Abstract
We propose a mathematical model for simulating the leading-edge dynamics of a migrating cell from the interplay among elastic properties, architecture of the actin cytoskeleton, and the mechanics of the membrane. Our approach is based on the description of the length and attachment dynamics of actin filaments in the lamellipodium network. It is used to determine the total force exerted on the membrane at each position along the leading edge and at each time step. The model reproduces the marked state switches in protrusion morphodynamics found experimentally between epithelial cells in control conditions and cells expressing constitutively active Rac, a signaling molecule involved in the regulation of lamellipodium network assembly. The model also suggests a mechanistic explanation of experimental distortions in protrusion morphodynamics induced by deregulation of Arp2/3 and cofilin activity.
Introduction
Recent high-resolution analysis by live cell microscopy has revealed distinct phenotypes in the morphodynamics of cell protrusion. The phenotypes change discretely upon manipulation of pathways that regulate the assembly of the actin cytoskeleton (1), the main driver of cell protrusion (2). Remarkably, the phenotypes are characteristic to cell type and mode of perturbation. They show little cell-to-cell variation despite significant shape heterogeneity between cells. Therefore, cell shape dynamics contain relevant information about cytoskeleton regulation. Finding a way to extract this information would open the possibility of studying the function of molecular regulators of motility by measuring the effect of their perturbation on cell morphodynamics. To this end, we have begun to develop a computational model to simulate the behavior of cell morphodynamics in response to force generation by the cytoskeleton, taking into account the elastic properties and dynamic geometry of the membrane as well as of the actin cytoskeleton.
The actin network juxtaposed to the plasma membrane forms a brush of unlinked filament tips undergoing thermal fluctuation. This causes an entropic force estimated to be strong enough to drive membrane protrusion (3,4). The efficiency of entropic force generation relies on the close apposition of filament ends to the membrane and the shortness of the free fluctuating length of filaments (5). The first condition is fulfilled in the protruding lamellipodium by persistent polymerization of G-actin at the leading edge (2). The rate of filament growth is by itself sensitive to load (4,6,7). The second condition is the result of the balance among filament nucleation, capping, cross-linking, and depolymerization. Moreover, experimental evidence suggests transient attachment of filaments to the membrane during the nucleation process (8–11) entailing a force-extension relation different from the one of freely fluctuating tips and the possibility of filaments pulling back the membrane (7,11). It is reasonable to assume that the duration of attachment also depends on the force between filament and membrane (7,12). The forces exerted by actin filaments on the membrane determine therefore not only how the membrane moves, but they feed-back onto the state of filament attachment and growth rate, resulting in spatiotemporal coupling of dynamics of cytoskeleton assembly, force production, and cell morphological responses. These coupled dynamics could be the cause of the complex shape changes, as observed, for example, with epithelial cells.
A model that accounts for both the length and attachment dynamics of an actin filament population was proposed recently (13). It can mimic both steady and saltatory motion of the obstacle, comparable to the behavior observed for Listeria, and allows the prediction of transitions between these motion types upon parameter changes (13,14).
Here we extend this model to soft obstacles with dynamic shapes like the plasma membrane limiting the lamellipodium, thereby including membrane tension. Furthermore, we adapt the geometry of the actin network considered in Gholami et al. (13) to the structures described for the lamellipodium (2). Together, this provides us with a computational tool to map the internal dynamics of the actin cytoskeleton and to relate them to the external shape dynamics of the membrane. We focus our analysis on the two regimes of protrusion dynamics characteristic to the regulation of lamellipodia in migrating epithelial cells which were denoted as I-state and V-state (1). In the V-state, localized, random bursts of protrusions initiate protrusion waves that propagate transversally along the edge. Whenever two propagating waves collide, they annihilate each other. The I-state consists of spatially homogeneous and synchronized oscillations between protrusion and retraction along the entire leading edge. Experimentally, the switch between V- and I-states has been associated with deregulation of Rac-activation (1).
Model
The assembly of the actin cytoskeleton in the lamellipodium is controlled by a network of signaling pathways (15) converging onto a few parameters such as, e.g., the force-independent free polymerization velocity (see below) or the binding rate of filaments to the membrane. Our model starts with these parameters. With this approach, we closely follow the modeling strategy of the tethered ratchet model (7,16), to which we add the dynamics of the free length of polymer in the brush.
Single filaments transfer mechanical momentum to the membrane only if they are anchored in a scaffold. We assume that the filament network itself, cross-linked and adherent to the substrate, provides this support. The freely fluctuating part of a filament measured from the point of the last bond to the tip is flexed by Brownian motion and can be characterized by its contour length l, the distance z between fixed point and membrane, and the angle θ between filament and normal to the membrane. If the filament is not attached to the membrane, the probability density distribution P(z) of the end-to-end distance defines a free energy F(z) = –kBT ln P(z), from which the average normal force on the membrane can be derived as (17)
The scale of this force is given by the Euler buckling force
where lp denotes the persistence length of the filament (4,5). In the following, we use the force dependence on contour length, distance to the membrane, and angle fd(l, z, θ) (see Supporting Material and Fig. 1 A) in the weakly bending rod approximation derived in Gholami et al. (5). The function fd(l, z, θ) is suitable for polymer stiffness ɛ = l/lp < 0.1 (5), characteristic to lamellipodial actin networks.
Figure 1.
