Abstract
The solution structure of (+)-spongistatin 1 (1) has been determined, via 1- and 2-D NMR techniques in conjunction with extensive in silico conformational analysis, to comprise a mixture of 4 major rapidly interconverting conformational families.
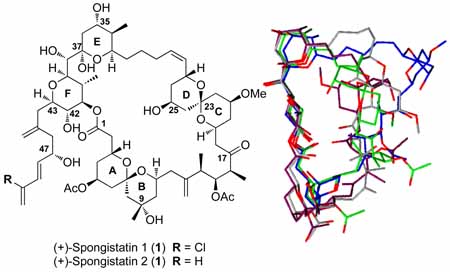
Spongistatins 1 and 2 [(+)-(1) and [(+)-(2)], principal members of the spongipyran family of highly cytotoxic marine metabolites, were isolated in 1993 by the research laboratories of Pettit,1 Kitagawa,2 and Fusetani.3 Initial reports on the structures were in agreement vis-à-vis the carbon skeleton, consisting of two [6.6] spiroketals and a bis-pyran moiety embedded within a 42-membered macrolide ring possesing an unsaturated side chain, and in the case of (+)-spongistatin 1 (1) decorated with a halogen. The complete stereochemical assignment of the 24 stereogenic centers was reported later in the same year by Kitagawa and coworkers.4 Assignment of the initial ambiguous stereochemistry was achieved by comparison of the NOE derived inter-hydrogen distances, with the average distances extracted from an ensemble of solution (+)-spongistatin 1 (1) conformers, the latter obtained by constrained molecular dynamics simulations of all possible diastereomers. Verification of the assigned structures of spongistatins 1 and 2 was subsequently achieved by total syntheses from the Evans and Kishi Laboratories.5
To explore the potential of using (+)-spongistatin 1 (1) as a chemotherapeutic agent, we initiated an analog program to identify the minimum structure required that would maintain a highly cytotoxic profile.6 Central to this study was the identification of the pharmacophore elements of (+)-1, in addition to defining the bioactive conformation. Due to the flexible structure of (+)-spongistatin 1, we reasoned that an increased understanding of solution conformations would be required before the design and synthesis of potentially bioactive analogs could be achieved. Herein we report the solution structure of (+)-spongistatin 1 (1), as well as an analysis of the structural elements responsible for the preferred solution conformations.
Spongistatin 1 [(+)-(1)] consists of a 42-membered macrolactone skeleton possessing 24 stereocenters, 18 rotatable σ bonds, one (Z)-olefinic linkage, two [6.6] spiroketals, one tetrahydropyran and a hemiketal tetrahydropyran moiety, in conjunction with an unsaturated side chain possesing a chlorine. For each of seven C(sp2)-C(sp3) and ten C(sp3)-C(sp3) linkages in the backbone, two and three different staggered conformations respectively are feasible, yielding, in theory, over 7,500,000 local minima. The actual number of possible minima however is significantly reduced due to the cyclic nature of the molecule in conjunction with the syn pentane interactions at tertiary ring junctions.
