Abstract
Double-stranded DNA viruses package their genomes into procapsids via an ATP-driven nanomotor. This ingenious motor configuration has inspired the development of biomimetics in nanotechnology. Bacteriophage ϕ29 DNA-packaging motor has been a popular tool in nanomedicine. To provide information for further motor modification, conjugation, labeling, and manufacturing, the connector protein gp10 of the ϕ29 DNA packaging motor was truncated, mutated, and extended. A 25-residue deletion or a 14-residue extension at the C terminus of gp10 did not affect procapsid assembly. A 42–amino acid extension at the N terminus did not interfere with the procapsid assembly but significantly decreased the DNA-packaging efficiency. DNA-packaging activity was restored upon protease cleavage of the extended region. Replacing the N-terminal peptide containing arginine and lysine with a histidine-rich peptide did not affect procapsid assembly but completely inhibited the packaging RNA (pRNA) binding to the connector and hindered subsequent DNA packaging. These results indicate that (1) the N-terminal arginine-lysine residues play a critical role in pRNA binding but are not essential for procapsid assembly; (2) the connector core, but not the flexible N- or C-terminal domains, is responsible for signaling the procapsid assembly; (3) pRNA binds to the connector as a result of electrostatic interactions between the polyanionic nature of nucleic acids and the cationic side groups of the amino acids, similar to RNA binding to Tat or polyArg.
Keywords: phi29, Connector, DNA packaging motor, Procapsid assembly
After synthesis by separate machinery, viral structural proteins and genome DNA interact to form DNA-filled capsids through a process referred to as DNA packaging.1–4 Most, if not all, double-stranded DNA (dsDNA) viruses, such as ϕ29,5 T4,6 SPP1,7 λ,8,9 T3 and T7,10,11 P22,12–14 P1,15 ψM2,16 Sfi21,17 herpes simplex virus,18–21 cytomegalovirus,22 adenovirus,23–25 and poxvirus,26,27 share common features in this late stage of the maturation process. In this group, their genomic DNA is packaged into a preformed procapsid. This entropically unfavorable DNA translocation process is accomplished by an ATP-driven nanomotor. In bacteriophages the motor involves DNA-packaging enzymes and a connector containing a central channel for DNA transport. This intriguing motor has provoked interest among virologists, bacteriologists, biochemists, biophysicists, chemists, structural biologists, and computational scientists alike. In the last decade extensive investigations28–30 have been carried out to unravel the motor mechanism in DNA packaging and viral assembly. Significant contributions have been made to the construction of bionanomotors,7,31,32 the development of therapeutic viral vectors, and the search for antiviral targets,33–36 as well as the exploration of their applications in nanobiotechnology.31,37–41
Bacteriophage ϕ29, of Bacillus subtilis, is a dsDNA virus with a 19,285–base pair linear genome.42 The infectious virion can be assembled in vitro in a defined biochemical system using eight purified components.43 Scaffold gp7, capsid gp8, head fiber gp8.5, and connector gp10 overexpressed in Escherichia coli from cloned genes self-assemble into empty procapsids. Its genomic DNA-gp3, a DNA with protein gp3 attached to each 5′ end, can be packaged into the procapsids with up to 90% efficiency with the aid of pRNA and the DNA-packaging protein gp16.44,45 Following the addition of the tail protein gp9, upper collar gp11, appendage gp12, and morphogenesis factor gp13, all of which are overexpressed in E. coli, up to 109 plaque-forming units/mL of infectious virion were assembled from the DNA-filled capsids.43 This in vitro assembly system makes ϕ29 a powerful model for the investigation of numerous biological processes.
The head-tail connector, also called portal vertex or DNA entrance vertex, plays a key role in the procapsid assembly and the DNA packaging. Structural analysis of the portal vertices of different phages reveals surprising structural similarities despite the lack of sequence homology. This structural similarity accounts for the common function in DNA translocation. A homo-dodecameric cone-shaped architecture with a central channel is shared by most bacteriophages including ϕ29, T3, T4, T7, λ, SPP1, and P22.46–55
