Abstract
Recent modeling of filopodia—the actin-based cell organelles employed for sensing and motility—reveals that one of the key limiting factors of filopodial length is diffusional transport of G-actin monomers to the polymerizing barbed ends. We have explored the possibility of active transport of G-actin by myosin motors, which would be an expected biological response to overcome the limitation of a diffusion-based process. We found that in a straightforward implementation of active transport the increase in length was unimpressive, ≤30%, due to sequestering of G-actin by freely diffusing motors. However, artificially removing motor sequestration reactions led to approximately threefold increases in filopodial length, with the transport being mainly limited by the motors failing to detach from the filaments near the tip, clogging the cooperative conveyer belt dynamics. Making motors sterically transparent led to a qualitative change of the dynamics to a different regime of steady growth without a stationary length. Having identified sequestration and clogging as ubiquitous constraints to motor-driven transport, we devised and tested a speculative means to sidestep these limitations in filopodia by employing cross-linking and putative scaffolding roles of Ena/VASP proteins. We conclude that a naïve design of molecular-motor-based active transport would almost always be inefficient—an intricately organized kinetic scheme, with finely tuned rate constants, is required to achieve high-flux transport.
Introduction
For processes including cancer metastasis (1), neuronal growth (2), wound healing (3), and embryonic development (4), cell motility is guided by the sensing function of fingerlike projections called filopodia. The final location of the cells is critical, so these cells must adeptly sense their environment to properly direct their movement. To shed light into these processes, the underlying physical and regulatory mechanisms of filopodial growth and retraction must be understood. In fact, the overall structure and function of filopodia is now largely known: G-actin monomers polymerize into F-actin filaments, which then bundle in parallel to protrude the cell membrane. Continued polymerization of G-actin at the barbed ends and depolymerization at the pointed ends results in treadmilling, one of the key processes in the dynamics of actin-based cellular structures (5,6).
In treadmilling, F-actin elongation results from polymerization at the barbed ends, whereas retraction results from depolymerization at the pointed ends, backward pushing from the stretched membrane, and ATP-driven pulling from the cell body (the latter two processes are termed retrograde flow (7,8)). Polymerization and retrograde flow are fast processes, and switching their equilibrium engenders complex and highly dynamical behavior, such as growth-retraction cycles and turnover process (9–13). These dynamics allow a filopodium to perform its role as a mechanochemical receptor to guide cell motility.
A more detailed understanding of filopodia, beyond this general picture of overall structure and function, is a challenging pursuit because of the complex interdependence of governing mechanical, chemical, and biological processes. Thus, the development of computational models which would produce quantitative and testable predictions can be an effective means toward gaining additional new insights.
Considerable efforts have been made in modeling filopodia and other organelles based on polymerizing bundles of F-actin, including stereocilia and microvilli (9,14–19). These models, some deterministic, some stochastic, suggest that the diffusional flux of G-actin to the polymerizing end, in the absence of other chemical or mechanical regulation, is a limiting factor determining the filopodial length (14,15). Although experimentally measured filopodia are often several microns in length, some reach 30, or even 100 μm (20–22). Interestingly, physiologically reasonable choices of parameters in the current computational models predict filopodial lengths which are many-fold shorter than the longest filopodia observed experimentally. In addition, the growth rates of the longest filopodia are ∼10 μm/min (22). This value is also many-fold higher than current models have reported. These discrepancies indicate that a fundamental mechanism of growth is currently unaccounted for. For this reason, we extended our previous model (9,14) to investigate how a hypothetical active transport of G-actin might influence filopodial length. ATP-driven molecular motors are employed for many transporting purposes in cells (23), providing a directed and faster alternative to diffusion. Will active transport of G-actin in filopodia sufficiently promote elongation to fill the gap between models and experimental observations?
This hypothesis of active transport goes against the grain of the long-held view of passive diffusion in the context of actin filament growth (24). Although polymerization is clearly diffusion-limited in vitro (25), an in vitro system necessarily omits much of the in vivo complexity, and mechanisms may vary across actin-based structures. In fact, an in vivo study tracking G-actin within a lamellipodium showed that G-actin migration is too fast to be explained by passive diffusion alone (26).
Molecular motors are a likely candidate for supplementing the passive diffusion of G-actin during filopodial elongation. Myosins, the family of proteins that can walk on actin filaments, have been detected in both filopodia and stereocilia. In fact, the importance of myosin X(M10) (27,28) in filopodial growth is well-supported experimentally (8,29), though the specific roles and cargo of various myosins remain unclear. As new myosin motors and their functions are still being identified (30), it may be valuable to use computational models to generate testable hypotheses for the role of these motors.
Myosin X has been identified at the filopodial tips (30), and its overexpression resulted in increased number and length of filopodia in motile cells (30). Furthermore, myosin X has been observed walking forward and moving rearward within filopodia (31). The forward motion suggests directed movement of motors along a filament that significantly outraces retrograde flow, and the slower rearward movement suggests stochastic periods without walking and backward movement due to retrograde flow.
Beyond the identification of such motors within filopodia, there is a variety of hypotheses for their actual role in filopodial growth. Although not mutually exclusive, they include a physical pushing against the membrane to effectively increase the polymerization rate (29), transport of integrin to form adhesive structures near the tip (32), and transport of regulatory proteins, such as Ena/VASP family members, toward the tip (33). The complete role of Ena/VASP in filopodial growth is not fully understood (34–36), although it has both G-actin and F-actin binding motifs (37), forms a tetramer to crosslink actin filaments (34,36–38), and is transported by myosin X (33).
We explore the hypothesis that myosin motors carry G-actin to the polymerizing barbed ends of the actin filaments, presumably widening the main bottleneck for the filopodial length. To our knowledge, only one experimental study suggested a mechanism of actin transport that goes beyond passive diffusion (26). In addition, a theoretical study, based on mean-field deterministic equations, investigated the distribution of various proteins along a stereocilium under the influence of motor proteins (17). This article represents the first filopodial model of active transport, building upon our previous work (9,14) to stochastically simulate individual motors walking on actin filaments and carrying G-actin inside filopodia.
