and the LP-derived CD103+CD8αlow/− DCs (Fig. S3, A and B). Further characterization of these subsets from WT mice revealed that only classical CD8α+ cDCs express Langerin, whereas both subsets express DEC205. Batf3−/− mice had a normal population of CD4+Foxp3+ T reg cells in both the LP and MLNs (Fig. 3, C and D; and Fig. S3 D), with no evidence of inflammatory bowel disease in either the small intestine or colon (Fig. 3 E). In addition, CD4 and CD8 T cells from the LP and MLNs of Batf3−/− mice showed normal expression of the gut-homing receptors α4β7 integrin and CCR9 (Fig. S4). There was no difference in the sensitivity of Batf3−/− and WT mice to the dextran sodium sulfate (DSS) model of induced colitis (Fig. S5; Brown et al., 2007).
Figure 3.
Batf3−/− mice lack intestinal CD103+CD11b− DCs. (A) Cell suspensions of LP from Batf3+/+ and Batf3−/− mice were stained for CD11c, I-Ab, CD11b, and CD103 (representative data from a single experiment involving two to three mice per group; two independent experiments were performed). Live CD11chighI-Ab+ cells were gated. Numbers represent the percentage of cells within the indicated gates. (B) Percentage of CD103+CD11b− LP DCs as gated in A. Horizontal lines represent the mean percentage. (C) Cell suspensions of LP from Batf3+/+ and Batf3−/− mice were stained for CD3e, CD4, CD8, and intracellular Foxp3 (representative data from a single experiment with three mice per group). Live CD3e+CD4+CD8− T cells were gated. Numbers represent the percentage of cells within the indicated gates. (D) Percentage of Foxp3+ LP T cells as gated in C. Horizontal lines represent the mean percentage. (D) Light microscopy of H&E-stained sections of the small intestine and colon from Batf3+/+ and Batf3−/− mice (data are representative of a single experiment involving two mice per group at 8–10 mo of age; a separate trial examined five mice per group at 3 mo of age with similar findings). Note the normal villous architecture and LP leukocytes, without evidence of inflammation. Bars: (left and right) 100 µm; (middle) 500 µm.
Batf3−/− mice lack dermal CD103+ DCs
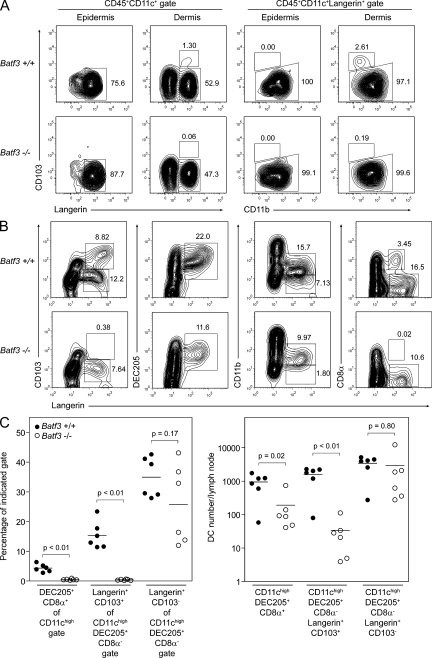
CD103+Langerin+ DCs have been recently described as a unique, blood-derived migratory DC subset present in the dermis of mouse skin, and have been shown to traffic to SDLNs both in the steady state and during inflammation (Bursch et al., 2007; Ginhoux et al., 2007; Poulin et al., 2007). These cells are distinct from CD103−Langerin+ Langerhans cells (LCs) of the epidermis and CD103−Langerin− dermal DCs (Merad et al., 2008). Migratory LCs themselves are also present in the dermis of mice but are thought to represent cells trafficking toward SDLNs. We performed flow cytometry on cell suspensions prepared from the epidermis and dermis of WT and Batf3−/− mice, and gated on DCs based on their expression of CD45 and CD11c (Fig. 4 A). Epidermal LCs were present normally in Batf3-deficient skin, whereas dermal preparations from these mice showed the absence of a small but distinct population of CD103+Langerin+ dermal DCs. Gating on all Langerin-expressing cells in each preparation showed the normal presence of epidermal and dermal CD103−CD11b+ LCs in WT and Batf3−/− mice, whereas the latter mice clearly lacked the dermal CD103+CD11blow/− DC subset.
Figure 4.
Batf3−/− mice lack dermal CD103+ DCs. (A) Cell suspensions of epidermal and dermal digests from Batf3+/+ and Batf3−/− mice were stained for CD45, CD11c, intracellular Langerin, CD103, and CD11b (representative data from a single experiment involving two mice per group). Live CD45+CD11c+ cells were initially gated (left), allowing for the discrimination of CD103−Langerin+ LCs and CD103+Langerin+ dermal DCs. Langerin-expressing cells were gated for further analysis (right), allowing good discrimination of these two populations based on their differential expression of CD11b. Numbers represent the percentage of cells within the indicated gates. (B) Cell suspensions of pooled SDLNs from Batf3+/+ and Batf3−/− mice were stained for CD11c, intracellular Langerin, CD103, DEC205, CD11b, and CD8α (representative data from a single experiment involving two mice per group; experiments with similar combinations of stains have been performed in two other independent trials involving at least one mouse per genotype in each trial). Live CD11chigh cells were gated. Note the complete absence of CD103+Langerin+ dermal DCs (far left) and CD8α+Langerinlow lymph node–resident DCs in Batf3−/− mice. Langerin-expressing cells present in Batf3−/− mice represent epidermal LCs. (C) Percentage and number of DCs per inguinal lymph node for three DC subsets. In this case, individual cell suspensions of inguinal lymph nodes (three mice per group, six lymph nodes total) were stained for CD11c, DEC205, CD8α, CD103, and intracellular Langerin. Live CD11chigh cells were initially gated. Lymph node–resident DEC205+CD8α+ cDCs were identified as a percentage of this gate. Migratory DCs were further gated as DEC205+CD8α− cells, and Langerin+CD103+ dermal DCs and Langerin+CD103− LCs were identified as percentages of this gate. Horizontal lines represent the mean percentage and cell number.
We next assessed SDLNs from WT and Batf3-deficient mice for the presence of CD103+Langerin+ dermal DCs and CD103−Langerin+ LCs, both known to express DEC205+ and lack expression of CD8α (Fig. 4, B and C). Batf3−/− mice again demonstrated the complete absence of CD103+ dermal DCs, with a normal population of LCs. As we have reported previously (Hildner et al., 2008), Batf3−/− mice also lacked LangerinlowCD8α+ cDCs in their SDLNs (Fig. 4 B). The loss of CD103+ dermal DCs in Batf3−/− mice was caused by a cell-intrinsic defect, based on analysis of mixed BM chimeras (Fig. S1 B).
