Abstract
FOXO transcription factors represent targets of the phosphatidylinositol 3-kinase/protein kinase B survival pathway controlling important biological processes, such as cell cycle progression, apoptosis, vascular remodeling, stress responses, and metabolism. Recent studies suggested the existence of alternative mechanisms of FOXO-dependent gene expression beyond classical binding to a FOXO-responsive DNA-binding element (FRE). Here we analyzed the relative contribution of those mechanisms to vascular function by comparing the transcriptional and cellular responses to conditional activation of FOXO3 and a corresponding FRE-binding mutant in human primary endothelial cells. We demonstrate that FOXO3 controls expression of vascular remodeling genes in an FRE-dependent manner. In contrast, FOXO3-induced cell cycle arrest and apoptosis occurs independently of FRE binding, albeit FRE-dependent gene expression augments the proapoptotic response. These findings are supported by bioinformatical analysis, which revealed a statistical overrepresentation of cell cycle regulators and apoptosis-related genes in the group of co-regulated genes. Molecular analysis of FOXO3-induced endothelial apoptosis excluded modulators of the extrinsic death receptor pathway and demonstrated important roles for the BCL-2 family members BIM and NOXA in this process. Although NOXA essentially contributed to FRE-dependent apoptosis, BIM was effectively induced in the absence of FRE-binding, and small interfering RNA-mediated BIM depletion could rescue apoptosis induced by both FOXO3 mutants. These data suggest BIM as a critical cell type-specific mediator of FOXO3-induced endothelial apoptosis, whereas NOXA functions as an amplifying factor. Our study provides the first comprehensive analysis of alternatively regulated FOXO3 targets in relevant primary cells and underscores the importance of such genes for endothelial function and integrity.
Keywords: Apoptosis, Cell Cycle, Chromatin Immunoprecipitation (ChiP), Endothelium, Microarray, Signal Transduction, BIM, Forkhead, FOXO, NOXA
Introduction
The FOXOs represent an evolutionarily conserved subfamily of forkhead transcription factors unified by the presence of a highly homologous DNA-binding domain termed the “forkhead box.” In contrast to invertebrates, which only express a single FOXO gene (dfoxo in Drosophila melanogaster and daf-16 in Caenorhabditis elegans) mammals contain four distinct isoforms: FOXO1, FOXO3, FOXO4 and the atypical FOXO6, which shows divergent modes of regulation and expression (1). FOXOs are regulated by various posttranslational modifications, including phosphorylation, acetylation, and ubiquitination. Among those, phosphorylation by the insulin receptor/phosphatidylinositol 3-kinase/protein kinase B cascade provides a master switch of FOXO activity. Growth factors or insulin stimulation leads to activation of this pathway and triggers protein kinase B-dependent FOXO phosphorylation at three distinct residues, followed by nuclear exclusion and proteasomal degradation (2). In contrast, the absence of growth factors or treatment with inhibitors of the phosphatidylinositol 3-kinase/protein kinase B pathway results in silencing of protein kinase B activity, promoting nuclear localization and transcriptional activation of FOXOs. This antagonism is corroborated by findings in C. elegans, in which inactivating mutations of the insulin receptor/phosphatidylinositol 3-kinase/protein kinase B pathway result in increased stress resistance and longevity, whereas DAF-16 deficiency reverts this phenotype (3, 4). At the cellular level, constitutive protein kinase B activation promotes proliferation, survival, and tumorigenesis, whereas expression of protein kinase B-insensitive variants of FOXO containing alanine replacements at the three protein kinase B phosphorylation sites (FOXO.A3) effectively antagonizes these effects and induces either cell cycle arrest (5–7) or apoptosis (8–10) in a cell type-specific manner. Over time, various functionally relevant FOXO targets have been identified. These include genes required for G1-S progression, such as the cyclin-dependent kinase inhibitor p27KIP1 and D-type cyclins (5–7); proapoptotic genes, such as the death ligands FASL (8) and TRAIL (10); or the proapoptotic Bcl-2 family member BIM (9, 11, 12) as well as stress resistance genes, such as superoxide dismutase (13) and GADD45 (14), which promote detoxification of reactive oxygen species and DNA damage repair, respectively. FOXOs furthermore modulate genes involved in energy metabolism and gluconeogenesis in the liver (15) and are key players in vascular development, remodeling, and angiogenesis (16–18). The latter is underscored by recent knock-out studies, which revealed that germ line deletion of FOXO1 results in embryonic lethality due to defects in blood vessel formation (16, 19). Consistently, combined somatic deletion of FOXO1 and at least one other FOXO gene leads to the development of hemangiomas (20). Although this supports the concept of functional redundancy of the different FOXOs, FOXO4-deficient mice are vital and do not show any overt phenotype (19). Likewise, FOXO3−/− mice reveal no apparent signs of endothelial dysfunction but suffer from ovarian infertility (19) and disturbed lymphocyte homeostasis (21). Although the absence of endothelial phenotypes may reflect a low or absent expression of FOXO3 and FOXO4 in embryonic vessels, recent in vitro data suggest that the two major FOXOs in adult vessels, FOXO1 and FOXO3, may have divergent targets in endothelial cells (ECs)3 and could fulfill alternative functions (18).
FOXOs classically regulate gene expression by binding of their forkhead boxes to a specific TTGTTTAC consensus sequence present in the promoter of their target genes (22, 23). Although the forkhead boxes of human FOXOs are almost identical, the isoforms vary in their transactivation domain and exhibit distinct and overlapping tissue distribution (19). Moreover, transcriptional regulation is not only promoted by binding to forkhead-responsive elements (FREs) but also alternatively by binding to unknown target motifs or interaction with other transcriptionally active mediators (24). We and others have previously reported that FOXOs can effectively repress transcription of D-type cyclins, which lack classical FREs in their promoter (6, 7). Furthermore, a mutant of FOXO1, which failed to bind to its established DNA consensus sequence, still efficiently inhibited cell cycle progression and cyclin D (CCND) expression in PTEN-negative renal carcinoma cells (6), suggesting the existence of alternative mechanisms of transcriptional regulation beyond classical DNA binding.
Here, we used functional assays, microarray analysis, and bioinformatic approaches to elucidate the endothelial responses elicited by conditional activation of FOXO3 or a corresponding FRE-binding mutant. We demonstrate that FRE-independent cell cycle regulation by FOXOs is not specific for tumor cells but also occurs by similar mechanisms in relevant primary cells. We further show for the first time that FOXO3-induced apoptosis as well as other responses can likewise occur in the absence of FRE binding and results from combined induction of FRE-dependent and FRE-independent FOXO3 targets. Our data provide evidence that FOXOs utilize both classical and alternative transcriptional mechanisms to control specific functions in primary human ECs.
EXPERIMENTAL PROCEDURES
Chemicals and Antibodies
The following commercially available antibodies were used: α-tubulin (T5168) and BIM (B7929) (Sigma); ERK1/2 (catalog number sc-154) (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)); and FOXO3 (catalog number 9467) and cleaved CASP-3 (caspase-3) (catalog number 9664) (Cell Signaling). p27KIP1 (catalog number 610241) and bromodeoxyuridine (BrdUrd)-FITC (catalog number 347583) were obtained from BD Biosciences; the monoclonal antibodies against cFLIP (NF6) and CASP-8 (caspase-8) (C15) are available from Enzo Life Sciences. 4-Hydroxy-(Z)-tamoxifen (4OHT) (Calbiochem catalog number 579002) and LY294002 (Sigma catalog number L-9908) were used at a concentration of 100 nm or 10 μm, respectively. Z-IETD-fluoromethyl ketone (caspase inhibitor II; catalog number 218759) was purchased from Calbiochem.
Culture of Primary Cells and Cell Lines
Primary human umbilical vein endothelial cells (HUVECs) were purchased from Lonza and cultured as described previously (25). Only cells from passage 3 were used for infection and maximally passaged once for the experiments.
Plasmids and Site-directed Mutagenesis
Plasmids for retroviral expression of conditionally active FOXO3.A3.ER (pBabe puro HA-FOXO3.A3.ER) and cFLIPL (PINCO-cFLIPL) have been described previously (26, 27). A corresponding DNA-binding mutant of FOXO3 (pBabe puro HA-FOXO3.A3.ER.H212R) was generated by using a site-directed mutagenesis kit (Stratagene) with the following primers: forward, 5′-GAACTCCATCCGGAGAAACCTGTCACTGCATAG-3′; reverse, 5-CTATGCAGTGACAGGTTTCTCCGGATGGAGTTC-3′. The 6×DBE-luc construct containing six copies of the DAF-16-binding motif and the BIM-luc construct containing the natural rat BIM promoter have been described earlier (12, 22).
Microarray and Statistical Analysis
HUVECs were infected in three independent experiments with either empty pBabe puro vector, pBP-HA-FOXO3.A3.ER, or pBP-HA-FOXO3.A3.ER.H212R in three consecutive rounds, and 72 h postinfection, cells were selected for puromycin resistance by adding 2 μg/ml puromycin overnight. Subsequently, cells were reseeded in puromycin-free medium, and total RNA from medium-stimulated controls or from cells treated for 12 h with 4OHT was isolated and individually processed for microarray hybridization using Affymetrix HG-U133 Plus 2.0 arrays according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). A Custom CDF Version 11 with Entrez-based gene definitions was used to annotate the arrays (28). Raw fluorescence intensity values were normalized, applying quantile normalization. Differential gene expression was analyzed based on log linear mixed model analysis of variance (29) using the commercial software package SAS JMP7 Genomics, version 3.1 from SAS (SAS Institute, Cary, NC). A false positive rate of α = 0.05 with Holm correction was taken as level of significance. To be significantly regulated, only genes with a -fold change of ≥1.5 and a p value less than or equal to the threshold estimated by the Holm multiple testing algorithm, as compared with the empty vector without 4OHT, were considered. Additionally, genes with a significantly different regulation between FOXO3.A3.ER and FOXO3.A3.ER.H212R according to Holm correction and a ≥2-fold differential regulation between FOXO3.A3.ER and FOXO3.A3.ER.H212R were resorted from the co-regulated group to FOXO3.A3.ER or FOXO3.A3.ER.H212R, respectively. Functional annotation clustering was performed, employing the Database for Annotation, Visualization, and Integrated Discovery (DAVID) with the parameters “Affymetrix HG-U133_Plus_2” as background, “H. sapiens” as species, “GOTERM_BP_ALL” for Gene Ontology, and “high” as level of classification stringency.