The stationary length in our previous model is set by a balance of fluxes: G-actin diffuses forward to the tip along its gradient; all of that flux is consumed for polymerization to F-actin; and as the F-actin filaments are pulled back by retrograde flow, an equal flux but opposite in direction returns actin to the cell bulk. Transport by motors adds to the forward flux: this addition is directional, as in retrograde flow, and not linearly decreasing with length as would diffusional flux (14,15,17). If motor-based flux dominates the diffusional flux, then motor-based transport of actin indeed may be visualized, similar to cartoons in biology textbooks, as a conveyor belt. Furthermore, if active transport flux exceeds the retrograde flow flux, there will be no stationary length.
Despite this possibility, straightforward introduction of motors did not lead to skyrocketing of the stationary filopodial length, let alone indefinite filopodial growth. In fact, freely diffusing motor proteins sequester G-actin previously available for polymerization. As a result, motors at large concentrations play a length-diminishing rather than a length-promoting role. In cases when simulated filopodia did grow longer, it was only a modest increase in length. Therefore, sequestration by motors can severely undermine active transport, and should be avoided in order to achieve a noticeable effect.
We repeated the simulations while disabling sequestration, by forbidding the freely diffusing motors (as opposed to motors bound to and walking along a filament) to load G-actin from the solution. This yielded a three-to-fivefold increase in filopodial length. Interestingly, we also noticed that the longest filopodia were observed in cases when motors had lower affinity to the filaments. This is a manifestation of clogging of the filament rails by the empty motors closer to the tip. We ran control simulations where motors loaded with actin are allowed to pass through empty motors: the most common result was large increase in stationary lengths, even in one case resulting in unsaturable linear growth.
Having identified limiting factors to motor-assisted filopodial growth, we explored a plausible biological means to side-step these limitations. In these simulations, myosin X does not load G-actin directly—instead loading Ena/VASP as a multisite adaptor, which in turn binds several G-actins. In this model, Ena/VASP is irreversibly consumed at the tip by a sink mimicking its cross-linking role (35) with simultaneous release of G-actin to the solution. Consequently, sinking of Ena/VASP for cross-linking results in decreased Ena/VASP concentration in solution, diminishing G-actin sequestration. The largest observed stationary lengths in this set were many-folds longer compared with filopodia produced using a naïve model of motor transport.
We summarize these findings as the rules of active transport: an effective transport mechanism must successfully overcome sequestration of the cargo and clean the clogged rails. It is likely that these are general principles that apply beyond the context of active transport in filopodia, where they have been deduced.
The results presented in this article were obtained from ∼8000 simulation trajectories, which took ∼100,000 CPU hours on the UNC Topsail supercomputer.
Methods
Here we report a stochastic treatment of filopodial dynamics based on the Gillespie algorithm (39) that builds on our previously developed model (14) to study motor-mediated active transport of G-actin monomers to the polymerizing end. Individual reactions in the Gillespie scheme included:
-
1.
Diffusion of G-actin along the length of the filopodium (rate kD),
-
2.
Polymerization and depolymerization at the barbed end, as affected by stochastic membrane force (rates k+ and k−),
-
3.
Motor loading and unloading of G- actin (rates kl and ku),
-
4.
Motor binding and unbinding to and from the filament (rates kfb and kfu), and
-
5.
Motor steps along F-actin filaments (rates k→ and k←).
In the simulations with Ena/VASP acting as an adaptor, the motor could be only loaded with an adaptor. The adaptor could be empty, or scaffolding one or two G-actin monomers. In yet another set of simulations, there was additional reaction, irreversible sinking of the adaptor with simultaneous release of G-actin (if it carries any) to the cytosol.
Actin bundle
Filopodia are formed by an F-actin bundle that extends the cell membrane, forming fingerlike protrusions along the leading edge of the cell (11,40,41). Typical extension lengths in real filopodia are several microns with growth and retraction rates of 0.1–0.2 μm/s (42). Emergence of a filopodium from the three-dimensional actin mesh of the cell or the lamellipodium is not simulated here. Instead, we start with a preformed filopodium of 80 to 600 nm length, the actual value having no effect on the steady state. Because there are typically 10–30 actin filaments per filopodium, we used 16 as the fixed number of filaments (15). Accordingly, filopodia tend to be 100–300 nm in width (16), so we used a width of 150 nm and assumed rapid mixing in the transverse direction. Longer filopodia may be thicker to maintain structural rigidity, although the steady-state effect of increased diffusing species would be canceled by the increased filament number. Having a persistence length of ∼10 μm, or longer if tightly cross-linked, the bundle of F-actin in our model was assumed straight for simplicity. Buckling and bending may be modeled in the future.
Diffusion
Given the linear dimension, molecules diffuse quickly with respect to molecule reaction times up to a certain length, called the Kuramoto length (43), which is ∼100 nm with physiological molecule concentrations. This length can be thought of as a mean free (i.e., without-reacting) path of a protein molecule. For stochastic treatment of diffusion, we divided the filopodium into compartments 50-nm in height, on the scale of Kuramoto length. This is to allow molecules to randomly hop in one dimension from one compartment to another at rates that correspond to typical diffusion rates (D = 5 μm2 s−1 diffusion coefficient, or 2000 s−1 hopping rate), although diffusion rates within filopodia have not been measured. Varying compartment height has been shown to have little effect on simulation results (14). The diffusion constant for all the diffusing species in our simulation was D = 5 μm2 s−1.
A boundary condition at the filopodial base maintains a G-actin concentration of 10 μM, and consumption of G-actin from barbed-end polymerization establishes a base-to-tip gradient. G-actin concentration at the tip can be very low (14), which is one of the motivations for stochastic simulation of filopodial dynamics. The rate of diffusion thus limits filopodial growth, which prompted us to investigate the effects of active transport of G-actin along the F-actin filaments by molecular motors.
Retrograde flow
Because the depolymerization rate at the pointed end is slow, the limitation of filopodial length is mainly controlled by retrograde flow, where the entire actin bundle moves backward at a constant velocity (11,15). The exact mechanism of retrograde flow remains speculative (44), but rates are likely subject to regulatory proteins. Here we did not consider variations in retrograde flow rates but instead used 70 nm/s in all simulations, whereas experimentally measured values are ∼10–200 nm/s (7,30,41,45). Technically, we drag all the filaments with everything bound to them toward the filopodial base with a constant velocity of vretr = 70 nm/s.