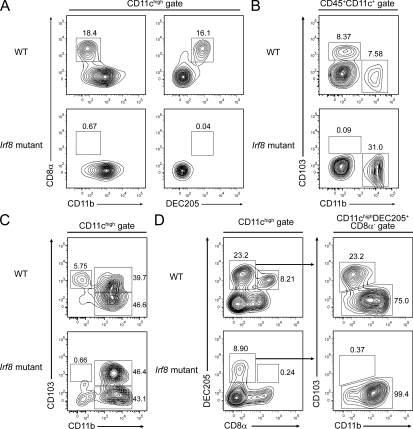
Irf8 mutant mice lack nonlymphoid CD103+ DCs
Irf8-deficient and BXH2 recombinant inbred mice, harboring a point mutation in the Irf8 gene, lack classical CD8α+ cDCs of the spleen (Aliberti et al., 2003; Tailor et al., 2008). These strains also have other immune and hematopoietic abnormalities because of the actions of Irf8 in other cell types and the presence of a replication-competent endogenous mouse leukemia virus in BXH2 mice (Holtschke et al., 1996; Turcotte et al., 2005; Turcotte et al., 2007). However, in the cDC compartment of both strains, there is a selective absence of CD8α+ DCs in vivo and the CD8α+ surrogate population in Flt3L-derived BM cultures in vitro. Because of overlapping effects of Irf8 and Batf3 deficiency, and high expression of Irf8 in CD103+ lung DCs (Fig. 2 A), we hypothesized that Irf8 mutant BXH2 mice would also lack development of nonlymphoid CD103+ DCs. Irf8 mutant mice lacked CD8α+ splenic cDCs (Fig. 5 A), consistent with an earlier report (Tailor et al., 2008). As we predicted, we found that Irf8 mutant mice lacked CD103+CD11b− lung DCs (Fig. 5 B). In addition, Irf8 mutant mice lacked LP CD103+CD11b− DCs (Fig. 5 C) and lacked both DEC205+CD8α+ cDCs and CD103+DEC205+CD8α− dermal DCs in SDLNs (Fig. 5 D). These findings show that nonlymphoid CD103+ DCs and CD8α+ splenic cDCs share a developmental dependence on both Irf8 and Batf3.
Figure 5.
Irf8 mutant mice lack nonlymphoid CD103+ DCs. (A) Cell suspensions of spleens from WT (B6C3HF1/J) and Irf8 mutant (BXH2) mice were stained for CD11c, CD11b, CD8α, and DEC205 (representative data from a single experiment involving two mice per group; a second independent experiment was performed with one mouse per group). Live CD11chigh cells were gated. Numbers represent the percentage of cells within the indicated gates. (B) Cell suspensions of lung digests from WT (B6C3HF1/J) and Irf8 mutant (BXH2) mice were stained for CD45, CD11c, CD11b, and CD103 (representative data from a single experiment involving two mice per group; two independent experiments were performed; a second independent experiment was performed with one mouse per group). Live CD45+CD11c+ cells, representing a mixture of lung macrophages and DCs, were gated. Numbers represent the percentage of cells within the indicated gates. (C) Cell suspensions of LP from WT (C57BL/6) and Irf8 mutant (BXH2) mice were stained for CD11c, CD11b, and CD103 (representative data from a single experiment involving two mice per group; a second independent experiment was performed with one mouse per group). Live CD11chigh cells were gated. Numbers represent the percentage of cells within the indicated gates. (D) Cell suspensions of pooled SDLNs from WT (B6C3HF1/J) and Irf8 mutant (BXH2) mice were stained for CD11c, CD8α, DEC205, CD11b, and CD103 (representative data from a single experiment involving two mice per group; a second independent experiment was performed with one mouse per group). Live CD11chigh cells were initially gated (left) to allow the identification of lymph node–resident DEC205+CD8α+ cDCs. Migratory DCs were further gated as DEC205+CD8α− cells and analyzed for their expression of CD11b and CD103. Numbers represent the percentage of cells within the indicated gates.
Batf3−/− mice display a normal CHS response
CHS is an inflammatory reaction of the skin mediated by hapten-specific T cells (Gocinski and Tigelaar, 1990). Several studies have examined the role of epidermal LCs in the CHS response (Bennett et al., 2005; Kaplan et al., 2005; Kissenpfennig et al., 2005). Each of these groups used a different mouse model lacking LCs, and each came to a different conclusion regarding the role of this DC subset in CHS. Two of these models used a diphtheria toxin receptor–based conditional depletion strategy to eliminate Langerin-expressing cells. However, it is now known that diphtheria toxin can deplete CD103+Langerin+ dermal DCs in addition to epidermal LCs in these mice (Bursch et al., 2007; Ginhoux et al., 2007; Poulin et al., 2007). Appropriate timing of diphtheria toxin treatment can allow for depletion of both Langerin-expressing cell types or the selective depletion of LCs (Bursch et al., 2007; Wang et al., 2008). Using this approach, CHS responses were found to be diminished upon depletion of both epidermal LCs and dermal CD103+ DCs, but were normal upon depletion of only epidermal LCs (Bursch et al., 2007; Wang et al., 2008). By inference, these studies suggested that that dermal CD103+ DCs were sufficient for mediating CHS. Because this conclusion was based on indirect evidence, we wished to test whether CHS responses were normal in Batf3-deficient mice, which lack CD103+ dermal DCs but have normal epidermal LCs.
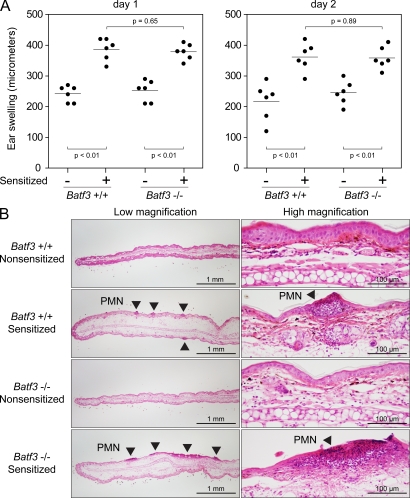
Mice were sensitized with 0.5% 1-fluoro-2,4-dinitrobenzene (DNFB) on their abdomens 5 and 4 d before the elicitation of an ear-swelling response (Fig. 6 A). Batf3−/− mice displayed normal hapten-specific ear swelling in sensitized mice, suggesting no absolute requirement for CD103+ dermal DCs in CHS. Histological examination of the ears on day 2 after elicitation showed significant edema and leukocyte infiltration in both WT and Batf3−/− mice (Fig. 6 B). To rule out the possibility that our CHS protocol may have overlooked a role for CD103+ dermal DCs caused by the dose and schedule of sensitization and elicitation, we performed a separate experiment in which three different protocols for DNFB-based CHS were used (Fig. S6). In each case, Batf3−/− mice displayed normal hapten-specific ear swelling in sensitized mice. In summary, our results together with previous studies indicate that epidermal LCs are sufficient to support CHS responses in the absence of CD103+ dermal DCs.
Figure 6.
