Abstract
Rationale
The intercalated disc (ID) is a highly specialized cell-cell contact structure that ensures mechanical and electrical coupling of contracting cardiomyocytes. Recently, the ID has been recognized to be a hot spot of cardiac disease, in particular inherited cardiomyopathy.
Objective
Given its complex structure and function we hypothesized that important molecular constituents of the ID still remain unknown.
Methods
Using a bioinformatic screen, we discovered and cloned a previously uncharacterized 54 kDa cardiac protein which we termed Myozap (Myocardium-enriched ZO-associated protein).
Results
Myozap is strongly expressed in the heart and lung. In cardiac tissue it localized to the ID and directly binds to desmoplakin and ZO-1. In a yeast-two hybrid screen for additional binding partners of Myozap we identified myosin phosphatase-RhoA interacting protein (MRIP), a negative regulator of Rho activity. Myozap, in turn, strongly activates SRF-dependent transcription through its ERM (Ezrin/radixin/moesin)-like domain in a Rho-dependent fashion. Finally, in vivo knockdown of the Myozap orthologue in zebrafish led to severe contractile dysfunction and cardiomyopathy.
Conclusions
Taken together, these findings reveal Myozap as a previously unrecognized component of a Rho-dependent signaling pathway that links the intercalated disc to cardiac gene regulation. Moreover, its subcellular localization and the observation of a severe cardiac phenotype in zebrafish, implicate Myozap in the pathogenesis of cardiomyopathy.
Keywords: myocytes, cardiac, cardiomyopathies, serum response factor
Introduction
A unique morphological feature of cardiac muscle is the intercalated disc (ID), a highly specialized cell-cell contact structure that connects individual cardiomyocytes. Several independent substructures of the ID have been distinguished, including (i) desmosomes, which link the intermediate filament apparatus of the cell, (ii) fasciae adhaerentes, to which the actomyosin filament bundles are attached, and (iii) gap junctions, which allow free movement of ions and small molecules between adjacent cardiomyocytes. Together, these structures form a network of proteins which ensure mechanical and electrical coupling of contracting cardiomyocytes (“functional syncytium”) (reviewed in1). Recently, immunoelectron microscopy and biochemical studies have revealed that major constituents of the desmosomes such as desmoplakin, plakophilin-2 and the cadherins can also be detected in adherens junctions2. Correspondingly, typical components of adherens and gap junctions were found to colocalize with desmosomal molecules3. Thus, it has been proposed that in the heart these specialized cell-cell contact structures are merged in an “area composita” (AC)2,4.
Given the critical importance of the intercalated disc for cardiac integrity and function, it is perhaps not surprising that the ID has become a hot spot of inherited cardiac disease. In particular, arrhythmogenic right ventricular cardiomyopathy (ARVC), has been coined a “desmosome cardiomyopathy”5, since the majority of the ARVC-associated genes encode for desmosomal ID components6. Yet, the complex interplay of the multiprotein network of the ID still remains poorly understood and it is likely that many critical ID components have still not been identified to date.
In an effort to identify such novel cardiac ID components, we explored expressed sequence tag (EST) databases for previously uncharacterized sequences with high abundance in cardiac cDNA libraries. With this approach, we discovered a novel open reading frame encoding for a 466 amino acid protein which we termed Myozap (Myocardium-enriched Zo-1-interacting protein). Here we show that this cardiac-enriched protein colocalizes with β-catenin, N-cadherin as well as plakophilin-2 at the intercalated disc and directly binds to ID components such as desmoplakin and ZO-1. In addition, Myozap promotes serum-response-factor (SRF)-signaling in a Rho-dependent fashion, while a newly identified Myozap binding partner, MRIP (Myosin phosphatase-RhoA interacting protein), inhibits this pathway. Finally, knockdown of myozap’s orthologue in zebrafish results in cardiomyopathy with severe contractile dysfunction.
Materials and Methods
Experimental procedures for cloning and the subsequent bioinformatics, Northern blot analysis and radioactive in situ hybridization, the generation of a Myozap-specific antiserum and western blot analyses, immunofluorescence and immunoelectron microscopy, yeast two hybrid assays, tissue culture, immunoprecipitations, reporter gene assays as well as zebrafish injection procedures, and fractional shortening measurement are provided in the online methods supplement.
Statistical analyses of the data were carried out using ANOVA followed by Student-Newman-Keuls post-hoc tests. If appropriate, Student’s t-test was employed (two sided, assuming similar variances). P values <0.05 were considered statistically significant.
The authors had full access to the data and take full responsibility for their integrity. All authors have read and agree to the manuscript as written.
Results
Myozap is a highly conserved cardiac protein
In order to identify novel cardiac-enriched genes, we searched the expressed sequence tag (EST) database for uncharacterized sequences predominantly found in cardiac cDNA libraries7. Several of these ESTs corresponded to the UNIGENE cluster Mm.27585, which we used to construct an open reading frame (ORF, GenBankTM accession number bankit1213482, FJ970029) encoding for a 466 amino acid polypeptide with a calculated molecular weight of 54.2 kDa (Figure 1A). A protein sequence alignment between the putative human, mouse, rat, xenopus, and zebrafish Myozap homologues revealed high evolutionary sequence conservation among mammalian species (Figure 1A). Molecular cloning of human, murine and rat myozap confirmed the predicted amino acid sequences.
Figure 1. Protein sequence alignment.
(A) Protein sequence alignment of human (NP_001018110), mouse (NP_001028380), rat (NP_001014233), Xenopus (XP_683507), and zebrafish (XP_683507) Myozap shows high conservation among species. Conserved amino acids are highlighted. h: human; b: bovine; m: mouse; r: rat, x: xenopus, d: zebrafish. (B) schematic structure of the Myozap protein revealing a central ERM-like domain, shown in part in an alignment with the known ERM protein Moesin.
According to the draft of the human genome, myozap maps to chromosome 15q21.3 and encompasses 13 exons, spanning a total of ~125.6 kb. Interestingly, the myozap gene is located in close proximity to a gene termed GRINL1A. In fact, Myozap’s sequence has been previously deposited in the NCBI databank under the denotation “Grinl1a isoform 7”. Moreover, it has been suggested that a common mRNA (Gcom) of Myozap/Grinl1a7 and a downstream part of the GRINL1A locus might exist8. However, utilizing rT-PCR experiments with several primers, we could only detect two distinct transcripts (online figure I), suggesting that Myozap and Gcom/Gdown are independent proteins. This notion is further supported by the finding that no overlapping ESTs between Myozap and the downstream gene Grinl1A exist in the database (online figure II). Taken together, these results make the existence of a common mRNA - at least at significant levels rather unlikely. Nevertheless, in order to acknowledge that Myozap´s sequence has been deposited as “Grinl1a7”, we subsequently used the gene name “Myozap/Grinl1a7”.