Procapsid morphogenesis is an intriguing topic for the study of macromolecular assembly. The connector of T4 bacteriophage (gp20) can initiate and induce the procapsid and core assembly.56 The connector core of T7 permits the incomplete capsid shells to assemble into a complete procapsid.57 In contrast, the ϕ29 connector has been shown to be a key component in regulating the shape and size of the procapsid during its assembly. During morphogenesis, scaffolding proteins interact with both the capsid protein and connector.58,59 The wider domain of the connector is encapsulated inside the procapsid.46,48,60 Structure determination by x-ray crystallography revealed that the narrow domain, also the N terminus (residues 1–10), of ϕ29 connector is flexible and structurally disordered.
The unique homo-dodecameric structure with an inner channel makes the connector a distinct and fascinating building block in nanotechnology and nanomedicine. Further understanding of the role of the amino acid sequence of the connector in procapsid assembly will facilitate motor or procapsid conjugation, labeling, modification, and manufacture. Using proteolytic cleavage we have demonstrated elsewhere that the N-terminal 14 residues of the connector are critical for RNA binding.61, 62 In this article we extend our analysis on the connector structure-function relationship by investigating the effect of N- and C-terminal mutations on procapsid assembly and DNA packaging.
Results
Effect of C-terminal extension on the procapsid assembly and DNA packaging
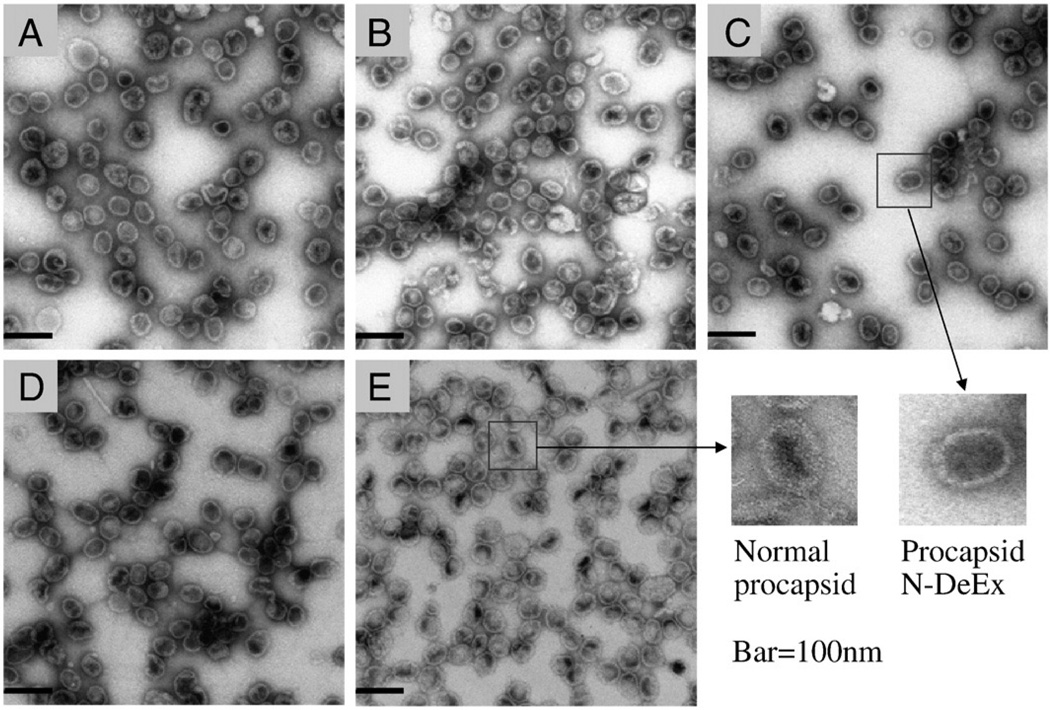
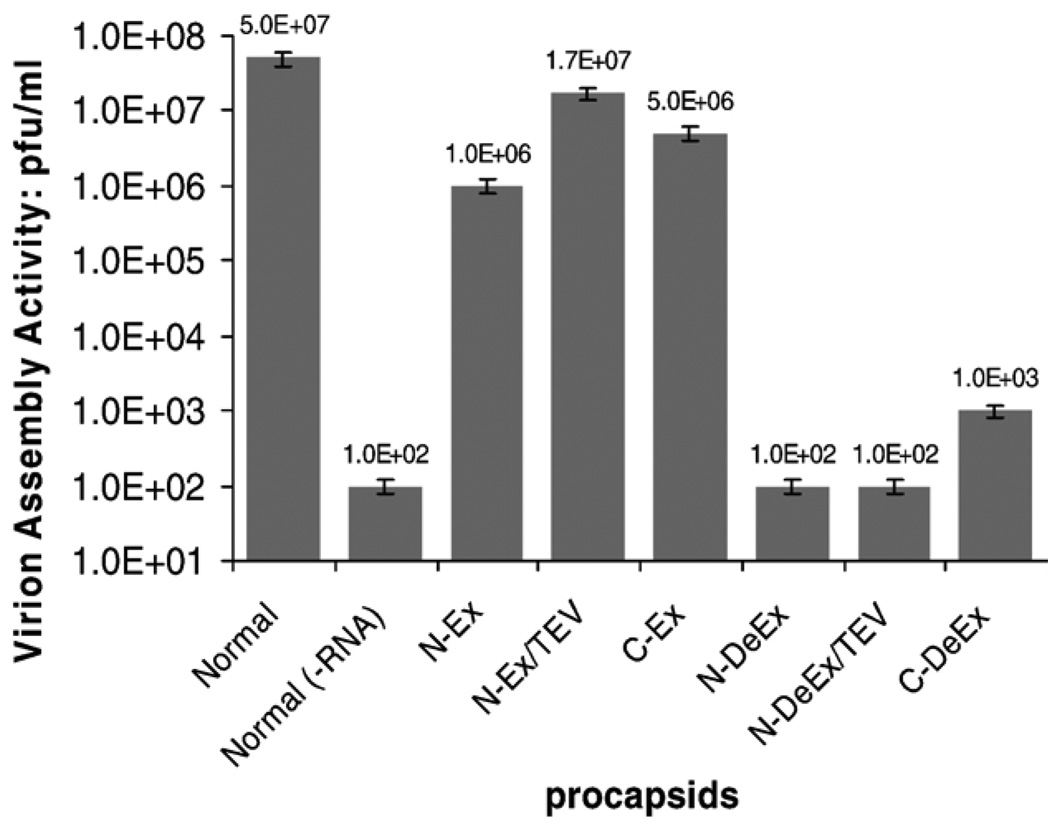
The C terminus of gp10 is located at the wide end of the connector, and residues 286–309 represent a flexible region embedded inside the procapsid.47,48 An extension of 14 residues to the C terminus of gp10 (C-Ex) was designed to investigate its influence in procapsid assembly and DNA packaging. Procapsid C-Ex (Figure 1) was constructed using mutant gp10 (Figure 2) with a C-terminal extension of (Gly)6 (GGGGGG) and a Strep-tag II (WSHPQFEK) (Table 1). The assembled procapsids were isolated using sucrose gradient sedimentation and imaged by transmission electron microscopy (TEM; Figure 3). The C-Ex procapsid (Figure 3, B) displayed a morphology similar to the unmodified procapsid (Figure 3, E). However, the biological activities of the mutant procapsid with C-terminal fusions were slightly altered (Figures 4 and 5). The activity of C-Ex procapsid in the DNA-packaging experiment decreased 10-fold (Figure 4, A and B, lane 5) as compared with normal procapsid (Figure 4, A and B, lane 2); the same was observed for its virion assembly activity (Figure 5).
Figure 1.
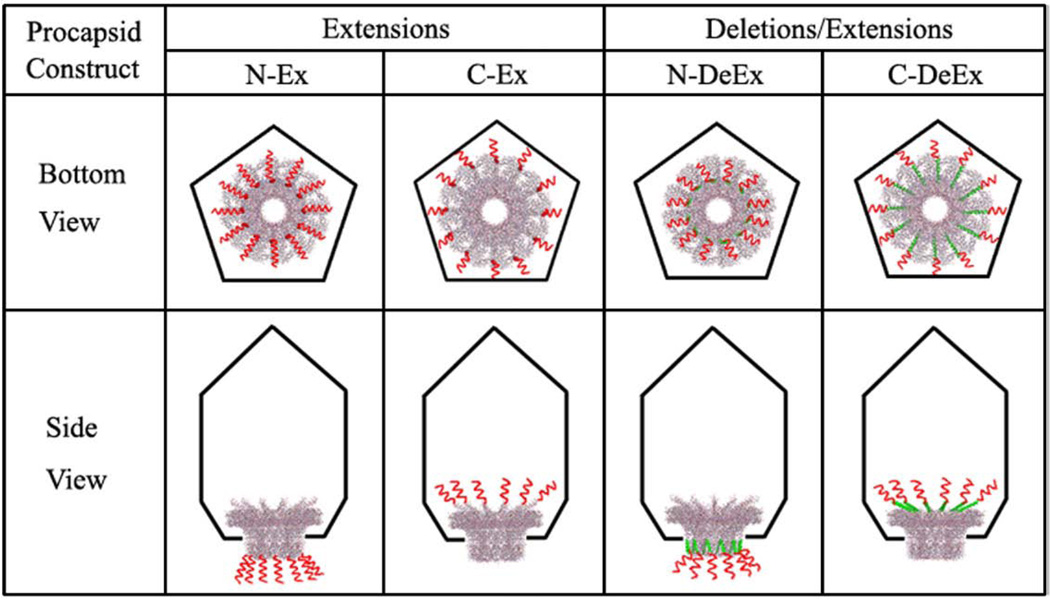
Illustration of different procapsid constructs (bottom and side view). The procapsid is shown as a black pentagon. The portal protein is shown in gray wireframe (PDB ID: 1h5w).47 The N- or C- terminal deletions, named as N-DeEx (deletion and extension both located at the N terminus) and C-DeEx (deletion and extension both located at the C terminus), are displayed as green ribbons. The N- or C-terminal extensions, N-Ex (extension only at the N terminus), C-Ex (extension only at the C terminus), N-DeEx and C-DeEx, are displayed as red curly lines but are not drawn to scale with the connector size.
Figure 2.
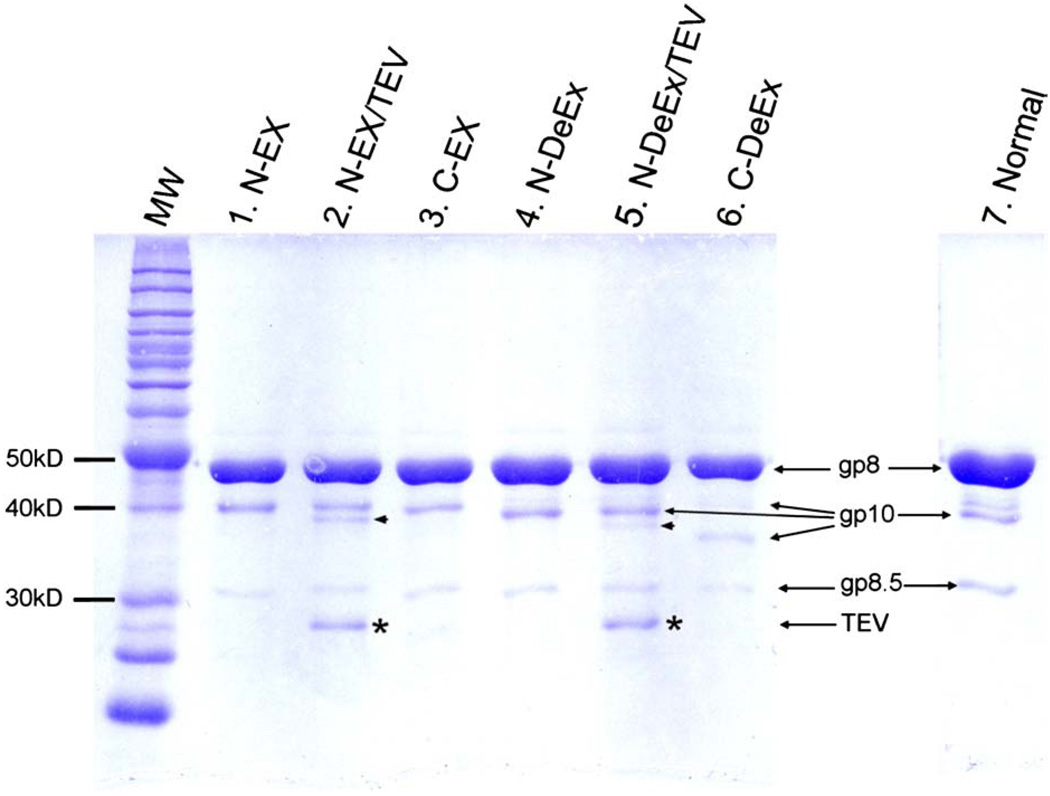
10% SDS-PAGE analysis of mutant procapsids. Lane 1, N-Ex procapsid, molecular weight (MW) of N-Ex gp10 was 40.2 kDa; lane 2, N-Ex procapsid cleaved by TEV. MW of treated N-Ex gp10 was 35.9 kDa; lane3, C-Ex procapsid. MW of C-Ex gp10 was 37.3 kDa; lane 4, N-DeEx procapsid. MW of N-DeEx gp10 was 36.8 kDa; lane 5, N-DeEx procapsid cleaved by TEV. MW of treated N-DeEx gp10 was 35.9 kDa; lane 6, C-DeEx procapsid. MW of C-DeEx gp10 was 34.3 kDa; lane 7, normal procapsid. MW of normal gp10 was 35.9 kDa. The small arrowhead refers to the mutated gp10, which has been treated by TEV, removing the N-terminus peptide tag. The star refers to the TEV protease.