Batf3−/− mice display a normal CHS response. (A) Sensitized and nonsensitized Batf3+/+ and Batf3−/− mice were treated on their ears with DNFB to elicit a hapten-specific CHS response. Ear swelling was measured at days 1 and 2 after elicitation. Each point represents an individual ear (three mice per group, six ears per group; Fig. S6 A shows an independent trial of this same protocol). Horizontal lines represent the mean. (B) Light microscopy of H&E-stained sections of treated ears from nonsensitized and sensitized Batf3+/+ and Batf3−/− mice. Sensitized ears demonstrate edema and focal areas of leukocyte infiltration (arrowheads) composed largely of PMNs. Bars: (left) 1 mm; (right) 100 µm.
DISCUSSION
CD103+ DCs constitute a unified DC subset
We present evidence for a common developmental pathway for peripheral CD103+ DCs in lung, gut, and skin- and lymphoid organ–resident CD8α+ cDCs. Conceivably, all peripheral CD103+ and lymphoid organ–resident CD8α+ cDCs constitute a single branch of cDC development sharing a common developmental precursor, developmental pathway, and overall function. Several papers over the last few years have collectively reported a pathway whereby BM common myeloid progenitors give rise to macrophage and DC precursors, which give rise to common DC precursors (CDPs; Fogg et al., 2006; Naik et al., 2007; Onai et al., 2007; Auffray et al., 2009a; Liu et al., 2009). In transfer experiments, CDPs are committed to give rise to pDCs, CD8α+ cDCs, and CD8α− cDCs. In the BM, spleen, and blood, an intermediate cell, the pre-cDC, has been identified that gives rise only to cDC subsets but not pDCs (Liu et al., 2009). In contrast, blood monocytes are not precursors for splenic cDCs or pDCs, but in some settings may generate inflammatory DCs, epidermal LCs, and lung and gut CD11b+ DCs (León and Ardavin, 2008; Auffray et al., 2009b; Bogunovic et al., 2009; Varol et al., 2009). Based on these studies and our results, we suggest that pre-cDCs may also be blood-circulating precursors for CD103+ DCs in peripheral tissues.
During the review of our manuscript, a separate report also identified the developmental relationship between peripheral CD103+ DCs and lymphoid organ–resident CD8α+ cDCs (Ginhoux et al., 2009). As in our experiments, Irf8 was found necessary for peripheral CD103+ DC development, but Batf3 was not examined. In addition, Id2 was required, in agreement with this factor’s role in CD8α+ cDC development (Hacker et al., 2003) and its higher expression in CD103+ DCs (Fig. 2 A). Using cell transfers, it was confirmed that pre-cDCs were in fact circulating precursors of peripheral tissue CD103+ DCs. In Batf3−/− mice, we find that the number of pre-cDCs is normal (Fig. S7, A–C).
Gene expression data (Fig. 2 and Fig. S7 D) suggest a model in which Irf8 and Batf3 cooperate functionally to allow development of both lymphoid organ–resident CD8α+ cDCs and peripheral tissue CD103+CD11b− DCs. These two subsets share many features, including patterns of gene expression and their role in priming CD8 T cell responses. Irf8 is expressed early, beginning at the CDP, and remains highly expressed in the pre-cDC and in terminally differentiated CD8α+ and peripheral CD103+ DC subsets. Importantly, its expression is extinguished in lymphoid organ–resident CD8α− cDCs, which also derive from the CDP/pre-cDC. In contrast, Batf3 becomes expressed only in the terminal stages of DC development, being highly expressed by both subsets of lymphoid organ–resident DCs and both subsets of peripheral DCs. However, the overlap of high Irf8 and Batf3 expression is only found within the CD8α+ and CD103+ subsets, suggesting cooperative interactions in their effects on functional development.
One report that is inconsistent with the hypothesis that pre-cDCs are the precursor for peripheral CD103+ DCs found that Ly6Chi monocytes may be the precursor for CD103+ lung DCs (Jakubzick et al., 2008). This study was based on the correlation of relative frequencies of blood Ly6Chigh and Ly6Clow monocytes with lung CD103+ and CD11b+ DCs, and used injected latex beads to identify blood-derived Ly6Chigh monocytes and trace their differentiation into lung DCs. However, no monocyte transfer experiments were performed. It is not clear at this time whether lung CD103+ DCs represent a special class of CD103+ DCs that may arise from blood monocytes, or whether the manipulations used resulted in additional pathways besides those operating in the steady state. Because Flt3 is expressed on CDPs and splenic cDCs but not blood monocytes (Kingston et al., 2009), it would be unexpected for monocytes to be the circulating precursor for CD103+ lung DCs, which themselves express high levels of Flt3 (Fig. 2 C). On the other hand, lung CD11b+ DCs and monocytes both express Csf1r (Fig. 2 C; Kingston et al., 2009), consistent with their developmental relationship.
Functions of peripheral CD103+ DCs
CD103+ DCs have been identified in individual anatomical locations. In the lung, CD103+ DCs were notable for their response to TLR3 stimulation and their ability to cross-present antigen to CD8+ T cells (Sung et al., 2006; del Rio et al., 2007). Our microarray analysis confirms selective expression of Tlr3 on lung CD103+ DCs and splenic CD8α+ cDCs. Some studies have suggested that lung CD103+ DCs preferentially prime CD8+ T responses to respiratory influenza infection (GeurtsvanKessel et al., 2008; Kim and Braciale, 2009). One study made use of Langerin–diphtheria toxin receptor mice for the selective depletion of Langerin+CD103+ lung DCs (GeurtsvanKessel et al., 2008), finding reduced viral clearance and virus-specific CD8+ T cell priming in their absence (GeurtsvanKessel et al., 2008). A second study examined the differential capacity of lung DC subsets to prime CTLs and cross-present antigens ex vivo (Kim and Braciale, 2009). In agreement, our results now show that CD8 T cell responses to SeV are reduced in the absence of pulmonary CD103+ DCs. In contrast, a recent report has claimed that CD103−CD11bhigh DCs of the lung were the dominant cell presenting antigen to CD8 T cells during influenza infection based on ex vivo analysis of their presentation capacity at the peak of infection, although both lung DC subsets presented antigen during early infection (Ballesteros-Tato et al., 2010). Further, CD70, which is selectively expressed by CD103−CD11bhigh DCs, was important for the expansion of influenza-specific CD8 T cells, but no direct assessment of the requirement for CD103+ DCs for in vivo CD8 T cell responses to influenza was made. Infection of Batf3−/− mice with influenza would help resolve this issue.