Myozap does not contain significant sequence homology to any other known protein. However, between amino acids 181 and 348, myozap contains an ERM (Ezrin, Radixin, and Moesin)-like domain (Figure 1B), which has been identified in several other proteins that link the cell membrane and the cytoskeleton9.
Myozap/Grinl1a7 is predominantly expressed in the heart
To determine myozap’s expression in different tissues, we performed multi-tissue Northern blot experiments using mRNA from various human and mouse tissues (Figure 2A). Human Myozap mRNA is predominantly expressed in the heart, and to a much lesser degree in skeletal muscle, placenta and lung. The murine Myozap/Grinl1a7 gene displayed a similar expression pattern, again with predominant expression in the myocardium. In order to be able to analyze myozap on the protein level, an antiserum was generated in rabbits utilizing a myozap-specific synthetic peptide. As myozap´s ERM-like domain suggested an association with the cell membrane, we performed Western blot experiments with membrane-rich fractions from various rat tissues. Myozap- and control-transfected HEK 293 cells served as positive and negative controls. These experiments revealed strong synthesis of myozap in the heart and in lung tissue at the predicted size of ~ 55 kDa, other tissues examined were completely negative (Figure 2B). As an additional control, the same tissues and cell culture extracts were subsequently probed with the antiserum in combination with the synthetic peptide the antiserum was generated against. No significant signals could be observed, confirming the specificity of the antiserum. To investigate myozap’s subcellular distribution, we treated the membrane rich fraction with a Triton-containing buffer, which yielded a Triton (detergent)-soluble fraction representing the membrane fraction and a Triton-insoluble fraction rich in cytoskeletal proteins10. In particular, the Triton-insoluble fraction yielded significant bands for myozap in heart and lung tissue (Figure 2C).
Figure 2. Expression patterns of Myozap.
(A) Northern blots from adult human and mouse tissues. Human Myozap is predominantly expressed in cardiac tissue, and to a lesser degree in skeletal muscle, placenta and lung. Mouse Myozap could not be detected in skeletal muscle. (B) A Myozap-specific antiserum was generated using a Myozap-derived synthetic peptide. A Western blot from membrane fractions from various rat tissues as well as a positive and negative controls (Myozap and empty vector transfected HEK 293 cells, respectively) revealed an expression of Myozap protein in the heart and in the lung. The same tissues and controls were probed with the antiserum plus the peptide. No significant signal could be observed, confirming the specificity of the antiserum. (C) For specification of Myozap’s subcellular distribution, the membrane rich fraction was treated with a Triton-containing buffer which yielded a Triton (detergent)-soluble fraction representing the membrane fraction and a Triton-insoluble fraction rich in cytoskeletal proteins. Particularly the Triton-insoluble fraction yielded significant bands for myozap in heart and lung tissue. (D) In situ hybridizations of adult murine cardiac tissue utilizing a Myozap probe. The antisense probed revealed an intense signal throughout the myocardium, whereas no significant signal could be observed with the sense probe. The coronary vasculature (v) is spared, suggesting a cardiomyocyte-specific expression pattern.
To further examine myozap’s expression in the heart, in situ hybridizations of adult murine cardiac tissue were performed which revealed an intense and specific signal throughout the myocardium (Figure 2D). The coronary vasculature (v) was spared, consistent with a cardiomyocyte-specific expression pattern.
Myozap expression is developmentally regulated
In order to investigate the developmental expression pattern of Myozap, we performed an additional series of in situ hybridization experiments of staged mouse embryos at embryonic days (ED) 8.0, 9.0, 11.5, 12.5, 15.5, as well as postnatal day (P) 1. At ED 8.0 of mouse embryonic development, Myozap/Grinl1a7 gene expression was confined to the embryonic vasculature (Figure 3A, dorsal aortae, da). At ED 9.0, the vasculature still shows a strong signal, including the dorsal aortae (da) and head veins (hv). In addition, the endocardium (ec) of the outflow tract (ot) and the primitive ventricle (pv), were intensely stained, whereas the myocardium was still negative at this timepoint (Figure 3B). At ED 11.5, not only the vasculature including the ductus cuvieri (which subsequently develops into the vena cava), but also the trabeculated myocardium (m) of the left ventricle revealed a strong hybridization signal (Figure 3C). One day later, at ED 12.5 (Figure 3D), an intense signal was observed throughout the entire myocardium (left ventricle, lv, and left atrium, la). Mesenchymal tissue within the lung (l) was also myozap-positive. At late embryonic stages (ED15.5) and postnatal day 1(P1), a strong hybridization signal of the heart and lung was maintained (Figure 3E&F), whereas expression in the vasculature had become undetectable. Taken together, these findings indicate that Myozap/Grinl1a7 expression undergoes developmental regulation with a shift from a vascular and endocardial pattern to a strong and specific myocardial staining.
Figure 3. Developmental regulation of Myozap expression.
(A) At embryonic day (ED) 8.0 of mouse development Myozap expression is confined to the embryonic vasculature (i.e. dorsal aortae) as revealed by in situ hybridizations. (B) At ED 9.0, the vasculature still displays a significant signal, including the dorsal aortae and head veins. In addition, the endocardium of the outflow tract and the primitive ventricle, but not the myocardium is strongly stained. (C) At ED 11.5, blood vessels such as the ductus cuvieri stain positive for Myozap. In addition, now the trabeculated myocardium of the left ventricle reveals a signal. (D) At ED 12.5, an intense signal is observed in the myocardium. Also, mesenchymal tissue within the lung is Myozap-positive. (E) and (F), a strong signal within the entire heart as well as lung is maintained throughout further development (ED 15.5 (E) and P1 (F)). Abbreviations used: da: dorsal aorta, hv: head vein, ec: endocardium, pv: primitive ventricle, ot: outflow tract, dc: ductus cuvieri, m: trabeculated myocardium, avc: atrio-ventricular cushion, la: left atrium, lv: left ventricle, l: lung, rv: right ventricle
Myozap is a novel component of the intercalated disc
Given myozap’s high protein levels in the myocardium, we next aimed to determine its subcellular localization in isolated adult murine cardiomyocytes.Using the myozap-specific antiserum, we detected a strong signal at the ID, as shown by colocalization with N-cadherin (Figure 4A). In addition, a weak staining of the sarcomeric Z-discs (as determined by colocalization with α-actinin) was observed (data not shown).