Table 1.
Gp10 sequence modification for the construction of mutant procapsids
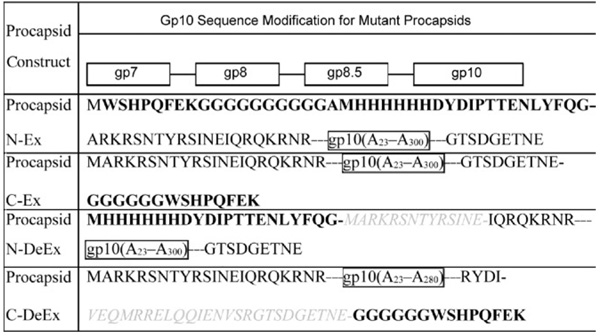 |
The extended sequence is shown in bold letters. The deleted sequence is shown in gray, italic. Letters within a border represent the sequence of the whole protein or protein fragment. Gp10 (A23 –A300 indicates the sequence of the 23–300 amino acids of gp10.).
Figure 3.
Transmission electron microscopy (TEM) images of procapsids. A, N-Ex procapsid; B, C-Ex procapsid; C, N-DeEx; D, C-DeEx procapsid; E, normal procapsid. Bar = 100 nm.
Figure 4.
DNA packaging activity of mutant procapsids with pRNA (A) Ii′ and (B) SphI Ii′. Same quantity of DNA (0.5 µg) as in lane 10, and same quantity of mutant procapsid proteins (1 µg) were used for each experiment. After DNA packaging, the mixtures excluding lane 10 were digested with DNase I to remove the unpackaged DNA. The packaged DNA was released from procapsid by heating at 75°C in the presence of 0.05 M EDTA, then treated with protease K before loading onto the gel. Lane 1: 1 kb DNA ladder; 2: normal procapsid; 3: N-Ex; 4: procapsid N-Ex + TEV; 5: C-Ex; 6: N-DeEx; 7: N-DeEx + TEV; 8: C-DeEx; 9: normal procapsid without ATP as negative control; 10: phi29 DNA without DNase treatment but digested with protease K used for the packaging experiment.
Figure 5.
Virion assembly activity of four mutant procapsids. Equal amount of procapsid protein was used for each test. “TEV” represents the procapsids being treated with TEV protease to remove the N-terminal extensions.
Effect of C-terminal deletion on procapsid assembly and DNA packaging
Procapsid C-terminal deletion (C-DeEx) was generated by replacing the 25 residues at the C-terminal flexible region of gp10 with a 14–amino acid peptide containing (Gly)6-Strep-tag II (Figure 1). As expected, the structure of the assembled procapsid was normal as revealed by TEM (Figure 3, D). This provides evidence that truncation and replacement of the last 25 residues of the C-terminal domain do not interfere with the procapsid assembly. It was interesting to find that the C-DeEx procapsid showed nonvisible DNA-packaging activity (Figure 4, A and B, lane 8). Results suggest that the 25-residue domain at the C terminus of gp10, which was found to be a structurally flexible region as revealed in x-ray crystallography, plays a key role in ϕ29 DNA packaging.
Effect of N-terminal extension on procapsid assembly and DNA packaging
A procapsid containing a connector with a 20-residue extension at its N terminus was shown elsewhere to retain the typical prolate shape of ϕ29 procapsid but showed diminished DNA-packaging activity.62 To further investigate the effect of N-terminal extensions on DNA packaging, we constructed a procapsid with a 40–amino acid addition to the N terminus of gp10 (N-Ex), which was located at the narrow domain of the connector. The extended sequence included a Strep-tag II (WSHPQFEK), a 10-glycine linker, a His6-tag, and a tobacco etch virus (TEV) protease recognition site (ENLYFQG) (Table 1). The TEV cleavage site permitted the removal of the extended region by TEV protease digestion if desired. The sensitivity to protease digestion (Figure 2, lane 2) of procapsid N-Ex also confirmed that the extended peptides protrude toward the outside of the procapsid (Figure 1).