CD103+ DCs of the gut have been shown to exist in both mouse and human (Jaensson et al., 2008). Initial interest in this subset of gut DCs came after the observation that host CD103 expression on a non–T or –B cell population was important for the ability of T reg cells to ameliorate disease in a T cell transfer model of colitis (a model of inflammatory bowel disease induced by the transfer of CD4+CD45RBhigh T cells into severe combined immunodeficiency or Rag-deficient recipients; Annacker et al., 2005). Because a subset of gut DCs was the main cell type in these immunodeficient recipients expressing CD103, these authors set out to characterize the function of this subset. These CD103+ DCs were reported to migrate in the steady state from the LP to the MLNs, and displayed the unique ability to induce the expression of the gut-homing receptors CCR9 and α4β7 integrin on T cells in vitro (Annacker et al., 2005; Johansson-Lindbom et al., 2005). Subsequent work also highlighted the unique ability of CD103+ DCs to induce the expression of the transcription factor Foxp3 in CD4+ T cells in vitro, a marker of T reg cell development (Coombes et al., 2007; Sun et al., 2007). CD103+ gut DCs were shown to selectively express genes necessary for the production of TGF-β and retinoic acid when they were compared with CD103− gut DCs, and CD103+ DC induction of Foxp3 expression was shown to be dependent on both of these endogenously produced factors. More recently, mice made deficient in all gut DCs (using a CD11c–diphtheria toxin receptor transgenic system) and then reconstituted with monocytes, allowing the selective repopulation of CD103−CD11b+ intestinal DCs, were found to be more susceptible to DSS-induced colitis, although no inflammation was observed in the absence of DSS treatment (Bogunovic et al., 2009; Varol et al., 2009). These data have led to the hypothesis that gut CD103+ DCs are important for the maintenance of self-tolerance in the intestine (Coombes and Powrie, 2008; Kelsall, 2008).
Our analysis of Batf3−/− mice revealed the absence of CD103+CD11b− DCs in both the LP and MLNs, and yet these mice displayed no evidence of spontaneous inflammation in their gastrointestinal tract and maintained a normal population of T reg cells in their LP and MLNs. Several possibilities might explain the lack of predicted gut inflammation in Batf3−/− mice lacking CD103+ DCs. The presence of the CD103+CD11b+ DP LP DC subset in these mice may be sufficient to maintain homeostasis. In addition, other APCs in the LP or MLNs may be able to generate T reg cells in the gut. Indeed, LP macrophages, rather than CD103+ DCs, were recently reported to induce CD4+ T cell expression of Foxp3 in vitro (Denning et al., 2007). The LP of Batf3−/− mice shows a normal population of macrophages. Thymically derived T reg cells may be sufficient for intestinal homeostasis, and we find no evidence of abnormal T reg cell development in Batf3−/− mice. Finally, Batf3−/− mice also lack classical CD8α+ DCs in their MLNs, which may be required to prime T cells responses against intestinal flora.
We show Batf3 and Irf8 to be transcription factors required for the development of CD103+ dermal DCs. CD103+ dermal DCs represent a distinct population of cells, sharing the expression of Langerin with epidermal LCs. These two cell types have been recognized to differ in their radiosensitivity, their repopulation kinetics after conditional depletion, and their developmental requirements for TGF-β (Bursch et al., 2007; Ginhoux et al., 2007; Poulin et al., 2007; Nagao et al., 2009). Both Batf3−/− and Irf8 mutant mice selectively lack this skin-resident DC subset, in addition to their lack of lymphoid organ–resident CD8α+ cDCs, while maintaining a normal epidermal LC population. CD103+ dermal DCs have been reported to play an important role in the presentation of antigen after DNA vaccination (Nagao et al., 2009), to be sufficient in the absence of LCs to support epicutaneous immunization to a foreign protein antigen, and to be efficient cross-presenting APCs for both viral and skin-derived self antigens (Wang et al., 2008; Bedoui et al., 2009). It will be important to examine these responses in Batf3−/− and Irf8−/− or Irf8 mutant mice.
Previous studies indicated that CD103+ dermal DCs can support a normal CHS response in the absence of LCs, but that CHS was abrogated in the absence of both cell types (Bursch et al., 2007; Wang et al., 2008). Our data using Batf3−/− mice represent the first CHS experiments in which CD103+ dermal DCs were selectively eliminated in the presence of a normal LC population. We show no absolute requirement for this dermal DC subset in this response, and suggest that LCs are sufficient in this regard. However, it is possible that in the constitutive absence of CD103+ dermal DCs in Batf3−/− mice, other cells not normally involved in CHS compensate for the loss of this subset.
Although Batf3 is highly expressed in lymphoid organ–resident CD8α+ and CD8α− cDCs (Hildner et al., 2008), it is not required for the development of the CD8α− cDC population. Likewise, Batf3 is expressed in both subsets of lung DCs but is only required for the development of the CD103+ subset. Thus, Batf3 is necessary but not sufficient for the development of CD8α+ lymphoid organ–resident and CD103+ peripheral DC subsets. The transcription factors Irf8 and Id2, required for CD8α+ cDC development (Hacker et al., 2003; Kusunoki et al., 2003), are more selectively expressed in CD8α+ and CD103+ DC subsets but are also more widely expressed in non-DC lineages compared with Batf3. Thus, the development of DC subsets appears to be based on intersections of both subset- and lineage-specific expression of transcription factors.
Our results reveal a developmental relationship between peripheral CD103+ DCs present in diverse anatomical locations, as well as a shared developmental relationship between these DCs and lymphoid organ–resident CD8α+ cDCs. We show that all of these DC subsets depend on both Batf3 and Irf8 for their development. Numerous experimental systems including diverse vaccination and immunization strategies, and models of infection, transplantation, tumor immunity, and autoimmunity have assigned important functions to CD8α+ cDCs (Heath et al., 2004; López-Bravo and Ardavin, 2008). Given that CD103+ DCs reside in virtually all peripheral organs, their presence and activity should also be considered in these models.
MATERIALS AND METHODS
Mice.
WT 129S6/SvEv, C57BL/6, and B6.SJL mice were purchased from Taconic. Batf3−/− mice on a 129S6/SvEv background were previously generated in our laboratory (Hildner et al., 2008). Batf3−/− mice were backcrossed six generations to the C57BL/6 background for use in mixed BM chimera experiments. BXH2/TyJ (Irf8 mutant) mice, carrying a point mutation in exon 7 of Irf8 resulting in an Arg to Cys change at amino acid 294 of the Irf8 protein (Turcotte et al., 2005), were purchased from the Jackson Laboratory. These mice are a recombinant inbred strain between C57BL/6 and C3H/HeJ mice, and therefore B6C3HF1/J mice (The Jackson Laboratory) were used as WT controls for BXH2 mice in most experiments (although in some cases C57BL/6 mice were used as controls). Experiments were generally performed with sex-matched mice at 6–12 wk of age. Mice were bred and maintained in our specific pathogen–free animal facility according to institutional guidelines with protocols approved by the Animal Studies Committee of Washington University.
Antibodies and flow cytometry.