Figure 4. Subcellular localization of Myozap within isolated cardiomyocytes and in the intact heart.
(A) Cardiomyocytes from adult mouse heart stained with a Myozap-specific antiserum reveal an intense signal at the intercalated disc, as shown by co-localization with cadherin. Also, a weak staining of the Z-discs is observed. Scalebar: 20 µm. (B) Sections of mouse heart showing a strong signal at the intercalated disc with Myozap antiserum, co-localizing with cadherin, plakoglobin as well as plakophilin-2. Scalebars: 20 µm. (C) Immunoelectron microscopy of a cryosection through a bovine heart utilizing the Myozap-antiserum. An intense signal is seen at the intercalated disc. Scalebar: 1.18 µm
In sections of intact mouse heart tissue myozap was again predominantly found at intercalated discs where it colocalized with N-cadherin (Figure 4B, upper panels). Myozap also colocalized with plakoglobin and plakophilin-2, two prototypical area composita proteins (Figure 4B, lower panels). These data were further corroborated by immunoelectron microscopy analyses on cryosections through bovine hearts. Again, the myozap-specific antiserum was used, followed by an incubation step using a nanogold-coupled secondary antibody. We observed an intense signal at the cell-cell junctions between adjacent cardiomyocytes, further supporting the notion that myozap is a novel component of the ID (Figure 4C).
Myozap interacts and colocalizes with desmoplakin
Given myozap’s ID localization, we examined whether it binds to other known ID proteins, such as desmoplakin, plakoglobin and plakophilin-2. Co-immunoprecipatitation experiments failed to demonstrate a direct interaction of myozap with plakophilin-2 and plakoglobin (data not shown). In contrast, experiments with transfected HEK293T cells revealed an intense co-precipitation of desmoplakin with myozap, while no specific band was observed with empty vector alone (Figure 5A). The positive co-immunoprecipitation could be confirmed with endogenous proteins derived from adult mouse heart lysates (Figure 5B). Moreover, these findings were further supported by the colocalization of both proteins at the ID in immunostainings of adult bovine myocardium (Figure 5C).
Figure 5. Myozap interacts and co-localizes with desmoplakin.
(A) A coimmunoprecipitation experiment with transfected HEK293T reveals interaction of desmoplakin with Myozap (lane 1), whereas no signal was observed with empty vector alone (lane 2). IP: immunoprecipitation, WB: Western blot, IgH: immunoglobulin heavy chain. (B) Co-IP of endogenous desmoplakin and myozap from neonatal mouse hearts with anti-desmoplakin and mouse control serum, respectively. Western blots on immunoprecipitates were probed with anti-myozap or anti-demosplakin antibodies as indicated. (C) An immunofluorescence experiment revealing co-localization of Myozap (red) with desmoplakin (green) in a cryosection through bovine heart utilizing Myozap- and desmoplakin-antisera. Scalebar: 50 µm
Myozap binds to ZO-1 (zonula occludens-1) and colocalizes with the actin cytoskeleton
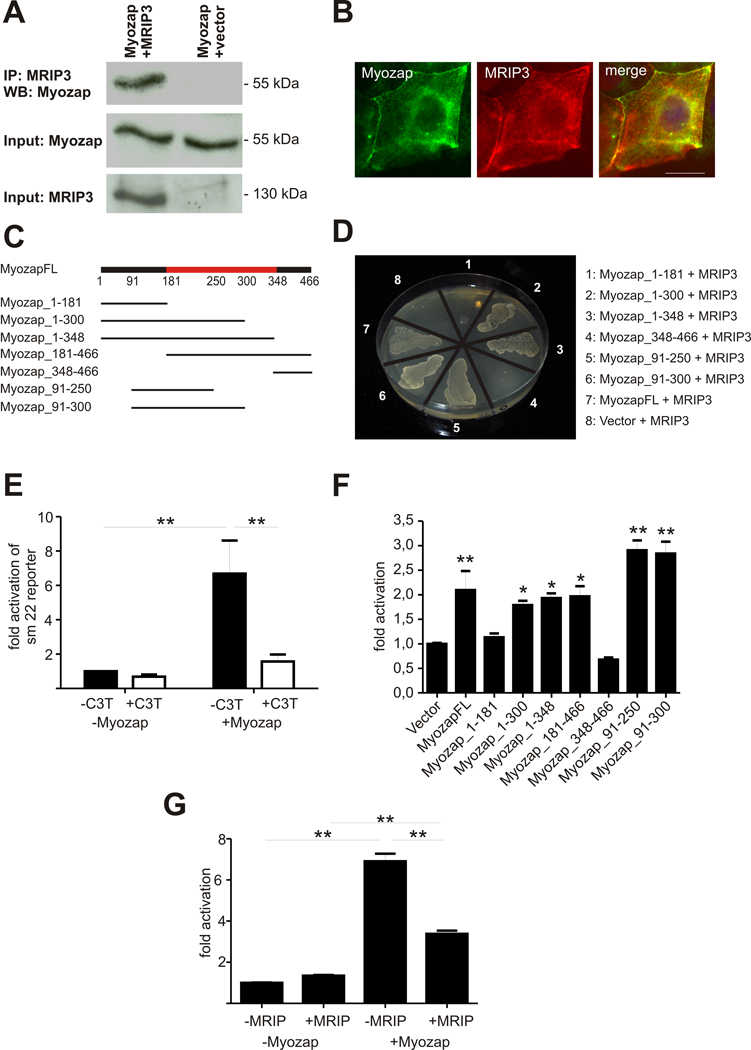
Next, we performed a yeast two-hybrid screening (Y2H) experiment to discover additional protein interaction partners of myozap. From this screen, several putative binding partners were identified, including zonula occludens-1 (ZO-1) and the Rho-inhibitor MRIP (Myosin phosphatase RhoA interacting protein). ZO-1 has been suggested to be a scaffold protein linking the actin cytoskeleton and structures of adherens and tight junctions11. In cardiomyocytes, ZO-1 has been shown to be part of the area composita of the intercalated disc3. In order to confirm the interaction with Myozap, immunoprecipitation experiments were performed. using a ZO-1 antibody and Myozap antiserum. The results reproduced the interaction between the two proteins (Figure 6A) and immunostaining of adult mouse heart cryosections revealed a colocalization of both proteins at the ID (Figure 6B).