Quantitative Real-time Reverse Transcription-PCR (qRT-PCR)
Total RNA was isolated by a spin column-based technique using an RNeasy minikit (Qiagen). RNA concentration was photometrically determined, and cDNA was synthesized from 1 μg of total RNA using the High Fidelity cDNA synthesis kit (Roche Applied Science). Primers and probes were obtained from Applied Biosystems for human NOXA (Hs00560402_m1), BIM (Hs01076940_m1), cFLIP (Hs800354474_m1), CCND1 (Hs99999004_m1), MEF2C (Hs00231149_m1), and GAPDH (Hs99999905_m1). Primer sequences for ID2, EGR1, SLIT2, INHBA, MET, PCNA, and IGFBP1 were taken from a public data base for primer and probe sequences used in real-time PCR assays (available at the RTPrimerDB Web site). qRT-PCR was performed using the primer probe method (TaqMan gene expression master mix; catalog number 4369016, Applied Biosystems) or SYBR Green (Brilliant II SYBR Green QPCR master mix; catalog number 600828, Stratagene) and raw data acquired with Stratagene's Mx3005P. Pipetting errors were corrected by passive 5-carboxy-X-rhodamine fluorescence. Gene expression was normalized to the housekeeping control gene GAPDH, and the relative expression of genes of interest was compared with the respective experimental control (empty vector 4OHT) calculated using the Comparative PCR Method MxPro-Mx3005 Version 4.01 software from Stratagene.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using the SimpleCHIP enzymatic chromatin immunoprecipitation kit (catalog number 9002) from Cell Signaling according to the manufacturer's protocol. Firefly, for one chromatin preparation, 10 × 106 cells were cross-linked with 1% (v/v) formaldehyde for 10 min at room temperature. Subsequently, nuclei were isolated by lysis of the cytoplasmic fraction, and chromatin was digested into fragments of 150–900 bp by micrococcal nuclease (400 gel units) for 20 min at 37 °C, followed by ultrasonic disruption of the nuclear membrane using a standard microtip and a Branson W250D Sonifier (four pulses, 60% amplitude, duty cycle 40%). For immunoprecipitation, 5–15 μg of total chromatin was incubated overnight at 4 °C with 5 μg of the respective antibodies: Abgent mouse α-HA-Tag Clone 12CA5 (AM1008a) and Cell Signaling Normal Rabbit IgG (catalog number 2729) as negative control. After incubation with 30 μl of ChIP grade protein G-agarose beads for 2 h at 4 °C, antibody-DNA complexes were eluted from the beads and digested by 40 μg of Proteinase K for 2 h at 65 °C, followed by spin column-based purification of the DNA. Transcription factor binding was finally assessed by qRT-PCR using the following primers: for the IGFBP1 promoter, 5′-CCTAACAACGGGACAAACAG-3′ (forward) and 5′-CTGCCAATCATTAACCTCCTG-3′ (reverse); for the BIM promoter, 5′-AGGCTAGGGTACACTTCG-3′ (forward) and 5′-AGGCTCGGACAGGTAAAG-3′ (reverse).
For quantification, the comparative cycle threshold (Ct) method was used. Briefly, the measured Ct values of each immunoprecipitation fraction were first normalized to the dilution-corrected Ct values of the respective input samples to obtain ΔCt values. Subsequently, differential site occupancy across samples (i.e. the enrichment of the analyzed promoter sequence relative to the respective experimental control) was determined according to the formula, 2−(ΔCtexperimental sample − ΔCtexperimental control). As additional controls, sequence enrichment relative to a nonspecific antibody control (i.e. -fold enrichment above background) and sequence enrichment relative to the total amount of input DNA (i.e. percentage of input) were determined.
Western Blot
For analysis of protein expression, cells were lysed in E1A lysis buffer (150 mm sodium chloride, 50 mm HEPES (pH 7.5), 5 mm EDTA, and 0.1% Nonidet P-40) freshly supplemented with 20 mm sodium glycerophosphate, 0.5 mm sodium orthovanadate, and Complete protease inhibitor mixture (Roche Applied Science), and proteins were detected by Western blot as described (7).
Cell Cycle Analysis
Apoptotic cells from supernatants were collected by centrifugation and combined with trypsinized adherent cells. Cells were fixed in 70% (v/v) ethanol, and RNA was degraded by 0.25 mg/ml RNase. Finally, cells were stained with 10 μg/ml propidium iodine, and cell cycle distribution and subdiploid DNA content were analyzed by flow cytometry using BD FACSDiva software, Version 6.1.1 (BD Biosciences).
BrdUrd Labeling
To determine S-phase distribution, HUVECs were incubated for 10 min with 1 μm BrdUrd and fixed in 70% ethanol overnight. Cells were then co-stained with fluorescein isothiocyanate-coupled anti-BrdUrd antibody and PI and analyzed by flow cytometry as described (7).
Colony Formation Assays
Control cells and cells expressing various transgenes were cultured until control cells reached confluence and then fixed by methanol (100%, v/v) and subsequently stained by 0.1% (m/v) crystal violet as described (7).
Cytotoxicity Assays
Viable cells expressing vector or the FOXO3 mutants were simultaneously fixed and stained with 10 g/liter crystal violet in 40% (v/v) methanol, washed, and air-dried. Remaining crystal violet was then extracted with methanol (100%, v/v), and dye intensity of the extracted fraction, indicating the relative amount of viable cells, was determined by measuring absorption at 570 nm using a spectrophotometer.
Retroviral Infections
Retroviral infections of HUVEC were done in three consecutive rounds as described (25), using stable ϕNX producer cells for pBabe puro empty vector, pBabe puro HA-FOXO3.A3.ER, pBabe puro HA-FOXO3.A3.ER.H212R, Pinco empty vector, or Pinco-cFLIPL, respectively.
Transfection of HUVECs with Small Interfering RNAs
For small interfering RNA (siRNA) transfection, HUVECs were seeded at a density of 12,000 cells/cm2 and siRNAs were transfected at a final concentration of 100 nm the following day by using Oligofectamine (Invitrogen) according to the manufacturer's protocol. Scrambled control siRNA (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) and siRNAs against NOXA (sense, 5′-CUUCCGGCAGAAACUUCUGTT-3′; antisense, 5′-CAGAAGUUUCUGCCGGAAGTT-3′) were synthesized by MWG Biotech AG and were described previously (30). Validated siRNA against BIM (sense, 5′-GAGACGAGUUUAACGCUUATT-3′; antisense, 5′-UAAGCGUUAAACUCGUCUCCG-3′) was purchased from Qiagen (catalog number SI2655359).
Luciferase Assays
HUVECs were retrovirally infected with pBP empty vector, pBP-HA-FOXO3.A3.ER, or pBP-HA-FOXO3.A3.ER.H212R and, after 72 h, selected for puromycin positivity overnight. Cells were reseeded at a density of 750,000 cells/10-cm dish and transfected with 5 μg of the respective firefly luciferase reporter constructs and 133 ng of ubiquitin-dependent Renilla luciferase as described (25). Cells were stimulated for 16 h with 4OHT, and luciferase activity was measured using the Dual-Glo luciferase assay system (Promega) according to the manufacturer's protocol. Firefly chemiluminescence values were each normalized to the co-transfected Renilla luciferase control reporter-generated chemiluminescence.
RESULTS
FOXO3 Induces Apoptosis by FRE-dependent and -independent Mechanisms
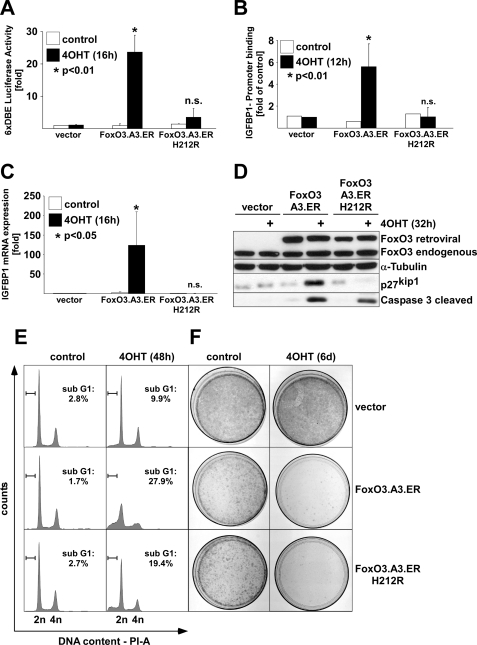
To study the impact of FOXO3 on ECs in vitro, we used a conditionally active FOXO3.A3 mutant, fused to a mutant form of the hormone-binding domain of the murine estrogen receptor, which renders this protein conditionally active by the addition of 4OHT to the culture medium (FOXO3.A3.ER) (26). We generated an FRE-binding mutant of FOXO3 by site-directed mutagenesis of histidine 212 in the third helix of the forkhead box to arginine (FOXO3.A3.ER.H212R). His212 was chosen because structural analysis revealed a direct interaction of this residue with bases of the FRE consensus site (23). Moreover, mutation of the corresponding His215 site in FOXO1 to arginine abolished FRE binding in bandshift assays and prevented recruitment to the promoter of IGFBP1 (insulin growth factor-binding protein-1), an established FOXO target gene containing a FRE sequence in its promoter region (6). Luciferase assays in various cells, including HUVECs, using a FOXO-responsive 6× DAF-16-binding element-driven luciferase reporter (6× DBE-luc) (22), confirmed the inability of FOXO3.13.ER.H212R to transactivate classical FOXO-responsive promoters upon the addition of 4OHT (Fig. 1A) (data not shown). In contrast, 4OHT treatment of HUVEC transfected with the parental FOXO3.A3.ER construct led to a 20–25-fold induction in this assay, suggesting a complete loss of FRE binding capacity of the H212R mutant. This loss of functionality was further supported by ChIP experiments, which confirmed its failure to bind to an established FRE-containing region of the IGFBP1 promoter in vivo (Fig. 1B) (6) (supplemental Fig. 1). Additionally, 4OHT stimulation of FOXO3.A3.ER.H212R-infected HUVEC had no effect on the levels of IGFBP1 mRNA or protein expression of p27KIP1 (Fig. 1, C and D), whereas both genes were strongly induced by conditional activation of FOXO.3.A3.ER. Remarkably, 4OHT treatment consistently triggered apoptosis in FOXO3.A3.ER.H212R-infected HUVECs, albeit less efficiently compared with FOXO3.A3.ER-infected cells. This was evident both by the presence of cleaved effector CASP-3 in Western blots (Fig. 1D) and by subdiploidy analysis of PI-stained cells (Fig. 1E), indicating apoptosis induction at 32 or 48 h after 4OHT treatment, respectively. Although apoptosis induction was less pronounced with the FRE-binding mutant as evident from statistical analysis of multiple experiments (supplemental Fig. 2A), prolonged activation of both mutants showed no major differences in colony formation assays and quantitative cytotoxicity assays (Fig. 1F and supplemental Fig. 2B), indicating that FOXO3 can equally affect EC proliferation and viability in the absence of a functional FRE-binding domain.
FIGURE 1.