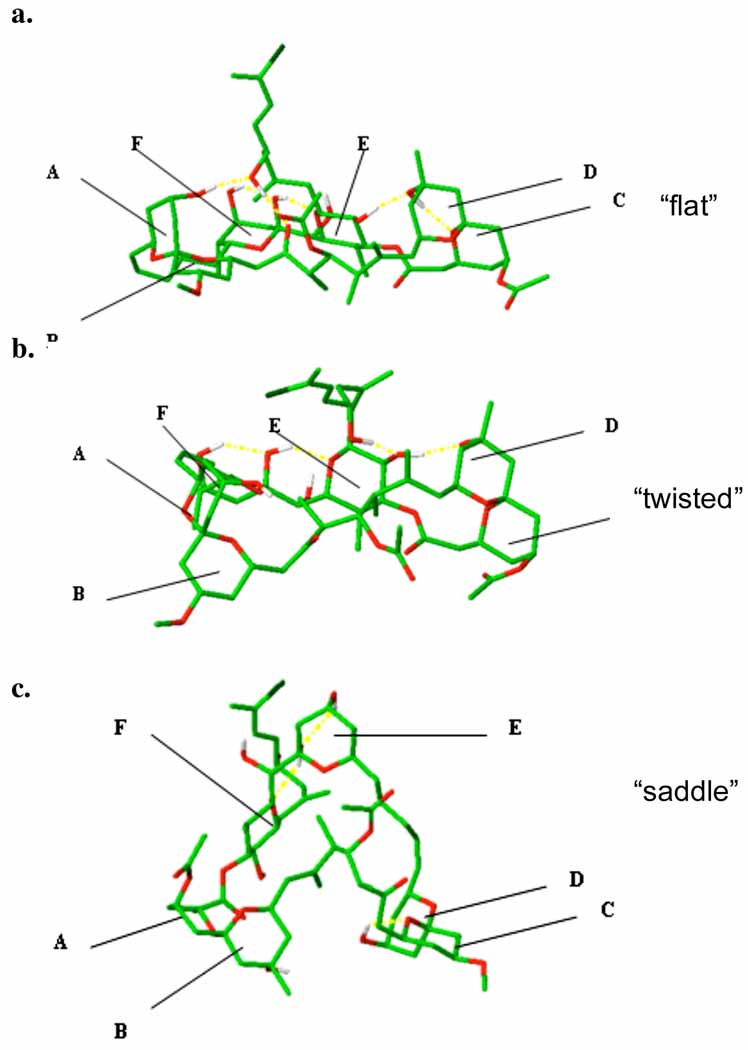
With the goal of identifying the predominant solution conformations, we initially conducted conformational searches using Macromodel 7.1. To cover the conformational space, 30,000-step Monte Carlo searches with the MMFF force field, in conjuction with generalized Born/surface area (GB/SA) solvation models for DMSO, acetonitrile, chloroform and water were performed. Calculations were repeated commencing from different initial geometries, until no additional distinct conformational families were obtained within a 100kJ/mol energy difference of the global minima. Clustering according to the backbone dihedral angles eliminated the redundant conformers, resulting in representative structures of distinct low energy conformers. Interestingly the calculated lowest energy conformations varied significantly in different solvents (Figure 1). In chloroform (+)-sponsigstatin 1 adapted a “flat” conformation in which the surface area of the molecule is maximized; four intramolecular hydrogen bonds were detected [C(9)-O→C(42)-OH, C(43)-O→C(37)-OH, C(37)-O→C(47)-OH and C(47)-O→C(25)-OH)]. In water a twisted or “infinity-like” (∞) conformer was preferred; three hydrogen bonds were detected [C(43)-O→C(37)-OH, C(37)-O→C(47)-OH, C(17)-O→C(25)-OH], while in DMSO and acetonitrile the lowest energy conformer comprised a “saddle” shape; three hydrogen bonds were detected [C(43)-O→C(37)-OH, C(37)-O→C(47)-OH, C(19)-O→C(25)-OH].
Figure 1.
Lowest energy conformations in (a) CHCl3 (flat shape) (b) water (∞-shape) (c) DMSO/acetonitrile (n-shape)
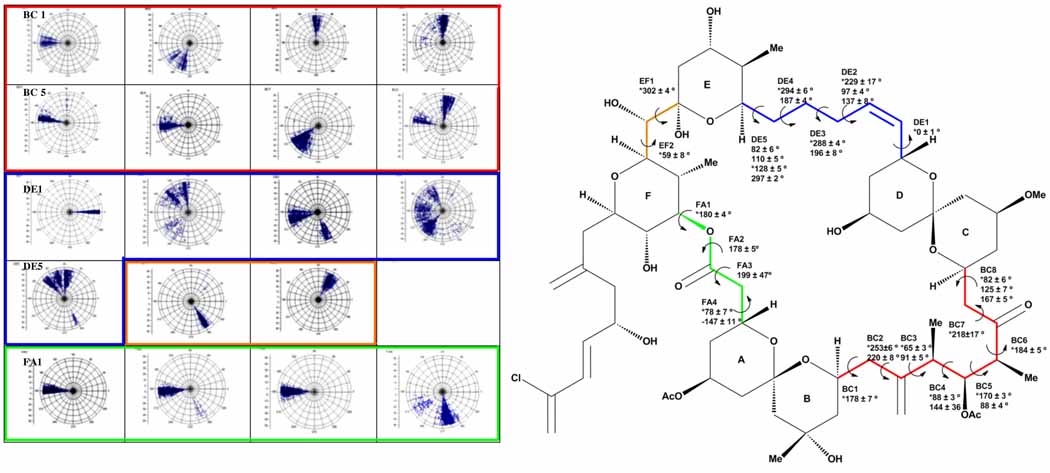
We focused first on the conformations in water, as initial calculations revealed (+)-spongistatin 1 has the highest flexibility in this medium. For the identification of the flexible portions of the molecule, we utilized a method involving polar coordinate maps (Figure 2), a tactic previously employed to great advantage by Taylor and Zajicek during their conformation-activity relationship studies of epothilone B.7 In a similar manner, we extracted the backbone torsional angles of each conformation and plotted the polar coordinate graphs.