Figure 6. Myozap interacts with ZO-1 and co-localizes with actin.
(A) HEK293T cells were transfected with plasmids encoding for HA-tagged myozap and ZO-1. Immunoprecipitation was performed using the ZO-1 antibody; the precipitate was immunoblotted with myozap antiserum. ZO-1 co-precipitated with myozap (lane 2), whereas no signal could be seen with empty vector (lane 1), confirming the interaction between the two proteins. (B) The immunostaining of an adult mouse cryosection with myozap- (Cy3) and ZO-1- (FITC) antibody reveals colocalization of the two proteins at the intercalated disc. (C) In order to determine which domain of myozap binds to ZO-1, several deletion variants were cloned. The red bar shows the ERM-like domain. (D) The ZO-1-binding domain was determined by performing a retransformation experiment. Therefore either one of the deletion variants, myozap full length or the pDest22 vector were transformed together with ZO-1 into a yeast strain. Only the variants myozap1-348, myozap91-250, myozap91-300 and myozap full length together with ZO-1 were able to grow on histidine lacking medium. The minimal myozap-domain necessary for binding ZO-1 is therefore the domain between amino acids 91 and 250. (E) To test whether myozap colocalizes with actin, COS7 cells were transfected with HA-tagged myozap. Immunostaining with phalloidin (red) and anti-myozap (green) reveals a localisation at the actin cytoskeleton. Scalebar: 10 µm. (F) In adult rat cardiomyocytes, a colocalization of myozap with actin is seen as well. Scalebar: 5 µm.
To further analyze which domains of myozap are sufficient to bind to ZO-1, we generated several deletion clones (Figure 6C) and again performed yeast two hybrid experiments to determine a potential interaction. The deletion variants, myozap full length or the empty pDest22 vector (activation domain) were transformed together with ZO-1 (cloned into the pDEST32 DNA-binding domain vector) into yeast. Only the deletion constructs myozap1-348, myozap91-250, myozap91-300 and myozap full length together with ZO-1 were able to grow on histidine lacking medium, suggesting that myozap’s amino acids 91 to 250 represent the minimal domain necessary to bind to ZO-1.
Given the ability of ERM proteins to bind to filamentous actin, we next tested whether full length Myozap colocalizes with actin. The use of an actin co-sedimentation assay as direct proof of a potential actin-myozap interaction was precluded due to spontaneous sedimentation of myozap during ultracentrifugation (data not shown). Therefore, COS7 cells were transiently transfected with HA-tagged myozap. Subsequent coimmunostaining with TRITC-labeled phalloidin for actin filaments and anti-myozap revealed colocalization of both proteins at the actin cytoskeleton (Figure 6E). To further corroborate these results, the experiment was repeated in isolated adult rat cardiomyocytes. Here, a colocalization of myozap with actin could be seen at ID where actin anchors at high densities to the cell membrane (Figure 6F).
Myozap induces Rho-dependent SRF signalling and binds to the Rho-Inhibitor MRIP
The Y2H screen also revealed a direct interaction between myozap and myosin phosphatase Rho-interacting protein (MRIP, its murine homologue is named p116Rip), which binds to Rho and inhibits Rho-dependent SRF signalling12. A coimmunoprecipitation experiment was performed in MDCK (Madin-Darby canine kidney) cells which again confirmed the interaction between the two proteins (Figure 7A). Moreover, both proteins colocalized at the cell membrane as well as with cortical actin as shown by immunostaining (Figure 7B). For further interaction domain mapping, we cloned several myozap deletion variants (Figure 7C). A yeast two hybrid retransformation interaction assay was performed by cotransforming either one of the deletion variants, myozap full length or empty vector together with MRIP3. The minimal domain of myozap necessary for binding MRIP3 was identified between amino acids 91 and 250 which includes parts of the ERM-like domain (Figure 7D).
Figure 7. Myozap interacts with MRIP and stimulates Rho-dependent signaling.
(A) A coimmunoprecipitation assay was performed by overexpressing Myozap and Myosin phosphatase Rho-interacting protein (MRIP) in MDCK cells. Using an anti-myc-antibody, Myozap could only be precipitated in the presence of MRIP (lane 1) but not in cells transfected with vector alone (lane 2), confirming the interaction between the two proteins. (B) MDCK cells were transfected with Myozap and MRIP. Immunostaining with Myozap- (FITC labeled secondary antibody) and MRIP- (Cy3 labeled secondary) antiserum reveals a colocalization at the cell membrane and the cytoskeleton. (C) In order to determine which domain of Myozap binds to MRIP, several deletion variants were cloned into the pDEST32 vector. The red bar shows the ERM-like domain. (D) An interaction assay was performed by transforming either one of the deletion variants, myozap full length or the pDest 32 vector together with MRIP3 (in pDest22) into yeast. The minimal domain of myozap necessary for binding MRIP3 was between amino acid 91 and 250 and includes the ERM-like domain. (E) A luciferase reporter assay was performed by overexpressing a luciferase reporter gene driven by the sm22-promoter together with Myozap in HEK293T cells. Myozap strongly activated the sm22-promoter. By adding C3-transferase (C3T), an inhibitor of RhoA, the activity could be significantly decreased, whereas C3-transferase did not significantly inhibit the sm22-promoter itself. **: p<0.01 (ANOVA, posthoc test: Student-Newman-Keuls). (F) In order to determine which domain of Myozap induces the sm22-promotor, a luciferase reporter assay using the sm22-promotor and myc-tagged full length Myozap (MyozapFL) and the myc-tagged deletion variants shown in Figure 7C was performed. All constructs except Myozap1-181 and Myozap348-466 significantly activated the sm22-promoter. * p<0.01 versus vector, ** p<0,001 versus vector (ANOVA, posthoc test: Student-Newman-Keuls). FL: full length. (G) HEK293T cells were transiently transfected with HA-tagged Myozap, myc-tagged MRIP and the sm22-luc reporter. Myozap activated the sm22-promoter, whereas MRIP did not induce the promoter. Coexpression of both, Myozap and MRIP, decreased the activation of the promoter compared to the activation of Myozap alone. **: p<0,001 (ANOVA, posthoc test: Student-Newman-Keuls). IP: Immunoprecipitation, WB: Western blot. Scalebar: 20 µm
Next, we investigated whether Myozap might have an impact on Rho-dependent signalling which has been shown to stimulate the transcriptional activity of SRF13,. Myozap and a luciferase reporter gene driven by the SRF-dependent sm22-promoter14,15, were overexpressed in HEK293T cells. Myozap strongly activated the sm22-promoter (~6.7 fold). Conversely, the addition of C3-transferase from Clostridium botulinum, which inhibits Rho via ADP-ribosylation of the GTPase, markedly attenuated sm22 activity (−76.7%, Figure 7E). In addition, HEK293T cells were cotransfected with myozap, MRIP and the sm22-luc reporter. Again, Myozap activated the sm22-promoter, whereas the addition of MRIP led to significant attenuation of sm22 activation (Figure 7F). Baseline promoter activity was not significantly affected by either C3-transferase or MRIP. Thus, myozap robustly induces SRF-dependent transcription, while its direct binding partner MRIP inhibits this activation.