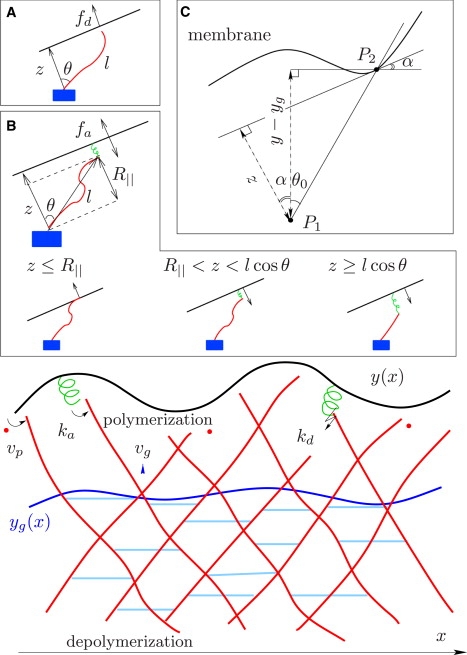
Interaction between membrane and actin network filaments at the leading edge. The lamellipodial actin network has two functionally different parts: a cross-linked part forming a gel, and a brush of free fluctuating polymer ends extending toward the cell membrane. The position of the membrane is described by the function y(x). The boundary between the brush and the cross-linked network region is described by the function yg(x). Filaments attach to the membrane at rate ka, and attached filaments detach at rate kd. Detached filaments elongate by polymerization with velocity vp. Cross-linkers continuously bind to the free polymers, so that the gel boundary yg(x) advances at velocity vg. (Inset A) Force fd exerted by detached filaments depends on the contour length l, distance to the membrane tangent z, and angle θ. (Inset B) Force fa exerted by attached filaments depends on the relation between the distance to the membrane z, the projection R‖ of the equilibrium end-to-end distance on the membrane normal, and the contour length l. (Inset C) Geometry of the problem: P1 is the fixed end of the filament and P1P2 is the grafting direction. The distance z from P1 to the local tangent to the membrane at P2 relates to y – yg by z/cos(θ0 + α) = (y – yg)/cos θ0, where α = arctan yx is the local slope of the membrane.
It is believed that the directionality of cell protrusions is maintained by directed growth (18). Whereas detached filaments always push the membrane, filaments can also exert a pulling force during attachment, depending on their length and position relative to the membrane. The molecular details of filament-membrane links are not yet fully understood. We therefore assume that single filaments can transiently attach to the membrane via linker proteins that behave like elastic springs. We distinguish three regimes for the force fa exerted by the serial arrangement of polymer and linker, depending on the relation between the distance to the membrane z, the projection R‖ of the equilibrium end-to-end distance onto the membrane normal, and the contour length l (see Fig. 1 B) (13):
The three cases correspond to
-
1.
The compressed filament pushes against the membrane.
-
2.
The filament and linker pull the membrane while being stretched together.
-
3.
A filament is fully stretched but the linker continues to pull the membrane by being stretched further.
We also assume that the linker can move freely in the membrane, so that the force exerted by attached filaments is normal to the membrane. Here, k‖, kl, and keff are the linear elastic coefficients of polymer, linker, and serial polymer-linker arrangement, respectively. For k‖ we use the linear response coefficient of a wormlike chain grafted at both ends (see Supporting Material) (19,20), itself a function of polymer stiffness and incidence angle (20).
Because the forces between membrane and single filaments are highly sensitive to the contour length and depend on the attachment state of the filament, the model has to include, as variables, the length and the attachment kinetics of the actin network. The free fluctuating length of a filament can change through elongation or shortening of both ends. For the sake of simplicity, we assume that depolymerization occurs only in the region where filaments are cross-linked. Thus, the dynamics of the filament length is controlled by two parameters—the rate of polymerization when the filament is detached from the membrane, and the rate of cross-linking (see Fig. 1).
According to Mogilner and Oster (4), the polymerization velocity of single filaments decreases exponentially with load
| (1) |
where fd is the force produced by a detached filament growing against the membrane, and δ is the size of an actin monomer. The free polymerization velocity depends on the G-actin concentration in the cell. We assume this to be a control parameter whose value is determined by the mechanisms of branching, capping, filament severing, ADP/ATP exchange, etc.
The rate of cross-linking, i.e., the probability for a filament to form new bonds with other filaments, increases with the available unlinked length of the filament and saturates at a value dependent on the available cross-linker concentration:
| (2) |
Here, is the maximum gel velocity and can be perceived as the width of the brush-gel transition region (see Fig. 1). We found that results only weakly depend on the choice of the value for (data not shown).
The transient attachment of filaments to the membrane is defined by the rate ka. We assume the force-dependent rate of detachment (12),
Here fa is the pulling force exerted on the membrane by an attached filament and denotes the rate of spontaneous detachment. We use again δ as a typical length for the dissociation process.
Our model actin network is polarized and grows in the direction of cell movement. This direction defines a symmetry axis. We consider four populations of filaments along the cell edge: attached (a) and detached (d) filaments, each of them divided into a subpopulation oriented to the right (+) and to the left (−) of the symmetry axis. These populations are described by their number density distributions (l, x, t) in dependence on the free contour length between gel and membrane l. The length distributions along the x axis obey
| (3) |
In addition, defines the local velocity of gelation, where θ0 is the angle between the grafting direction and the cross-linking velocity vg, which points in the direction of the symmetry axis y (see Fig. 1).
The total normal force density exerted by the actin cytoskeleton on the plasma membrane can be computed as
where z± = (y − yg) cos(θ±)/ cos(θ0) denotes the distance between the fixed end of the filament and the tangent to the membrane at the point closest to the free end, and θ± = ±θ0 + arctan(yx) is the angle between grafting direction and membrane normal (see Fig. 1 C).
We assume that the membrane is under constant tension S. This results in a resistance force to bending. The linear force density is given by
Ftension(x, t) points in the direction of the local normal to the membrane. Finally, the moving membrane is under the influence of the drag force of the surrounding intracellular fluid and other viscous forces, e.g., flow of membrane. The evolution of the membrane position is then defined by the force balance at the membrane,
| (4) |
where η is an effective viscous drag coefficient.