Motors
To investigate active transport of G-actin, we incorporated directed motors into our model. They can load and unload a G-actin monomer and step along the actin filaments. To model such a step in the Gillespie scheme, a motor on a filament reacts with an actin monomer 32.4 nm (this is the size of a motor step) away from its starting location along the filament at rates of k→ = 50 s−1 in the forward direction and k← = 5 s−1 in the reverse direction. Not knowing the real biological values and effects of unloading rates (ku), we explored rates ranging from 1 to 3000 s−1. Similarly, we explored motor-filament detachment rates (kfu from 0.1 to 300 s−1). Loading and attachment were assumed to be diffusion-limited (10 μM−1 s−1).
Several different scenarios may arise for motors during the simulation: Motors near the tip (closer to the barbed end than 32.4 nm) can no longer step forward, but can (un)load actin (with rates ku and kl), detach from the filament (with rate kfu), or step backward (with rate k←). Motors that are not attached to any filament also diffuse back and forth within the filopodial cytoplasm with D = 5 μm2 s−1. Those that are bound to a filament are dragged backward by retrograde flow, so they are in essence walking along a treadmilling rope. Thus, the retrograde flow speed is effectively subtracted from average motor walking speed (found from reaction rates and the step size).
Under more realistic consideration, the treadmilling speed of a filament depends on the elongation rate, and so does the retrograde flow. Most of the simulations reported here were carried out with constant retrograde flow rates, thus neglecting this coupling. However, as elaborated in Supporting Material, when we explicitly introduce the coupling between polymerization and retrograde flow processes, the same qualitative conclusions on the rules of active transport are reached as without coupling.
Ena/VASP
Ena/VASP are transported by myosin X (33) and have G-actin binding sites, therefore they might serve as adaptors/scaffolds for the myosin-based G-actin transport. In our scheme, an Ena/VASP molecule can bind one or two G-actins (with the diffusion-limited rate, 10 μM−1 s−1, one by one), release either one of bound G-actins (with the rate ks, that we explored in 0.1–300 s−1 range), be loaded to a motor (diffusion-limited rate), or unloaded from a motor with the rate ku.
The irreversible sinking of Ena/VASP at the tip, representing the filament cross-linking role of Ena/VASP, was also diffusion-limited.
Polymerization, depolymerization
Actin filaments are polar in that polymerization occurs predominantly at the tip, the barbed end, whereas depolymerization occurs predominantly at the base, the pointed end. For actin bundles that are linked to fixed substrate or extracellular matrix, this treadmilling is responsible for motility by extension of filopodia, and cytoskeletal dynamics in general (5,6). Whether protrusion or retraction occurs depends on the equilibrium between polymerization and depolymerization rates. G-actin polymerization rates are increased when bound to ATP, depolymerization rates are increased when bound to ADP, and F-actin:ATP hydrolyzes to F-actin:ADP, leading to aging (46). In our model, we assumed that only G-actin:ATP is polymerized at the tip and only G-actin:ADP is depolymerized at the base, allowing us to account for effects of hydrolysis solely via rates of polymerization (k+ = 11.6 μM−1 s−1) and depolymerization (k− = 1.4 s−1).
Polymerization and depolymerization were treated stochastically in the Gillespie scheme, and the depolymerization rate was slow compared to polymerization. One (de)polymerization event changed the filament length by 2.7 nm, although the diameter of G-actin is 5.4 nm, because F-actin consists of two protofilaments in a right-handed helix. Thus, we modeled F-actin as one protofilament with a polymerization step of 2.7 nm. The motor step of 32.4 nm, therefore, spanned 12 actin units.
Polymerization required a free G-actin monomer, so, before one could be incorporated into the filament, it had to have been unloaded from the motor or Ena/VASP.
Membrane force
Although depolymerization is unaffected by membrane force, polymerization rate decreases with membrane tension (47). The membrane height must be large enough to sterically accommodate a G-actin monomer at the barbed end, so membrane force and its effect on polymerization rate were derived from membrane height. A Gaussian distribution of membrane height with respect to filament tip position was used with a square root of variance of 20 nm (14). At the average height, the typical force was 10 pN. The height with respect to each of the 16 filaments was recalculated after each reaction because membrane fluctuations are on the micro- to millisecond timescale, rapid compared to growth dynamics on the second scale (48). Longer filaments experience stronger membrane force and polymerize more slowly than shorter filaments, on average. This negative feedback diminishes the heterogeneity in filament lengths (14). It should be noted that a second mechanism of negative feedback is the gradient of G-actin concentration along the filopodial tube, providing higher availability of G-actin for shorter filaments (14).
Simulation scheme
In this work we have used the spatial extension of the Gillespie algorithm (14). In the Gillespie algorithm, a simulation step requires two random numbers. The first determines the time step as influenced by the aggregate rate of all possible reactions in the scheme (diffusion of various proteins, (de)polymerization reactions, motor (un)loading of G-actin, and motor (de)attachment from F-actin). The second random number determines which of these reactions will occur, as affected by the rates. After an event, the following are updated: time, species in each compartment, filament length (always affected by retrograde flow, affected by (de)polymerization if the event occurred), and membrane force on each filament. Thus, the Gillespie method allows for simulation and evolution of reactions over continuous time while accounting for effects of molecular noise (49–58). In summary, we modeled stochastically the effects of motor transport of G-actin on filopodial dynamics, with molecular level spatial resolution for motors walking on actin filaments.
Results
The basic components of our filopodia model include polymerization, depolymerization, G-actin diffusion, retrograde flow, and membrane force (14). In the first set of simulations reported here, we added motor molecules that can diffuse, load actin, attach to the filaments, walk on filaments, detach, and unload actin (Fig. 1). Bulk G-actin concentration influences steady-state length (14), thus it was kept consistent between simulations. Accordingly, we placed corresponding boundary conditions at the bottom of a filopodium (where it emerges from the cell's leading edge), assuming equilibrium with the motor-loading reaction (M10+G-actin) in the cell bulk, such that the sum of freely diffusing G-actin and G-actin loaded on freely diffusing motors was kept constant at CA = 10 μM. G-actin binding to motors allows transport but can also result in sequestration, leaving little free G-actin available to polymerization at the filament barbed-end if motor-actin affinity is too high. Thus, rate of actin dissociation from motors (ku) is one key parameter we explore. We also explore the rate of motor dissociation from filaments (kfu) and motor concentration [M].
Figure 1.