In order to determine which domain of myozap induces the sm22-promotor, we used the same deletion fragments shown in Figure 7C as well as full length Myozap (MyozapFL). Myozap or its deletion variants were co-transfected with the sm22-promoter luciferase reporter. All constructs except Myozap1-181 and Myozap348-466 significantly activated the sm22-promoter (Figure 7G). Interestingly, the minimal activation domain localizes to the center of the ERM-like domain. Moreover, Myozap appears to specifically target SRF-dependent signalling, since MEF2, another basic loop helix transcription factor, was not significantly activated (online figure II).
Knockdown of Myozap in zebrafish causes cardiomyopathy
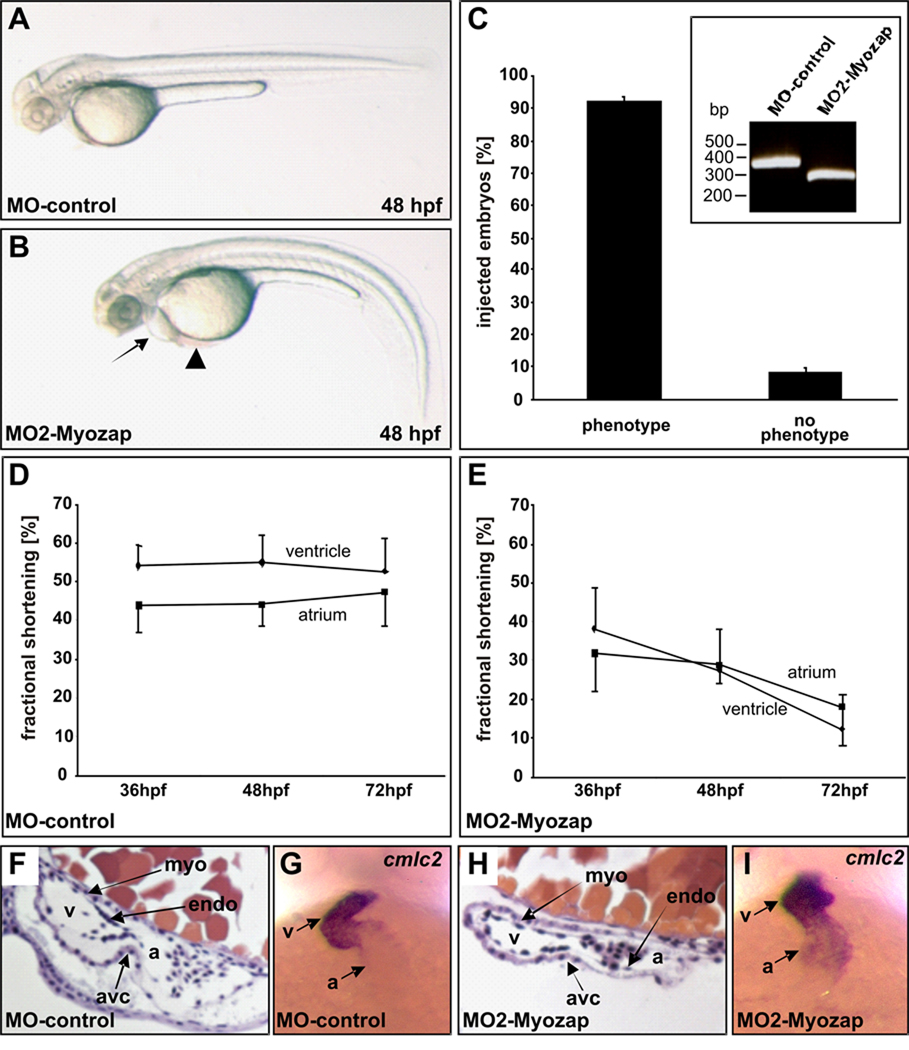
To assess the role of Myozap in vivo, we performed morpholino-modified (MO) antisense oligonucleotide-mediated knockdown experiments of the Myozap orthologue in zebrafish. We characterized embryos injected with MO1-control and MO2-myozap, the latter of which leads to an abnormal splice product (exon skipping, 95bp) and thus to premature termination of translation of zebrafish Myozap (zMyozap). MO-control injected zebrafish displayed normal morphology at 48 hpf (hours post fertilization) (Figure 8A), whereas MO2-Myozap injected embryos developed significant cardiac pathology (Figure 8B): The morphants showed pericardial edema as a sign of cardiomyopathy as well as blood pooling at the sinus venosus suggestive for low contractile performance. Efficacy of the MO2-myozap morpholino was tested by PCR, which showed that the majority of zMyozap mRNA was incorrectly spliced. By injecting 2.5 ng of MO2-myozap, 92% of the injected embryos (n=370) developed similar phenotypical characteristics including contractile dysfunction. Of note, the MO2-myozap injected embryos also displayed severe skeletal muscle dysfunction (online videos I&II). Repetitive measurements of atrial and ventricular fractional shortening of MO-control and MO2-Myozap injected embryos at 36, 48 and 72 hpf revealed a progressive loss of contractility (Figure 8D&E, and online videos III&IV). Despite the functional impairment, Myozap morphant hearts displayed normal morphology. Endocardial and myocardial cell layers as well as the atrioventricular canal (AVC) appeared unaltered in MO2-myozap injected embryos after 48hpf (Figure 8F&H). At 48hpf, the expression of cardiac myosin light chain 2, investigated by whole mount antisense RNA in situ hybridization, was similar in MO-control and MO2-Myozap injected embryos, indicating no severe changes in cardiac development and differentiation (Figure 8G&I).