The evolution of the actin gel boundary (see Fig. 1) can be described by the local average cross-linking velocity subtracted by the velocity vr of the retrograde flow of the cross-linked gel:
| (5) |
The retrograde flow arises from contraction of the gel by myosin motors and the resultant force naFa + ndFd exerted by the brush on the gel boundary. Whereas the magnitude of forces acting from the cell body on leading-edge membrane determines retrograde flow in general, changes of the resultant force arising from increases in leading-edge velocity occurring during V- and I-states modulate retrograde flow in the range 15–25% of the protrusion velocity (21). The relatively small flow variations during protrusion events have been ascribed to a tight feedback between increases in boundary force and increases in adhesion forces subadjacent to the cell edge (22). Furthermore, we neglect in our model the effect of variable contraction, as it has been shown experimentally that lamellipodium-driven morphodynamics in epithelial cells is only weakly dependent on the activity of myosin motors (21). Hence, whereas gel properties set the average velocity of the lamellipodium, at the timescales of the V- and I-states, these parameters have little impact on the model behavior and we set vr constant. In the following, ∂tyg(x, t) is referred to as the gel velocity.
The length distributions quickly collapse into narrow distributions around mean values determined by the length for which the advection velocity in the dynamics of is equal to zero (13). Therefore, in Eqs. 3–5, we approximate the mean values of forces, cross-linking velocity and polymerization velocity, with the values these functions assume at the mean lengths and obtain (see (13,14))
| (6) |
Here, are the total linear densities of the different filament populations. The results presented in the following are obtained from the expressions in Eq. 6.
Results
Our model implies a shape-mediated, spatial coupling of neighboring points on the membrane. First, the forces exerted by attached and detached filaments depend on the angle between filament and membrane normal, and on the distance from the fixed end of the filament to the membrane tangent. Both change when the local orientation of the membrane changes (see Fig. 1). Second, membrane tension depends on the local curvature. This means that the temporal evolution of a membrane point x depends not only on the position of the membrane y(x), but also on the positions of its neighbors. Accordingly, different morphodynamic patterns are being formed in dependence on the regime of local dynamics. We review first the local behavior, and then derive from it the behavior of the spatially coupled system.
Local dynamics
Local dynamics is described by the time evolution of a single point of the membrane in the absence of any coupling to neighboring points. Depending on parameter values, the system either is in steady motion, or shows velocity oscillations (13). The dynamic regime changes upon parameter variation due to a Hopf bifurcation (Fig. 2 D). It occurs because the system becomes unstable with respect to perturbations of na away from the stationary state. (In mathematical terms: the unstable eigenvector of the Jacobi matrix is almost parallel to the na-axis in phase space.) More generally, it becomes unstable with respect to all perturbations which entail a perturbation in na. For example, perturbations in the distance between membrane and gel boundary, which we applied in the simulations of Fig. 2, shift the force balance and dissociation rate, and therefore also change na. Decreasing na leads to stronger forces on the bonds of the remaining attached filaments, resulting in their detachment from the membrane. On the stable side of the bifurcation, na-perturbations are amplified, if they exceed a threshold value (Fig. 2 B). Perturbations smaller than that value, decay (Fig. 2 A). The existence of such a perturbation threshold above which perturbations are amplified is called excitable dynamics.
Figure 2.
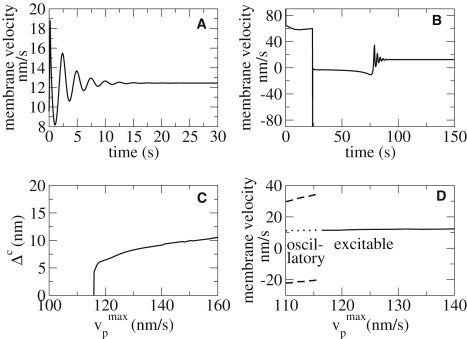
Excitability. The system is prepared in the stable stationary state (y = y0). At t = 0, the membrane position is perturbed (y(0) = y0 + Δ), whereas all other variables remain unperturbed. For Δ < Δc, a small-scale decaying response of the membrane is observed, shown here by decaying oscillations of the membrane velocity (A). For perturbations greater than a threshold Δc, the system undergoes large-scale nonlinear responses before returning to steady state (B). The value of the threshold Δc increases with the distance to the bifurcation in the polymerization velocity (C). (D) For values of larger than the value of the Hopf bifurcation, the system exhibits a stable stationary state (solid line); below that value, it oscillates with the amplitudes shown by the dashed lines, and the stationary state is unstable (dotted line). The amplitude jumps to large values immediately at the Hopf bifurcation due to a canard explosion (14). The dynamics are excitable for larger than and close to the Hopf bifurcation value. Parameter values are given in Table 1 and = 150 nm s−1 in panels A and B.
On the unstable side of the bifurcation, all na-perturbations grow until they turn into oscillations. The oscillation involves periodic attachment and detachment of filaments to the membrane and consists of two phases (see Fig. 3):
-
1.
A compression phase (e.g., between 1.0 and 1.6 min), where the gel boundary advances faster than the membrane. In this phase, the fraction of attached filaments is high.
-
2.
A relaxation phase (e.g., between 1.6 and 2.5 min), where the membrane moves faster than the gel and a large fraction of filaments is detached.
Figure 3.
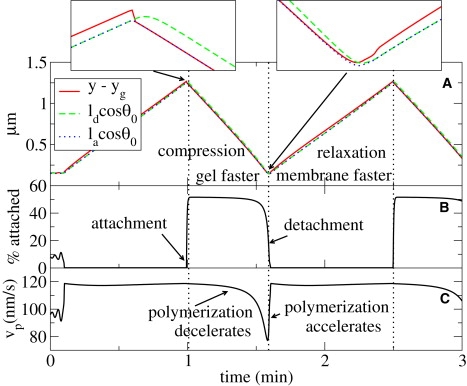
Different phases of motion during a protrusion cycle. (A) Time evolution of the distance y – yg between membrane and gel as well as the projections ld cos θ0 and la cos θ0 of the polymer lengths onto the direction of motion. The two insets show enlarged details of the maxima and minima of the curves to emphasize the difference between the two phases of the oscillation (see text): When gelation velocity exceeds membrane velocity, detached filaments are compressed (ld cos θ0 > y − yg ≈ lacosθ0); when membrane velocity exceeds gelation velocity, both detached and attached filaments are relaxed (y − yg > ld cos θ0 ≈ la cos θ0.) (B) Time evolution of the fraction of attached filaments. (C) Time evolution of the polymerization velocity.