Conditional FOXO3 activation induces apoptosis in primary ECs irrespective of direct FRE-binding. HUVECs were retrovirally infected with either empty vector, FOXO3.A3.ER or FOXO3.A3.ER.H212R, selected by puromycin, and reseeded. A, luciferase assay demonstrating the incapability of the H212R mutant to transactivate a forkhead-responsive 6×DBE-luc reporter upon 16-h treatment with 4OHT. Data represent -fold average luciferase activation ± S.D. of a 6× DBE-luc reporter transfected into the differently infected cells. Luciferase values were each normalized to activity of a co-transfected Renilla luciferase reporter and related to unstimulated empty vector-infected cells. B, HA tag-specific chromatin immunoprecipitation after 12 h of 4OHT treatment showing binding of HA-FOXO3.A3.ER but not HA-FOXO3.A3.ER.H212R to an established FOXO-responsive region (6) in the IGFBP1 promoter. Data are represented as -fold enrichment of binding related to 4OHT-treated vector controls. C, qRT-PCR demonstrating expression of IGFBP1, normalized to GAPDH expression after 12 h of 4OHT treatment. Data display average -fold regulation ± S.D. related to unstimulated vector. D, Western blot, showing expression of p27Kip1 and cleavage of caspase-3 32 h after 4OHT treatment. E, cell cycle profiles, displaying the percentage of subdiploid DNA content assessed by flow cytometric analysis of PI-stained cells 48 h after 4OHT treatment. F, colony formation assay after 6 days (6d) of 4OHT treatment. All data are derived from at least three independent experiments. Statistical significance was calculated by Student's t test.
FOXO3 Regulates Gene Expression Both by Classical and Alternative Mechanisms
To elucidate classical (i.e. targets that require FRE binding) and potentially existing alternatively regulated FOXO3 targets in ECs, which mediate the observed effects, we performed microarray analysis using Affymetrix HG-U133 Plus 2.0 chips, which cover 47,000 transcripts and 6,500 additional genes of the human genome. For this, HUVECs were retrovirally infected with either empty vector, FOXO3.A3.ER, or FOXO3.A3.ER.H212R, respectively. Cells were stimulated with 4OHT for 12 h, and total RNA from three independent experiments was individually processed for DNA microarray hybridization. An early time point for analysis was chosen to prevent secondary effects on gene expression. The raw and normalized data are deposited in the Gene Expression Omnibus (GEO) data base (accession number GSE-16573). Correlation heat mapping (supplemental Fig. 3A) and three-dimensional principal component analysis (supplemental Fig. 3B) were used to illustrate differential gene expression patterns between FOXO3.A3.ER and FOXO3.A3.ER.H212R and their controls, respectively. The two analyses concordantly show that 4OHT treatment of both FOXO3 mutants elicits gene expression patterns that are distinct from each other and additionally different from the gene expression of the controls. Only genes with a -fold change of ≥1.5 and a p value below the threshold estimated by the Holm multiple testing algorithm compared with the untreated empty vector were considered as significantly regulated. According to these criteria, overall, we identified 579 genes regulated by FOXO3.A3.ER alone and 348 genes that were co-regulated by the two mutants, demonstrating the potential of FOXO3 to regulate gene expression by alternative mechanisms. No genes were regulated by the addition of 4OHT in vector-infected cells, and only 13 genes were nonspecifically regulated by introduction of the two FOXO3.ER mutants. For statistical reasons, we also found a class of genes regulated by FOXO3.A3.ER.H212R alone, which probably resulted from an increased availability of this mutant for binding partners due to its deficiency to bind classical FRE. This group primarily contained nonspecific gene classes (data not shown) and thus was excluded from further analysis.
FOXO3 Regulates Distinct Endothelial Functions in the Absence of Direct FRE Binding
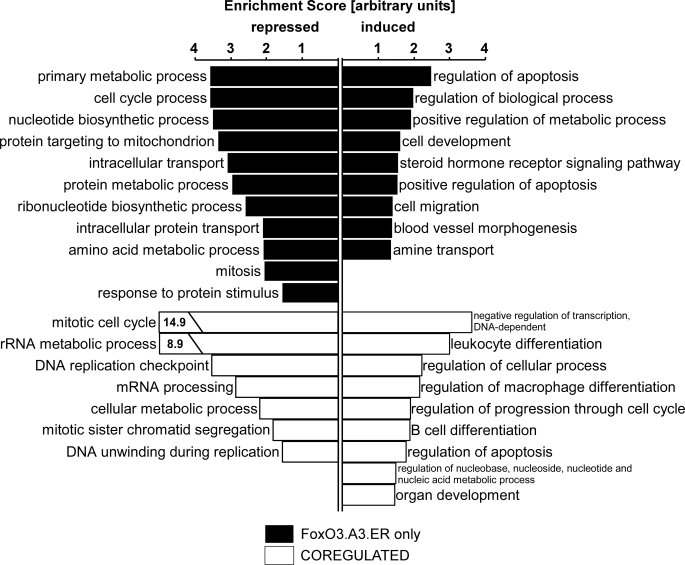
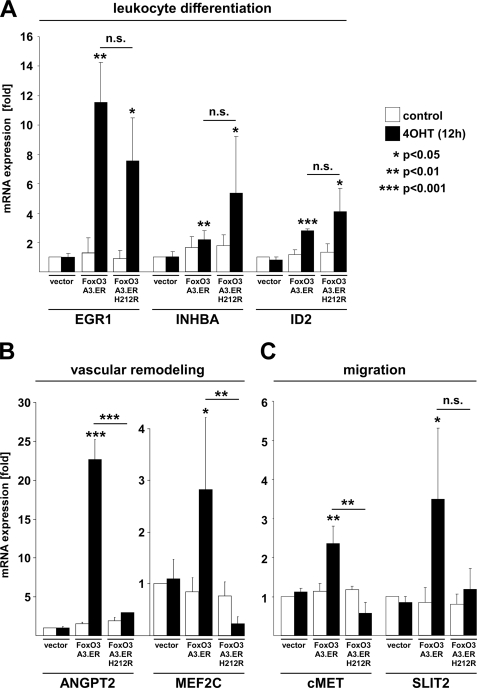
We then performed functional annotation clustering, using a bioinformatics tool provided by DAVID (31) to identify classes of functionally related genes regulated by the two FOXO variants. This tool integrates functionally related gene ontology (GO) groups and ranks these functional clusters accordingly to their statistical significance of overrepresentation by calculating an enrichment score. Interestingly, this analysis revealed the existence of various functional gene clusters that are not regulated classically by binding to FREs (Fig. 2). These include multiple clusters related to cell cycle regulation, which were statistically overrepresented within the group of co-regulated transcripts compared with the total amount of genes present on the chip. In agreement with an established inhibitory effect of FOXOs on proliferation, cell cycle-related clusters were particularly found in the co-repressed group of transcripts. In addition, DAVID analysis revealed the overrepresentation of functional annotation clusters related to the immune system, such as “leukocyte differentiation,” “regulation of macrophage differentiation,” and “B-cell differentiation,” among the co-up-regulated group. This included genes such as ID2 (inhibitor of DNA binding 2), a repressor of helix-loop-helix proteins and established regulator of B-cell differentiation and granulopoiesis (32, 33), the transforming growth factor β family member INHBA (inhibin βA), an established inhibitor of follicle-stimulating hormone secretion and regulator of hemopoiesis (34, 35), as well as the crucial mediator of thymocyte development EGR1 (early growth response-1) (36, 37) (supplemental Table 1). FRE-independent regulation of all of these genes was independently verified by qRT- PCR (Fig. 3A), confirming the reliability of our microarray data. Clustering of the genes up-regulated by 4OHT-induced activation of FOXO3.A3.ER alone (below referred to as the “FOXO3.A3.ER only” group) uncovered a direct role of FOXO3 in the regulation of cell development, cell migration, and blood vessel morphogenesis. Genes involved in these functional clusters comprised various well known modulators of blood vessel morphogenesis, such as the previously described FOXO1 target ANGPT2 (17) and MEF2C (myocyte enhancer factor 2C) as well as migration-associated genes, such as cMET and SLIT2 (supplemental Table 2). FRE-dependent expression of all of these genes again was independently confirmed by qRT-PCR (Fig. 3, B and C).
FIGURE 2.
Functional annotation clustering reveals the presence of classically and alternatively regulated functional gene clusters. HUVECs were retrovirally infected with either empty vector, FOXO3.A3.ER, or FOXO3.A3.ER.H212R and reseeded after puromycin selection. Total RNA from three independent experiments was isolated after 12 h of 4OHT treatment, and global gene expression was determined by oligonucleotide microarray analysis, as described under “Experimental Procedures.” Gene profiles were analyzed using DAVID as described under “Experimental Procedures.” The enrichment score on the x axis ranks the overrepresentation of a cluster composed of functionally similar GO groups in relation to the total number of genes on the chip. Functional annotation clusters were assigned by the subset GO group designation with the highest significant p value.
FIGURE 3.
FOXO3 induces genes involved in migration, vascular remodeling, and leukocyte differentiation. HUVECs were retrovirally infected with the indicated constructs and puromycin-selected prior to reseeding. A–C, qRT-PCR showing expression of genes involved in migration, vascular remodeling, and leukocyte differentiation by classical and alternative mechanisms after 12 h of 4OHT treatment. Gene expression was normalized to GAPDH expression, and data are presented as average -fold regulation ± S.D. as compared with unstimulated vector controls and are derived from at least three independent experiments. Statistical significances were calculated by Student's t test. n.s., not significant.