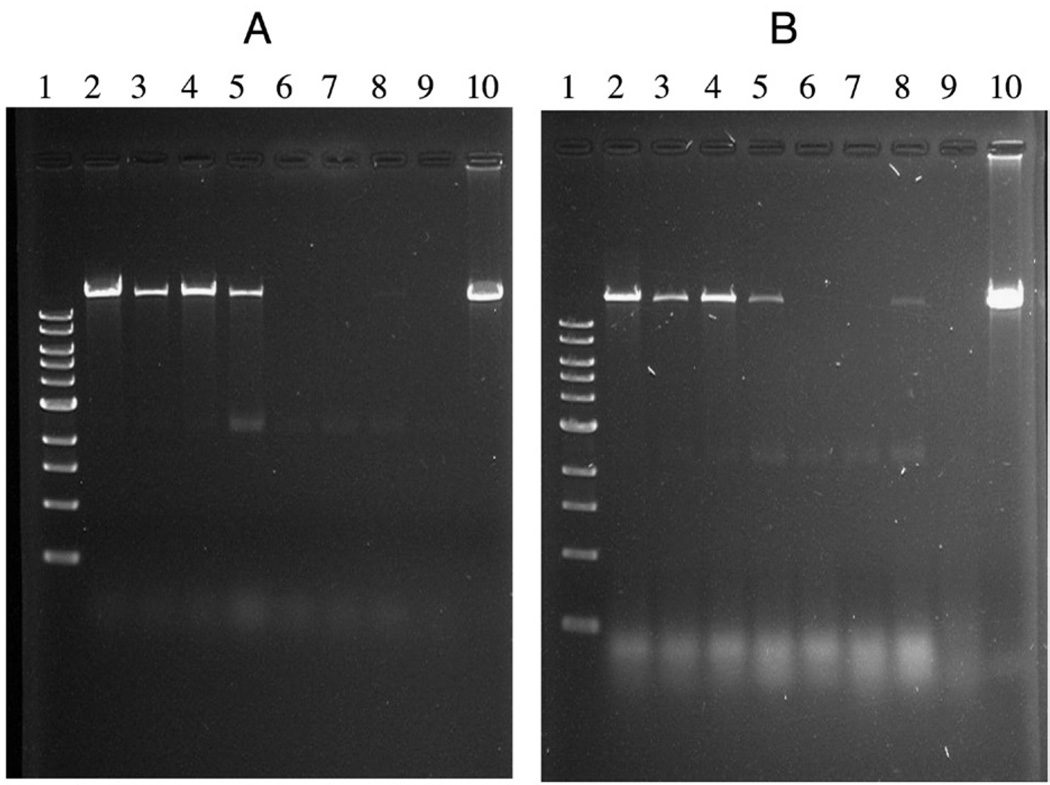
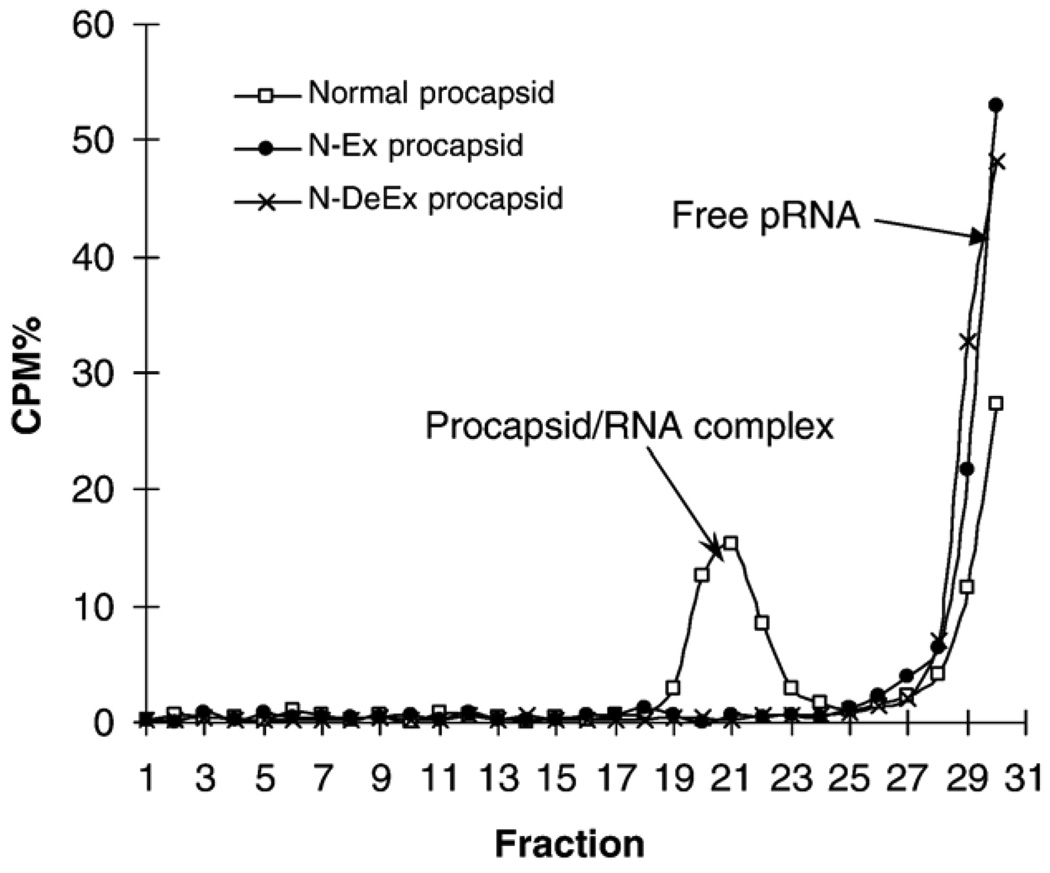
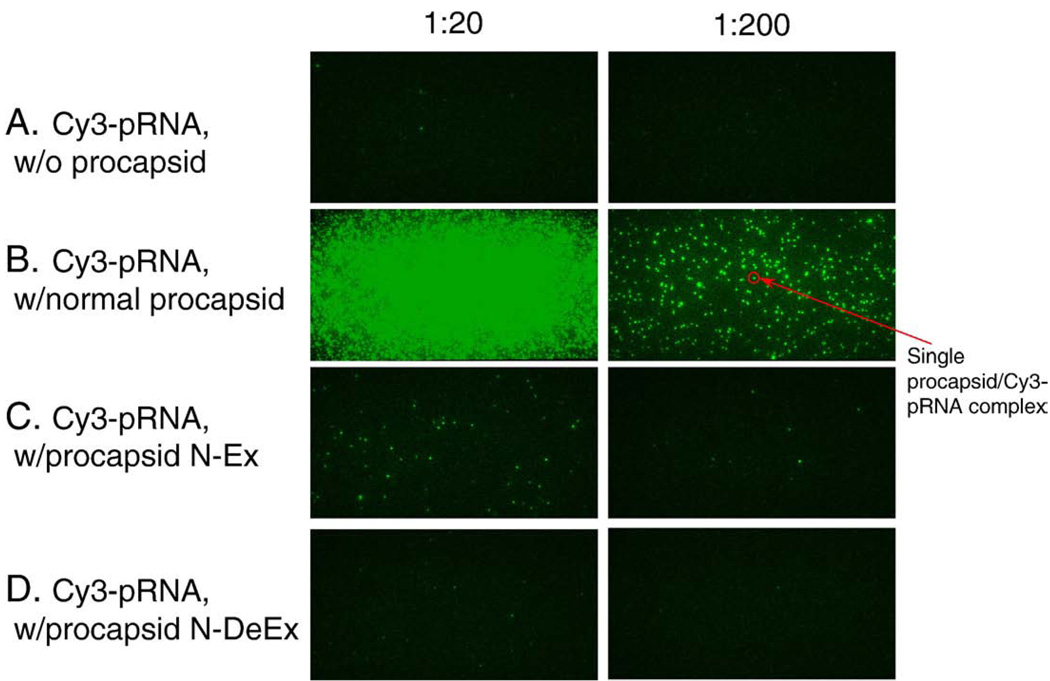
TEM images of the N-Ex procapsid (Figure 3, A) were indistinguishable, in terms of size and structure, from those of normal procapsids (Figure 3, E), suggesting that the N-terminal fusions oriented toward the outside of the procapsid and did not interfere with procapsid morphogenesis. However, the biological functions of the N-Ex procapsid were affected. A 10-fold decrease in DNA-packaging activity was observed when using either pRNA Ii′ (Figure 4, A, lane 3) or pRNA SphI Ii′ (Figure 4, B, lane 3). The 100-fold reduction in phage assembly activity of N-Ex as compared with the wild-type procapsid provides further evidence that the biological activity is impaired by the N-terminal extension (Figure 5). No binding of 3H-labeled pRNA to procapsid N-Ex could be detected by 5–20% sucrose gradient sedimentation analysis (Figure 6). Such data was consistent with the previous report that the binding of pRNA to gp10 was sterically hindered when a His6-tag was attached to the N terminus of the protein.62 The binding ability of N-Ex procapsid with pRNA was further investigated by single-molecule fluorescent imaging, in which each pRNA molecule was labeled with one Cy3 fluorophore45. In this experiment, individual normal procapsid–Cy3-pRNA and N-Ex procapsid–Cy3-pRNA complexes, immobilized on the surface through the anti-procapsid IgG, appeared as distinct bright spots under the microscope after laser excitation (Figures 7 and 8). The single-molecule imaging result indicated that the pRNA-binding activity of N-Ex procapsid was about 100-fold (Figure 8, C) lower than that of a normal procapsid (Figure 8, B), which agreed with the finding in sucrose gradient sedimentation where the binding of 3H-labeled pRNA to N-Ex procapsid was undetectable (Figure 6). The N-terminal extension was susceptible to TEV protease, although the cleavage was incomplete (Figure 2, lane 2). Removal of the extended sequence restored the DNA-packaging activity (Figure 4, A and B, lane 4) and the virion assembly activity of infectious ϕ29 virions (Figure 5). Therefore, it was further proved that the N-terminal extension did not disrupt the morphogenesis of ϕ29 procapsid but instead altered the pRNA binding ability and the DNA-packaging efficiency.62
Figure 6.
5–20% Sucrose gradient sedimentation used to detect the binding of N-terminus mutant procapsid to 3H-labeled pRNA.
Figure 7.