Figure 2.
Left Polar coordinate graphs for each backbone dihedral angle (color code/order is given right side). Each point represents a dihedral angle of a conformation where the distance from the origin is the relative energy of that conformer to the global minimum. An enlarged graphic can be found in the supporting information. Right Dihedral angle preferences of the low energy conformations (10 kcal/mol energy window) of (+)-spongistatin 1 in water.
As summarized in Figure 2, the western hemisphere of (+)-spongistatin 1 (1) has a preferred conformation where there is a hydrogen bond between the C(43) oxygen and the C(37)-hydroxyl; this arrangement is seen in a majority of the conformers. In addition, the tether (green) between the AB and F rings adapts a conformation to minimize the steric interactions, as opposed to the regions adjacent to the CD ring (blue and red) which remain quite flexible, enabling the molecule to adjust into a series of conformers according to solvent polarity.
In addition to these studies, a five ns molecular dynamics simulation in water was performed permitting backbone dihedral angles to be extracted. Principal component analysis revealed that while the molecule adapts to different conformations, several bonds have coupled conformational changes. Importantly strong correlations were observed between changes in dihedral angles BC2 and BC8, and DE5 and BC4, with weak correlations for variations at BC3 and DE3, and BC3 and BC4. These calculations, together with the conformational searches, reveal that the CD ring portion of the molecule to be flexible in solution moving up and down with coupled movements in the C(11)-C(19) and C(27)-C(33) tethers, in contrast to the western hemisphere which is held more rigid with a C(43)-O→C(37)-OH hydrogen bond preserved in many of the conformations.
Having identified the flexible components of the molecule, we next turned attention to the solution structure in DMSO, the solvent employed by Kitagawa et al. 4 Here our calculated lowest energy conformation did not conform with the reported structure, with the reported structure being some 47 kcal/mol higher in energy after minimization employing the PM3 method. This result was not totally surprising give that literature data suggests that both conformational searches and MD simulations with NOE distance constraints can generate high energy average conformations if the molecule is interconverting between different conformations in solution. We therefore envisioned that (+)-spongistatin 1, with flexible CD ring orientations available, would be better represented as an ensemble of conformations for validation of both NOESY derived inter-hydrogen distances and 1H NMR derived coupling constants. To date several strategies have been developed to estimate the solution distribution of conformations based on NMR observables. We selected the DISCO (Distribution of Conformations) tactic from among the different strategies,8a,b an in house written software similar to the NAMFIS method.8c
We initiated our NMR studies with the assignment of all non-overlapping protons in the 1H spectra, including the four hydroxyl hydrogens, via a combination of COSY,9 TOCSY,10 HSQC11 and HMBC12 NMR techniques at 600 MHz, employing DMSO and acetonitrile as solvents. The coupling constants for adjacent protons were obtained from spectra in acetonitrile, when the corresponding resonances in DMSO could not be resolved. Due to the similarity in the polarities of DMSO and acetonitrile, a similar conformational preference would be anticipated. This hypothesis was confirmed by comparison of the 1H NMR coupling constants and by conformational search calculations in the two solvents, which yielded the same low energy conformational families. Overall, out of 60 resolved coupling constants, eleven belonging to the 1H couplings around single bonds were selected for DISCO calculations (See supporting information).
We next performed NOESY experiments in DMSO at various mixing times. Buildup curves revealed a mixing time optimum of 600 ms. Out of 314 NOESY cross-peaks, 15 peaks belonging to non-adjacent protons were selected (see supporting information). The short range NOEs were excluded as they were common to all conformations. The NOE peak volumes were integrated and normalized by diagonal peaks. Calibrations were performed for each proton independently according to the known distances in the spiroketals. Strong correlations between protons of the C(14)-C(16) and C(38)-C(40) regions were observed in agreement with the Kitagawa structure.4
Combining the results of the conformational searches in the different solvents, in conjunction with elimination of the redundant conformers, provided 220 distinct conformations. The distances and coupling constants corresponding to the NMR data were then extracted from each conformer employing the Janacchio software,13 and in turn subjected to clustering and deconvolution by DISCO.