Staining was performed at 4°C in the presence of Fc Block (clone 2.4G2; BD) in FACS buffer (Dulbecco’s PBS [DPBS] + 0.5% BSA + 2 mm EDTA + 0.02% sodium azide). The following antibodies and secondary reagents were purchased from BD: biotin–anti–c-Kit (2B8), biotin–anti-CD3e (2C11), FITC–anti-CD3e (2C11), FITC–anti-Mac3 (M3/84), FITC–anti-Ly6C (AL-21), FITC–anti-B220 (RA3-6B2), FITC–anti–I-Ab (25-9-17), FITC–anti-Gr1 (RB6-8C5), PE–anti-Flt3 (A2F10.1), PE–anti–LPAM-1 (α4β7 integrin, DATK32), PE–anti-CD43 (S7), PE–anti–I-Ab (AF6-120.1), PE–anti-CD11c (HL3), PE– and PE-Cy7–anti-CD11b (M1/70), PE-Cy7–anti-CD4 (RM4-5), PE-Cy7–anti-CD8α (53-6.7), PerCP Cy5.5–anti-CD45.1 (A20), allophycocyanin–anti-SIRPα (P84), allophycocyanin–anti-CD8α (53-6.7), Alexa Fluor 700–anti-CD45.2 (104), streptavidin-PerCP, streptavidin–PE-Cy7, and streptavidin-allophycocyanin. The following antibodies and secondary reagents were purchased form eBioscience: biotin–anti-B220 (RA3-6B2), biotin–anti-CD49b (DX5), biotin–anti–Thy-1.2 (53-2.1), biotin–anti-Ter119 (Ter119), biotin–anti-CD103 (2E7), FITC–anti-CD19 (ebio1D3), PE–anti-Flt3 (A2F10), PE–anti-CD103 (2E7), PE–anti-Foxp3 (FKJ-16s), PE-Cy7–anti–c-Kit (2B8), allophycocyanin–anti-CD115 (AFS98), allophycocyanin–anti-CD45.2 (104), allophycocyanin Alexa Fluor 750–anti-CD11c (N418), Alexa Fluor 700–anti–Sca-1 (D7), allophycocyanin Alexa Fluor 750–anti-CD8a (53-6.72), allophycocyanin eFluor 780–anti-CD11c (N418), and streptavidin–allophycocyanin eFluor 780. Alexa Fluor 488–anti-CD207/Langerin (929F3.01) was purchased from Imgenex. FITC–anti-CD8α (5H10), FITC–anti-CD4 (CT-CD4), allophycocyanin–anti-CD8α (5H10), and streptavidin–allophycocyanin-Cy7 were purchased from Invitrogen. Allophycocyanin–anti-CD205/DEC205 (NLDC-145) was purchased from Miltenyi Biotec. FITC–anti-CCR9 (242503) was purchased from R&D Systems. The following antibodies were maintained as hybridomas, with purified antibody conjugated to Pacific blue succinimidyl ester (Invitrogen): anti-CD11b (M1/70), anti-B220 (6B2), anti-Gr1 (RB6-8C5), anti-Ter119 (Ter119), anti-CD3e (2C11), anti-CD4 (GK1.5), and anti-CD8α (53-6.7). Kb-SeV NP 324–332 (FAPGNYPAL) MHC pentamers were purchased from Proimmune.
For lung DC staining, samples were fixed in 4% formaldehyde for 15 min at room temperature after surface antibody staining and before use of streptavidin reagents. For all experiments involving intracellular anti-Langerin staining, cells were surface antibody stained before fixation in 4% formaldehyde for 15 min at room temperature. Cells were subsequently permeabilized (DPBS + 0.1% BSA + 0.5% saponin) for 5 min at 4°C and stained in the same buffer with anti-Langerin and steptavidin reagents. Cells were washed twice in permeabilization buffer before returning them to FACS buffer. For intracellular Foxp3 staining, cells were fixed and permeabilized with buffers purchased as part of the Foxp3 staining buffer set (eBioscience). Flow cytometry was performed on a FACSCalibur, FACSCanto II, or LSR II (BD).
DC preparation and cell sorting.
Lungs were perfused with 10 ml DPBS via injection into the right ventricle after transection of the lower aorta. Dissected lungs were minced and digested in 5 ml of Iscove’s modified Dulbecco’s media + 10% FCS (cIMDM) with 4 mg/ml collagenase D (Roche) for 1 h at 37°C with stirring. EDTA (5-mM final concentration) was added to cell suspensions, and cells were incubated on ice for 5 min. Cells were passed through a 40-µm strainer before red blood cells lysis with ACK lysis buffer. Cells were counted on an analyzer (Vi-CELL; Beckman Coulter), and 2–3 × 106 cells were used per antibody staining reaction.
Spleens, MLNs, and SDLNs (typically pooled cervical, brachial, axillary, and inguinal nodes, although in some cases inguinal nodes were analyzed separately) were digested in 5 ml cIMDM with 250 µg/ml collagenase B (Roche) and 30 U/ml DNase I (Sigma-Aldrich) for 1 h at 37°C with stirring. EDTA (5-mM final concentration) was added to cell suspensions, and cells were incubated on ice for 5 min. Cells were passed through a 40-µm strainer before red blood cells lysis with ACK lysis buffer. Cells were counted on a Vi-CELL analyzer, and 2–3 × 106 cells were used per antibody staining reaction.
Small intestinal LP cells were prepared from dissected small intestines after removal of Peyer’s patches and residual fat. Small (∼5-mm) pieces of opened intestine were washed well in DPBS, incubated in 40 ml cIMDM for 45 min at 37°C with shaking at 250 rpm, vortexed, and washed again in cIMDM. Pieces were digested in 30 ml cIMDM with 250 µg/ml collagenase B and 30 U/ml DNase I for 1 h at 37°C with stirring. Cells were passed through a 40-µm strainer before resuspension in 4 ml of 40% Percoll (Sigma-Aldrich). This suspension was overlaid with 4 ml of 70% Percoll and centrifuged at room temperature for 20 min at 900 g without braking. Cells were collected from the interface, washed with buffer (DPBS + 3% FCS + 0.02% sodium azide), counted, and stained for flow cytometry.
Dermal and epidermal cell suspensions were prepared from mouse ear skin. Dorsal and ventral ear flaps were teased apart with gentle traction, and individual flaps were digested in 3 ml cIMDM with 2 mg/ml Dispase II (Roche) for 2.5 h at 37°C with slow rotation in 6-well plates. Epidermal sheets generally “floated” off of the underlying dermis, although in some cases forceps were used to fully separate the skin layers. Epidermal and dermal sheets were separately digested in a fashion identical to that described for lymph nodes to result in cell suspensions for antibody staining and flow cytometry.
Monocytes, pre-cDCs, and mature cDCs were isolated on a cell sorter (MoFlo; Dako). BM monocytes were identified as CD11b+Ly6Chigh cells, excluding CD11b+Ly6Cint neutrophils. Pre-cDCs from BM and spleen were identified as CD11c+CD43+SirpαintI-Ab−CD11b−B220−Gr1−CD4−CD8− cells after enrichment of CD11c+ cells by magnetic bead–based positive selection with anti-CD11c beads (Miltenyi Biotec). Mature cDCs from spleens were identified as either CD11c+CD8α+DEC205+CD4−CD11b−B220−DX5−Thy1.2−Gr1−Ter119− or CD11c+CD4+CD11b+CD8α−DEC205−B220−DX5−Thy1.2−Gr1−Ter119− cells after enrichment of CD11c+ cells by magnetic bead–based positive selection using anti-CD11c beads. RNA from these sorted cell populations was purified using RNeasy kits (QIAGEN).