FOXO3-induced Cell Cycle Arrest Occurs Independently of FRE Binding
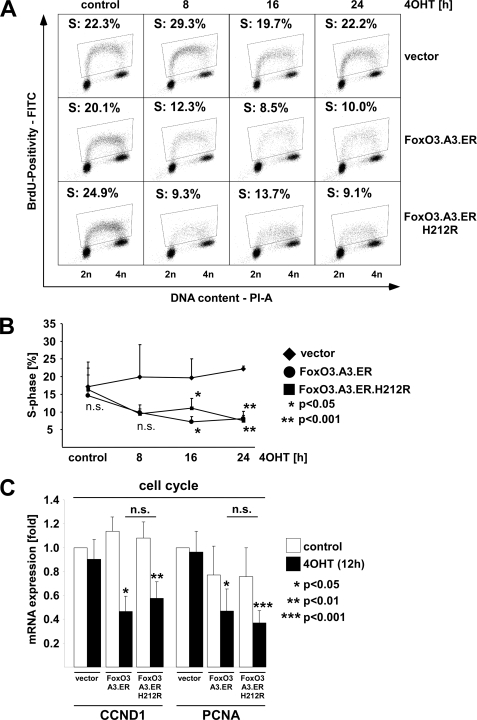
An immediate prediction emerging from the DAVID functional cluster analysis was that FOXO3 should induce cell cycle arrest independently of FRE binding. Such a finding would corroborate earlier findings in a cancer cell line, in which a FRE-binding mutant of FOXO1 induced a G1-S cell cycle arrest by repression of CCND1 and CCND2 (6). Remarkably, the majority of the genes in the cell cycle-associated functional clusters of the co-repressed group had predicted or demonstrated functions in replication/replication initiation (e.g. PCNA, CDC6, and MCM2, -4, and -7) or G2-M progression (CCNB2, AURKA (Aurora kinase A), BUB1B/BUBR1, CDC20, and ZWINT; see Table 3) rather than in G1-S transition, suggesting that in ECs, FOXO3 might affect cell cycle progression at different and/or multiple stages. To investigate at which stage FOXO3 activation predominantly inhibits cell cycle progression in ECs, we stably expressed FOXO3.A3.ER or FOXO3.A3.ER.H212R in HUVECs. Subsequently, we performed DNA profiling and BrdUrd incorporation experiments at different time points after 4OHT treatment to assess their effect on cell cycle progression. Supplemental Fig. 4 and Fig. 4A illustrate that activation of both mutants resulted in accumulation of cells in G1 and decreased BrdUrd incorporation as early as 8 h after 4OHT treatment. Fig. 4B shows a statistical summary of three independent BrdUrd incorporation experiments, which confirms a visible but non-significant decrease of BrdUrd incorporation by both activated mutants after 8 h of 4OHT treatment, which becomes statistically significant at 16–24 h. We next examined the list of genes regulated by activation of the two FOXO variants for potential mediators of the observed G1-S cell cycle arrest. Although we identified the established cyclin-dependent kinase inhibitor p27KIP1 as a FRE-dependent FOXO3 target gene, which could account for the observed G1 cell cycle arrest upon 4OHT treatment of FOXO3.A3.ER-infected cells, we failed to detect the major D-type cyclins, CCND1 and CCND2, in the list of genes in the microarray and merely found CCND3 as a jointly down-regulated transcript (Table 3). However, closer examination of the microarray data revealed repression of CCND1 just below the threshold of 1.5-fold in the co-regulated group (data not shown) as well as a repression of the DNA replication cofactor PCNA. To evaluate CCND1 and PCNA repression as potential mechanisms for the observed G1-S cell cycle arrest with the FRE-binding mutant, we retrovirally expressed both FOXO3.ER variants in HUVECs and analyzed CCND1 and PCNA expression by quantitative RT-PCR 12 h after 4OHT treatment. Concordantly, we observed repression of CCND1 and PCNA mRNA by FOXO3.A3.ER and FOXO3.A3.ER.H212R (Fig. 4C). Thus, CCND1 and PCNA are regulated independently of FRE binding and represent valid candidates to mediate FOXO3-induced cell cycle arrest in the absence of a functional FRE-binding domain.
TABLE 3.
Genes involved in proliferation and cell cycle regulation that are repressed by FOXO3.A3.ER and FOXO3.A3.ER.H212R as identified by functional annotation clustering
| Entrez Gene ID | Gene symbol | Full name | -Fold change |
Othersa | |||
|---|---|---|---|---|---|---|---|
| HUVEC FoxO3 (this work) |
786-O renal carcinoma FoxO1 (6) |
||||||
| A3.ER | A3.ER.H212R | A3 | A3.H215R | ||||
| -fold | -fold | ||||||
| Mitotic cell cycle | |||||||
| 6790 | AURKA | Aurora kinase A | 0.52 | 0.55 | Ref. 44 | ||
| 332 | BIRC5 | Baculoviral IAP repeat-containing 5 | 0.61 | 0.62 | |||
| 701 | BUB1B (BUBR1) | BUB1 budding Uninhibited by benzimidazol | 0.58 | 0.62 | |||
| 220134 | C18orf24 | Chromosome 18 open reading frame 24 | 0.66 | 0.62 | |||
| 8900 | CCNA1 | Cyclin A1 | 0.48 | 0.45 | Refs. 17 and 18 | ||
| 9133 | CCNB2 | Cyclin B2 | 0.56 | 0.51 | |||
| 991 | CDC20 | Cell division cycle 20 homolog (Saccharomyces cerevisiae) | 0.32 | 0.33 | Refs. 17 and 18 | ||
| 990 | CDC6 | Cell division cycle 6 homolog (S. cerevisiae) | 0.51 | 0.50 | |||
| 55143 | CDCA8 | Cell division cycle-associated 8 | 0.56 | 0.62 | |||
| 1019 | CDK4 | Cyclin-dependent kinase 4 | 0.43 | 0.43 | 0.49 | 0.26 | |
| 1033 | CDKN3 | Cyclin-dependent kinase inhibitor 3 | 0.55 | 0.56 | |||
| 81620 | CDT1 | Chromatin licensing and DNA replication | 0.66 | 0.63 | |||
| 55165 | CEP55 | Centrosomal protein of 55 kDa | 0.51 | 0.57 | |||
| 9787 | DLG7 | Discs, large homolog 7 (Drosophila) | 0.50 | 0.47 | |||
| 9156 | EXO1 | Exonuclease 1 | 0.63 | 0.60 | |||
| 348235 | FAM33A | Family with sequence similarity 33, member A | 0.57 | 0.53 | |||
| 26271 | FBXO5 | F-box protein 5 | 0.57 | 0.55 | |||
| 56992 | KIF15 | Kinesin family member 15 | 0.61 | 0.58 | |||
| 11004 | KIF2C | Kinesin family member 2C | 0.63 | 0.59 | |||
| 9918 | NCAPD2 | Non-SMC condensin I complex, subunit D2 | 0.64 | 0.58 | |||
| 54892 | NCAPG2 | Non-SMC condensin II complex, subunit G2 | 0.70 | 0.66 | |||
| 9221 | NOLC1 | Nucleolar and coiled body phosphoprotein | 0.61 | 0.64 | |||
| 55872 | PBK | PDZ binding kinase | 0.46 | 0.42 | |||
| 23082 | PPRC1 | Peroxisome proliferator-activated receptor | 0.66 | 0.80 | |||
| 9232 | PTTG1 | Pituitary tumor-transforming 1 | 0.57 | 0.53 | |||
| 8607 | RUVBL1 | RuvB-like 1 (Escherichia coli) | 0.64 | 0.67 | |||
| 29901 | SAC3D1 | SAC3 domain-containing 1 | 0.58 | 0.58 | |||
| 6502 | SKP2 | S-phase kinase-associated protein 2 (p45) | 0.66 | 0.62 | |||
| 10615 | SPAG5 | Sperm-associated antigen 5 | 0.57 | 0.63 | |||
| 7027 | TFDP1 | Transcription factor Dp-1 | 0.66 | 0.64 | |||
| 54962 | TIPIN | TIMELESS-interacting protein | 0.53 | 0.50 | |||
| 22974 | TPX2 | TPX2, microtubule-associated | 0.49 | 0.51 | |||
| 11065 | UBE2C | Ubiquitin-conjugating enzyme E2C | 0.51 | 0.50 | Refs. 17 and 46 | ||
| 11130 | ZWINT | ZW10 interactor | 0.58 | 0.54 | Ref. 46 | ||
| DNA replication checkpoint | |||||||
| 8318 | CDC45L | CDC45 cell division cycle 45-like (S. cerevisiae) | 0.63 | 0.62 | 0.5 | 0.48 | |
| 990 | CDC6 | Cell division cycle 6 homolog (S. cerevisiae) | 0.51 | 0.50 | |||
| 81620 | CDT1 | Chromatin licensing and DNA replication | 0.66 | 0.63 | |||
| 51053 | GMNN | Geminin, DNA replication inhibitor | 0.49 | 0.49 | |||
| 5111 | PCNA | Proliferating cell nuclear antigen | 0.50 | 0.46 | 0.24 | 0.22 | |
| 54962 | TIPIN | TIMELESS-interacting protein | 0.53 | 0.50 | |||
| Mitotic sister chromatid segregation | |||||||
| 9787 | DLG7 | Discs, large homolog 7 (Drosophila) | 0.50 | 0.47 | |||
| 9918 | NCAPD2 | Non-SMC condensin I complex, subunit D2 | 0.64 | 0.58 | |||
| 54892 | NCAPG2 | Non-SMC condensin II complex, subunit G2 | 0.70 | 0.66 | |||
| 9232 | PTTG1 | Pituitary tumor-transforming 1 | 0.57 | 0.53 | |||
| 11130 | ZWINT | ZW10 interactor | 0.58 | 0.54 | |||
| DNA unwinding during replication | |||||||
| 4171 | MCM2 | Minichromosome maintenance complex component 2 | 0.49 | 0.58 | Ref. 18 | ||
| 4173 | MCM4 | Minichromosome maintenance complex component 4 | 0.54 | 0.57 | |||
| 4176 | MCM7 | Minichromosome maintenance complex component 7 | 0.53 | 0.51 | |||
| Non-clustered cell cycle regulators | |||||||
| 896 | CCND3 | Cyclin D3 | 0.65 | 0.66 | |||
FIGURE 4.
Conditional activation of FOXO3. H212R.ER induces a G1 cell cycle arrest in primary human ECs. HUVECs were retrovirally infected with either empty vector, FOXO3.A3.ER or FOXO3.A3.ER.H212R, selected by puromycin, and reseeded. A, flow cytometric quantification of BrdUrd positivity after stimulation with 4OHT for the indicated times. The percentage of BrdUrd-positive cells (i.e. cells undergoing replication) is indicated. B, statistical analysis of BrdUrd-positive cells after activation of FOXO3. Data represent mean values of BrdUrd positivity ± S.D. and are derived from three independent experiments. C, qRT-PCRs, showing repression of endothelial CCND1 and PCNA mRNA 12 h after 4OHT treatment. Gene expression was normalized to GAPDH expression and is presented as average -fold expression ± S.D. of three independent experiments in relation to unstimulated vector controls. Statistical significances were determined using Student's t test. n.s., not significant.
FOXO-induced Regulation of Endothelial Apoptosis
A special case among the overrepresented functional groups was the clusters associated with apoptosis. Although “regulation of apoptosis” was the top cluster with the highest enrichment score among the “FOXO3.A3.ER only” group, it was also statistically significantly represented in the co-up-regulated genes. However, in the “FOXO3.A3.ER only” group, two clusters of apoptosis-related genes with high enrichment scores were found as compared with only one apoptosis cluster with a lower enrichment score present in the co-up-regulated group. Furthermore, the GO groups constituting the apoptosis-related functional cluster of the co-up-regulated transcripts in general showed less significant (higher) p value scores (Table 1).
TABLE 1.