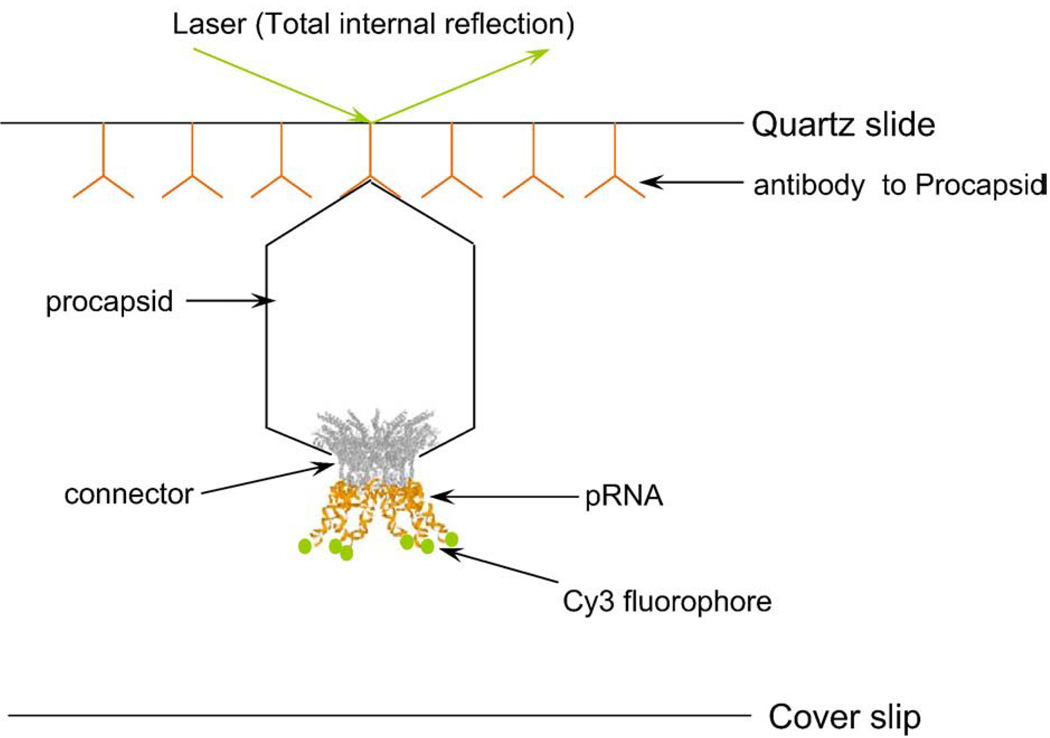
Schematic drawing of single-molecule imaging design. The surface of the quartz slide of the perfusion chamber was coated with anti-procapsid IgG. Each pRNA was labeled by one Cy3 fluorophore at the 5′ end. The procapsid–Cy3-pRNA complex, immobilized to the quartz surface by the antibody to procapsid, was imaged through a total internal reflection.
Figure 8.
Single-molecule fluorescence microscopy used to visualize the relative binding affinity of mutant procapsids to Cy3-pRNA. Individual mutant procapsids with Cy3-pRNA were incubated at room temperature for 30 minutes. The mixtures were then diluted 20 (left column) or 200 times (right column) before applying to the quartz slide surface. Imaging was carried out using a single-molecule dual-viewing system with a 60× objective at 1.6× magnification.45 A, Cy3-pRNA without procapsid. B, Normal procapsid with Cy3-pRNA. C, Procapsid N-Ex with Cy3-pRNA. D, Procapsid N-DeEx with Cy3-pRNA. The green color was presented as the pseudo color, with each spot representing one procapsid–pRNA complex.
Effect of N-terminal mutation on procapsid assembly and DNA packaging
It was reported that the flexible region of residues 1–10 protrudes out of the connector’s N-terminal narrow domain, followed by residues 11–14, which act as the distal end for one of the five α-helices of the gp10 subunit.47 Many RNA-binding proteins, such as Tat peptide63,64 or polyArg,65 use the cationic side groups of arginine or lysine to interact with polyanionic nucleic acids. Thus, it is reasonable to assume that the basic residues at the N-terminal domain of gp10, especially the three consecutive basic residues Arg-Lys-Arg, are essential for pRNA binding. To confirm this assumption, we deleted the 14–amino acid sequence at the N-terminal domain of gp10 and replaced it with a peptide that does not contain arginine or lysine but histidine and other uncharged amino acids. This mutant procapsid, N-DeEx (Figure 1), displayed the typical morphology of ϕ29 procapsid as revealed by TEM (Figure 3, C). The result suggests that the flexible region at the N-terminal domain and the distal α-helical region are dispensable for procapsid assembly.
To further investigate the role of these 14 residues in ϕ29 motor function, the DNA-packaging activity of the resulting mutant procapsid was tested (Figure 4) and was found to be inactive in DNA packaging (Figure 4, A and B, lane 6). This implies that the 14–amino acid sequences play a critical role in DNA packaging. In vitro ϕ29 virion assembly assay revealed that no virion was produced when procapsid N-DeEx was used (Figure 5). Removal of the extended peptide by TEV protease cleavage did not rescue its DNA-packaging activity (Figure 4, A and B, lane 7) or virion assembly activity (Figure 5), further supporting the conclusion that the sequences containing arginine and lysine are essential for DNA packaging and cannot be replaced with a peptide not containing arginine or lysine.
The procapsid-RNA binding assays were performed to further elucidate the mechanisms responsible for the loss of DNA-packaging function of the mutant procapsid. It was shown that no binding between mutant procapsid N-DeEx and pRNA could be detected either by 5–20% sucrose gradient sedimentation analysis (Figure 6) or single-molecule fluorescence imaging (Figure 8, D). Because the total number of amino acids in gp10 of mutant procapsid N-Ex was similar to that of normal procapsid, the difference of activity in DNA packaging and phage assembly implied that the loss of activity in N-DeEx was a result of the deletion of Arg-Lys-Arg. It was concluded that the Arg-Lys-Arg sequence in the N-terminal 14 amino acids is essential for pRNA binding and cannot be replaced by the histidine-containing sequence. The loss of DNA-packaging activity and/or virion assembly is the consequence of the failure of pRNA binding to the N terminus of gp10.
Discussion
Four different kinds of mutant procapsids, with modification on either the C or N terminus of gp10, were constructed. No significant negative effect on procapsid assembly was observed for both N- and C-terminal extensions. Structural studies have revealed that both the N- and C-terminal domains are structurally flexible.47,48 We speculate that as long as the modified gp10 proteins can self-associate into the connector core, they will interact with the scaffolding protein gp7 and the capsid protein gp10 to guide the assembly of the procapsid into the correct prolate particle. Truncation of up to 14 residues at the N terminus or up to 25 residues at the C terminus did not affect procapsid assembly. However, truncation of more than 35 residues at the C terminus or more than 37 residues at the N terminus resulted in a gp10 mutant that was insoluble and unable to assemble into the connector (unpublished data).
The detectable sequential steps in the assay for the morphogenesis of ϕ29 virion are: binding of pRNA to the connector of the procapsid → DNA packaging → virion assembly assayed by a plaque-forming unit. Although the binding of pRNA to procapsid is a prerequisite in DNA packaging and virion assembly, it was interesting to discover that the pRNA binding activity of some of the procapsid mutants with partial activity in DNA packaging and/or virion assembly could not be detected by sucrose gradient sedimentation. For example, procapsid N-Ex might have low pRNA-binding affinity that leads to the formation of unstable pRNA/procapsid complexes that could not be detected with our techniques. The complexes would fall apart when using sucrose gradient sedimentation or gel shift assay. The DNA-packaging activity could still be observed, because it occurred in solution.
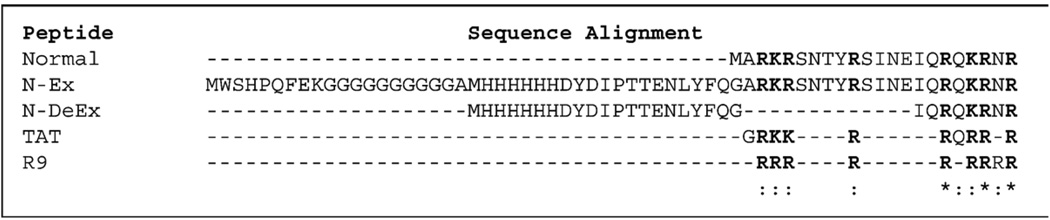
Various basic peptides have the ability to bind to nucleic acids. For example, HIV-1 Tat protein contains a highly basic region responsible for RNA binding and import. This cationic core of the peptide (Tat48–57) is rich in arginine and lysine residues and is often referred to as the Tat peptide.63,64 An oligomer of arginine, (Rn, n = 5–9) is also frequently designed for the purpose of RNA/DNA binding, membrane translocation, cellular uptake, etc.65 Based on their common function of binding to nucleic acids, we assume that the N-terminal domain of the connector binds to pRNA in a similar fashion. The sequence homologies among the N-terminal residues of the connector in normal procapsid, mutants N-Ex, N-DeEx, Tat peptide (Tat48–57), and polyArg indicate that the eight basic amino acids (arginine or lysine) in the N-terminal 22 residues of the normal connector protein display surprising similarities to the Tat peptide, which also contains eight basic amino acids (Figure 9). The pRNA-connector interactions could be described by a mechanism similar to that of the RNA and polyArg or Tat interactions. This finding is in concurrence with previous studies on cleavage,61 extension,62 and mutagenesis66 of the N terminus of connector protein gp10. Here we demonstrate that the gp10 N terminus, which is extended from the narrow domain and oriented to the outside of the procapsid, contributes to the binding of pRNA and subsequently to the binding of gp16 to activate the DNA-packaging motor. Because pRNA binding is critical for the function of the ϕ29 DNA-packaging motor, we believe that these basic amino acids contribute to the pRNA binding and thus are essential for the motor function. Deletion or replacement of the N-terminal 14 residues resulted in the loss of RNA-binding ability;61,62 therefore, the DNA-packaging activity was lost for procapsid N-DeEx. However, extension on the N terminus of the connector with a 40-residue peptide significantly reduced the binding ability of pRNA to the procapsid N-Ex (Figures 6 and 8, C) but only negligibly altered its biological activities (Figure 4, A and B, lane 3; Figure 5). The DNA-packaging activity of the procapsid was rescued following the cleavage of the extended sequence, which confirmed the previous report that the pRNA binding to the connector might be physically hindered by the sequence extension.65 We further speculate that the connector C-terminal region localized inside the procapsid might play a key role in the DNA packaging. A 105-fold higher virion assembly activity for procapsid C-Ex as compared with procapsid C-DeEx has been observed (Figure 5). Both procapsids contained the same extended sequence, except that procapsid C-DeEx included an additional C-terminal deletion. The data suggests that the sequence of the C-terminal structurally flexible 25 residues is involved in DNA packaging or ejection. Because the C terminus is embedded inside the procapsid, its interaction with DNA may play a mechanical role in the processes of DNA entering or exiting the portal vertex.