Figure 8. Knockdown of the Myozap/Grinl1A7 orthologue leads to cardiomyopathy in zebrafish.
(A) MO-control injected zebrafish at 48 hpf (hours post fertilization) displaying normal morphology. (B) MO2-Myozap injected embryo at 48 hpf showing cardiac pathology. Arrow: heart/pericardial edema; Arrowhead: blood pool at sinus venosus. (C) Knockdown-efficacy. 92% knockdown phenotype (n=370; two independent experiments). Inset: effectiveness of MO2-Myozap on mRNA splicing. Injection of MO2-Myozap results in an abnormal splice product (exon skipping, 95bp), leading to premature termination of translation of zebrafish Myozap.
(D & E): Fractional shortening of the atrial and ventricular chamber of MO-control and MO2-Myozap injected embryos at 36, 48 and 72 hpf. Atrial as well as ventricular fractional shortening of MO2-myozap morphants decreases over time consistent with cardiomyopathy. (F & H) Myozap morphant hearts display normal morphology. Endocardial and myocardial cell layers as well as the atrioventricular canal (AVC) are unaltered in MO2-myozap injected embryos after 48hpf. (G & I) At 48hpf, the expression of myosin light chain 2, investigated by whole mount antisense RNA in situ hybridization, is similar in MO-control and MO2-Myozap injected embryos indicating no severe changes in cardiac development.
Discussion
We describe a previously uncharacterized cardiac protein, Myozap, which localizes to the ID and binds to several members of the area composita, the predominant cell-cell-contact structure of the mature cardiomyocyte. Since the actin myofilament cytoskeleton anchors at fascia adhaerens and area composita, the ID is directly involved in the versatile role of actin signaling and actomyosin contractility. The actin cytoskeleton acts in a variety of cellular motility processes, including contractility, mitosis, cytokinesis, secretion, and endocytosis. Moreover, transcription and gene expression is regulated by processes involving actin either directly through an association with nuclear chromatin remodeling proteins16 or indirectly through changes of actin dynamics17. Specifically, monomeric actin serves as an upstream regulator of SRF (serum response factor)-dependent signaling (for review see18). SRF is a MADS-box transcription factor that controls the expression of a wide range of muscle-specific genes18.
In this context, it is noteworthy that myozap contains an ERM-like domain, since ERM proteins have important roles in adherens junction formation and maintenance via binding and stabilization of the actin cytoskeleton9. Of note, ERM proteins are also involved in the regulation the activity of the small GTPase Rho (for review see13). The small GTPases of the Rho family, including Rho, Rac, and Cdc42, have been shown to function as major activators of SRF-dependent transcriptional activity via changes in actin dynamics17,19,20. Rho GTPases thereby serve as molecular switches in a multitude of signaling pathways especially those involving actin polymerization and stress fiber formation21. Myozap, which colocalizes with filamentous actin, activates SRF signaling as shown by the robust activation of a sm22-promoter driven reporter. This reporter gene activation could be inhibited by cotransfection of C3-transferase, which irreversibly blocks Rho activation, suggesting that Myozap uses a Rho-dependent mechanism of SRF activation. Mechanistically, most ERM proteins positively modulate Rho9 via binding of RhoGDI, a negative regulator of Rho activity. This in turn releases inactive Rho, allowing its subsequent activation22. However, ERM domains may also function downstream of Rho, where ERM proteins are activated by Rho-dependent threonine phosphorylation. Both mechanisms together form a positive feedback loop23.
In cardiomyocytes, signaling from the small GTPase Rho and Rac diverges into several downstream targets which include SRF, other transcription factors such as NF-κB, as well as myofilament proteins, and ion channels24. Myozap’s direct interaction with MRIP, an endogenous inhibitor of Rho-mediated SRF signaling11, adds another layer of complexity. The addition of MRIP in a SRF-dependent reporter assay significantly blunts myozap’s capability of Rho-dependent SRF activation. Consistently, MRIP´s murine homologue, p116RIP, has been shown to interfere with RhoA-mediated transcription through its ability to disassemble the actomyosin cytoskeleton downstream of RhoA25.
An important hallmark of SRF is its ability to orchestrate the activity of numerous upstream signaling events26. The tight control of SRF activity is essential for maintenance of cardiac contractility and the heart’s ability to respond to stress and diverse disease stimuli. In this regard, it is worth noting that overexpression of the cardiac SRF activator STARS leads to an increased sensitivity to hemodynamic stress27. These findings are also in line with data derived from mice with cardiac-specific overexpression of SRF, which develop severe dilated cardiomyopathy28. Conversely, inactivation of SRF in the adult mouse heart has been reported to lead to impaired left ventricular function and rapid progression to dilated cardiomyopathy mediated by a progressive loss of contractile proteins29. Further support for the essential role of SRF in cardiac integrity comes from the results of embryonic deletion of SRF, which results in cardiovascular defects and intrauterine death30,31. Moreover, recently the microRNAs 133a-1 and -2 have been shown to be critical components of an SRF-dependent myogenic transcriptional circuit32. Our data suggest that myozap might represent another novel regulator of myocardial SRF signaling. Consistently, knockdown of myozap in zebrafish results in severe cardiomyopathy.
In addition to its effects on cardiac signal transduction, myozap’s localization at the ID junctions implies a function in the mechanical and/or electrical coupling of cardiomyocytes. The ID is known as a hot spot of inherited cardiac disease, which include arrhythmogenic right ventricular cardiomyopathy (ARVC) and dilated cardiomyopathy. The disruption of desmosomal integrity appears to be a key factor in mediating the development of ARVC, resulting in defective mechanics, abnormal localization of cell-cell-adhesion proteins and gap junction remodeling33. It will thus be interesting to see whether myozap also plays a role in the etiology of ARVC. In addition, the ID has also been implicated into the pathogenesis of dilated cardiomyopathy (DCM)34. This notion is supported by data from mouse models for DCM including the muscle LIM protein knockout mouse35 and the tropomodulin overexpressing mouse36, which showed a marked upregulation of adherens junction-associated proteins including N-cadherin, α- and β-catenin, plakoglobin, N-RAP, and vinculin37. In our zebrafish knockdown model, Myozap deficiency leads to severe contractile dysfunction without disturbing cardiac development. Contractile dysfunction is a hallmark of DCM suggesting a potential role for myozap in the pathogenesis of cardiomyopathy.