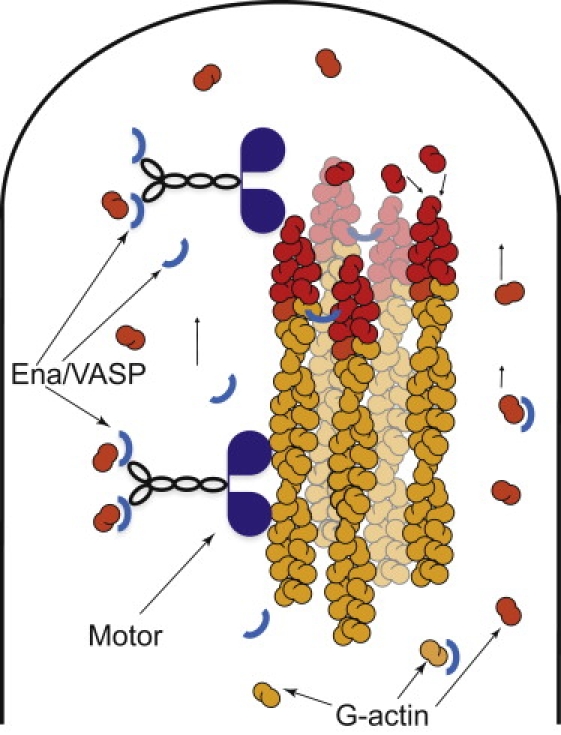
A schematic representation of the filopodial tip in the model is shown. A bundle of polymerizing actin filaments is enveloped by membrane which affects polymerization rates. Transported G-actin must dissociate before polymerization. Retrograde flow pulls filaments back with constant velocity. Myosin X motors travel the filaments in a directed fashion toward the barbed ends at the filopodial tip. Ena/VASP serves as a scaffold between G-actin monomers and motor molecules, and it is consumed near the tip due to cross-linking of the filaments.
Exploring the three-parameter space (ku, kfu, [M]; see Table 1 for other parameters) via 8 × 8 × 8 logarithmic grid (that is, each parameter was scanned on a logarithmic scale, e.g., [M] = 0.1, 0.3, 1, 3, 10, 30, 100, 300 μM), we covered the biologically plausible ranges for these parameters. Intuitively, very low [M] or very high ku are equivalent to the absence of any motors. In the latter case, motors are present, but do not carry actin. Thus, ku determines the fraction of motors that do carry G-actin. For this reason, the stationary length depends mostly on the ratio of ku and [M] (as can be seen by partial collapse of curves in Fig. 2): very high motor concentrations or low ku result in sequestration of G-actin and filopodia do not grow. Therefore, in many cases the stationary length turned out to be shorter than without motors. In a few cases, we observed ∼30% increase in stationary length with the intermediate parameter values (Fig. 2). In Fig. 3 the three-parameter space is given in three two-dimensional projections and the bottom-right diagram in Fig. 3 includes all three parameters. The regions where filopodia do grow longer than they would without motors are marked to provide a phase-diagram-like comprehensive representation. These turn out to be quite localized regions, with low motor concentration. Counterintuitively, upregulating the motors will not lead to increase in transport efficacy because of sequestration. Moreover, motors accumulate at the tip, in qualitative agreement with previous mean-field calculations in stereocilia (17), strongly sequestering G-actin in the location where G-actin's concentration is initially low and where it is most needed for polymerization. In the stationary-state diffusional backward flux of the motors due to concentration, the gradient is equal to the flux of their directed motion forward.
Table 1.
Model variables and parameters
| Mechanics | |
|---|---|
| Half actin monomer size | δ = 2.7 nm |
| Number of filaments | N = 16 |
| Thermal energy | kBT = 4.1 pN nm |
| Membrane force | f = 10 pN |
| Diffusion rates (all species) | kD = 5 μm2 s−1 (2000 s−1) |
| Membrane fluctuation | σd = 20 nm |
| Retrograde flow speed | vretr = 70 nm/s |
| Chemical reaction rates | |
| Polymerization | k+ = 11.6 μM−1 s−1 (21.8 s−1) |
| Depolymerization | k− = 1.4 s−1 |
| Motor loading | kl = 10 μM−1 s−1 (18.8 s−1) |
| Motor unloading | ku = 1–3000 s−1 |
| Filament-binding | kfb = 10 μM−1 s−1 (18.8 s−1) |
| Filament-unbinding | kfu = 0.1 – 300 s−1 |
| Step forward | k→ = 50 s−1 |
| Step back | k← = 5 s−1 |
| Adaptor taking G-actin | ks+ = 10 μM−1 s−1 (18.8 s−1) |
| Adaptor releasing G-actin | ks− = 0.1–300 s−1 |
| Bulk concentrations | |
| Actin | CA = 10 μM |
| Myosin X | [M] = 0.1–300 μM |
| Ena/VASP | [V] = 0.1–300 μM |
The compartment volume was fixed in our computations, with compartment length of lD = 50 nm and filopodial diameter of 150 nm. Based on the compartment volume, the corresponding reaction rates and the protein diffusion rates are also given in units of seconds in parentheses.
Figure 2.
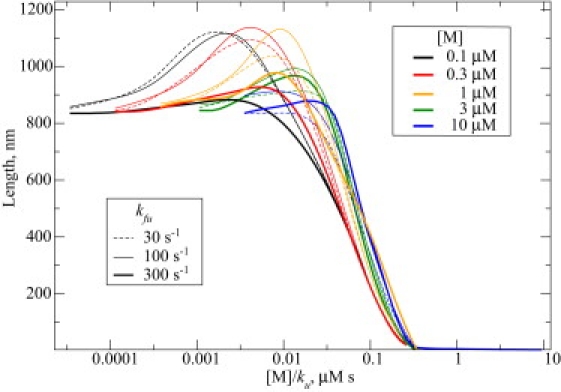
Filopodial stationary length, as a function of model motor-related parameters, is shown. Bulk motor concentration is color-coded. Filament unbinding rate for the motors is equal to 300 s−1 (thick solid lines), 100 s−1 (thin solid lines), and 30 s−1 (dashed lines). As the motor unloading rate ku essentially defines which fraction of motors carry actin, the length dependence comes mostly from the ratio of motor concentration to unloading rate. For this reason, the latter ratio is used as the variable on x axis.
Figure 3.