CDPs from BM were isolated on a cell sorter (FACSAria II; BD) as c-KitintSca-1−Flt3+CD115+CD11b−CD11c−B220−Gr1−Ter119−CD3−CD4−CD8− cells. Cells were sorted directly into 1 ml of TRIzol reagent (Invitrogen), and 25 µl of linear acrylamide (Applied Biosystems) was added as a coprecipitant. The aqueous phase was extracted with chloroform, and RNA was precipitated with 2-propanol, washed once with 75% ethanol, and resuspended in RNase-free water before quantitative real-time RT-PCR analysis.
Mixed BM chimeras.
BM cells from femurs and tibias were collected from B6.SJL (CD45.1), C57BL/6 (CD45.2), and Batf3−/− (CD45.2) mice (backcrossed six generations to the C57BL/6 background). Two mixtures of BM cells were prepared at a 1:1 ratio: B6.SJL cells mixed with C57BL/6 cells, and B6.SJL cells mixed with Batf3−/− cells. 20 million total BM cells were transplanted by intravenous injection into B6.SJL mice that had received 1,200 rads of whole body irradiation on the day of transplant. Mixed chimeras were analyzed at 6 wk after transplant for the presence of DCs in the lungs and SDLNs, using antibodies specific for CD45.1 and CD45.2 to identify the donor cell of origin.
SeV infection.
Sendai/52 virus (Fushimi strain) was purchased from the American Type Culture Collection and subjected to two rounds of in vitro plaque purification on Vero E6 cells. Afterward, a single clone was subjected to a round of amplification in embryonated chicken eggs. SeV was isolated from the allantoic fluid and diluted in PBS to generate a viral stock. Calculation of plaque-forming units was performed by standard plaque assay using either Vero E6 or LLC-MK2 cells. Mice were infected with 100 PFU of virus instilled intranasally. Mice were weighed at baseline and daily for 19 d. At the time points indicated in the figures, lungs were harvested and portions were fixed for histology. Remaining portions were digested for leukocyte preparation, and cells were stained and assayed by flow cytometry.
DSS-induced colitis.
Mice were allowed free access to filtered drinking water containing 5% DSS (TDB Consultancy). Mice were weighed at baseline and daily for 1 wk.
CHS.
Three separate protocols were used for CHS. A 2 × 2 cm area of abdominal skin was shaved in those groups of mice receiving sensitization. Control mice received no treatment to their abdomens. Sensitization in our basic protocol (Fig. 6; and Fig. S6, protocol A) involved two applications of 50 µl of 0.5% DNFB (Fluka) in a 1:4 olive oil/acetone solution on days −5 and −4. Protocol B differed from protocol A in that only a single sensitizing dose was applied to the abdomen at day −5. Protocol C (a “low-dose” protocol, similar to that used in Bursch et al. [2007] and Wang et al. [2008]) involved only a single sensitizing dose of 25 µl of 0.3% DNFB in a 1:4 olive oil/acetone solution on day −5. In Protocols A and B, elicitation of CHS was performed on day 0 by the application of 20 µl of 0.2% DNFB in a 1:4 olive oil/acetone solution to each ear after measurement of baseline thickness with a micrometer. In Protocol C, elicitation was performed on day 0 by the application of 10 µl of 0.15% DNFB in a 1:4 olive oil/acetone solution to each ear after measurement of baseline thickness. Ears were measured on days 1–3, and ear swelling was calculated as the difference between baseline ear thickness and the measured value.
Histology.
Small pieces of lung, small intestine, colon, and ear were fixed in 10% buffered formalin and embedded in paraffin. 5-µm sections were stained with hematoxylin and eosin (H&E). For analysis of intestinal histology in the experiment involving DSS treatment, small intestines, cecums, and colons were harvested on day 7 and fixed in Bouin’s fixative and pinned on black wax for 3 h at 4°C. After washing with 70% ethanol, strips of tissue were embedded in 2% agar for routine paraffin processing, cut in serial sections, and stained with H&E. Morphometric analysis was performed on the distal rectum of these mice. Areas of atrophic crypts were defined as those with <20 epithelial cells observed on cross section through well-oriented crypts, with no cells in M phase present. Areas of intact crypts were defined as those with >30 epithelial cells observed on cross section through well-oriented crypts, with cells in M phase present.
Expression microarray analysis.
Expression microarrays (available from the Gene Expression Omnibus under accession no. GSE17322) for lung DC subsets were originally published in Sung et al. (2006). Expression microarrays (available from the Gene Expression Omnibus under accession no. GSE6259) for splenic cDC subsets were originally published in Dudziak et al. (2007). Data were normalized and expression values were modeled using DNA-Chip Analyzer software (dChip; Li and Wong, 2001a,b).
Quantitative RT-PCR.
cDNA was prepared using SuperScript III reverse transcriptase (Invitrogen). For real-time PCR, the relative standard curve method, SYBR Green PCR Master Mix, and a StepOnePlus Real-Time PCR System (Applied Biosystems) were used according to the manufacturer’s instructions. PCR conditions were 1 min at 95°C, followed by 45 two-step cycles consisting of 15 s at 95°C and 1 min at 50°C, followed by a melting curve analysis to determine product purity. The values were normalized to that of Hprt1 by dividing the mean copy number of the respective transcript sample. Primers used were as follows: Batf3 forward, 5′-CAGACCCAGAAGGCTGACAAG-3′; Batf3 reverse, 5′-CTGCGCAGCACAGAGTTCTC-3′; Irf8 forward, 5′-TGCCACTGGTGACCGGATAT-3′; Irf8 reverse, 5′-GACCATCTGGGAGAAAGCTGAA-3′; Hprt1 forward, 5′-TCAGTCAACGGGGGACATAAA-3′; and Hprt1 reverse, 5′-GGGGCTGTACTGCTTAACCAG-3′.
Statistical analysis.
Differences between groups were analyzed by an unpaired, two-tailed Student’s t test, with P ≤ 0.05 considered significant (Prism; GraphPad Software, Inc.).
Online supplemental material.
