Abstract
The accurate identification of Leishmania species is important for the treatment of infected patients. Molecular methods offer an alternative to time consuming traditional laboratory techniques for species determination. We redesigned a 7SL rRNA gene based PCR and sequence assay for increased species identification. DNA extracted from 17 reference strains and 10 cultured clinical isolates was examined. Sequence comparison was used successfully to identify organisms to the complex level with intercomplex similarity ranging from 77.5% to 98.4%. Many species within each complex were discriminated accurately by this method including: L. major, L. tropica, L. aethiopica, L. guyanensis, and the previously indistinguishable L. brasiliensis and L. panamensis. The L. donovani complex members remain indistinguishable by this method, as are the representatives of L. amazonensis/L. garnhami and L. mexicana/L. pifanoi.
Leishmaniasis is a vector borne disease caused by protozoan parasites belonging to the genus Leishmania. Over 20 species of Leishmania are found in 88 countries worldwide; causing approximately 2 million new reported infections per year (WHO Report 2004). Due to the large number of infective species and variability in host genetics and immune status a wide spectrum of clinical presentations may occur and species determination may be complicated ( Romero, Vinitius De Farias Guerra et al. 2001; Vega-Lopez 2003; Reithinger, Dujardin et al. 2007).
Research suggests that antiparasitic agents used for the treatment of Leishmaniasis demonstrates species dependent efficacy. Romero (2001) reported that meglumine antimoniate may show species dependent treatment success with L. braziliensis compared to L. guyanensis (Romero, Vinitius De Farias Guerra et al. 2001); and, Navin (1992) demonstrated that sodium stibogluconate produced a higher cure rate in patients with L. braziliensis lesions (96%) compared to those with L. mexicana lesions (57%) (Navin, Arana et al. 1992). Soto et. al established that miltefosine was effective against L. panamensis (>90% cure) but was not effective against L. braziliensis (Soto, Toledo et al. 2002). Additionally, L. braziliensis can be naturally azole resistant (Saenz, Paz et al. 1990; Navin, Arana et al. 1002; Rangel, Dagger et al. 1996). Traditional laboratory diagnostic methods for leishmaniasis include direct microscopy of skin sections and lengthy and technically demanding culture followed by analysis with multi-locus enzyme electrophoresis (MLEE). To provide more timely identification we previously reported on the use of the partial 7SL ribosomal RNA gene sequence, present in single copy, for complex identification (Zelazny, Fedorko et al. 2005). While the previous assay was useful for many species, it was unable to differentiate L. braziliensis from L. panamensis. Additionally, it was not tested in a real time format that allows for melting peak visualization rather than agarose gel electrophoresis to confirm product production. Through the use of novel primers, we extended the previously described 7SL sequenced region to include a small segment of a putative tRNA gene followed by a 98bp spacer region and a large segment of the 7SL rRNA gene. While still employing single PCR and sequencing reactions, we gain a larger sequenced area and enhanced species identification capabilities. Interestingly, the spacer region between the two genes contains conserved base differences that were especially important for distinguishing between the L. braziliensis and L. guyanensis complexes.
Strains of Leishmania used in this work were cultured and DNA was extracted as previously described (Zelazny, Fedorko et al. 2005). PCR was carried out with M13 tailed primers (LeishFW- 5′GTAAAACGACGGCCAGCATCCGTGACAGGATTCGAACC-3′) corresponding to sequence ~200bp upstream from the putative 7SL gene start sequence and (LeishRV 5′-CAGGAAACAGCTATGACCGTGGGGCTCAAGTGCGGACATG-3′) corresponding to sequence at position 36 bp upstream from the end of the putative 7SL gene sequence. Real-Time PCR amplification was carried out with a reaction mixture containing 50ng of extracted DNA, 0.4μM LeishFW primer, 0.4μM LeishRV primer, 1X QuantiTect SYBR Green PCR Mastermix (Qiagen, Valencia, Ca), and 0.5 units UNG Uracil-DNA-Glycosylase (Roche Applied Science, Indianapolis, IN). The reaction was brought to 20ul total volume with PCR grade water. Real Time PCR was carried out in a Rotor-Gene RG-3000 (Corbett Research, San Francisco, CA) with a program consisting of a warming to 50°C for 2 minutes, initial denaturation at 95°C for 15 minutes, followed by 45 cycles of 95°C for 15 seconds, 65°C for 30 seconds, 72°C for 30 seconds resulting in a ~465 bp amplification product. Products used for sequencing reactions were purified using Microcon YM-100 Centrifugal Filter Devices (Millipore, Billerica, MA) following manufacture recommendations. Sequencing and analysis was carried out as previously described except using The Lasergene program suite (version 7.1.0; DNASTAR Inc. Madison, WI). Leishmania spp. sequenced regions, presented in this paper, were deposited in GenBank under accession numbers: FJ525405, FJ525406, FJ525407, FJ525408, FJ525409, FJ525410, FJ525411, FJ525412, FJ525413, FJ525414, FJ525415, FJ525416, FJ525417, FJ525418, FJ525419, FJ525420.