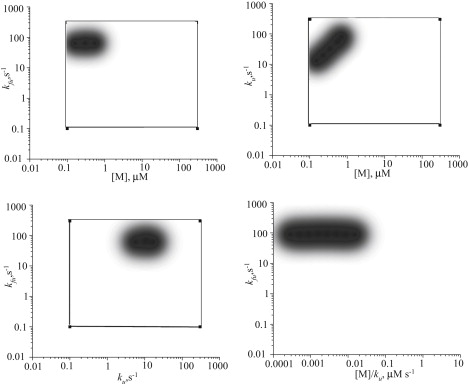
Only in specific local regions of the parameter space do motors provide an increase in filopodial length. These regions are always characterized by low motor concentrations. High motor concentration area does not lead to stable filopodia as most G-actin required for growth is sequestered.
Realizing that sequestration limits length, we ran another set of simulations within the same parameter space, but with actin-loading of diffusing motors (as opposed to filament-bound motors) turned off. Conceptually, motors falling off filaments (accumulating at the tip) did not sequester actin. The goal of these simulations was to confirm the limiting role of sequestration and to find out what would limit the length in its absence. In addition, because binding of G-actin by myosin X has not been observed experimentally, this scheme is not necessarily unrealistic.
The longest filopodia observed in this set of simulations were almost 6 μm, >7 times longer than those that grow in a model without active transport. However, the increase could have been merely an effect of increased presence of G-actin in the filopodial tube. In addition to those molecules that freely diffuse in cytosol (maintained to be 10 μM at the bottom), some are carried by the motors attached to the filaments. To verify that directed transport was responsible for this length increase, we ran two control simulations: In one, we set the motor backward step-rate k← to be equal to forward step-rate k→ = 50 s−1, making the motors nondirectional, random walkers. The resulting filopodial lengths were back to those without motors. In a second control simulation, we set k← = k→ = 2000 s−1, similar to diffusion rate, in which case the increase in length was minuscule compared to directional motors even at these unrealistically high motor speeds.
The largest lengths in the nonsequestering motors model were achieved at the kfu values on the higher end of the parameter space (30 s−1, 100 s−1, 300 s−1), corresponding to lower affinity between motors and F-actin. On one hand, high kfu helps to clear the filaments from the motors that have delivered cargo at the tip; on the other hand, if very few motors stay on the filaments the transport is inefficient. Our results show that the former outweighs the latter: the high values of kfu for the longest filopodia indicate that the clogging of filaments by empty motors represents yet another key bottleneck for achieving long stationary lengths.
To gauge the influence of this bottleneck, we created an artificial setup where loaded motors could step forward even if the site was occupied by an empty motor (i.e., pass through each other). In one case (ku = 3 s−1, kfu = 300 s−1, [M] = 1 μM) such a scheme results in a qualitatively new growing regime. In the new conditions the forward directional actin flux overcame the backward directional flux due to retrograde flow, providing the unquenchable source of actin for the polymerizing barbed ends (see Fig. 4, juxtaposing the two regimes). In these simulations, filopodia continued to grow linearly for >100 s up to >6 μM, without any indication of growth saturation, where the latter has been found in all other cases discussed here or in prior works (14,15).
Figure 4.
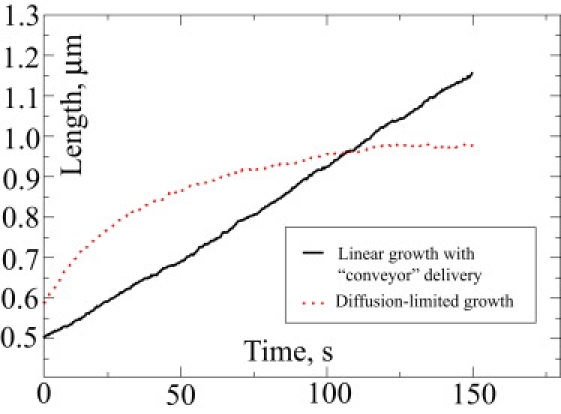
Comparison between diffusion-limited and linear growth regimes is shown. These are two simulations with different parameters from the Ena/VASP set.
Thus, we have learned that sequestration and clogging can stunt motor-assisted filopodial growth. If active transport is indeed employed in living organisms for G-actin supply of the polymerizing barbed ends, there has to be a biological mechanism for disabling these limitations. In search of a plausible biological mechanism, we turned to Ena/VASP, which has traditionally been implicated in anticapping activity but actually has additional functions (34). Ena/VASP can bind G-actin (37,59), and it has also been observed as myosin X cargo (33).
However, from ideas in our previous works (9,14), we conclude that a protein only needs to be transported actively if it is consumed during the growth, like G-actin. Regulatory molecules, like capping or anticapping proteins that only interact with the filament ends, are required in only very limited amounts, and at realistic growth speeds diffusion will always provide sufficient flux. Therefore, we suggest that active transport of Ena/VASP indicates that it may be continuously consumed, most likely to cross-link the filaments (35). There can be other reasons to be motor-transported. For instance, as we investigate here, Ena/VASP might serve as a scaffold, or an adaptor, between myosin X and actin monomers. In this scenario of a sinking adaptor, motors would not bind and sequester G-actin when the adaptor is absent (and consumption due to the cross-linking will diminish adaptor concentration), although clogging would not necessarily be avoided.
To explore this sinking adaptor possibility, we allowed Ena/VASP to bind up to two G-actin monomers and/or be loaded to a motor. We simulate a maximum of two G-actin molecules transported per motor for simplicity, although one motor could carry up to eight G-actin molecules: Ena/VASP can form tetramers (37) where each tetramer has four G-actin binding sites. Myosin X is also a dimer with two heads and two tails (28,60,61), and each tail could bind an Ena/VASP tetramer as cargo (not considered in our scheme for simplicity). This 8:1 stoichiometry could lead to efficient transport, providing that sequestration is avoided.
In the first set of Ena/VASP simulations, a cross-linking sink was not incorporated. Sequestration still occurred, and filopodia lengths were similar to those with naïve motors. In the second set of Ena/VASP simulations, a cross-linking sink was incorporated. Ena/VASP does have a bundling role (35) and is localized near the tip (34) (where sequestration is most deleterious), so it is possibly consumed there for cross-linking, releasing actin for polymerization. Bundling would provide structural rigidity for long filopodia, but because we assume straight filopodia, the desired scenario is simply achieved by reactions of Ena/VASP irreversible sinking near the tip with simultaneous release of any G-actin it had carried.