When the clustering level was set to 25 structures, the ensemble generated lower violations to the NMR observables compared to the lowest energy single conformation, as achieved in the Kitagawa study.
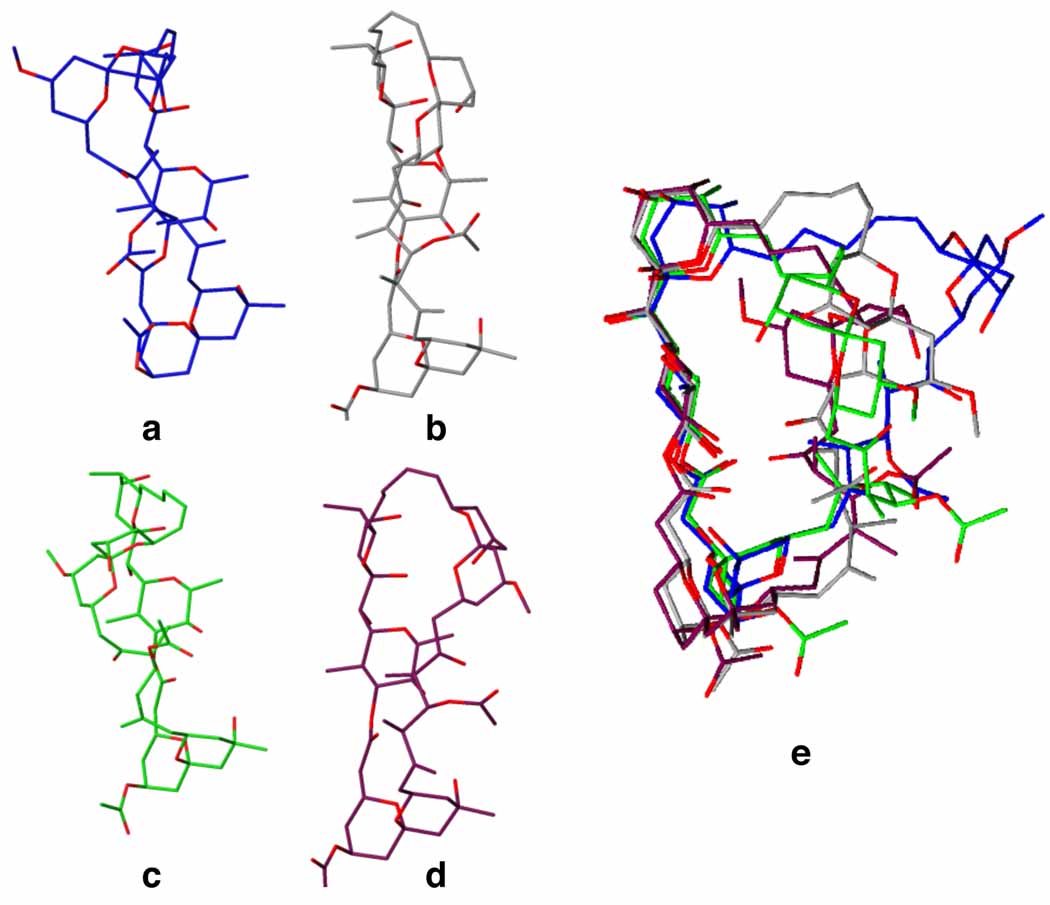
Taken together these studies revealed 4 major conformational families (Figure 3), that in combination fulfilled the constraints obtained from the NMR experiment. The major conformational family (57 %) comprises a minimum energy conformation family found in water (i.e., twisted). When separate calculations were run with only J value constraints, DISCO provided the same conformation in 53%, whereas when only NOE constraints were employed this conformation comprised 66%. These observations indicate that the calculations were not over parameterized. Minor conformations as illustrated in Figure 3b–d (4–13%) can be described as a “linear” conformation along the ABEF ring perimeter, whereas the rest of the molecule is able to occupy different conformations. The remaining conformations, including those having different ABEF ring alignments, are not found to be significant. These results, in agreement with prior studies, reveal (+)-spongistatin 1 has a preferred linear alignment for the ABEF ring perimeter (Figure 4e). The CD ring portion, as well as the chlorodiene sidechain are considerably more mobile and are able to adapt many different conformations.