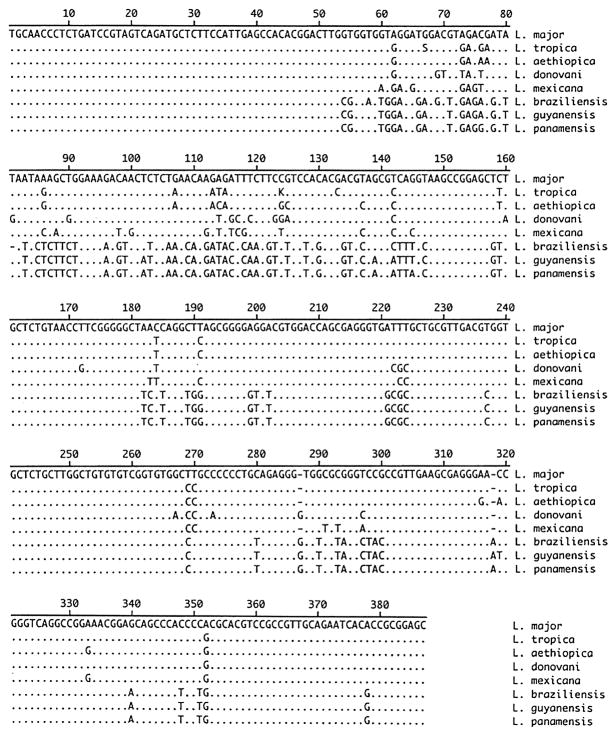
Seventeen Leishmania reference strains and 10 clinical isolates were examined in this study. (Table 1) These organisms represent the most commonly isolated species of Leishmania causing human disease (Jeronimo S., et al (2006). Nucleotide sequence information for each isolate was compared pairwise to determine percent similarity. After primer removal, a 385–387bp amplified product was seen (Figure 1). Some strains containing insertions, as compared to L. major, have been previously described(Zelazny, Fedorko et al. 2005).
Figure 1.
Alignment of PCR amplified region from Leishmania spp. Representative sequence information from one isolate of each species is shown. Dots indicate identity with Leishmania major sequence.
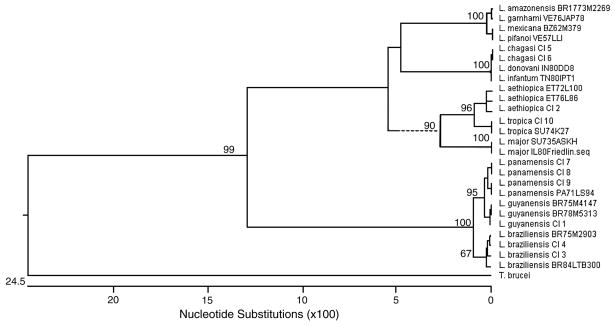
The sequencing strategy used was able to clearly differentiate the tested strains to the complex level (L. major, L. tropica. L. aethiopica, L. braziliensis, L. guyanensis, L. mexicana, L. donovani) (Figure 1). Intercomplex similarity ranged from 77.5 % (L. guyanensis v. L. donovani) to 98.4 % (L. braziliensis v. L. guyanensis). In accordance with other sequence regions examined for use in identification schemes, intraspecies polymorphisms were present (Foulet, Botterel et al. 2007). When multiple isolates of a species were examined, intraspecies similarity ranged from 99.2 to 100%. Of four L. aethopica isolates tested, two isolates were conserved and two exhibited 1 or 2 bp changes. Within the four L. braziliensis strains examined, L. braziliensis MHOM/BR/84/LTB300 contained a change within the upstream putative noncoding region; and, L. braziliensis MHOM/BR/75/M2903 contained a deletion within this region. Of four L. panamensis isolates, two examined have a 1bp change. However, these polymorphisms did not affect the ability to identify species and all are appropriately grouped on the phylogenetic tree (Figure 2).
Figure 2.
Phylogenetic tree of 27 reference and clinical isolates of Leishmania spp.. constructed by the neighbor-joining method, using Trypanosoma brucei strain 427 homologous sequence as the outgroup. Numbers on branches represent the percentage of 1,000 bootstrap samples supporting the branch. Only values >50 are shown. CI=clinical isolate.