Apoptosis-related gene ontology groups induced by FoxO3.A3.ER and FoxO3.A3.ER.H212R
| Gene ontology | FoxO3.A3.ER |
FoxO3.A3.ER.H212R |
||||
|---|---|---|---|---|---|---|
| Position of functional annotation cluster | Gene count | p value | Position of functional annotation cluster | Gene count | p value | |
| GO:0042981, regulation of apoptosis | 1 | 24 | 7.70E−04 | 7 | 10 | 8.6E−03 |
| GO:0043067, regulation of programmed cell death | 1 | 24 | 8.90E−04 | 7 | 10 | 9.3E−03 |
| GO:0006915, apoptosis | 1 | 29 | 3.50E−03 | 7 | 12 | 1.4E−02 |
| GO:0012501, programmed cell death | 1 | 29 | 4.00E−03 | 7 | 12 | 1.5E−02 |
| GO:0016265, death | 1 | 30 | 4.34E−03 | 7 | 12 | 2.1E−02 |
| GO:0008219, cell death | 1 | 30 | 4.34E−03 | 7 | 12 | 2.1E−02 |
| GO:0048468, cell development | 1 | 38 | 1.43E−02 | 7 | 15 | 3.1E−02 |
| GO:0043065, positive regulation of apoptosis | 6 | 12 | 1.95E−02 | |||
| GO:0043068, positive regulation of programmed cell death | 6 | 12 | 2.05E−02 | |||
| GO:0006917, induction of apoptosis | 6 | 10 | 3.77E−02 | |||
| GO:0012502, induction of programmed cell death | 6 | 10 | 3.85E−02 | |||
The Extrinsic Death Receptor Pathway Is Not Involved in FOXO3-mediated Apoptosis
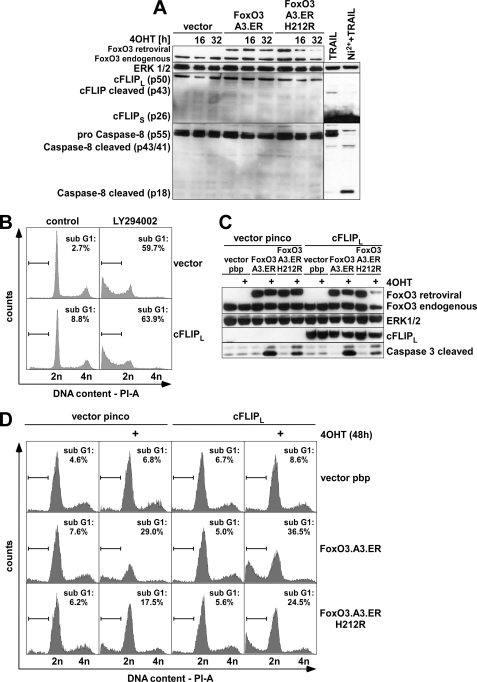
To better understand how exactly FOXO3 mediates apoptosis in ECs, we summarized all apoptosis-related genes of the overrepresented functional clusters in Table 2. Interestingly, both genes from the intrinsic mitochondrial pathway, such as NOXA and BIM, and genes of the extrinsic death receptor pathway, such as TRAIL, were found among the FOXO3-regulated genes. To determine the relative contribution of genes from the extrinsic death receptor pathway and the intrinsic mitochondrial pathway to FOXO3-mediated apoptosis, we investigated cleavage of initiator CASP-8, which is commonly activated during death receptor-mediated apoptosis. Time course experiments revealed no obvious CASP-8 cleavage even at late time points (Fig. 5A), at which apoptosis induction had already commenced as judged by the presence of cleaved CASP-3 p17/19 (compare Fig. 1A). In contrast to a recent report (38), we also did not find any indication of altered transcript (supplemental Fig. 5A) or protein levels of the CASP-8 antagonist cFLIPL (Fig. 5A); nor did we detect cleavage to cFLIP-p43, which indicates successful receptor ligation (39) and is readily detectable upon treatment of primary ECs with sublethal doses of recombinant TRAIL that only triggers partial CASP-8 cleavage to p41/p43 (Fig. 5A) and no major CASP-3 cleavage and subdiploidy induction (supplemental Fig. 6, A and B). By contrast, co-treatment of TRAIL with nickel chloride (Ni2+), an established repressor of cFLIP and sensitizer for TRAIL-induced apoptosis in ECs (39), allowed full CASP-8 cleavage to its active p18 fragment (Fig. 5A). This correlated with CASP-3 cleavage and subdiploidy induction (supplemental Fig. 6, A and B), confirming full functionality of the TRAIL death receptor pathway in ECs and its responsiveness to cFLIP modulation. To further validate that CASP-8 was not involved in FOXO-induced apoptosis, we additionally employed a CASP-8-specific inhibitor, Z-IETD-fluoromethyl ketone. Supplemental Fig. 6C illustrates that Z-IETD-fluoromethyl ketone treatment did not reduce FOXO3-induced subdiploidy at doses that significantly decreased CASP-8 and CASP-3 cleavage as well as subdiploidy induction by Ni2+ and TRAIL in parallel experiments (supplemental Fig. 6, A and B). We ultimately excluded a causal involvement of the extrinsic death receptor pathway by retroviral expression of cFLIPL, which can antagonize CASP-8-induced apoptosis in various cells (40, 41), including HUVECs (39). Neither the proapoptotic effects of endogenous FOXO activation by prolonged treatment with the phosphatidylinositol 3-kinase inhibitor LY294002 (Fig. 5B; statistics provided in supplemental Fig. 5B) nor apoptosis induction by 4OHT treatment of cells expressing either one of the two FOXO3.ER mutants could be reversed by cFLIPL overexpression (Fig. 5, C and D, and supplemental Fig. 5C), indicating that in ECs, FOXO3-induced apoptosis is triggered by activation of the intrinsic pathway.
TABLE 2.
Genes involved in apoptosis that are induced by FOXO3.A3.ER and FOXO3.A3.ER.H212R as identified by functional annotation clustering
| Entrez Gene ID | Gene symbol | Full name | -Fold change |
Othersa | |||
|---|---|---|---|---|---|---|---|
| HUVEC FoxO3 (this work) |
786-O renal carcinoma FoxO1 (6) |
||||||
| A3.ER | A3.ER.H212R | A3 | A3.H215R | ||||
| -fold | -fold | ||||||
| 90 | ACVR1 | Activin A receptor, type I | 2.10 | 1.91 | |||
| 196 | AHR | Aryl hydrocarbon receptor | 3.20 | ||||
| 11199 | ANXA1 | Annexin A1 | 1.84 | ||||
| 10018 | BCL2L11/BIM | BCL2-like 11 (apoptosis facilitator) | 1.61 | 2.19 | |||
| 23786 | BCL2L13 | BCL2-like 13 (apoptosis facilitator) | 1.63 | ||||
| 604 | BCL6 | B-cell CLL/lymphoma 6 | 2.00 | 2.55 | 3.10 | 3.40 | Refs. 17, 18, and 46 |
| 650 | BMP2 | Bone morphogenetic protein 2 | 2.76 | 3.23 | |||
| 663 | BNIP2 | BCL2/adenovirus E1B 19-kDa interacting protein 2 | 1.59 | ||||
| 664 | BNIP3 | BCL2/adenovirus E1B 19-kDa interacting protein 3 | 1.50 | ||||
| 669 | BPGM | 2,3-Bisphosphoglycerate mutase | 1.70 | 1.85 | |||
| 694 | BTG1 | B-cell translocation gene 1, anti-proliferative | 4.48 | 4.20 | 3.30 | ||
| 114926 | C8orf4 | Chromosome 8 open reading frame 4 | 2.01 | 1.94 | |||
| 120329 | CASP12 | Caspase-12 (gene/pseudogene) | 2.07 | 1.71 | |||
| 837 | CASP4 | Caspase-4, apoptosis-related cysteine peptidase | 2.01 | ||||
| 1027 | CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 2.33 | ||||
| 114769 | COP1 | Caspase recruitment domain family, member 16 | 3.07 | ||||
| 1459 | CSNK2A2 | Casein kinase 2, α prime polypeptide | 1.57 | 2.72 | |||
| 1508 | CTSB | Cathepsin B | 1.76 | Ref. 46 | |||
| 7852 | CXCR4 | Chemokine (CXC motif) receptor 4 | 2.23 | Refs. 17 and 18 | |||
| 10059 | DNM1L | Dynamin 1-like | 1.54 | ||||
| 2296 | FOXC1 | Forkhead box C1 | 1.51 | ||||
| 1647 | GADD45A | Growth arrest and DNA damage-inducible, α | 2.05 | 2.37 | Ref. 46 | ||
| 2729 | GCLC | Glutamate-cysteine ligase, catalytic subunit | 1.57 | 2.19 | 5.60 | 16.90 | |
| 3267 | HRB | HIV-1 Rev-binding protein | 2.22 | ||||
| 10553 | HTATIP2 | HIV-1 Tat-interactive protein of 2.30 kDa | 1.75 | 1.54 | |||
| 64135 | IFIH1 | Interferon induced with helicase C domain 1 | 1.71 | ||||
| 3480 | IGF1R | Insulin-like growth factor 1 receptor | 2.18 | Ref. 46 | |||
| 51447 | IHPK2 | Inositol hexaphosphate kinase 2 | 1.81 | ||||
| 3592 | IL12A | Interleukin 12A | 2.49 | ||||
| 3624 | INHBA | Inhibin, βA | 1.62 | 3.13 | |||
| 4154 | MBNL1 | Muscleblind-like (Drosophila) | 1.71 | 3.90 | 2.20 | ||
| 4233 | MET | MET met proto-oncogene (hepatocyte growth factor receptor) | 1.61 | ||||
| 64112 | MOAP1 | Modulator of apoptosis 1 | 1.64 | ||||
| 4646 | MYO6 | Myosin VI | 2.78 | 4.90 | 0.80 | ||
| 59277 | NTN4 | Netrin 4 | 2.40 | ||||
| 26471 | NUPR1 | Nuclear protein 1 | 3.86 | ||||
| 10133 | OPTN | Optineurin | 1.57 | ||||
| 734 | OSGIN2 | Oxidative stress-induced growth inhibitor family member 2 | 2.70 | 3.60 | 0.70 | ||
| 5074 | PAWR | PRKC, apoptosis, WT1, regulator | 2.25 | 2.16 | |||
| 10015 | PDCD6IP | Programmed cell death 6-interacting protein | 1.52 | ||||
| 5290 | PIK3CA | Phosphoinositide-3-kinase, catalytic, α-polypeptide | 3.27 | Ref. 46 | |||
| 5366 | PMAIP1/NOXA | Phorbol 12-myristate 13-acetate-induced protein 1 | 3.43 | ||||
| 5921 | RASA1 | RAS p21 protein activator (GTPase-activating protein) 1 | 2.13 | 1.75 | |||
| 10313 | RTN3 | Reticulon 3 | 3.52 | 5.60 | 2.00 | ||
| 57142 | RTN4 | Reticulon 4 | 1.70 | ||||
| 51100 | SH3GLB1 | Src homology 3 domain GRB2-like endophilin B1 | 1.57 | ||||
| 51804 | SIX4 | SIX homeobox 4 | 1.77 | ||||
| 9353 | SLIT2 | Slit homolog 2 (Drosophila) | 4.57 | 1.91 | Refs. 17 and 18 | ||
| 6648 | SOD2 | Superoxide dismutase 2, mitochondrial | 4.19 | 2.93 | 7.90 | 3.50 | |
| 6789 | STK4 | Serine/threonine kinase 4 | 1.64 | Ref. 17 | |||
| 8743 | TNFSF10/TRAIL | Tumor necrosis factor (ligand) superfamily, member 10 | 3.70 | Refs. 17 and 18 | |||
| 94241 | TP53INP1 | Tumor protein p53-inducible nuclear protein 1 | 2.71 | 1.80 | 2.00 | ||
FIGURE 5.