During the compression phase, filaments shorten as the distance between membrane and gel decreases. Both Euler buckling force (∼1/l2) and the spring constant (∼1/l4) of the filament are strongly length-dependent. Consequently, the magnitude of forces exerted by both attached and detached filaments increases when filaments shorten. Due to the force dependence of the polymerization velocity, filament growth slows down even further. As a result, the magnitude of pulling forces increases superlinearly, which leads to explosive filament detachment (at t ≈ 1.6 min). The pushing force exerted by short detached filaments is strong and initiates accelerated membrane protrusion. The membrane advances faster than the gel, inducing relaxation of the filaments. In this phase, filaments grow at the maximum polymerization velocity, and initially do not stay attached to the membrane because detachment rate is still very high. With progression to longer filaments, the forces on the membrane begin to decrease, causing a reduced detachment rate (at t ≈ 2.5 min). An increased number of attached filaments slows down the membrane below the velocity of gelation. This condition initiates a new cycle of protrusion.
Membrane oscillations occur within certain ranges of polymerization, attachment, detachment, and cross-linking rates (see (14) for a detailed analysis). Very fast polymerization velocities lead to very high forces exerted by detached filaments. As a result, even when filaments attach, the membrane movement never falls below the gel growth velocity. Filaments do not reach the shortening phase and the membrane ends up moving steadily at the velocity of gelation. On the other hand, very slow polymerization velocities decrease the force of detached filaments below the critical value necessary to propel the membrane faster than the gel, as required for oscillations. Steady motion is reached also in this case.
Spatial coupling of membrane dynamics in asymmetric networks
The dependence of the entropic forces on the angle between filament and membrane (Fig. 4 A) induces a strongly nonlinear force response to a perturbation in the membrane shape. Filaments oriented closer to the membrane normal exert higher forces than filaments oriented with a larger angle to it (Fig. 4, B and C). Therefore, the force distribution over a shape perturbation is asymmetric, causing a one-sided propagation of the perturbation along the membrane. In the example of Fig. 4 D, most filaments are oriented to the right. The forces on the membrane are weaker where the membrane has a positive slope and stronger where the membrane has a negative slope. Accordingly, both shoulders of the shape perturbation propagate in the direction of the predominant filament orientation.
Figure 4.
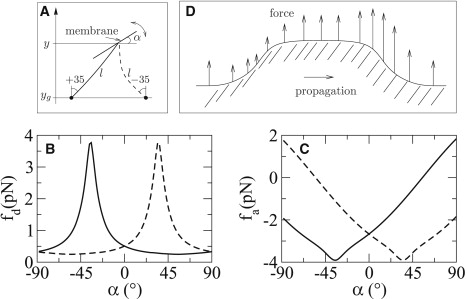
Dependence of the forces on the local slope of the membrane. (A) Definition of geometry. Force dependence of (B) detached filaments (see also Gholami et al. (5), their Fig. 15); and (C) attached filaments. Solid and dashed curves correspond to the filament configuration illustrated in panel A. For all calculations, filament length and distance between fixed point yg and membrane position y are kept constant at values la = 192 nm, ld = 200 nm, and y − yg = 161 nm, corresponding to the parameter set producing the stable yet excitable steady state of the system discussed in Fig. 2. The magnitude of forces is maximal when filaments are normal to the membrane. (D) The dependence of forces on the angle between filament and membrane tangent leads to one-sided propagation of a local perturbation for asymmetric filament networks.
Spatial coupling of membrane dynamics in symmetric networks
Traveling waves
Although the asymmetry of filament orientation may alone define a mechanism for the transversal propagation of protrusion events, there is little evidence from electron micrographs for such an arrangement in lamellipodial actin networks (23,24). Therefore, we consider the effect of spatial coupling via tension and angular dependence of forces in shape perturbations with symmetric networks. We assume right- and left-oriented filaments with identical density n/2 at an angle of ±35°. To explore the dynamics under these conditions we excite the system with variations in the polymerization velocity associated with biochemical noise:
Here, ξ denotes the space- and time-dependent Gaussian white noise (with the properties 〈ξ(x, t)〉 = 0, 〈ξ(x, t)ξ(x′, t′)〉 = δ(x − x′)δ(t − t′)). The factor Q controls the noise amplitude.
Propagation of protrusions in the symmetric case is based on the excitability of the system. Excitability does not require filaments to be tilted with respect to the membrane and works in a large range of the angle θ (14). Protrusion waves can be initiated by perturbations in polymerization velocity, leading to perturbations of na. If a local perturbation exceeds the excitability threshold, it will be amplified (see Fig. 2 C). The membrane quickly moves forward due to the decrease of naFa. The resulting protrusion pulls on the bonds of attached filaments in the immediate neighborhood, which increases the dissociation rate there. As a result, the neighboring regions are perturbed above the excitability threshold causing the propagation of protrusion pulses in both directions along the membrane. When two pulses traveling in opposite directions meet, they annihilate. This is due to the general property of excitable systems that, once excited, each point is insensitive to further perturbation for some time, before it returns to equilibrium. In our model, the refractory period corresponds to the recovery of the slow variables la and ld to their stationary values.