Figure 9.
Sequence alignment for the N-terminal sequence of the connector in normal procapsid, procapsid N-Ex, procapsid N-DeEx with the Tat peptide (Tat48–57), and polyArg R9.
Methods
Construction of plasmids for the expression of four procapsids
Four mutant procapsids have been constructed: (1) N-Ex, N-terminal extension of 40 residues to the connector; (2) C-Ex, C-terminal extension of 14 residues to the connector; (3) N-DeEx, N-terminal deletion of 14 residues and extension of 21 residues to the connector; (4) C-DeEx, C-terminal deletion of 25 residues and extension of 14 residues to the connector (Figure 1, Table 1).
The plasmid vectors for the production of four procapsids were constructed using the same plasmid pARgp7–8–8.5 and four corresponding plasmid coding for gp10 plasmid DNA. gp10 N-Ex was generated by inserting the annealed primer pair F1-R1 into the NdeI site of vector pHis-gp10.67 gp10 C-Ex, gp10 N-DeEx, and gp10 C-DeEx were constructed from vector pET-21a(+) (EMB Biosciences, Madison, Wisconsin) with a two-step polymerase chain reaction (PCR). First primer pair F1-R1 was used to amplify the gp10 gene from the ϕ29 genomic DNA-gp3; its product was then used as a template for the subsequent PCR with the primer pair F2-R2 to incorporate restriction sites and affinity tags. The second PCR product was digested with NdeI-XhoI and ligated into the NdeI-XhoI sites of the vector pET-21a(+). The plasmid pARgp7–8–8.5 was digested with BamHI and BglII, generating a DNA fragment containing gp7–8–8.5 with two cohesive ends, both compatible with BglII. Four gp10 plasmid DNAs were digested with BglII and dephosphorylated with shrimp alkaline phosphatase (USB, Cleveland, Ohio). Ligation of the gp7–8–8.5 fragment with gp10 vectors digested with BglII generated the four plasmids desired for the expression of the four procapsids. The nucleotide sequence of the primers used in the procapsid construction is given in Tables 2 and 3.
Table 2.
Sequence of the PCR primers used for the plasmid construction of mutant procapsids*
| Procapsid | Primers | Sequence (5′–3′) |
|---|---|---|
| Procapsid N-Ex | F1 | TAATGTGGAGCCACCCGCAGTTCGAAAAGGGAGGTGGTGGAGGAGGTGGAGGTGGTGGAGC |
| R1 | TACACCTCGGTGGGCGTCAAGCTTTTCCCTCCACCACCTCCTCCACCTCCACCACCTCGAT | |
| Procapsid C-Ex | F1, F2 | CGCAGCTGGCATATGGCACGTAAACGCAGTAAC |
| R1 | GGATGACTCCAACCTCCTCCACCACCTCCCTCATTTGTTTCACCGT | |
| R2 | ATAATGTTCTCGAGCTACTTTTCGAACTGCGGATGACTCCAACCTC | |
| Procapsid N-DeEx | F1 | GTAACCTGCATATGCACCATCACCATCACCATGATTACGATATCCCAACGACTGA |
| F2 | CGATATCCCAACGACTGAAAACCTGTACTTCCAGGGAATACAGCGTCAAAAACGG | |
| R1, R2 | CTAGCTATCTCGAGTTAGATGTCATATCTGAATTTAACTT | |
| Procapsid C-DeEx | F1, F2 | CGCAGCTGGCATATGGCACGTAAACGCAGTAAC |
| R1 | GGATGACTCCAACCTCCTCCACCACCTCCGATGTCATATCTGAATT | |
| R2 | ATAATGTTCTCGAGCTACTTTTCGAACTGCGGATGACTCCAACCTC |
F1/R1 and F2/R2 were the first and second pair of primers, respectively.
Table 3.
Effect of N- and C-terminal extensions with/without truncations on the procapsid structure and function*
| Procapsid mutant |
Procapsid morphology |
DNA packaging |
Virion assembly |
|---|---|---|---|
| N-Ex | Normal | ↓ 10-fold | ↓ 100-fold |
| C-Ex | Normal | ↓ 10-fold | ↓ 10-fold |
| N-DeEx | Normal | ~0 | ~0 |
| C-DeEx | Normal | ↓ 100-fold | ~0 |
The DNA packaging activity and phage assembly activity were given relative to those of the wild-type procapsid.
Expression and purification of procapsids
The constructed plasmid vectors were transformed into the E. coli strain HMS174 (DE3), respectively, for protein expression. A volume of 10 mL cell culture was grown overnight at 37°C in a Luria-Bertani medium containing 100 µg/mL ampicillin and periodically shaken at 250 rpm. One percent inoculation (v/v) was used for 500 mL culture, and 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) induction was applied when the optical density at 600 nm (OD600) reached 0.5–0.6. Cells were harvested 3 hours after induction by centrifugation at 5000 g for 20 minutes in a Beckman JS-7.5 rotor, and then stored at −70°C before purification. His- and Strep II–tagged connector proteins were purified by His- or Strep-affinity chromatography, respectively, whereas the assembled mutant procapsids were isolated using sucrose gradient sedimentation as described.62 Gel analysis (10% SDS-PAGE) of the purified mutant procapsids is shown in Figure 2.
Tobacco etch virus protease cleavage of N-terminal tags
The N-terminal tags of gp10 can be cleaved by TEV protease. The reaction was performed by incubating 1 mg of purified His–TEV protease with 20 mg of gp10 protein in the presence of 1 mM dithiothreitol at 4°C overnight. Removal of the N-terminal tag was verified by 10% SDS-PAGE gel electrophoresis (Figure 2).
DNA packaging and in vitro virion assembly assay
The procedures for DNA packaging and in vitro virion assembly assay have been described.43 Wild-type pRNA Ii′68 and SphI Ii′, a 26-base extension at the 3′ end of wild-type Ii′ pRNA,61,62 were used in comparing the DNA-packaging activity of different procapsid constructs. For in vitro virion assembly assay, pRNA Ii′ was included.
Binding of 3H-labeled pRNA to procapsid detected by 5–20% sucrose gradient sedimentation
Binding ability of pRNA Aa′69 to the mutant procapsids N-Ex and N-DeEx was tested through 5–20% sucrose gradient sedimentation as reported elsewhere.61,62
Transmission electron microscopy imaging
The TEM grids were coated with 400-mesh formvar and carbon and glow-discharged before use to increase their hydrophilicity. The purified procapsid protein was dialyzed and diluted when necessary before being negatively stained with 2% uranyl acetate. Samples were imaged on a Philips CM-100 transmission electron microscope with a 200- and 45-µm condenser and objective aperture, respectively, operated at 80 kV.