The extrinsic death receptor pathway is not involved in FOXO3-induced apoptosis. HUVECs were retrovirally infected with the indicated constructs, puromycin-selected, and reseeded for the respective experiments. A, Western blot, showing cFLIP and caspase-8 levels after exposure of cells to medium or 4OHT for 16 or 32 h. Additionally, stimulations with TRAIL (100 ng/ml, 3 h) or TRAIL + NiCl2 (Ni) (1.5 mm) are shown as positive control for cFLIP or caspase-8 cleavage, respectively. B, cell cycle profiles of HUVECs, infected as indicated and treated with 10 μm LY392002 for 48 h. The percentage of subdiploid cells was determined by flow cytometric analysis of PI-stained cells. C, Western blot showing expression of FOXO3, cFLIPL, and cleaved caspase-3 after 32 h of 4OHT treatment. D, cell cycle profiles after 48 h of 4OHT treatment. The percentage of PI-stained cells with a subdiploid DNA content was assessed by flow cytometry. All Western blots and cell cycle profiles are representative of three independent experiments.
BIM Is Regulated by Classical and Alternative Mechanisms and Critically Contributes to FOXO3-induced Apoptosis
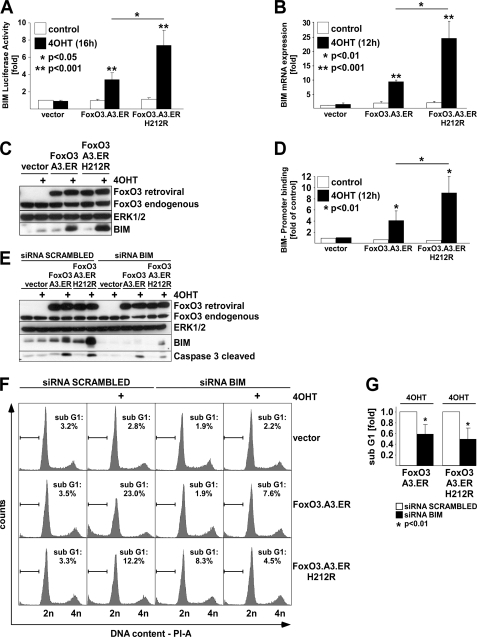
Analysis of the apoptosis-related genes regulated by FOXO3 (Table 2) for modulators of the intrinsic apoptosis pathway suggested the established proapoptotic FOXO target BIM as a potential mediator of FRE-independent apoptosis induction because it was found in the class of co-up-regulated genes (Table 2). To consolidate this finding, we first tested the effect of conditional activation of both FOXO3.A3.ER and FOXO3.A3.ER.H212R on BIM promoter activity using a BIM promoter-driven luciferase reporter (BIM-Luc) (12). Both 4OHT-mediated activation of FOXO3 and its corresponding FRE-binding mutant were sufficient to transactivate the BIM promoter construct (Fig. 6A). Likewise, qRT-PCR for BIM mRNA (Fig. 6B) and Western analysis of BIM protein (Fig. 6C) confirmed regulation of BIM by the FRE-binding mutant. Despite the failure of FOXO3.A3.ER.H212R to bind and transactivate classical FRE-containing promoters (Fig. 1, compare A and B), ChIP analysis revealed 4OHT-induced recruitment of both proteins to a conserved region of the BIM promoter (Fig. 6D) encompassing its previously reported FRE-related sequence (12) (supplemental Fig. 7). Similar results were obtained using an anti-ER antibody, excluding an antibody artifact (data not shown). Thus, recruitment of FOXO3 to this region may obviously also occur via alternative mechanisms. Notably, both BIM expression and promoter recruitment appeared to be stronger upon activation of FOXO3.A3.ER.H212R than FOXO3.A3.ER. To evaluate the exact contribution of BIM to FOXO3-mediated apoptosis, we performed knockdown experiments using siRNA against BIM. Introduction of BIM siRNA into HUVECs stably expressing either FOXO3.A3.ER or FOXO3.A3.ER.H212R strongly reduced FOXO3-induced BIM protein levels and resulted in a reduced cleavage of CASP-3 in Western blot (Fig. 6E). Furthermore, it significantly decreased the number of apoptotic cells in DNA-profiling experiments (Fig. 6F). Summarizing three independent cell cycle profiling experiments (Fig. 6G), BIM siRNA suppressed apoptosis induced by both mutants by ∼50% as compared with cells transfected with control siRNA. Thus, BIM clearly contributes to FOXO3-induced apoptosis and is regulated by classical and alternative mechanisms.
FIGURE 6.
The Bcl-2 family member BIM contributes to FOXO3-induced apoptosis. HUVECs were retrovirally transduced with either empty vector, FOXO3.A3.ER, or FOXO3.A3.ER.H212R and puromycin-selected prior to reseeding. A, luciferase assay, showing BIM promoter activity normalized to Renilla activity after co-transfection of BIM- and Renilla-luciferase reporter gene constructs and 4OHT treatment for 16 h. B, qRT-PCR demonstrating BIM expression, normalized to GAPDH expression after 12 h of 4OHT treatment. C, Western blot, indicating expression of BIM 32h after 4OHT treatment. D, HA tag-specific chromatin immunoprecipitation after 12 h of 4OHT treatment showing binding of HA-FOXO3.A3.ER and HA-FOXO3.A3.ER.H212R to a conserved region in the BIM promoter containing a reported FRE-related sequence (12). E, Western blot, showing knockdown levels of BIM protein and cleavage of caspase-3 protein in response to transfection with either siRNA against BIM or scrambled control siRNA and 4OHT treatment for 24 h. F, cell cycle profiles, displaying the percentage of cells with subdiploid DNA content after transfection of siRNA against BIM or scrambled siRNA followed by 48-h treatment with 4OHT. G, statistical analysis of apoptosis suppression upon knockdown of BIM. Relative levels of 4OHT-induced apoptosis were calculated in relation to subdiploidy values of the individual scrambled siRNA-transfected cells expressing the indicated FOXO mutants (arbitrarily set to 1). Statistical data in A and B are presented as mean -fold regulation ± S.D. in relation to unstimulated vector. In D, OHT-treated vector-infected cells served as reference. All data are derived from at least three independent experiments. Significances were calculated by Student's t test.
NOXA Partially Contributes to FOXO3- induced Apoptosis
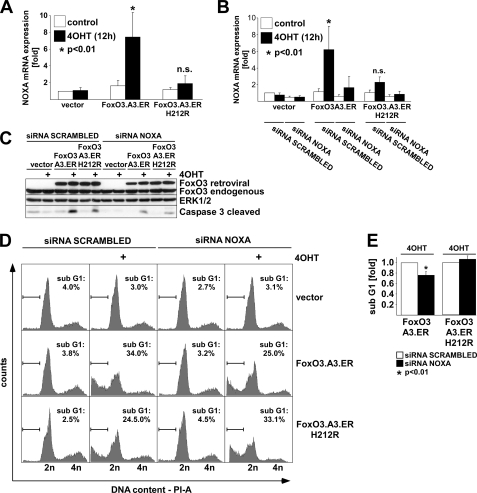
Considering that FOXO3.A3.ER.H212R induced BIM expression to higher levels than FOXO3.A3.ER, whereas apoptosis induction was less pronounced, we concluded that BIM induction alone is unlikely to be the only mediator of FOXO3-induced apoptosis in ECs. Hence, we postulated the existence of at least one other target, which is regulated by direct FRE binding. Among the potential candidates listed in Table 2, another proapoptotic Bcl-2-like family member, NOXA, fulfilled both criteria of being (a) an inducer of the intrinsic apoptotic pathway and (b) a FRE-dependently (i.e. a FOXO3.A3.ER only) regulated gene. qRT-PCR of samples from FOXO3-infected HUVECs validated NOXA as a classically regulated gene (Fig. 7A). To confirm the role of NOXA in FOXO3-induced apoptosis, we overexpressed the two FOXO3 mutants in HUVECs, followed by transfection of NOXA siRNA and treatment with 4OHT. NOXA siRNA introduction resulted in efficient knockdown of FOXO3-induced NOXA mRNA levels in qRT-PCR analysis (Fig. 7B) and decreased cleavage of CASP-3 (Fig. 7C) and subdiploid DNA content (Fig. 7D). On average, NOXA siRNA reduced FOXO3.A3.ER-induced subdiploidy by about 25% in three independent experiments (Fig. 7E), whereas FOXO3.A3.ER.H212R-dependent subdiploidy was not significantly altered (p > 0.05). Hence, NOXA critically contributes to FOXO3-induced apoptosis via classical but not alternative mechanisms.
FIGURE 7.
The Bcl-2 family member NOXA contributes to FOXO3-induced apoptosis. HUVECs were retrovirally infected with the indicated constructs and reseeded after puromycin selection. A, qRT-PCR showing NOXA expression, normalized to GAPDH expression after 12 h of 4OHT treatment. B, qRT-PCR demonstrating knockdown of NOXA mRNA after transfection of either siRNA against NOXA or scrambled siRNA and 4OHT treatment for 12 h. Data in A and B are presented as average -fold regulation ± S.D. related to unstimulated vector controls and are derived from at least three independent experiments. C, Western blot, displaying cleavage of caspase-3 upon transfection with either siRNA against NOXA or scrambled siRNA, followed by 4OHT treatment for 24 h. D, cell cycle profiles, showing the amount of PI-stained cells with subdiploid DNA content as determined by flow cytometry of the differently infected and siRNA-transfected cells 48 h after treatment with 4OHT. E, statistical analysis of apoptosis suppression by NOXA siRNA. Relative apoptosis was calculated as described in the legend to Fig. 6G. Statistical significances were calculated using Student's t test.
DISCUSSION
Our knowledge of FOXO-dependent processes has tremendously increased with the availability of knock-out mice for the individual FOXO factors and the discovery of FRE-independent gene regulation in tumor cells. However, studies that investigate the relevance of this mechanism for FOXO-mediated gene expression and cell type-specific functions in primary human cells are still not available.