The morphodynamics resembles the V-state identified experimentally for PtK1 epithelial cells (1): Most spontaneous protrusions split and propagate in both directions. They always terminate upon collision as waves in an excitable system do. The normal velocity of the membrane switches in this case between three phases (see Fig. 5, A–C): the base level (green), corresponding in our model to the steady state without noise; fast protrusion (red), corresponding to emerging protrusions; and fast retraction (blue) after every protrusion. Model parameters were tuned to obtain a pattern that mimics the normal velocity maps in Fig. 12 of Machacek and Danuser (1): The ratio ka to was used to adjust the period of the I-state to measured values; the membrane tension S adjusts the propagation velocity of laterally traveling waves; and, essentially, − vr sets the scale of the normal velocity map. The values of ka to , S and − vr obtained that way are close to measured values or values used by other models also (see references in Table 1). Characteristics of the simulated waves resulting from these parameter choices also agree very well with the ones observed in experiments: The velocity amplitudes are ∼30 nm s−1 in experiments and 40 nm s−1 in simulations (see Fig. 5 and Fig. 3) and the width of the fast protruding region is ∼5 μm in simulations and 4–12 μm in experiments. The lateral velocity of the protrusions along the leading edge is independent of the migration velocity of the cell, which is also in agreement with experiments (1).
Figure 5.
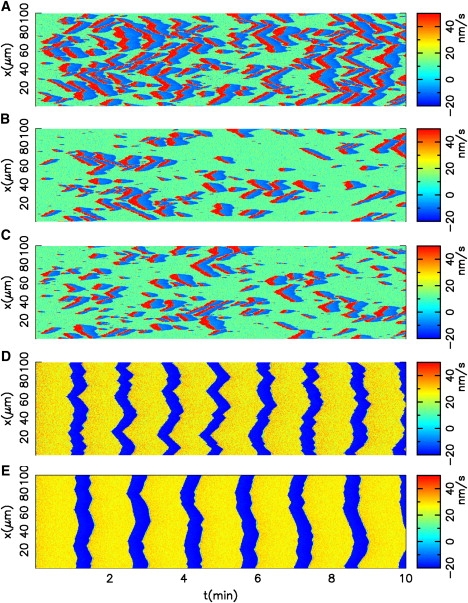
Simulated normal velocity maps of the membrane using different values for the polymerization velocity and noise level. Columns of the maps indicate the velocity values along the membrane for one time point. Rows indicate the velocity values for one point on the membrane over time. (A) = 140 nm s−1, Q = 0.3 nm s−1; (B) = 140 nm s−1, Q = 0.25 nm s−1; (C) = 141 nm s−1, Q = 0.3 nm s−1; (D) = 110 nm s−1, Q = 0.3 nm s−1; and (E) = 110 nm s−1, Q = 0.1 nm s−1. All other parameters are fixed as listed in Table 1. (A–C) Defined conditions for the V-state; (D and E) conditions for the I-state.
Table 1.
Model parameter values
| Parameter | Value | Remark | ||
|---|---|---|---|---|
| Actin monomer radius | δ | 2.7 nm | (16) | |
| Persistence length of actin | lp | 15 μm | (49) | |
| Attachment rate | ka | 2.16 s−1 | Assumed∗ 10 s−1 in Shaevitz and Fletcher (50) | |
| Detachment constant | 2 s−1 | Assumed∗ 0.5 s−1 in Mogilner and Oster (7) | ||
| Saturation value of cross-linking velocity | 76 nm s−1 | Assumed∗† (1) | ||
| Saturation length of cross-linking velocity | 100 nm | Assumed | ||
| Saturation value of polymerization velocity | 110–150 nm s−1 | (51–53) | ||
| Total filament density | n | 100 μm−1 | (11)‡ | |
| Orientation angle | θ0 | 35° | (23,24) | |
| Spring constant of linker | kl | 0.7 pN nm−1 | (7, 54) | |
| Effective drag coefficient | η | 2 pN μm−2 | (55)‡ | |
| Membrane tension | S | 10 pN | Assumed∗‡§ (52) | |
| Retrograde flow | vr | 60 nm s−1 | Assumed∗†(1) |
The ratio ka/, S, and − vr were chosen to fit the normal velocity maps in Machacek and Danuser (1). However, the obtained values are close to values given in the above references.
Results depend essentially on − vr only, because vg(l) (Eq. 2) is almost always in the saturation range.
Results depend only on n/η and S/η, not on the absolute values of these parameters.
Occurrence and persistence of the waves depend strongly on the relationship between noise level and excitability threshold: Decrease of the noise level (Fig. 5, A and B) or increase of the threshold (Fig. 5, A–C) lead to decreased induction of V-events. However, noise does not affect propagation velocity or amplitude of the waves.
Synchronous oscillations
In addition to the propagation of transversal waves in a V-state, Machacek and Danuser (1) described morphodynamic patterns where long sectors of the membrane are synchronized in cycles of protrusion and retraction. This behavior was referred to as the I-state, observed in PtK1 cells expressing constitutively active Rac1, as well as in newt lung epithelial cells. In our model, I-state behavior is reproduced when polymerization velocity is reduced, so that the local dynamics of the system becomes oscillatory. Here, the spatial coupling leads to a fast synchronization of the different points of the membrane (Fig. 5, D and E). Noise affects the regularity of the pattern. Decreased noise levels (Fig. 5, D and E) yield longer synchronized sectors of the membrane. The period of oscillations is comparable to the period of the V-events in the stable system, in accordance with Machacek and Danuser (1). In addition, our simulations show no difference in the average velocity of V- and I-state, which is also in line with experiments. Based on our model, we reason this is because these two events occur on two sides of a Hopf bifurcation of the brush dynamics (14).
Discussion
Morphodynamic profiling of PtK1 epithelial cells identified two protrusion phenotypes by manipulating the pathways of actin assembly (1): Control cells are characterized by the V-state whereas upregulation of Rac1 activity induced a switch to the I-state. Inhibition of the filament nucleator Arp2/3 maintains the V-state but leads to fewer and less persistent waves. We have therefore analyzed if and how the change of the different control parameters can induce the same transitions in our model.