L. panamensis and the closely related L. guyanensis species isolates are distinguished by 2 conserved basepair differences and there are 5 conserved basepair changes between L. braziliensis and L. panamensis isolates allowing for a clear division in the phylogenetic tree (Figure 1, Figure 2). Additionally, while only previously separated by a single base, L. mexicana and L. amazonensis are separated by 2bp differences. Due to the small number of base changes between these species we suggest that bidirectional sequencing and a comparison of both sequences always be performed. The new assay did not allow for separation of L. amazonensis and L. garnhami or L. mexicana and L. pifanoi. As expected, the sequenced region was unable to differentiate among members of the L. donovani complex (L. donovani, L. infantum, L. chagasi). Previous work suggests that L. infantum and L. chagasi are generally considered to be genetically identical (Mauricio, Stothard et al. 2000) and all three species of the complex have been shown to be extremely similar (Kuhls, Mauricio et al. 2005; Mauricio, Yeo et al. 2006). Species dependent treatment schemes are not yet being employed for the L. donovani complex, lessening the need for species level discrimination ( Chappuis, Sundar et al. 2007).
Here, we evaluated a Real-Time PCR assay followed by a single sequencing reaction for the identification of Leishmania spp. All complexes are clearly differentiated and L. braziliensis and L. panamensis are clearly separated, allowing for the option of species-specific treatment. Additionally, the primers utilized show no similarity to human DNA and no amplification was observed when the assay was preformed with DNA extracted from human material (blood, CSF, skin biopsy, joint fluid). When the PCR assay was carried out with a known positive human skin biopsy sample we were able to identify the infecting species accurately and further work will be carried out to evaluate the use of this assay for clinical specimens.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chappuis F, Sundar, et al. Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat Rev Microbiol. 2007;5(11):873–82. doi: 10.1038/nrmicro1748. [DOI] [PubMed] [Google Scholar]
- Foulet F, Botterel, et al. Detection and identification of Leishmania species from clinical specimens by using a real-time PCR assay and sequencing of the cytochrome B gene. J Clin Microbiol. 2007;45(7):2110–5. doi: 10.1128/JCM.02555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo SB, et al. Leishmaniasis. In: Guerrant RL, Walker DH, Weller PF, editors. Tropical Infectious Diseases Principles, Pathogens, and Practice. 2. Philadelphia: Elsevier; 2006. pp. 1095–1113. [Google Scholar]
- Kuhls K, I, Mauricio L, et al. Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes Infect. 2005;7(11–12):1224–34. doi: 10.1016/j.micinf.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Lukes J, I, Mauricio L, et al. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U S A. 2007;104(22):9375–80. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio IL, Stothard JR, et al. The strange case of Leishmania chagasi. Parasitol Today. 2000;16(5):188–9. doi: 10.1016/s0169-4758(00)01637-9. [DOI] [PubMed] [Google Scholar]
- Mauricio IL, Yeo M, et al. Towards multilocus sequence typing of the Leishmania donovani complex: resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD) Int J Parasitol. 2006;36(7):757–69. doi: 10.1016/j.ijpara.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Navin TR, Arana BA, et al. Placebo-controlled clinical trial of sodium stibogluconate (Pentostam) versus ketoconazole for treating cutaneous leishmaniasis in Guatemala. J Infect Dis. 1992;165(3):528–34. doi: 10.1093/infdis/165.3.528. [DOI] [PubMed] [Google Scholar]
- Rangel, Dagger HF, et al. Naturally azole-resistant Leishmania braziliensis promastigotes are rendered susceptible in the presence of terbinafine: comparative study with azole-susceptible Leishmania mexicana promastigotes. Antimicrob Agents Chemother. 1996;40(12):2785–91. doi: 10.1128/aac.40.12.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reithinger R, Dujardin JC, et al. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7(9):581–96. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- Romero GA, Vinitius De Farias Guerra M, et al. Comparison of cutaneous leishmaniasis due to Leishmania (Viannia) braziliensis and L. (V.) guyanensis in Brazil: clinical findings and diagnostic approach. Clin Infect Dis. 2001;32(9):1304–12. doi: 10.1086/319990. [DOI] [PubMed] [Google Scholar]
- Saenz RE, Paz H, et al. Efficacy of ketoconazole against Leishmania braziliensis panamensis cutaneous leishmaniasis. Am J Med. 1990;89(2):147–55. doi: 10.1016/0002-9343(90)90292-l. [DOI] [PubMed] [Google Scholar]
- Soto JM, Toledo JT, et al. Treatment of cutaneous leishmaniasis with a topical antileishmanial drug (WR279396): phase 2 pilot study. Am J Trop Med Hyg. 2002;66(2):147–51. doi: 10.4269/ajtmh.2002.66.147. [DOI] [PubMed] [Google Scholar]
- Vega-Lopez F. Diagnosis of cutaneous leishmaniasis. Curr Opin Infect Dis. 2003;16(2):97–101. doi: 10.1097/00001432-200304000-00006. [DOI] [PubMed] [Google Scholar]
- Zelazny AM, Fedorko DP, et al. Evaluation of 7SL RNA gene sequences for the identification of Leishmania spp. Am J Trop Med Hyg. 2005;72(4):415–20. [PubMed] [Google Scholar]