Figure 3.
Representative solution conformations of (+)-spongistatin 1 in DMSO (estimated solution distribution by DISCO). a. Twisted (57%) b. Flat (13%) c. Flat conformation where the CD ring is flipped up (8%) d. Flat conformation where the CD ring is flipped down (4%) e. Overlay of the found conformations. The chlorodiene sidechains have been omitted for a clearer picture.
Supplementary Material
Acknowledgment
Financial support was provided by the NIH (NCI) through Grant No. CA-70329.
Footnotes
Supporting Information Available: Spectroscopic and analytical data and computational procedures and results of calculations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Pettit GA, Cichacz ZA, Gao F, Herald CL, Boyd MR. J. Chem. Soc., Chem. Commun. 1993:1166. [Google Scholar]; (b) Pettit GR, Cichacz ZA, Gao F, Herald CL, Boyd MR. J. Chem. Soc., Chem. Commun. 1993:1805. [Google Scholar]
- 2.Kobayashi M, Aoki S, Sakai H, Kawazoe K, Kihara N, Sasaki T, Kitagawa I. Tetrahedron Lett. 1993;34:2795. [Google Scholar]
- 3.Fusetani N, Shinoda K, Matsunaga S. J. Am. Chem. Soc. 1993;115:3977. [Google Scholar]
- 4.Kobayashi M, Aoki S, Sakai H, Kihara N, Sasaki T, Kitigawa I. Chem. Pharm. Bull. 1993;41:989. doi: 10.1248/cpb.41.989. [DOI] [PubMed] [Google Scholar]
- 5.(a) Guo J, Duffy KJ, Stevens KL, Dalko PI, Roth RM, Hayward MM, Kishi Y. Angew. Chem., Int. Ed. 1998;37:187. [Google Scholar]; (b) Hayward MM, Roth RM, Duffy KJ, Dalko PI, Stevens KL, Guo J, Kishi Y. Angew. Chem., Int. Ed. 1998;37:192. [Google Scholar]; (c) Evans DA, Coleman PJ, Dias LC. Angew. Chem., Int. Ed. Engl. 1997;36:2738. [Google Scholar]; (d) Evans DA, Trotter BW, Côté B, Coleman PJ. Angew. Chem., Int. Ed. 1997;36:2741. [Google Scholar]; (e) Evans DA, Trotter BW, Côté B, Coleman PJ, Dias LC, Tyler AN. Angew. Chem., Int. Ed. 1997;36:2744. [Google Scholar]
- 6.(a) Bai R, Cichacz ZA, Herald CL, Pettit GR, Hamel E. Mol. Pharmacol. 1993;44:757. [PubMed] [Google Scholar]; (b) Bai R, Taylor GF, Cichacz ZA, Herald CL, Kepler JA, Pettit GR, Hamel E. Biochemistry. 1995;34:9714. doi: 10.1021/bi00030a009. [DOI] [PubMed] [Google Scholar]
- 7.Taylor RE, Zajicek J. J. Org. Chem. 1999;64:7224–7228. [Google Scholar]
- 8. Landis C, Allured VS. J. Am. Chem. Soc. 1991;113:9493. Cicero DO, Barbato G, Bazzo R. J. Am. Chem. Soc. 1995;117:1027. (c) Software sourcecode, binaries of DISCO as well as benchmark studies can be downloaded from Sourceforge (http://discoupenn.sourceforge.net/).
- 9.Rance M, Sorensen OW, Bodenhausen G, Wagner G, Ernst RR, Wuthrich K. Biochem. Biophys. Res. Commun. 1983;117:479. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 10.Bax A, Davis DG. J. Magn. Reson. 1985;65:355. [Google Scholar]
- 11.Bayer RD, Johnson R, Krishanamurthy K. J. Magn. Reson. 2003;165:253. doi: 10.1016/j.jmr.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Bax A, Summers MF. J. Am. Chem. Soc. 1986;108:2093. [Google Scholar]
- 13.Evans DA, Bodkin MJ, Baker SR, Sharman GJ. Magn. Reson. Chem. 2007;45:595. doi: 10.1002/mrc.2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.