Some of the simulations with this scheme did show significantly increased stationary lengths compared to simulation of naïve motors. In the most interesting case (ku = 30 s−1, ks = 30 s−1, kfu = 100 s−1, [M] = 1 μM), the growth seemed to be linear for >300 s, although with a very gradual slope change. This corresponds to a small difference between retrograde flow actin flux and motor-transported actin flux, with the former slightly larger, so that it dominates until the filopodium becomes very long. The stationary length here reached 3.5 μM, severalfold higher than when using naïve motors.
To summarize our results for various active transport designs, a comparative chart for the maximal observed lengths is given in Fig. 5.
Figure 5.
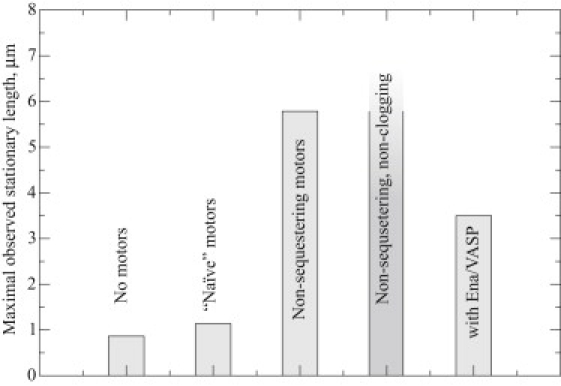
The largest observed stationary length in each set of simulations is shown. In the case of simulations with artificial conditions in which motors do not sequester actin and do not clog the filaments (inasmuch as they are sterically transparent to each other), a linear growth regime was observed that did not reach a stationary length.
Discussion
When considering biological active transport realized by molecular motors instead of passive diffusion, one typically envisions a conveyorlike delivery of the materials to the construction site. Without active transport, as diffusional flux and the resulting elongation rate decrease with the length, retrograde flow plus depolymerization equalizes polymerization at the stationary length, and filopodial elongation stops (14). With active transport, there is the possibility that forward flux exceeds that of retrograde flow, in which case there is no steady state, and conveyorlike delivery with linear growth would last indefinitely.
Even when the motor-based actin forward flux is barely below the retrograde actin flux, it will still dominate the dynamics, which will exhibit a phase of linear growth for quite a long time (see Fig. 4). Diffusional flux will be gradually decreasing until the total flux becomes equal to retrograde flux, which will happen when the filopodium has already grown long, as in the presented case with Ena/VASP.
If one were to write a diffusion equation for G-actin concentration (15), an addition of motor-based flux would be represented as a convectional term. From the arguments above, the relative magnitude of this term with respect to retrograde flow is crucial, as it sets the asymptotic growth regime.
Our findings indicate that for a conveyorlike delivery achieved by an active transport term of sufficient magnitude, the process of transport has to be organized quite intricately. Even for a modest increase in stationary length, certain rules have to be observed. These rules are quite general, so one can think of them as the rules for effective active transport in polymerization-based protrusions. To follow the rules, 1), motors should be kept from sequestering freely diffusing cargo; and 2), rails of the carriers should be kept clear of clogging by empty motors.
In our simulations, the directional flux due to active transport was almost always less than backward flux due to retrograde flow; therefore, diffusion still set the stationary length. This is when the diffusional flux thins out such that, in sum with the motor-based flux, it balances the retrograde flow. Surprisingly, it was only a narrow area of parameter space that yielded the conveyor picture of delivery, even in the artificial control simulations with sequestration and clogging being turned off. Perhaps this regime of transport would have been enhanced by simulating transport stoichiometry of eight G-actins per motor instead of two, leading to flux amplification. Nevertheless, we did not observe a perpetual conveyor in our speculative, yet plausible scheme with Ena/VASP. However, even achieving a transient, yet prolonged conveyorlike behavior required intricacy in that Ena/VASP needs to have both bundling and scaffolding (adaptor) functions. Interestingly, Ena/VASP is known to bundle filaments, could possibly act as a scaffold via its G-actin binding domains, and has been observed as myosin X cargo.
Thus, the balance between active transport-based flux and backward flux due to retrograde flow can dramatically affect filopodial growth. In this article, we considered a hypothetical active transport of G-actin to promote elongation. To the same purpose, retrograde flow might serve as another convenient way for a cell to regulate the filopodial length, switching between different growth regimes. Reducing the retrograde flow severalfold will increase the filopodial stationary length (14). In some cases, the reported values of bulk G-actin concentrations are much higher, of ∼100 μM (62); however, in vitro, 100 μM pure G-actin polymerizes, leaving only 0.1–1 μM monomeric G-actin (63). To maintain a pool of unpolymerized actin in vivo, most of monomeric actin is typically sequestered by special proteins such as thymosin-β4 (64). Prior experimental and computational analysis found that the concentration for nonsequestered G-actin is ∼10–50 μM (65) (see also the Supporting Material for a brief review of experimental measurements of actin concentrations in various cell types). Even at the upper limits of polymerizable G-actin concentrations, this will only lead to several-fold-larger filopodial stationary lengths (14). However, in combination with another severalfold increase that we observed when using the postulated Ena/VASP based scheme of G-actin transport, this can take filopodia from submicron lengths to lengths of ∼10 μM. The same factors also increase growth speeds. Decreasing retrograde flow rate would increase the growth speed ≤4 μm/min. The combination of the latter effect with enhanced G-actin flux due to active transport and high G-actin concentration, may be enough to result in the high growth speeds of ∼10 μm/min observed in some experiments (22). Thus, the cells that grow extremely long filopodia perhaps use multiple facilities to achieve this, which would include the downregulation of retrograde flow, upregulation of actin concentration, and possibly active transport, as explored in this work.
It would be interesting to test experimentally whether G-actin is transported actively with myosin X and/or Ena/VASP utilized as an adaptor. For example, one might mutate the G-actin binding domain of Ena/VASP and monitor filopodial length. However, Ena/VASP needs to bind G-actin for its anticapping activity, so mutation may diminish filopodial length through disabling anticapping, thus obscuring any conclusions regarding transport of G-actin. Another possibility is to fluorescently label G-actin: to avoid an intense background of glowing F-actin, one could consider labeling a small fraction of actin, or more attractively, labeling DnaseI, which binds to G-actin but not F-actin (66). Concurrent labeling of myosin X or Ena/VASP with a second fluorophore might reveal colocalization with G-actin along the length of the filopodia, or perhaps even FRET-based spectral changes, offering support for the active transport of G-actin. Such a colocalization study might rely on techniques used in recent work that demonstrates espin1 and myosin IIIa are cotransported along the length of a stereocilium (67).