Effects of Rac1 activation on Arp2/3- and cofilin-mediated actin assembly
Increased Rac1 levels lead to increased activation of Arp2/3 and inhibition of cofilin via PAK (25,26). Arp2/3 nucleates filaments off existing filaments (27,28), whereas cofilin severs filaments and promotes their depolymerization (29–31). As a result of expressing constitutively active Rac1, branching is therefore intensified whereas depolymerization is weakened. Both effects change density and total length of filaments and affect the concentration of the actin monomers available for polymerization, G, to which growth velocity is proportional (32). Therefore, the model parameters most likely to depend on Rac1 activity are the total filament density n and the polymerization velocity . Simple calculations based on the model presented in Carlsson et al. (33) yield the following conditions as to how n and G vary with the amount of cofilin C and the total amount of active Arp2/3-complex A0 (see Supporting Material):
An increase of activated Arp2/3 and a decrease of activated cofilin both result in less G-actin. Consequently, expression of constitutively active Rac1 corresponds in our model to a decrease of the polymerization velocity, which is proportional to the amount of polymerizable G-actin.
Rac1-dependent transition between traveling waves and synchronous oscillations
Fig. 6 shows the transition lines between V and I phenotypes in different parameter planes as they result from numerical simulations of the expressions in Eq. 6. In our model, a transition between V- and I-states cannot be induced by changing the total filament density while keeping G constant (top left). Instead, the V-state is turned into the I-state by decreasing the polymerization velocity alone. Indeed, experimentally it was observed that expression of constitutively active Rac1, which does decrease the polymerization velocity (see above), does induce a V- to I-state transition. Constitutively active Rac1 may decrease polymerization velocity by promotion of branching, or by inhibition of cofilin. Both mechanisms lead to a depletion of the monomeric actin pool. The change in morphodynamics observed under inhibition of Arp2/3 can be used to distinguish these scenarios quantitatively: Direct inhibition of Arp2/3 does not change the phenotype, but leads only to decreased incidence and persistence of the waves (1). Such a change in morphodynamics can be achieved in the model by increasing polymerization velocity inside the excitable domain, and therefore increasing the excitability threshold. Interestingly, only a relatively minor change of the polymerization velocity is required to reproduce this change in the model: Changes from 140 nm s−1 to 141 nm s−1 cause the transition from Fig. 5, A–C. In contrast, a larger change of the polymerization velocity (from 140 nm s−1 in A to 110 nm s−1 in D) is necessary to reproduce the effect of excess Rac1 activation with the model. Association of the pattern in Fig. 5 A with wild-type cells, of the pattern in Fig. 5 C with Arp2/3-inhibited cells, and of the pattern in Fig. 5 D with Rac1-activated cells, suggests that downregulation of Arp2/3 activation must have a smaller effect on the polymerization velocity than downregulation of cofilin. Therefore, our model predicts that inhibition of cofilin, and not activation of Arp2/3, is the dominant mechanism to decrease the pool of monomeric actin under expression of constitutively active Rac1 responsible for the switch V- to I-state morphodynamics.
Figure 6.
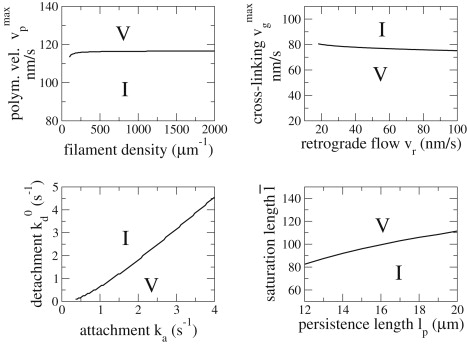
Transition lines between the V- and I-states in different parameter spaces. All other parameters are fixed as listed in Table 1 and = 110 nm s−1.
The ratio between the variations of the monomer concentration with the amount of Arp2/3 and cofilin is given by (see Supporting Material and (34))
Our results therefore imply that the concentration C of active cofilin is much smaller than the concentration of Arp2/3 bound to filaments in PtK1 cells denoted by A0 − A.
Alternative mechanisms for a transition between traveling waves and synchronous oscillations
Transitions from a V- to an I-state can be induced also by variation of attachment (ka) and detachment (kd0) rates, as well as by variation of the saturation value of the cross-linking velocity () and its saturation length () (Fig. 6). In contrast, variations in retrograde flow (vr, top right) and persistence length (lp, bottom right) have almost no effect on the morphodynamic pattern. This suggests that manipulations of a PtK1 cell that would affect the binding or cross-linking processes could lead to shape dynamics similar to the dynamics of cells expressing constitutively active Rac1. In contrast to the activation of Rac1, Arp2/3, and cofilin, these parameters are currently impossible to manipulate.
Relation to other models of cell protrusion dynamics
In our model, different dynamic regimes emerge from the changes of the free polymer length in the filament brush growing toward the plasma membrane and interactions between filament tips and membrane. These aspects are distinct from other models linking actin cytoskeleton dynamics to cell shape (35–41). Besides the field tests of the model based on our own, existing morphodynamic profiles, the importance of la and ld as dynamic variables is also supported by experiments which changed the free filament length by perturbation of Ena/VASP (42). In agreement with our model, long free lengths yield slow edge velocities because filaments are too floppy to exert a strong pushing force, whereas short free lengths yield slow velocities due to the polymerization rate limitation by strong force (42). Similar observations were made by Koestler et al. (11).
There are a variety of studies describing the shape dynamics of whole cells, in particular of keratocytes (43,44). These studies describe a correlation between smooth leading-edge shape and high steady migration velocity and more dynamic and irregular leading-edge shape at low velocities. The change of leading-edge dynamics is linked to a change of migration velocity. We propose that the acceleration of migration corresponds to an increase of away from the Hopf bifurcation into the excitable regime, leading to a transition from irregular patterns to steady homogenous protrusion. Therefore, our model adds to these earlier models a mechanism for the transition between oscillating and persistent protrusion observed between different cell types.