Sample preparation for single-molecule measurements
A perfusion chamber was constructed from a quartz slide with two holes and a glass coverslip held together with double-sided adhesive tape. A channel was cut into the tape connecting the two holes in the quartz slide before the assembly of the perfusion chamber. The procapsid mutants– Cy3-pRNA complexes were immobilized to the surface of the quartz slide via specific binding to anti-procapsid IgG (Proteintech Group, Chicago, Illinois). The surface of the quartz slide was first coated with IgG by incubation overnight at 4°C with phosphate-buffered saline containing 0.1 mg/mL IgG. The unbound IgG was rinsed with phosphate-buffered saline, and the quartz slide was blocked with 10 mg/mL ultrapure bovine serum albumin (Ambion, Austin, Texas) by incubation for 1 hour at 20°C. The flow chamber was rinsed again with TMS buffer (100 mM Tris-HCl, pH 8, 100 mM NaCl, 10 mM MgCl2) and then flushed with procapsid–Cy3-pRNA mixture (Figure 7).45 Samples were prepared by incubating for 30 minutes at room temperature with 0.1 µg individual mutant procapsids and 20 ng Cy3–pRNA bearing matching loops. A dilution factor of 20 or 200 from the stock solution was applied before applying the samples to the flow chamber for another 30 minutes of incubation. After removing the excess procapsid–Cy3-pRNA by TMS buffer, the measurements were performed in the presence of an oxygen-depleting solution containing glucose oxidase, catalase, β-d-glucose, and 2-mercaptoethanol.70
Total internal fluorescence microscope experimental setup
Individual procapsid–Cy3-pRNA complexes were imaged using a home-built, prism-type, dual-view total Single Molecule Dual-View Total Internal Reflection Fluorescence System (SMDV-TIRF).45 Briefly, the system was constructed based on an inverted microscope Olympus IX71 (Olympus, Center Valley, Pennsylvania) combined with a Dual-View imager (Mag Biosystems, Tucson, Arizona). The 532-nm laser (Crystalaser, Reno, Nevada) was used to excite the Cy3 flourescence. The angle of incidence for the excitation light was adjusted using a series of mirrors and a 45-degree quartz prism. Fluorescence was collected via a 60× oil-immersion objective, numerical aperture = 1.4 (Plan Apo, San Marcos, California) and detected by an iXon 887 V EMCCD camera (Andor Technology, South Windsor, Connecticut) after passing through the Dual-View imager. Data analysis was performed using the Andor IQ software provided with the camera. A more detailed description of the SMDV-TIRF has been given elsewhere.45
Acknowledgments
The authors would like to thank Karen Rufus from Vanderbilt University for providing the strain containing the plasmid for the expression of TEV protease; Nicola Stone-house from University of Leeds for providing the plasmids for the expression of gp10-Nhis and gp10-Chis; Debra Sherman for help in TEM imaging; Hui Zhang for helping with single-molecule imaging.
This research was supported by National Institutes of Health grant R01-EB03730.
References
- 1.Guo P, Grimes S, Anderson D. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage ϕ29. Proc Natl Acad Sci U S A. 1986;83:3505–3509. doi: 10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hohn B, Wurtz M, Klein B, Lustig A, Hohn T. Phage lamda DNA packaging in vitro. J Supramol Struct. 1974;2:302–317. doi: 10.1002/jss.400020220. [DOI] [PubMed] [Google Scholar]
- 3.Black LW. In vitro packaging of bacteriophage T4 DNA. Virology. 1981;113:336–344. doi: 10.1016/0042-6822(81)90160-4. [DOI] [PubMed] [Google Scholar]
- 4.Strobel E, Behnish W, Schmieger H. In vitro packaging of mature phage DNA by Salmonella phage P22. Virology. 1984;133:158–165. doi: 10.1016/0042-6822(84)90434-3. [DOI] [PubMed] [Google Scholar]
- 5.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage ϕ29. J Mol Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 6.Rao VB, Black LW. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4: construction of a defined in vitro DNA packaging system using purified terminase proteins. J Mol Biol. 1988;200:475–488. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- 7.Oliveira L, Alonso JC, Tavares P. A defined in vitro system for DNA packaging by the bacteriophage SPP1: insights into the headful packaging mechanism. J Mol Biol. 2005;353:529–539. doi: 10.1016/j.jmb.2005.08.063. [DOI] [PubMed] [Google Scholar]
- 8.Becker A, Gold M. Prediction of an ATP reactive center in the small subunit, gpNul of phage λ terminase enzyme. J Mol Biol. 1988;199:219–222. doi: 10.1016/0022-2836(88)90391-9. [DOI] [PubMed] [Google Scholar]
- 9.Catalano CE, Cue D, Feiss M. Virus DNA packaging: the strategy used by phage λ. Mol Microbiol. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 10.Yamada M, Fujisawa H, Kato H, Hamada K, Minagawa T. Cloning and sequencing of the genetic right end of bacteriophage T3 DNA. Virology. 1986;151:350–361. doi: 10.1016/0042-6822(86)90055-3. [DOI] [PubMed] [Google Scholar]
- 11.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 12.Casjens S, Huang WM, Hayden M, Parr R. Initiation of bacteriophage P22 DNA packaging series: analysis of a mutant which alters the DNA target specificity of the packaging apparatus. J Mol Biol. 1987;194:411–422. doi: 10.1016/0022-2836(87)90671-1. [DOI] [PubMed] [Google Scholar]
- 13.Prasad BVV, Prevelige PE, Marietta E, Chen RO, Thomas D, King J, et al. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- 14.Galisteo M, King J. Conformational transformations in the protein lattice of phage P22 procapsids. Biophys J. 1993;65:227–235. doi: 10.1016/S0006-3495(93)81073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skorupski K, Pierce JC, Sauer B, Sternberg N. Bacteriophage P1 genes involved in the recognition and cleavage of the phage packaging site (pac) J Mol Biol. 1992;223:977–989. doi: 10.1016/0022-2836(92)90256-j. [DOI] [PubMed] [Google Scholar]
- 16.Pfister P, Wasserfallen A, Stettler R, Leisinger T. Molecular analysis of Methanobacterium phage ψM2. Mol Microbiol. 1998;30:233–244. doi: 10.1046/j.1365-2958.1998.01073.x. [DOI] [PubMed] [Google Scholar]
- 17.Desiere F, Lucchini S, Brussow H. Evolution of Streptococcus thermophilus bacteriophage genomes by modular exchanges followed by point mutations and small deletions and insertions. Virology. 1998;241:345–356. doi: 10.1006/viro.1997.8959. [DOI] [PubMed] [Google Scholar]
- 18.Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, et al. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu D, Weller SK. Generic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology. 1998;243:32–44. doi: 10.1006/viro.1998.9041. [DOI] [PubMed] [Google Scholar]
- 20.Salmon B, Baines JD. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U(L)15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes. J Virol. 1998;72:3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng B, O’Connor CM, Kedes DH, Zhou ZH. Direct visualization of the putative portal in the Kaposi’s sarcoma-associated herpesvirus capsid by cryoelectron tomography. J Virol. 2007;81:3640–3644. doi: 10.1128/JVI.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheffczik H, Savva CG, Holzenburg A, Kolesnikova L, Bogner E. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 2002;30:1695–1703. doi: 10.1093/nar/30.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid SI, Hearing P. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J Virol. 1997;71:3375–3384. doi: 10.1128/jvi.71.5.3375-3384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bett A, Prevec L, Graham F. Packaging capacity and stability of human adenovirus type 5 vectors. J Virol. 1993;67:5911–5921. doi: 10.1128/jvi.67.10.5911-5921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Low JA, Christensen JB, Imperiale MJ. Role for the adenovirus IVa2 protein in packaging of viral DNA. J Virol. 2001;75:10446–10454. doi: 10.1128/JVI.75.21.10446-10454.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin EV, Senkevich VI, Chernos VI. Gene A32 product of vaccinia virus may be an ATPase involved in viral DNA packaging as indicated by sequence comparisons with other putative viral ATPases. Virus Genes. 1993;7:89–94. doi: 10.1007/BF01702351. [DOI] [PubMed] [Google Scholar]