Fig. S1 demonstrates that the defect in the development of peripheral CD103+CD11b− DCs is cell intrinsic in Batf3−/− mice. Fig. S2 demonstrates increased weight loss and pulmonary inflammation in Batf3−/− mice infected with SeV. Fig. S3 shows an analysis of DCs and T reg cells from the MLNs of Batf3−/− mice. Fig. S4 shows an analysis of homing receptor expression on intestinal T cells from Batf3−/− mice. Fig. S5 demonstrates normal sensitivity to DSS-induced colitis in Batf3−/− mice. Fig. S6 tests alternative protocols for the induction of CHS. Fig. S7 shows an analysis of pre-cDCs from Batf3−/− mice, and shows quantitative RT-PCR data for the expression of Batf3 and Irf8 in developing and mature DC populations. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091627/DC1.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute (K.M. Murphy), the Burroughs Wellcome Fund Career Award for Medical Scientists (B.T. Edelson), the Jack H. Ladenson Fellowship in Experimental Clinical Pathology at Washington University School of Medicine (B.T. Edelson), the Emmy Noether Program of the German Research Foundation (K. Hildner), and the National Institutes of Health (K.M. Murphy, P.A. Klekotka, D. Bhattacharya, M.J. Holtzman, and S.-S.J. Sung).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- cDC
- conventional DC
- CDP
- common DC precursor
- CHS
- contact hypersensitivity
- DNFB
- 1-fluoro-2,4-dinitrobenzene
- DP
- double positive
- DSS
- dextran sodium sulfate
- LC
- Langerhans cell
- LP
- lamina propria
- MLN
- mesenteric lymph node
- pDC
- plasmacytoid DC
- SDLN
- skin-draining lymph node
- SeV
- Sendai virus
References
- Aliberti J., Schulz O., Pennington D.J., Tsujimura H., Reis e Sousa C., Ozato K., Sher A. 2003. Essential role for ICSBP in the in vivo development of murine CD8alpha+ dendritic cells. Blood. 101:305–310 10.1182/blood-2002-04-1088 [DOI] [PubMed] [Google Scholar]
- Annacker O., Coombes J.L., Malmstrom V., Uhlig H.H., Bourne T., Johansson-Lindbom B., Agace W.W., Parker C.M., Powrie F. 2005. Essential role for CD103 in the T cell–mediated regulation of experimental colitis. J. Exp. Med. 202:1051–1061 10.1084/jem.20040662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C., Fogg D.K., Narni-Mancinelli E., Senechal B., Trouillet C., Saederup N., Leemput J., Bigot K., Campisi L., Abitbol M., et al. 2009a. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J. Exp. Med. 206:595–606 10.1084/jem.20081385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C., Sieweke M.H., Geissmann F. 2009b. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 27:669–692 10.1146/annurev.immunol.021908.132557 [DOI] [PubMed] [Google Scholar]
- Ballesteros-Tato A., León B., Lund F.E., Randall T.D. 2010. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8(+) T cell responses to influenza. Nat. Immunol. 11:216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoui S., Whitney P.G., Waithman J., Eidsmo L., Wakim L., Caminschi I., Allan R.S., Wojtasiak M., Shortman K., Carbone F.R., et al. 2009. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat. Immunol. 10:488–495 10.1038/ni.1724 [DOI] [PubMed] [Google Scholar]
- Bennett C.L., van Rijn E., Jung S., Inaba K., Steinman R.M., Kapsenberg M.L., Clausen B.E. 2005. Inducible ablation of mouse Langerhans cells diminishes but fails to abrogate contact hypersensitivity. J. Cell Biol. 169:569–576 10.1083/jcb.200501071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., et al. 2009. Origin of the lamina propria dendritic cell network. Immunity. 31:513–525 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.L., Riehl T.E., Walker M.R., Geske M.J., Doherty J.M., Stenson W.F., Stappenbeck T.S. 2007. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J. Clin. Invest. 117:258–269 10.1172/JCI29159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch L.S., Wang L., Igyarto B., Kissenpfennig A., Malissen B., Kaplan D.H., Hogquist K.A. 2007. Identification of a novel population of Langerin+ dendritic cells. J. Exp. Med. 204:3147–3156 10.1084/jem.20071966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G.A., Hogg T.L., Woodland D.L. 1994. The MHC class I-restricted T cell response to Sendai virus infection in C57BL/6 mice: a single immunodominant epitope elicits an extremely diverse repertoire of T cells. Int. Immunol. 6:1767–1775 10.1093/intimm/6.11.1767 [DOI] [PubMed] [Google Scholar]
- Coombes J.L., Powrie F. 2008. Dendritic cells in intestinal immune regulation. Nat. Rev. Immunol. 8:435–446 10.1038/nri2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes J.L., Siddiqui K.R., Arancibia-Cárcamo C.V., Hall J., Sun C.M., Belkaid Y., Powrie F. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β– and retinoic acid–dependent mechanism. J. Exp. Med. 204:1757–1764 10.1084/jem.20070590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio M.L., Rodriguez-Barbosa J.I., Kremmer E., Förster R. 2007. CD103− and CD103+ bronchial lymph node dendritic cells are specialized in presenting and cross-presenting innocuous antigen to CD4+ and CD8+ T cells. J. Immunol. 178:6861–6866 [DOI] [PubMed] [Google Scholar]
- Denning T.L., Wang Y.C., Patel S.R., Williams I.R., Pulendran B. 2007. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 8:1086–1094 10.1038/ni1511 [DOI] [PubMed] [Google Scholar]
- Dudziak D., Kamphorst A.O., Heidkamp G.F., Buchholz V.R., Trumpfheller C., Yamazaki S., Cheong C., Liu K., Lee H.W., Park C.G., et al. 2007. Differential antigen processing by dendritic cell subsets in vivo. Science. 315:107–111 10.1126/science.1136080 [DOI] [PubMed] [Google Scholar]
- Fogg D.K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D.R., Cumano A., Geissmann F. 2006. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 311:83–87 10.1126/science.1117729 [DOI] [PubMed] [Google Scholar]
- Gabriele L., Ozato K. 2007. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 18:503–510 10.1016/j.cytogfr.2007.06.008 [DOI] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Lambrecht B.N. 2008. Division of labor between dendritic cell subsets of the lung. Mucosal Immunol. 1:442–450 10.1038/mi.2008.39 [DOI] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Willart M.A., van Rijt L.S., Muskens F., Kool M., Baas C., Thielemans K., Bennett C., Clausen B.E., Hoogsteden H.C., et al. 2008. Clearance of influenza virus from the lung depends on migratory langerin+CD11b− but not plasmacytoid dendritic cells. J. Exp. Med. 205:1621–1634 10.1084/jem.20071365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Collin M.P., Bogunovic M., Abel M., Leboeuf M., Helft J., Ochando J., Kissenpfennig A., Malissen B., Grisotto M., et al. 2007. Blood-derived dermal langerin+ dendritic cells survey the skin in the steady state. J. Exp. Med. 204:3133–3146 10.1084/jem.20071733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., et al. 2009. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206:3115–3130 10.1084/jem.20091756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocinski B.L., Tigelaar R.E. 1990. Roles of CD4+ and CD8+ T cells in murine contact sensitivity revealed by in vivo monoclonal antibody depletion. J. Immunol. 144:4121–4128 [PubMed] [Google Scholar]
- Hacker C., Kirsch R.D., Ju X.S., Hieronymus T., Gust T.C., Kuhl C., Jorgas T., Kurz S.M., Rose-John S., Yokota Y., Zenke M. 2003. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 4:380–386 10.1038/ni903 [DOI] [PubMed] [Google Scholar]