Shlomovitz and Gov (45) and Kuusela and Alt (46) have both proposed models that also explain shape fluctuations, including the lateral propagation of protrusion waves. In contrast to our experimental data from polarized epithelial cells, which suggest that dynamics of lamellipodia is myosin-independent (1), these models require motor-driven contraction. Experimental data supporting the notion that actomyosin contractility plays an important role in cell shape dynamics was published by the Sheetz lab (47). It should be noted that these experiments investigated the molecular mechanisms of cell spreading before cell polarization and directed cell protrusion. Whereas, in our model, I-states arise with a rapid synchronization of the lamellipodium network assembly along the edge of a polarized epithelial cell, the I-state during initial cell spreading originates from the periodic contraction-relaxation of an unpolarized lamellar actin network. Intriguingly, Sheetz and colleagues (48) noted in their experimental system a phase transition from I-state to V-state behavior at later stages of the spreading process. It is tempting to speculate that this transition is associated with the formation of a noncontractile lamellipodium in front of the contractile lamella, accompanied by a gradual increase of the contributions of a brush-driven to a contraction-driven pulsation mechanism. Future versions of our model will therefore add gel dynamics to brush-driven mechanisms to account for pulsatile retrograde flow. However, such a model will be significantly more complex, as it needs to incorporate a description of the dynamic interactions between lamellipodium and lamella, much of which remain to be explored experimentally.
Conclusion
In summary, we propose here a model for the spatial coupling of protrusion dynamics via the mechanical interaction of actin filaments with the plasma membrane. Our model has demonstrated capability of explaining key switches in the morphodynamic behaviors of epithelial cells induced by molecular manipulations of actin assembly, quantified in Machacek and Danuser (1). It also explains a large body of further, more-qualitative observations of protrusion behaviors. Future extensions of the model may consider explicitly processes that are currently lumped in phenomenological parameters. For example, retrograde flow is likely to depend on the forces generated by interactions between membrane and actin filaments as well as the dynamic activity of myosin motors. Moreover, the restriction to a constant total barbed-end density could be replaced by inclusion of nucleation and capping as control mechanisms of free barbed-end dynamics.
In many cases, the effects of manipulations of actin cytoskeleton dynamics are often difficult to observe directly. Hence, linking molecular functions to morphodynamic responses remains challenging. The model presented here presents a first cornerstone of a quantitative bridge between the modes of action of an experimental perturbation at the molecular scale and the morphodynamics of the cell edge. Given the ease with which both perturbation experiments and morphodynamic measurements can be conducted, this model will become a powerful tool in defining regulatory pathways of cell morphology.
Supporting Material
One table and six figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00012-3.
Supporting Material
Acknowledgments
This work was funded by the National Institutes of Health grant No. R01 GM071868 (to G.D.).
Footnotes
Gaudenz Danuser's present address is Harvard Medical School, Boston, Massachusetts.
References
- 1.Machacek M., Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophys. J. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard T.D. The cytoskeleton, cellular motility and the reductionist agenda. Nature. 2003;422:741–745. doi: 10.1038/nature01598. [DOI] [PubMed] [Google Scholar]
- 3.Peskin C.S., Odell G.M., Oster G.F. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mogilner A., Oster G. Cell motility driven by actin polymerization. Biophys. J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gholami A., Wilhelm J., Frey E. Entropic forces generated by grafted semiflexible polymers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;74:041803. doi: 10.1103/PhysRevE.74.041803. [DOI] [PubMed] [Google Scholar]
- 6.Hill T.L. Microfilament or microtubule assembly or disassembly against a force. Proc. Natl. Acad. Sci. USA. 1981;78:5613–5617. doi: 10.1073/pnas.78.9.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mogilner A., Oster G. Force generation by actin polymerization II: the elastic ratchet and tethered filaments. Biophys. J. 2003;84:1591–1605. doi: 10.1016/S0006-3495(03)74969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Co C., Wong D.T., Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svitkina T. N-WASP generates a buzz at membranes on the move. Cell. 2007;128:828–830. doi: 10.1016/j.cell.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 10.Upadhyaya A., van Oudenaarden A. Biomimetic systems for studying actin-based motility. Curr. Biol. 2003;13:R724–R744. doi: 10.1016/j.cub.2003.08.051. [DOI] [PubMed] [Google Scholar]
- 11.Koestler S.A., Auinger S., Small J.V. Differentially oriented populations of actin filaments generated in lamellipodia collaborate in pushing and pausing at the cell front. Nat. Cell Biol. 2008;10:306–313. doi: 10.1038/ncb1692. [DOI] [PubMed] [Google Scholar]
- 12.Evans E., Ritchie K. Strength of a weak bond connecting flexible polymer chains. Biophys. J. 1999;76:2439–2447. doi: 10.1016/S0006-3495(99)77399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gholami A., Falcke M., Frey E. Velocity oscillations in actin-based motility. N. J. Phys. 2008;10:033022. [Google Scholar]
- 14.Enculescu M., Gholami A., Falcke M. Dynamic regimes and bifurcations in a model of actin-based motility. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2008;78:031915. doi: 10.1103/PhysRevE.78.031915. [DOI] [PubMed] [Google Scholar]
- 15.Burridge K., Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 16.Mogilner A. Mathematics of cell motility: have we got its number? J. Math. Biol. 2009;58:105–134. doi: 10.1007/s00285-008-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frey E., Kroy K., Sackmann E. Springer Verlag; Berlin, Germany: 1998. Dynamical Networks in Physics and Biology. [Google Scholar]