Here we provide the first study analyzing FRE-dependent and -independent effects of FOXO3 in relevant primary human cells (i.e. ECs). Our bioinformatical analysis of the functional gene clusters regulated by FOXO3.A3.ER and its corresponding FRE-binding mutant revealed several overrepresented clusters regulated by classical and alternative mechanisms. The list of classically regulated functional groups primarily contained clusters related to metabolism, cell migration, and blood vessel morphogenesis. Intriguingly, the presence of the cluster “blood vessel morphogenesis” implicates a direct effect of FOXO3 on vascular remodeling. This cluster contains genes such as ANGPT2, which antagonizes the vessel-stabilizing effects of ANGPT1 (42), and MEF2C, an important factor required for early cardiac development and vascular remodeling (43, 44), which both were confirmed as FRE-dependent targets by qRT-PCR. Although a recent study reported exclusive dependence of ANGPT2 expression on FOXO1 but not FOXO3 in ECs (18), implicating non-redundant regulation of ANGPT2, our experiments clearly demonstrate FRE-dependent regulation of ANGPT2 by activated FOXO3. These findings are corroborated by data of Daly et al. (17), which show direct regulation of ANGPT2 by activated FOXO1 but not by a FRE-binding mutant in human ECs, supporting a functional redundancy of FOXOs in vascular remodeling.
Remarkably, our study revealed that the two most extensively studied responses to FOXO activation, induction of cell cycle arrest and apoptosis, do not require FRE-dependent gene expression. Although the former confirms the observation of FRE-independent cell cycle regulation by expression of a corresponding FOXO1 mutant in tumor cells (6), this early study did not report a proapoptotic effect of the FOXO1-FRE mutant. This suggests that the FRE-independent antiproliferative effect appears to be mediated by a conserved mechanism, whereas FRE-independent apoptosis induction is cell type-specific. Both the study by Ramaswamy et al. (6) and our own observations previously implicated down-regulation of D-type cyclins as an important mechanism of FRE- independent cell cycle arrest by FOXOs in other cells. The data presented here suggest that a similar mechanism also contributes to FRE-independent cell cycle arrest in primary ECs. This is particularly evident from our qRT-PCR experiments, which confirmed FRE-independent repression of CCND1. Remarkably, we also found multiple genes related to replication or G2-M transition in the group of co-repressed genes, which is also reflected by the distinct overrepresentation of replication- and G2-M-related gene clusters identified by the DAVID analysis. It is tempting to speculate that FOXO3 thus might inhibit cell cycle progression at multiple levels. Indeed, our finding that FOXO3 can repress expression of the essential S-phase regulator PCNA supports this notion. However, it is unlikely that repression of S-phase regulators and G2-M genes is causal for the observed FRE-independent cell cycle arrest in ECs because our time course experiments demonstrate that the majority of cells arrest in G1-S. Nevertheless, FOXO3 has previously been implicated with regulation of G2-M progression in cancer cells (45, 46). Thus, we formally cannot exclude inhibition of G2-M progression as an additional mechanism of FOXO3-induced cell cycle arrest. Future experiments are mandatory to clarify this issue.
Regarding the mechanism of FRE-independent apoptosis induction, our data strongly suggest BIM as the crucial tissue-specific proapoptotic factor in ECs. This was surprising because the BIM promoter contains conserved albeit not perfectly matching FREs (compare supplemental Fig. 7) that can directly bind FOXO3 in bandshift assays (12). Furthermore, these were reported to participate in FOXO3-induced transactivation of a BIM promoter-driven luciferase reporter in neuronal cells (12). However, mutation of these FRE sites did not completely abolish FOXO3-mediated promoter activation, indicating that effective BIM expression may depend on the presence of additional tissue-specific factors. Consistently, BIM was not found as a regulated gene in the aforementioned FOXO1 study (6); nor was it identified as a FOXO3 target in a recent microarray study performed in DLD1 colon carcinoma cells, which undergo cell cycle arrest but not apoptosis upon FOXO3 activation (46). In contrast, BIM represents an established FOXO3 target in various cells that show a proapoptotic response to FOXO3 (47), supporting the view that tissue-specific factors cooperate with FOXO3 to induce BIM. Remarkably, our ChIP experiments revealed that the FOXO3.A3.ER.H212R mutant efficiently bound to a region of the BIM promoter containing one of the aforementioned imperfect FREs. At the same time, it failed to bind to the IGFBP1 promoter, which contains a perfectly matching FRE. An attractive explanation for this discrepancy could be recruitment via interaction with a (tissue-specific) interaction partner. Although it is presently unknown how exactly FOXO3.A3.ER.H212R regulates BIM in ECs, a recent report demonstrated physical interaction of FOXO3 with RUNX transcription factors, which were required to completely activate BIM expression in different cell types (48).
Although we demonstrate that FRE-independent gene expression is sufficient for apoptosis induction, our data suggest that a FRE-dependent mechanism greatly amplifies this process. For one, this is implicated by our bioinformatical analysis, which revealed a more prominent overrepresentation of apoptosis-associated GO groups among the FRE-dependent FOXO3 targets. Second, we consistently observed higher induction levels of BIM with the FRE-binding mutant despite decreased CASP-3 cleavage and subdiploidy. Although both the higher BIM expression and promoter binding may perhaps reflect an increased availability of the FRE-mutant for binding partners due to its deficiency in binding classical FREs, BIM up-regulation alone cannot sufficiently explain the proapoptotic effect of FOXO3. Hence, one or several FRE-dependent targets are required for effective apoptosis induction by FOXO3 in ECs. We definitively can exclude a death receptor-dependent mechanism (e.g. via repression of the CASP-8 antagonist cFLIP (38) or induction of TRAIL (10)) because neither cFLIPL expression nor incubation with a CASP-8 inhibitor affected FOXO3-induced apoptosis. Moreover, primary human ECs are highly resistant to death ligand-induced apoptosis and survive a stimulation of up to 3 mg/ml TRAIL in vitro (39). BIM is also usually bound at the cytoskeleton and first has to be released to exert its proapoptotic effect (49). Its release has recently been found to be mediated by GADD45, an established FOXO3 target and DNA damage response gene (14), which likewise was present in our list of genes co-induced by the two FOXO mutants (data not shown). Thus, participation of another regulator of the intrinsic pathway is the most likely explanation for our observation. Among those, NOXA is the most promising candidate. NOXA has recently been described as a FOXO3 target (50), and here we provide the first evidence that its regulation is FRE-dependent. Normally, apoptosis initiation is inhibited by antiapoptotic BCL-2 family members, which prevent mitochondrial BAX/BAK activation (51). Unlike BIM and other proapoptotic BCL-2 family members that trigger apoptosis by their ability to neutralize these antiapoptotic proteins, NOXA can only engage a subset of them (52). Thus, NOXA requires the aid of other proapoptotic BCL-2 proteins and/or simultaneous loss of antiapoptotic BCL-2 proteins to influence apoptosis. NOXA therefore may merely act as an amplifier of the process, whereas tissue-specific expression of FRE-independent targets, such as BIM, is required to determine the apoptotic fate. Consistently, NOXA depletion reduced FOXO3.A3.ER-induced apoptosis in our experiments but failed to reduce apoptosis induction by the FRE mutant. In contrast, BIM siRNA efficiently suppressed apoptosis induced by the two FOXO3 mutants, suggesting a potential cooperation of both factors in FOXO3-induced apoptosis. It remains to be elucidated whether such cooperation can sufficiently explain the mystery of cell type-specific apoptosis by FOXO3, but at least this is an attractive hypothesis.
Besides cell cycle and apoptosis regulation, we also found several functional annotation clusters related to leukocyte differentiation overrepresented among the co-regulated genes. Particularly in view of the FOXO3 knock-out phenotype, which exhibits defects in lymphocyte homeostasis in the T-cell compartment (21), this is an interesting finding. A potential candidate gene contributing to this imbalance is the transcription factor EGR1. EGR1-deficient mice have previously been shown to contain increased numbers of T-cells (36), and haplodeficiency of EGR1 leads to development of T-cell lymphomas (37), the second prevalent tumor type induced by combined somatic deletion of multiple FOXO factors besides hemangiomas (20). It is noteworthy that EGR1 is induced by vascular injury (53) and promotes FGF-dependent angiogenesis during tumor growth upon transient activation (54), whereas long term EGR1 expression blocks tumor angiogenesis (55). Loss of EGR1 thus might contribute to hemangioma formation in FOXO1/3 double deficient mice (20) and partially account for the antiangiogenic properties of FOXO1 and FOXO3 in ECs (18) even more because EGR1 is also a FOXO1 target gene in human ECs (17). The exact roles of individual targets of these leukocyte-related clusters remain to be determined, but in any case, the high prevalence of such clusters in the group of co-induced genes implicates a potentially important role of FRE-independent FOXO3 targets in the regulation of cell type-specific functions, such as lymphocyte homeostasis, inflammation, and tumor angiogenesis.
Although both the divergent knock-out phenotypes (19) and the reported gene expression differences upon knockdown of FOXO1 and -3 in human ECs (18) suggest a non-redundant function of FOXOs in ECs, the tumor-suppressive function appears to be conserved because combined deletion of FOXO1, -3, and -4 greatly enhanced hemangioma formation (20). At first glance, this appears to be at odds with the finding that FOXO-induced cell cycle arrest does not require FRE-dependent gene expression. However, both FOXO3.A3.ER.H212R (this study) and FRE binding-efficient FOXO1 (6), can repress D-type cyclins. Two scenarios could explain these results. First, FOXO1 and FOXO3 could interact with common binding partners to regulate CCND expression. Alternatively, both FRE-binding mutants could interact with similar alternative binding sites at the CCND1 promoter. Both possibilities suggest a participation of either the highly homologous forkhead box or other conserved motifs. Whereas it is unclear which mechanism is operative for CCND1, it is well known that some binding partners, such as nuclear hormone receptors, bind to FOXOs via a conserved LXXLL motif C-terminal of the forkhead box (56). Interestingly, comparison of our target genes with those of FOXO1 in ECs (17, 18) and of FOXO1 and FOXO3 in other cell systems (6, 46) reveals co-regulation of various alternative FOXO3 targets (compare Tables 2 and 3 and supplemental Tables 1 and 2), implicating a substantial overlap in FRE-independent gene expression. Thus, FRE-independent gene regulation does not generally exclude functional redundancy and does not necessarily implicate cell type-specific regulation. On the other hand, we also found cell type-specific genes among the classical target genes, as exemplified by ANGPT2, demonstrating tissue-specific regulation of classical target genes as well. This probably occurs due to posttranslational modulation of FOXOs, such as acetylation or phosphorylation, which have previously been shown to be important for the regulation of direct FOXO targets, such as FasL (57). Thus, the picture emerges that post-translational modifications of FOXOs in combination with interaction with other transcriptional mediators can turn FOXO responses in a certain direction. Therefore, the outcome of FOXO activation is probably dependent on tissue-specific expression of the different FOXOs as well as on the presence and activation status of cofactors and posttranslational modification enzymes, which together determine the cell type-specific responses to FOXO factors. Future comparative studies of FRE-dependent and -independent gene expression in a single cell type are required to dissect the relative contributions of these different mechanisms in individual cells.