Moreover, a critical feature in end stage heart failure is the progressive loss and altered distribution of connexin 43 (Cx43). This process, referred to as “gap junction remodeling”, may lead to electrophysiological perturbations and contractile dysfunction, as recently reviewed38. In this context, myozap′s binding partner ZO-1 seems to play an essential role since it directly interacts with Cx43 at the ID where it controls gap junction size. Recent data suggest that the binding of ZO-1 to Cx43 is increased in heart failure leading to an altered gap junction architecture (please see39 and references quoted herein). It remains to be seen whether the direct interaction of myozap with ZO-1 modulates this process and thus plays a role in these fundamental remodeling processes.
In summary, we have discovered a previously uncharacterized, cardiac-enriched protein, termed myozap (myocardium-enriched ZO-1 associated protein). Myozap acts as an activator of Rho-dependent SRF signaling, which in turn is controlled by a direct interaction with MRIP, a molecule that can attenuate myozap-dependent SRF activation. Interestingly, myozap is localized at the ID where it interacts with desmoplakin and ZO-1, two proteins that have been implicated in the pathogenesis of human heart disease. The knockdown of Myozap in zebrafish leads to severe contractile dysfunction. Further in vivo gain and loss-of-function animal experiments as well as the careful analyses of the Myozap/Grinl1A7 gene in ARVC patients, will help to clarify whether myozap also participates in the pathogenesis of cardiomyopathy.
Novelty and Significance
What is known?
The intercalated disc (ID) is an essential cell contact structure ensuring mechanical and electrical coupling between cardiomyocytes.
The importance of the ID is further underscored by the findings that many proteins involved in this complex contribute to the pathogenesis of inherited cardiac disease.
What new information does this article contribute?
Using a bioinformatic screen we were able to identify a previously uncharacterized protein termed Myozap which localizes to the intercalated disc
Myozap regulates signaling pathways activated by the serum response factor”Knockdown” of Myozap in zebrafish causes severe impairment of cardiac contractility.
Summary
Given the complexity and the importance of the intercalated disc, we hypothesized that there may be ID components that are still unknown. In bioinformatic screening experiments, we identified an ID protein termed Myozap which colocalizes and interacts with several other ID proteins, including desmoplakin and ZO-1. Moreover, we found that Myozap is a positive regulator of serum-response-factor signaling. When downregulated in vivo, Myozap deficiency caused severe cardiomyopathy in zebrafish. Myozap may be an important component — -molecular pathways that regulate normal cardiac function and may be involved in the pathogenesis of cardiomyopathies.
Supplementary Material
Acknowledgements
We thank Ulrike Oehl and Jutta Krebs for excellent technical assistance. Furthermore, we are grateful to Niels Weinhold for his essential help with the MDCK cells. We are indebted to Kathleen J. Green for the kind gift of the desmoplakin cDNA.
Sources of funding
This work has been supported by the Landesstiftung Baden-Württemberg as well as by a grant of the Bundesministerium für Bildung und Forschung, Germany (Nationales Genomforschungsnetz: NGFNplus) to N.F. E.N.O was supported by grants from the NIH, the Donald W. Reynolds Center for Clinical Cardiovascular Research and the Robert A. Welch Foundation.
Abbreviations
- BSA
bovine serum albumin
- ERM
Ezrin, Radixin, and Moesin
- MRIP
Myosin phosphatase Rho interacting protein
- Myozap
Myocardium-enriched Zo-1-interacting protein
- ORF
open reading frame
- PFA
paraformaldehyde
- ZO-1
zonula occludens 1
- ID
Intercalated Disc
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None.
Subject Codes: [137, 138, 143]
References
- 1.Noorman M, van der Heyden MA, van Veen TA, Cox MG, Hauer RN, de Bakker JM, van Rijen HV. Cardiac cell-cell junctions in health and disease: Electrical versus mechanical coupling. J Mol Cell Cardiol. 2009 doi: 10.1016/j.yjmcc.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Franke WW, Borrmann CM, Grund C, Pieperhoff S. The area composita of adhering junctions connecting heart muscle cells of vertebrates. I. Molecular definition in intercalated disks of cardiomyocytes by immunoelectron microscopy of desmosomal proteins. Eur J Cell Biol. 2006;85:69–82. doi: 10.1016/j.ejcb.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Borrmann CM, Grund C, Kuhn C, Hofmann I, Pieperhoff S, Franke WW. The area composita of adhering junctions connecting heart muscle cells of vertebrates. II. Colocalizations of desmosomal and fascia adhaerens molecules in the intercalated disk. Eur J Cell Biol. 2006;85:469–485. doi: 10.1016/j.ejcb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Goossens S, Janssens B, Bonne S, De Rycke R, Braet F, van Hengel J, van Roy F. A unique and specific interaction between alphaT-catenin and plakophilin-2 in the area composita, the mixed-type junctional structure of cardiac intercalated discs. J Cell Sci. 2007;120:2126–2136. doi: 10.1242/jcs.004713. [DOI] [PubMed] [Google Scholar]
- 5.van Tintelen JP, Hofstra RM, Wiesfeld AC, van den Berg MP, Hauer RN, Jongbloed JD. Molecular genetics of arrhythmogenic right ventricular cardiomyopathy: emerging horizon? Curr Opin Cardiol. 2007;22:185–192. doi: 10.1097/HCO.0b013e3280d942c4. [DOI] [PubMed] [Google Scholar]