- 18.Carlier M.F., Pantaloni D. Control of actin assembly dynamics in cell motility. J. Biol. Chem. 2007;282:23005–23009. doi: 10.1074/jbc.R700020200. [DOI] [PubMed] [Google Scholar]
- 19.Kroy K., Frey E. Force-extension relation and plateau modulus for wormlike chains. Phys. Rev. Lett. 1996;77:306–309. doi: 10.1103/PhysRevLett.77.306. [DOI] [PubMed] [Google Scholar]
- 20.Kroy K. Hieronymus; Munich, Germany: 1998. Viscoelasticity of solutions of semi-flexible polymers. [Google Scholar]
- 21.Ponti A., Machacek M., Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 22.Ji L., Lim J., Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat. Cell Biol. 2008;10:1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svitkina T.M., Verkhovsky A.B., Borisy G.G. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verkhovsky A.B., Chaga O.Y., Borisy G.G. Orientational order of the lamellipodial actin network as demonstrated in living motile cells. Mol. Biol. Cell. 2003;14:4667–4675. doi: 10.1091/mbc.E02-10-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eden S., Rohatgi R., Kirschner M.W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 26.Huang T.Y., DerMardirossian C., Bokoch G.M. Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Stradal T.E., Scita G. Protein complexes regulating Arp2/3-mediated actin assembly. Curr. Opin. Cell Biol. 2006;18:4–10. doi: 10.1016/j.ceb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Higgs H.N., Pollard T.D. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 2001;70:649–676. doi: 10.1146/annurev.biochem.70.1.649. [DOI] [PubMed] [Google Scholar]
- 29.Bamburg J.R. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 30.Carlier M.-F., Ressad F., Pantaloni D. Control of actin dynamics in cell motility. Role of ADF/cofilin. J. Biol. Chem. 1999;274:33827–33830. doi: 10.1074/jbc.274.48.33827. [DOI] [PubMed] [Google Scholar]
- 31.Lai F.P.L., Szczodrak M., Rottner K. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delorme V., Machacek M., Bokoch G.M. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev. Cell. 2007;13:646–662. doi: 10.1016/j.devcel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlsson A.E., Wear M.A., Cooper J.A. End versus side branching by Arp2/3 complex. Biophys. J. 2004;86:1074–1081. doi: 10.1016/S0006-3495(04)74182-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chauvière A., Preziosi L., Verdier C., editors. Cell Mechanics: From Single Scale Based Models to Multiscale Modeling. Taylor & Francis; New York: 2009. [Google Scholar]
- 35.Kruse K., Joanny J.F., Prost J. Contractility and retrograde flow in lamellipodium motion. Phys. Biol. 2006;3:130–137. doi: 10.1088/1478-3975/3/2/005. [DOI] [PubMed] [Google Scholar]
- 36.Larripa K., Mogilner A. Transport of a 1D viscoelastic actin-myosin strip of gel as a model of a crawling cell. Physica A. 2006;372:113–123. doi: 10.1016/j.physa.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawes A.T., Edelstein-Keshet L. Phosphoinositides and Rho proteins spatially regulate actin polymerization to initiate and maintain directed movement in a one-dimensional model of a motile cell. Biophys. J. 2007;92:744–768. doi: 10.1529/biophysj.106.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawes A.T., Bard Ermentrout G., Edelstein-Keshet L. Actin filament branching and protrusion velocity in a simple 1D model of a motile cell. J. Theor. Biol. 2006;242:265–279. doi: 10.1016/j.jtbi.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura S.I., Ueda M., Sasai M. Cortical factor feedback model for cellular locomotion and cytofission. PLOS Comput. Biol. 2009;5:e1000310. doi: 10.1371/journal.pcbi.1000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alt W., Dembo M. Cytoplasm dynamics and cell motion: two-phase flow models. Math. Biosci. 1999;156:207–228. doi: 10.1016/s0025-5564(98)10067-6. [DOI] [PubMed] [Google Scholar]
- 41.Alt W., Bock M., Möhl C. Taylor & Francis; New York: 2009. Coupling of Cytoplasm and Adhesion Dynamics Determines Cell Polarization and Locomotion. [Google Scholar]
- 42.Bear J.E., Svitkina T.M., Gertler F.B. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 43.Lacayo C., Pincus Z., Theriot J. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 2007;5:2035–2052. doi: 10.1371/journal.pbio.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keren K., Pincus Z., Theriot J.A. Mechanism of shape determination in motile cells. Nature. 2008;453:475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shlomovitz R., Gov N.S. Membrane waves driven by actin and myosin. Phys. Rev. Lett. 2007;98:168103. doi: 10.1103/PhysRevLett.98.168103. [DOI] [PubMed] [Google Scholar]
- 46.Kuusela E., Alt W. Continuum model of cell adhesion and migration. J. Math. Biol. 2009;58:135–161. doi: 10.1007/s00285-008-0179-x. [DOI] [PubMed] [Google Scholar]
- 47.Giannone G., Dubin-Thaler B.J., Sheetz M.P. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 48.Döbereiner H.-G., Dubin-Thaler B., Sheetz M.P. Dynamic phase transitions in cell spreading. Phys. Rev. Lett. 2004;93:108105. doi: 10.1103/PhysRevLett.93.108105. [DOI] [PubMed] [Google Scholar]
- 49.Le Goff L., Hallatschek O., Amblard F. Tracer studies on F-actin fluctuations. Phys. Rev. Lett. 2002;89:258101. doi: 10.1103/PhysRevLett.89.258101. [DOI] [PubMed] [Google Scholar]
- 50.Shaevitz J.W., Fletcher D.A. Load fluctuations drive actin network growth. Proc. Natl. Acad. Sci. USA. 2007;104:15688–15692. doi: 10.1073/pnas.0702601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abraham V.C., Krishnamurthi V., Lanni F. The actin-based nanomachine at the leading edge of migrating cells. Biophys. J. 1999;77:1721–1732. doi: 10.1016/S0006-3495(99)77018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mogilner A., Oster G. The physics of lamellipodial protrusion. Eur. Biophys. J. 1996;25:47–53. [Google Scholar]
- 53.Novak I.L., Slepchenko B.M., Mogilner A. Quantitative analysis of G-actin transport in motile cells. Biophys. J. 2008;95:1627–1638. doi: 10.1529/biophysj.108.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans E. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 55.Berg H. Princeton University Press; Princeton, NJ: 1983. Random Walks in Biology. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