Supplementary Material
Acknowledgments
We thank N. Schmidt and A. Huss for excellent technical assistance and B. Burgering (Utrecht) for critically reading the manuscript.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft “International Research Training Group Vascular Medicine” (Graduiertenkolleg 880/2) (to M. S. and M. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–7.
- EC
- endothelial cell
- FRE
- FOXO-responsive DNA-binding element
- BrdUrd
- bromodeoxyuridine
- Z
- benzyloxycarbonyl
- HUVEC
- human umbilical vein endothelial cell
- qRT
- quantitative real-time reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- PCNA
- proliferating cell nuclear antigen
- ChIP
- chromatin immunoprecipitation
- PI
- propidium iodide
- GO
- gene ontology
- HIV-1
- human immunodeficiency virus, type 1.
REFERENCES
- 1.Greer E. L., Brunet A. (2005) Oncogene 24, 7410–7425 [DOI] [PubMed] [Google Scholar]
- 2.Dansen T. B., Burgering B. M. (2008) Trends Cell Biol. 18, 421–429 [DOI] [PubMed] [Google Scholar]
- 3.Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., Ruvkun G. (1997) Nature 389, 994–999 [DOI] [PubMed] [Google Scholar]
- 4.Lee S. S., Kennedy S., Tolonen A. C., Ruvkun G. (2003) Science 300, 644–647 [DOI] [PubMed] [Google Scholar]
- 5.Medema R. H., Kops G. J., Bos J. L., Burgering B. M. (2000) Nature 404, 782–787 [DOI] [PubMed] [Google Scholar]
- 6.Ramaswamy S., Nakamura N., Sansal I., Bergeron L., Sellers W. R. (2002) Cancer Cell 2, 81–91 [DOI] [PubMed] [Google Scholar]
- 7.Schmidt M., Fernandez de Mattos S., van der Horst A., Klompmaker R., Kops G. J., Lam E. W., Burgering B. M., Medema R. H. (2002) Mol. Cell. Biol. 22, 7842–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 9.Dijkers P. F., Medema R. H., Lammers J. W., Koenderman L., Coffer P. J. (2000) Curr. Biol. 10, 1201–1204 [DOI] [PubMed] [Google Scholar]
- 10.Modur V., Nagarajan R., Evers B. M., Milbrandt J. (2002) J. Biol. Chem. 277, 47928–47937 [DOI] [PubMed] [Google Scholar]
- 11.Dijkers P. F., Birkenkamp K. U., Lam E. W., Thomas N. S., Lammers J. W., Koenderman L., Coffer P. J. (2002) J. Cell Biol. 156, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilley J., Coffer P. J., Ham J. (2003) J. Cell Biol. 162, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W., Coffer P. J., Huang T. T., Bos J. L., Medema R. H., Burgering B. M. (2002) Nature 419, 316–321 [DOI] [PubMed] [Google Scholar]
- 14.Tran H., Brunet A., Grenier J. M., Datta S. R., Fornace A. J., Jr., DiStefano P. S., Chiang L. W., Greenberg M. E. (2002) Science 296, 530–534 [DOI] [PubMed] [Google Scholar]
- 15.Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 16.Furuyama T., Kitayama K., Shimoda Y., Ogawa M., Sone K., Yoshida-Araki K., Hisatsune H., Nishikawa S., Nakayama K., Nakayama K., Ikeda K., Motoyama N., Mori N. (2004) J. Biol. Chem. 279, 34741–34749 [DOI] [PubMed] [Google Scholar]
- 17.Daly C., Wong V., Burova E., Wei Y., Zabski S., Griffiths J., Lai K. M., Lin H. C., Ioffe E., Yancopoulos G. D., Rudge J. S. (2004) Genes Dev. 18, 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Potente M., Urbich C., Sasaki K., Hofmann W. K., Heeschen C., Aicher A., Kollipara R., DePinho R. A., Zeiher A. M., Dimmeler S. (2005) J. Clin. Invest. 115, 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosaka T., Biggs W. H., 3rd, Tieu D., Boyer A. D., Varki N. M., Cavenee W. K., Arden K. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paik J. H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J. W., Carrasco D. R., Jiang S., Gilliland D. G., Chin L., Wong W. H., Castrillon D. H., DePinho R. A. (2007) Cell 128, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin L., Hron J. D., Peng S. L. (2004) Immunity 21, 203–213 [DOI] [PubMed] [Google Scholar]
- 22.Furuyama T., Nakazawa T., Nakano I., Mori N. (2000) Biochem. J. 349, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai K. L., Sun Y. J., Huang C. Y., Yang J. Y., Hung M. C., Hsiao C. D. (2007) Nucleic Acids Res. 35, 6984–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Vos K. E., Coffer P. J. (2008) Oncogene 27, 2289–2299 [DOI] [PubMed] [Google Scholar]
- 25.Viemann D., Schmidt M., Tenbrock K., Schmid S., Müller V., Klimmek K., Ludwig S., Roth J., Goebeler M. (2007) J. Immunol. 178, 3198–3207 [DOI] [PubMed] [Google Scholar]
- 26.Dijkers P. F., Medema R. H., Pals C., Banerji L., Thomas N. S., Lam E. W., Burgering B. M., Raaijmakers J. A., Lammers J. W., Koenderman L., Coffer P. J. (2000) Mol. Cell. Biol. 20, 9138–9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stassi G., Di Liberto D., Todaro M., Zeuner A., Ricci-Vitiani L., Stoppacciaro A., Ruco L., Farina F., Zummo G., De Maria R. (2000) Nat. Immunol. 1, 483–488 [DOI] [PubMed] [Google Scholar]
- 28.Dai M., Wang P., Boyd A. D., Kostov G., Athey B., Jones E. G., Bunney W. E., Myers R. M., Speed T. P., Akil H., Watson S. J., Meng F. (2005) Nucleic Acids Res. 33, e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh W. P., Chu T. M., Wolfinger R. D., Gibson G. (2003) Genetics 165, 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng Y., Tang W., Dai Y., Wu X., Liu M., Ji Q., Ji M., Pienta K., Lawrence T., Xu L. (2008) Mol. Cancer Ther. 7, 2192–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 32.Ji M., Li H., Suh H. C., Klarmann K. D., Yokota Y., Keller J. R. (2008) Blood 112, 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buitenhuis M., van Deutekom H. W., Verhagen L. P., Castor A., Jacobsen S. E., Lammers J. W., Koenderman L., Coffer P. J. (2005) Blood 105, 4272–4281 [DOI] [PubMed] [Google Scholar]
- 34.Bernard D. J., Chapman S. C., Woodruff T. K. (2001) Recent Prog. Horm. Res. 56, 417–450 [DOI] [PubMed] [Google Scholar]
- 35.Shav-Tal Y., Zipori D. (2002) Stem Cells 20, 493–500 [DOI] [PubMed] [Google Scholar]
- 36.Bettini M., Xi H., Milbrandt J., Kersh G. J. (2002) J. Immunol. 169, 1713–1720 [DOI] [PubMed] [Google Scholar]
- 37.Joslin J. M., Fernald A. A., Tennant T. R., Davis E. M., Kogan S. C., Anastasi J., Crispino J. D., Le Beau M. M. (2007) Blood 110, 719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skurk C., Maatz H., Kim H. S., Yang J., Abid M. R., Aird W. C., Walsh K. (2004) J. Biol. Chem. 279, 1513–1525 [DOI] [PubMed] [Google Scholar]
- 39.Schmidt M., Hupe M., Endres N., Raghavan B., Kavuri S., Geserick P., Goebeler M., Leverkus M. (June16, 2009) J. Cell. Mol. Med. 10.1111/j.1582-4934.2009.00823.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irmler M., Thome M., Hahne M., Schneider P., Hofmann K., Steiner V., Bodmer J. L., Schröter M., Burns K., Mattmann C., Rimoldi D., French L. E., Tschopp J. (1997) Nature 388, 190–195 [DOI] [PubMed] [Google Scholar]
- 41.Scaffidi C., Schmitz I., Krammer P. H., Peter M. E. (1999) J. Biol. Chem. 274, 1541–1548 [DOI] [PubMed] [Google Scholar]
- 42.Augustin H. G., Koh G. Y., Thurston G., Alitalo K. (2009) Nat. Rev. Mol. Cell Biol. 10, 165–177 [DOI] [PubMed] [Google Scholar]
- 43.Bi W., Drake C. J., Schwarz J. J. (1999) Dev. Biol. 211, 255–267 [DOI] [PubMed] [Google Scholar]
- 44.Lin Q., Lu J., Yanagisawa H., Webb R., Lyons G. E., Richardson J. A., Olson E. N. (1998) Development 125, 4565–4574 [DOI] [PubMed] [Google Scholar]
- 45.Alvarez B., Martínez-A C., Burgering B. M., Carrera A. C. (2001) Nature 413, 744–747 [DOI] [PubMed] [Google Scholar]
- 46.Delpuech O., Griffiths B., East P., Essafi A., Lam E. W., Burgering B., Downward J., Schulze A. (2007) Mol. Cell. Biol. 27, 4917–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Z., Tindall D. J. (2008) Oncogene 27, 2312–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamura Y., Lee W. L., Inoue K., Ida H., Ito Y. (2006) J. Biol. Chem. 281, 5267–5276 [DOI] [PubMed] [Google Scholar]
- 49.Puthalakath H., Villunger A., O'Reilly L. A., Beaumont J. G., Coultas L., Cheney R. E., Huang D. C., Strasser A. (2001) Science 293, 1829–1832 [DOI] [PubMed] [Google Scholar]
- 50.Obexer P., Geiger K., Ambros P. F., Meister B., Ausserlechner M. J. (2007) Cell Death Differ. 14, 534–547 [DOI] [PubMed] [Google Scholar]
- 51.Adams J. M., Cory S. (2007) Oncogene 26, 1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. (2005) Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 53.Khachigian L. M., Lindner V., Williams A. J., Collins T. (1996) Science 271, 1427–1431 [DOI] [PubMed] [Google Scholar]
- 54.Fahmy R. G., Dass C. R., Sun L. Q., Chesterman C. N., Khachigian L. M. (2003) Nat. Med. 9, 1026–1032 [DOI] [PubMed] [Google Scholar]
- 55.Lucerna M., Pomyje J., Mechtcheriakova D., Kadl A., Gruber F., Bilban M., Sobanov Y., Schabbauer G., Breuss J., Wagner O., Bischoff M., Clauss M., Binder B. R., Hofer E. (2006) Cancer Res. 66, 6708–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao H. H., Herrera R. E., Coronado-Heinsohn E., Yang M. C., Ludes-Meyers J. H., Seybold-Tilson K. J., Nawaz Z., Yee D., Barr F. G., Diab S. G., Brown P. H., Fuqua S. A., Osborne C. K. (2001) J. Biol. Chem. 276, 27907–27912 [DOI] [PubMed] [Google Scholar]
- 57.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.