- 6.Herren T, Gerber PA, Duru F. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: a not so rare "disease of the desmosome" with multiple clinical presentations. Clin Res Cardiol. 2009;98:141–158. doi: 10.1007/s00392-009-0751-4. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 8.Roginski RS, Mohan Raj BK, Birditt B, Rowen L. The human GRINL1A gene defines a complex transcription unit, an unusual form of gene organization in eukaryotes. Genomics. 2004;84:265–276. doi: 10.1016/j.ygeno.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 10.Hunter AW, Barker RJ, Zhu C, Gourdie RG. Zonula occludens-1 alters connexin-43 gap junction size and organization by influencing channel accretion. Mol Biol Cell. 2005;16:5686–5689. doi: 10.1091/mbc.E05-08-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. 2008;1778:660–669. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surks HK, Richards CT, Mendelsohn ME. Myosin phosphatase-Rho interacting protein.A new member of the myosin phosphatase complex that directly binds RhoA. J Biol Chem. 2003;278:51484–51493. doi: 10.1074/jbc.M305622200. [DOI] [PubMed] [Google Scholar]
- 13.Hughes SC, Fehon RG. Understanding ERM proteins--the awesome power of genetics finally brought to bear. Curr Opin Cell Biol. 2007;19:51–56. doi: 10.1016/j.ceb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Liu Z, Mercer B, Overbeek P, Olson EN. Evidence for serum response factor-mediated regulatory networks governing SM22alpha transcription in smooth, skeletal, and cardiac muscle cells. Dev Biol. 1997;187:311–321. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Ip HS, Lu MM, Clendenin C, Parmacek MS. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol Cell Biol. 1997;17:2266–2278. doi: 10.1128/mcb.17.4.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rando OJ, Zhao K, Crabtree GR. Searching for a function for nuclear actin. Trends Cell Biol. 2000;10:92–97. doi: 10.1016/s0962-8924(99)01713-4. [DOI] [PubMed] [Google Scholar]
- 17.Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 18.Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Mack CP, Somlyo AV, Hautmann M, Somlyo AP, Owens GK. Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J Biol Chem. 2001;276:341–347. doi: 10.1074/jbc.M005505200. [DOI] [PubMed] [Google Scholar]
- 20.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 21.Pellegrin S, Mellor H. Actin stress fibres. J Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K, Sasaki T, Mammoto A, Takaishi K, Kameyama T, Tsukita S, Takai Y. Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J Biol Chem. 1997;272:23371–23375. doi: 10.1074/jbc.272.37.23371. [DOI] [PubMed] [Google Scholar]
- 23.Tsukita S, Yonemura S. Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J Biol Chem. 1999;274:34507–34510. doi: 10.1074/jbc.274.49.34507. [DOI] [PubMed] [Google Scholar]
- 24.Brown JH, Del Re DP, Sussman MA. The Rac and Rho hall of fame: a decade of hypertrophic signaling hits. Circ Res. 2006;98:730–742. doi: 10.1161/01.RES.0000216039.75913.9e. [DOI] [PubMed] [Google Scholar]
- 25.Mulder J, Ariaens A, van Horck FP, Moolenaar WH. Inhibition of RhoA-mediated SRF activation by p116Rip. FEBS Lett. 2005;579:6121–6127. doi: 10.1016/j.febslet.2005.09.083. [DOI] [PubMed] [Google Scholar]
- 26.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 27.Kuwahara K, Teg Pipes GC, McAnally J, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. Modulation of adverse cardiac remodeling by STARS, a mediator of MEF2 signaling and SRF activity. J Clin Invest. 2007;117:1324–1334. doi: 10.1172/JCI31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Azhar G, Chai J, Sheridan P, Nagano K, Brown T, Yang J, Khrapko K, Borras AM, Lawitts J, Misra RP, Wei JY. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am J Physiol Heart Circ Physiol. 2001;280:H1782–H1792. doi: 10.1152/ajpheart.2001.280.4.H1782. [DOI] [PubMed] [Google Scholar]
- 29.Parlakian A, Charvet C, Escoubet B, Mericskay M, Molkentin JD, Gary-Bobo G, De Windt LJ, Ludosky MA, Paulin D, Daegelen D, Tuil D, Li Z. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation. 2005;112:2930–2939. doi: 10.1161/CIRCULATIONAHA.105.533778. [DOI] [PubMed] [Google Scholar]
- 30.Parlakian A, Tuil D, Hamard G, Tavernier G, Hentzen D, Concordet JP, Paulin D, Li Z, Daegelen D. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol Cell Biol. 2004;24:5281–5289. doi: 10.1128/MCB.24.12.5281-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miano JM, Ramanan N, Georger MA, de Mesy Bentley KL, Emerson RL, Balza RO, Jr, Xiao Q, Weiler H, Ginty DD, Misra RP. Restricted inactivation of serum response factor to the cardiovascular system. Proc Natl Acad Sci U S A. 2004;101:17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z, Bowles NE, Scherer SE, Taylor MD, Kearney DL, Ge S, Nadvoredskiy VV, DeFreitas G, Carabello B, Brandon LI, Godsel LM, Green KJ, Saffitz JE, Li H, Danieli GA, Calkins H, Marcus F, Towbin JA. Desmosomal dysfuntion due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ Res. 2006;99:646–655. doi: 10.1161/01.RES.0000241482.19382.c6. [DOI] [PubMed] [Google Scholar]
- 34.Perriard JC, Hirschy A, Ehler E. Dilated cardiomyopathy: a disease of the intercalated disc? Trends Cardiovasc Med. 2003;13:30–38. doi: 10.1016/s1050-1738(02)00209-8. [DOI] [PubMed] [Google Scholar]
- 35.Arber S, Hunter JJ, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 36.Sussman MA, Welch S, Cambon N, Klevitsky R, Hewett TE, Price R, Witt SA, Kimball TR. Myofibril degeneration caused by tropomodulin overexpression leads to dilated cardiomyopathy in juvenile mice. J Clin Invest. 1998;101:51–61. doi: 10.1172/JCI1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehler E, Horowits R, Zuppinger C, Price RL, Perriard E, Leu M, Caroni P, Sussman M, Eppenberger HM, Perriard JC. Alterations at the intercalated disk associated with the absence of muscle LIM protein. J Cell Biol. 2001;153:763–772. doi: 10.1083/jcb.153.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Severs NJ, Bruce AF, Dupont E, Rothery S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc Res. 2008;80:9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruce AF, Rothery S, Dupont E, Severs NJ. Gap junction remodelling in human heart failure is associated with increased interaction of connexin43 with ZO-1. Cardiovasc Res. 2008;77:757–765. doi: 10.1093/cvr/cvm083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.