- 27.Cassetti MC, Merchlinsky M, Wolffe EJ, Weiserg AS, Moss B. DNA package mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graeble M, Hearing P. Cis and trans requirements for the selective packaging of adenovirus type 5 DNA. J Virol. 1992;66:723–731. doi: 10.1128/jvi.66.2.723-731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins RR, Becker A. Interaction of terminase, the DNA packaging enzyme of phage λ, with its cos DNA substrate. J Mol Biol. 1995;252:31–45. doi: 10.1006/jmbi.1995.0473. [DOI] [PubMed] [Google Scholar]
- 30.Hang JQ, Catalano CE, Feiss M. The functional asymmetry of cosN, the nicking site for bacteriophage λDNA packaging, is dependent on the terminase binding site, cosB. Biochemistry. 2001;40:13370–13377. doi: 10.1021/bi011126r. [DOI] [PubMed] [Google Scholar]
- 31.Guo P. Bacterial virus phi29 DNA-packaging motor and its potential applications in gene therapy and nanotechnology. Methods Mol Biol. 2005;300:285–324. doi: 10.1385/1-59259-858-7:285. [DOI] [PubMed] [Google Scholar]
- 32.Hwang Y, Feiss M. A defined system for in vitro λ-DNA packaging. Virology. 1997;211:367–376. doi: 10.1006/viro.1995.1419. [DOI] [PubMed] [Google Scholar]
- 33.Zhang CL, Garver K, Guo P. Inhibition of phage ϕ29 assembly by antisense oligonucleotides targeting viral pRNA essential for DNA packaging. Virology. 1995;211:568–576. doi: 10.1006/viro.1995.1439. [DOI] [PubMed] [Google Scholar]
- 34.Trottier M, Zhang CL, Guo P. Complete inhibition of virion assembly in vivo with mutant pRNA essential for phage ϕ29 DNA packaging. J Virol. 1996;70:55–61. doi: 10.1128/jvi.70.1.55-61.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogner E. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev Med Virol. 2002;12:115–127. doi: 10.1002/rmv.344. [DOI] [PubMed] [Google Scholar]
- 36.Visalli RJ, van Zeijl M. DNA encapsidation as a target for anti-herpesvirus drug therapy. Antiviral Res. 2003;59:73–87. doi: 10.1016/s0166-3542(03)00108-6. [DOI] [PubMed] [Google Scholar]
- 37.Shu D, Huang L, Hoeprich S, Guo P. Construction of phi29 DNA-packaging RNA (pRNA) monomers, dimers and trimers with variable sizes and shapes as potential parts for nano-devices. J Nanosci Nanotechnol. 2003;3:295–302. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 38.Hess H, Bachand GD, Vogel V. Powering nanodevices with biomolecular motors. Chemistry. 2004;10:2110–2116. doi: 10.1002/chem.200305712. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt JJ, Montemagno CD. Bionanomechanical systems. Annu Rev Mater Res. 2004;34:315–337. [Google Scholar]
- 40.Shu D, Moll D, Deng Z, Mao C, Guo P. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett. 2004;4:1717–1724. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo P. RNA nanotechnology: engineering, assembly and applications in detection, gene delivery and therapy. J Nanosci Nanotechnol. 2005;5:1964–1982. doi: 10.1166/jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meifer WJJ, Horcajadas JA, Salas M. Phi29 family of phages. Microbiol Mol Biol Rev. 2001;65:261–287. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CS, Guo P. In vitro assembly of infectious virions of ds-DNA phage ϕ29 from cloned gene products and synthetic nucleic acids. J Virol. 1995;69:5018–5023. doi: 10.1128/jvi.69.8.5018-5023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage ϕ29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 45.Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guasch A, Pous J, Ibarra B, Gomis-Rüth FX, Valpuesta JM, Sousa N. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 47.Orlova EV, Gowen B, Dröge A, Stiege A, Weise F, Lurz R, et al. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Badasso MO, Leiman PG, Tao Y, He Y, Ohlendorf DH, Rossman MG, et al. Purification, crystallization and initial X-ray analysis of the head-tail connector of bacteriophage phi29. Acta Crystallogr D. 2000;56:1187–1190. doi: 10.1107/s0907444900009239. [DOI] [PubMed] [Google Scholar]
- 49.Doan DN, Dokland T. The gpQ portal protein of bacteriophage P2 forms dodecameric connectors in crystals. J Struct Biol. 2007;157:432–436. doi: 10.1016/j.jsb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Carazo JM, Fujisawa H, Nakasu S, Carrascosa JL. Bacteriophage T3 gene 8 product oligomer structure. J Ultrastruct Mol Struct Res. 1986;94:105–113. doi: 10.1016/0889-1605(86)90056-x. [DOI] [PubMed] [Google Scholar]
- 51.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78:12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cerritelli ME, Studier FW. Purification and characterization of T7 head-tail connectors expressed from the cloned gene. J Mol Biol. 1996;285:299–307. doi: 10.1006/jmbi.1996.0251. [DOI] [PubMed] [Google Scholar]
- 53.Kochan J, Carrascosa JL, Murialdo H. Bacteriophage λ preconnectors: purification and structure. J Mol Biol. 1984;174:433–447. doi: 10.1016/0022-2836(84)90330-9. [DOI] [PubMed] [Google Scholar]
- 54.Agirrezabala X, Martin-Benito J, Valle M, Gonzáles JM, Valencìa A, Valpuesta JM, et al. Structure of the connector of bacteriophage T7 at 8 Å resolution: structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005;347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Cingolani G, Moore SD, Prevelige PE, Jr, Johnson JE. Preliminary crystallographic analysis of the bacteriophage P22 portal protein. J Struct Biol. 2002;139:46–54. doi: 10.1016/s1047-8477(02)00512-9. [DOI] [PubMed] [Google Scholar]
- 56.Kato H, Baschong C. Isolation of a gp20-complex and its role in in vitro assembly of both prohead and core of bacteriophage T4. Virology. 1997;227:400–408. doi: 10.1006/viro.1996.8322. [DOI] [PubMed] [Google Scholar]
- 57.Cerritelli ME, Studier FW. Assembly of T7 capsids from independently expressed and purified head protein and scaffolding protein. J Mol Biol. 1996;258:286–298. doi: 10.1006/jmbi.1996.0250. [DOI] [PubMed] [Google Scholar]
- 58.Guo P, Erickson S, Xu W, Olson N, Baker TS, Anderson D. Regulation of the phage ϕ29 prohead shape and size by the portal vertex. Virology. 1991;183:366–373. doi: 10.1016/0042-6822(91)90149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu CY, Morais MC, Battisti AJ, Rossmann MG, Prevelige PE., Jr Molecular dissection of ϕ29 scaffolding protein function in an in vitro assembly system. J Mol Biol. 2007;366:1161–1173. doi: 10.1016/j.jmb.2006.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, et al. Structure determination of the head-tail connector of bacteriophage phi29. Acta Crystallogr D Biol Crystallogr. 2001;57:1260–1269. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- 61.Xiao F, Moll D, Guo S, Guo P. Binding of pRNA to the N-terminal 14 amino acids of connector protein of bacterial phage ϕ29. Nucleic Acids Res. 2005;33:2640–2649. doi: 10.1093/nar/gki554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun J, Cai Y, Moll WD, Guo P. Controlling bacteriophage phi29 DNA-packaging motor by addition or discharge of a peptide at N-terminus of connector protein that interacts with pRNA. Nucleic Acids Res. 2006;34:5482–5490. doi: 10.1093/nar/gkl701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;20:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z, Wang X, Rana TM. Protein orientation in the Tat-TAR complex determined by psoralen photocross-linking. J Biol Chem. 1996;19:16995–16998. doi: 10.1074/jbc.271.29.16995. [DOI] [PubMed] [Google Scholar]
- 65.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc Natl Acad Sci U S A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atz R, Ma S, Gao J, Anderson DL, Grimes S. Alanine scanning and Fe-BABE probing of the bacteriophage ϕ29 prohead RNA-connector interaction. J Mol Biol. 2007;369:239–248. doi: 10.1016/j.jmb.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson MA, Wood JP, Capaldi SA, Baron AJ, Gell C, Smith DA, et al. Affinity of molecular interactions in the bacteriophage ϕ29 DNA packaging motor. Nucleic Acids Res. 2006;34:2698–2709. doi: 10.1093/nar/gkl318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C, Sheng S, Shao Z, Guo P. A dimer as a building block in assembling RNA. A hexamer that gears bacterial virus ϕ29 DNA-translocating machinery. J Biol Chem. 2000;275:17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 69.Chen C, Zhang C, Guo P. Sequence requirement for hand-in-hand interaction in formation of pRNA dimers and hexamers to gear ϕ29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]