- Heath W.R., Belz G.T., Behrens G.M., Smith C.M., Forehan S.P., Parish I.A., Davey G.M., Wilson N.S., Carbone F.R., Villadangos J.A. 2004. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol. Rev. 199:9–26 10.1111/j.0105-2896.2004.00142.x [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtschke T., Löhler J., Kanno Y., Fehr T., Giese N., Rosenbauer F., Lou J., Knobeloch K.P., Gabriele L., Waring J.F., et al. 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 87:307–317 10.1016/S0092-8674(00)81348-3 [DOI] [PubMed] [Google Scholar]
- Jaensson E., Uronen-Hansson H., Pabst O., Eksteen B., Tian J., Coombes J.L., Berg P.L., Davidsson T., Powrie F., Johansson-Lindbom B., Agace W.W. 2008. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J. Exp. Med. 205:2139–2149 10.1084/jem.20080414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C., Tacke F., Ginhoux F., Wagers A.J., van Rooijen N., Mack M., Merad M., Randolph G.J. 2008. Blood monocyte subsets differentially give rise to CD103+ and CD103− pulmonary dendritic cell populations. J. Immunol. 180:3019–3027 [DOI] [PubMed] [Google Scholar]
- Johansson-Lindbom B., Svensson M., Pabst O., Palmqvist C., Marquez G., Förster R., Agace W.W. 2005. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J. Exp. Med. 202:1063–1073 10.1084/jem.20051100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.H., Jenison M.C., Saeland S., Shlomchik W.D., Shlomchik M.J. 2005. Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity. 23:611–620 10.1016/j.immuni.2005.10.008 [DOI] [PubMed] [Google Scholar]
- Kelsall B. 2008. Recent progress in understanding the phenotype and function of intestinal dendritic cells and macrophages. Mucosal Immunol. 1:460–469 10.1038/mi.2008.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Braciale T.J. 2009. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS One. 4:e4204 10.1371/journal.pone.0004204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston D., Schmid M.A., Onai N., Obata-Onai A., Baumjohann D., Manz M.G. 2009. The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood. 114:835–843 [DOI] [PubMed] [Google Scholar]
- Kissenpfennig A., Henri S., Dubois B., Laplace-Builhé C., Perrin P., Romani N., Tripp C.H., Douillard P., Leserman L., Kaiserlian D., et al. 2005. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity. 22:643–654 10.1016/ j.immuni.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Kusunoki T., Sugai M., Katakai T., Omatsu Y., Iyoda T., Inaba K., Nakahata T., Shimizu A., Yokota Y. 2003. TH2 dominance and defective development of a CD8+ dendritic cell subset in Id2-deficient mice. J. Allergy Clin. Immunol. 111:136–142 10.1067/mai.2003.29 [DOI] [PubMed] [Google Scholar]
- León B., Ardavín C. 2008. Monocyte-derived dendritic cells in innate and adaptive immunity. Immunol. Cell Biol. 86:320–324 10.1038/icb.2008.14 [DOI] [PubMed] [Google Scholar]
- Li C., Wong W.H. 2001a. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 98:31–36 10.1073/pnas.011404098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wong W.H. 2001b. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:research0032.1–research0032.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., Nussenzweig M. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bravo M., Ardavín C. 2008. In vivo induction of immune responses to pathogens by conventional dendritic cells. Immunity. 29:343–351 10.1016/j.immuni.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Merad M., Manz M.G. 2009. Dendritic cell homeostasis. Blood. 113:3418–3427 10.1182/blood-2008-12-180646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Ginhoux F., Collin M. 2008. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol. 8:935–947 10.1038/nri2455 [DOI] [PubMed] [Google Scholar]
- Nagao K., Ginhoux F., Leitner W.W., Motegi S., Bennett C.L., Clausen B.E., Merad M., Udey M.C. 2009. Murine epidermal Langerhans cells and langerin-expressing dermal dendritic cells are unrelated and exhibit distinct functions. Proc. Natl. Acad. Sci. USA. 106:3312–3317 10.1073/pnas.0807126106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S.H. 2008. Demystifying the development of dendritic cell subtypes, a little. Immunol. Cell Biol. 86:439–452 10.1038/icb.2008.28 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., Manz M.G. 2007. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8:1207–1216 [DOI] [PubMed] [Google Scholar]
- Poulin L.F., Henri S., de Bovis B., Devilard E., Kissenpfennig A., Malissen B. 2007. The dermis contains langerin+ dendritic cells that develop and function independently of epidermal Langerhans cells. J. Exp. Med. 204:3119–3131 10.1084/jem.20071724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman R.M. 2007. Lasker Basic Medical Research Award. Dendritic cells: versatile controllers of the immune system. Nat. Med. 13:1155–1159 10.1038/nm1643 [DOI] [PubMed] [Google Scholar]
- Sun C.M., Hall J.A., Blank R.B., Bouladoux N., Oukka M., Mora J.R., Belkaid Y. 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204:1775–1785 10.1084/jem.20070602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S.S., Fu S.M., Rose C.E., Jr., Gaskin F., Ju S.T., Beaty S.R. 2006. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J. Immunol. 176:2161–2172 [DOI] [PubMed] [Google Scholar]
- Tailor P., Tamura T., Morse H.C., III, Ozato K. 2008. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 111:1942–1945 10.1182/blood-2007-07-100750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura H., Tamura T., Gongora C., Aliberti J., Reis e Sousa C., Sher A., Ozato K. 2003. ICSBP/IRF-8 retrovirus transduction rescues dendritic cell development in vitro. Blood. 101:961–969 10.1182/blood-2002-05-1327 [DOI] [PubMed] [Google Scholar]
- Turcotte K., Gauthier S., Tuite A., Mullick A., Malo D., Gros P. 2005. A mutation in the Icsbp1 gene causes susceptibility to infection and a chronic myeloid leukemia–like syndrome in BXH-2 mice. J. Exp. Med. 201:881–890 10.1084/jem.20042170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte K., Gauthier S., Malo D., Tam M.F., Stevenson M.M., Gros P. 2007. Icsbp1/IRF-8 is required for innate and adaptive immune responses against intracellular pathogens. J. Immunol. 179:2467–2476 [DOI] [PubMed] [Google Scholar]
- Udey M.C., Nagao K. 2008. Characteristics and functions of murine cutaneous dendritic cells: a synopsis of recent developments. Mucosal Immunol. 1:470–474 10.1038/mi.2008.37 [DOI] [PubMed] [Google Scholar]
- Varol C., Vallon-Eberhard A., Elinav E., Aychek T., Shapira Y., Luche H., Fehling H.J., Hardt W.D., Shakhar G., Jung S. 2009. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 31:502–512 10.1016/j.immuni.2009.06.025 [DOI] [PubMed] [Google Scholar]
- Wang L., Bursch L.S., Kissenpfennig A., Malissen B., Jameson S.C., Hogquist K.A. 2008. Langerin expressing cells promote skin immune responses under defined conditions. J. Immunol. 180:4722–4727 [DOI] [PubMed] [Google Scholar]
- Wu L., D’Amico A., Winkel K.D., Suter M., Lo D., Shortman K. 1998. RelB is essential for the development of myeloid-related CD8alpha− dendritic cells but not of lymphoid-related CD8alpha+ dendritic cells. Immunity. 9:839–847 10.1016/S1074-7613(00)80649-4 [DOI] [PubMed] [Google Scholar]