Compared to filopodia, the requirement for active transport in other parallel actin-based structures may not be acute. For instance, stereocilia are maintained at very definite lengths, where this fine-tuning is indicative of regulation by a possible signaling subnetwork. Despite the fact that elongation rates in stereocilia are much lower than in filopodia (17), motors that have been characterized in stereocilia (67) might potentially carry G-actin. Motors are also found in microvilli; however, their lengths are short and diffusional transport should suffice. (More details on the role of motors in these organelles are provided in the Supporting Material.) Our model is general in its treatment of motors, hence, analogous active transport schemes could be constructed for microvilli and stereocilia, although one needs to take into account the mechanical and structural differences among these organelles.
Supporting Material
One table, one equation, one reaction set, two figures, and comments are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00097-4.
Supporting Material
Acknowledgments
We are grateful to Dr. Richard Cheney and his research group members for helpful discussions about possible roles of Myosin X and Ena/VASP in filopodia.
We acknowledge financial support from the National Science Foundation CAREER Award No. CHE-0846701. All calculations were carried out using the University of North Carolina Topsail supercomputer at Chapel Hill.
References
- 1.Lorenz M., Yamaguchi H., Condeelis J. Imaging sites of N-wasp activity in lamellipodia and invadopodia of carcinoma cells. Curr. Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Dent E.W., Gertler F.B. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- 3.Noselli S. Drosophila, actin and videotape—new insights in wound healing. Nat. Cell Biol. 2002;4:E251–E253. doi: 10.1038/ncb1102-e251b. [DOI] [PubMed] [Google Scholar]
- 4.Lawson N.D., Weinstein B.M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 5.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 6.Pantaloni D., Le Clainche C., Carlier M.F. Mechanism of actin-based motility. Science. 2001;292:1502–1506. doi: 10.1126/science.1059975. [DOI] [PubMed] [Google Scholar]
- 7.Kovar D.R. Intracellular motility: myosin and tropomyosin in actin cable flow. Curr. Biol. 2007;17:R244–R247. doi: 10.1016/j.cub.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Lin C.H., Espreafico E.M., Forscher P. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhuravlev P.I., Papoian G.A. Molecular noise of capping protein binding induces macroscopic instability in filopodial dynamics. Proc. Natl. Acad. Sci. USA. 2009;106:11570–11575. doi: 10.1073/pnas.0812746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellor H. The role of formins in filopodia formation. Biochim. Biophys. Acta. 2009 doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Faix J., Rottner K. The making of filopodia. Curr. Opin. Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Chang S., De Camilli P. Glutamate regulates actin-based motility in axonal filopodia. Nat. Neurosci. 2001;4:787–793. doi: 10.1038/90489. [DOI] [PubMed] [Google Scholar]
- 13.Wood W., Martin P. Structures in focus—filopodia. Int. J. Biochem. Cell Biol. 2002;34:726–730. doi: 10.1016/s1357-2725(01)00172-8. [DOI] [PubMed] [Google Scholar]
- 14.Lan Y., Papoian G.A. The stochastic dynamics of filopodial growth. Biophys. J. 2008;94:3839–3852. doi: 10.1529/biophysj.107.123778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mogilner A., Rubinstein B. The physics of filopodial protrusion. Biophys. J. 2005;89:782–795. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atilgan E., Wirtz D., Sun S.X. Mechanics and dynamics of actin-driven thin membrane protrusions. Biophys. J. 2006;90:65–76. doi: 10.1529/biophysj.105.071480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naoz M., Manor U., Gov N.S. Protein localization by actin treadmilling and molecular motors regulates stereocilia shape and treadmilling rate. Biophys. J. 2008;95:5706–5718. doi: 10.1529/biophysj.108.143453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prost J., Barbetta C., Joanny J.-F. Dynamical control of the shape and size of stereocilia and microvilli. Biophys. J. 2007;93:1124–1133. doi: 10.1529/biophysj.106.098038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gov N.S. Dynamics and morphology of microvilli driven by actin polymerization. Phys. Rev. Lett. 2006;97:018101. doi: 10.1103/PhysRevLett.97.018101. [DOI] [PubMed] [Google Scholar]
- 20.Rifkin J.L., Speisman R.A. Filamentous extensions of vegetative amoebae of the cellular slime mold Dictyostelium. Trans. Am. Microsc. Soc. 1976;95:165–173. [PubMed] [Google Scholar]
- 21.Gustafson T., Wolpert L. Studies on the cellular basis of morphogenesis in the sea urchin embryo. Directed movements of primary mesenchvme cells in normal and vegetalized larvae. Exp. Cell Res. 1961;24:64–79. doi: 10.1016/0014-4827(61)90248-8. [DOI] [PubMed] [Google Scholar]
- 22.Miller J., Fraser S.E., McClay D. Dynamics of thin filopodia during sea urchin gastrulation. Development. 1995;121:2501–2511. doi: 10.1242/dev.121.8.2501. [DOI] [PubMed] [Google Scholar]
- 23.Berg J.S., Powell B.C., Cheney R.E. A millennial myosin census. Mol. Biol. Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollard T.D., Blanchoin L., Mullins R.D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 25.Drenckhahn D., Pollard T.D. Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J. Biol. Chem. 1986;261:12754–12758. [PubMed] [Google Scholar]
- 26.Zicha D., Dobbie I.M., Dunn G.A. Rapid actin transport during cell protrusion. Science. 2003;300:142–145. doi: 10.1126/science.1082026. [DOI] [PubMed] [Google Scholar]
- 27.Berg J.S., Derfler B.H., Cheney R.E. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J. Cell Sci. 2000;113:3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 28.Bohil A.B., Robertson B.W., Cheney R.E. Myosin-X is a molecular motor that functions in filopodia formation. Proc. Natl. Acad. Sci. USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheetz M.P., Wayne D.B., Pearlman A.L. Extension of filopodia by motor-dependent actin assembly. Cell Motil. Cytoskeleton. 1992;22:160–169. doi: 10.1002/cm.970220303. [DOI] [PubMed] [Google Scholar]
- 30.Berg J.S., Cheney R.E. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 31.Kerber M.L., Jacobs D.T., Cheney R.E. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr. Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H., Berg J.S., Strömblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 33.Tokuo H., Ikebe M. Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 2004;319:214–220. doi: 10.1016/j.bbrc.2004.04.167. [DOI] [PubMed] [Google Scholar]
- 34.Applewhite D.A., Barzik M., Borisy G.G. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol. Biol. Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schirenbeck A., Arasada R., Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc. Natl. Acad. Sci. USA. 2006;103:7694–7699. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barzik M., Kotova T.I., Schafer D.A. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krause M., Dent E.W., Gertler F.B. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell Dev. Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 38.Hüttelmaier S., Harbeck B., Jockusch B.M. Characterization of the actin binding properties of the vasodilator-stimulated phosphoprotein VASP. FEBS Lett. 1999;451:68–74. doi: 10.1016/s0014-5793(99)00546-3. [DOI] [PubMed] [Google Scholar]
- 39.Gillespie D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. [Google Scholar]
- 40.Wicki A., Lehembre F., Christofori G. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell. 2006;9:261–272. doi: 10.1016/j.ccr.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Lidke D.S., Lidke K.A., Arndt-Jovin D.J. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J. Cell Biol. 2005;170:619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Argiro V., Bunge M.B., Johnson M.I. A quantitative study of growth cone filopodial extension. J. Neurosci. Res. 1985;13:149–162. doi: 10.1002/jnr.490130111. [DOI] [PubMed] [Google Scholar]
- 43.Kuramoto Y. Springer-Verlag; Berlin, Heidelberg, New York, Tokyo: 1984. Chemical Oscillations, Waves and Turbulence. [Google Scholar]
- 44.Heid P.J., Geiger J., Soll D.R. Computer-assisted analysis of filopod formation and the role of myosin II heavy chain phosphorylation in Dictyostelium. J. Cell Sci. 2005;118:2225–2237. doi: 10.1242/jcs.02342. [DOI] [PubMed] [Google Scholar]
- 45.Brown M.E., Bridgman P.C. Retrograde flow rate is increased in growth cones from myosin IIB knockout mice. J. Cell Sci. 2003;116:1087–1094. doi: 10.1242/jcs.00335. [DOI] [PubMed] [Google Scholar]
- 46.Stukalin E.B., Kolomeisky A.B. ATP hydrolysis stimulates large length fluctuations in single actin filaments. Biophys. J. 2006;90:2673–2685. doi: 10.1529/biophysj.105.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raucher D., Sheetz M.P. Cell spreading and lamellipodial extension rate is regulated by membrane tension. J. Cell Biol. 2000;148:127–136. doi: 10.1083/jcb.148.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin L.C.-L., Brown F.L.H. Brownian dynamics in Fourier space: membrane simulations over long length and time scales. Phys. Rev. Lett. 2004;93:256001. doi: 10.1103/PhysRevLett.93.256001. [DOI] [PubMed] [Google Scholar]
- 49.Kepler T.B., Elston T.C. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys. J. 2001;81:3116–3136. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasai M., Wolynes P.G. Stochastic gene expression as a many-body problem. Proc. Natl. Acad. Sci. USA. 2003;100:2374–2379. doi: 10.1073/pnas.2627987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korobkova E., Emonet T., Cluzel P. From molecular noise to behavioral variability in a single bacterium. Nature. 2004;428:574–578. doi: 10.1038/nature02404. [DOI] [PubMed] [Google Scholar]
- 52.Walczak A.M., Onuchic J.N., Wolynes P.G. Absolute rate theories of epigenetic stability. Proc. Natl. Acad. Sci. USA. 2005;102:18926–18931. doi: 10.1073/pnas.0509547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinberger L.S., Burnett J.C., Schaffer D.V. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Thattai M., van Oudenaarden A. Stochastic gene expression in fluctuating environments. Genetics. 2004;167:523–530. doi: 10.1534/genetics.167.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lan Y., Papoian G.A. Stochastic resonant signaling in enzyme cascades. Phys. Rev. Lett. 2007;98:228301. doi: 10.1103/PhysRevLett.98.228301. [DOI] [PubMed] [Google Scholar]
- 56.Lan Y., Papoian G.A. The interplay between discrete noise and nonlinear chemical kinetics in a signal amplification cascade. J. Chem. Phys. 2006;125:154901. doi: 10.1063/1.2358342. [DOI] [PubMed] [Google Scholar]
- 57.Lan Y., Wolynes P.G., Papoian G.A. A variational approach to the stochastic aspects of cellular signal transduction. J. Chem. Phys. 2006;125:124106. doi: 10.1063/1.2353835. [DOI] [PubMed] [Google Scholar]
- 58.Artyomov M.N., Das J., Chakraborty A.K. Purely stochastic binary decisions in cell signaling models without underlying deterministic bistabilities. Proc. Natl. Acad. Sci. USA. 2007;104:18958–18963. doi: 10.1073/pnas.0706110104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chereau D., Dominguez R. Understanding the role of the G-actin-binding domain of Ena/VASP in actin assembly. J. Struct. Biol. 2006;155:195–201. doi: 10.1016/j.jsb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 60.Knight P.J., Thirumurugan K., Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 61.Nagy S., Ricca B.L., Rock R.S. A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. USA. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koestler S.A., Rottner K., Small J.V. F- and G-actin concentrations in lamellipodia of moving cells. PLoS One. 2009;4:e4810. doi: 10.1371/journal.pone.0004810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pollard T.D. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber A., Nachmias V.T., Safer D. Interaction of thymosin β 4 with muscle and platelet actin: implications for actin sequestration in resting platelets. Biochemistry. 1992;31:6179–6185. doi: 10.1021/bi00142a002. [DOI] [PubMed] [Google Scholar]
- 65.Novak I.L., Slepchenko B.M., Mogilner A. Quantitative analysis of G-actin transport in motile cells. Biophys. J. 2008;95:1627–1638. doi: 10.1529/biophysj.108.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cramer L.P., Briggs L.J., Dawe H.R. Use of fluorescently labeled deoxyribonuclease I to spatially measure G-actin levels in migrating and non-migrating cells. Cell Motil. Cytoskeleton. 2002;51:27–38. doi: 10.1002/cm.10013. [DOI] [PubMed] [Google Scholar]
- 67.Salles F.T., Merritt R.C., Kachar B. Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nat. Cell Biol. 2009;11:443–450. doi: 10.1038/ncb1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


