Abstract
α-Secretase cleavage of the amyloid precursor protein (APP) is of great interest because it prevents the formation of the Alzheimer-linked amyloid-β peptide. APP belongs to a conserved gene family including the two paralogues APP-like protein (APLP) 1 and 2. Insulin-like growth factor-1 (IGF-1) stimulates the shedding of all three proteins. IGF-1-induced shedding of both APP and APLP1 is dependent on phosphatidylinositol 3-kinase (PI3-K), whereas APLP2 shedding is independent of this signaling pathway. Here, we used human neuroblastoma SH-SY5Y cells to investigate the involvement of protein kinase C (PKC) in the proteolytic processing of endogenously expressed members of the APP family. Processing was induced by IGF-1 or retinoic acid, another known stimulator of APP α-secretase shedding. Our results show that stimulation of APP and APLP1 processing involves multiple signaling pathways, whereas APLP2 processing is mainly dependent on PKC. Next, we wanted to investigate whether the difference in the regulation of APLP2 shedding compared with APP shedding could be due to involvement of different processing enzymes. We focused on the two major α-secretase candidates ADAM10 and TACE, which both are members of the ADAM (a disintegrin and metalloprotease) family. Shedding was analyzed in the presence of the ADAM10 inhibitor GI254023X, or after transfection with small interfering RNAs targeted against TACE. The results clearly demonstrate that different α-secretases are involved in IGF-1-induced processing. APP is mainly cleaved by ADAM10, whereas APLP2 processing is mediated by TACE. Finally, we also show that IGF-1 induces PKC-dependent phosphorylation of TACE.
Keywords: Diseases/Amyloid, Protease/Adam/Adamts, Proteases/Secretases, Insulin-like Growth Factor (Igf), Protein Kinase C (Pkc), Signal Transduction, Adam10, Aplp-2, App Family, Tace
Introduction
Alzheimer disease (AD)2 is histopathologically characterized by the presence of amyloid plaques in the brain parenchyma. The major constituent of the plaques is the amyloid-β (Aβ) peptide, which is derived from the Aβ precursor protein (APP) by β- and γ-secretase cleavage in the amyloidogenic pathway (1). In addition to Aβ, a larger secreted fragment, sAPPβ, and an intracellular fragment (APP intracellular domain) are formed. Proteolytic processing of APP occurs mainly by an alternative pathway. In this nonamyloidogenic pathway, Aβ formation is precluded because α-secretase cleaves APP in the middle of the Aβ region. In addition to sAPPα, a small secreted peptide, p3, and APP intracellular domain are generated by subsequent γ-secretase cleavage.
APP belongs to a conserved gene family including the two mammalian paralogues amyloid precursor-like protein 1 and 2 (APLP1 and APLP2). The exact biological function of APP and its paralogues is still unknown, although several different studies have shown involvement of APP in cell adhesion, neurite outgrowth, synaptogenesis, modulation of synaptic plasticity, and neuroprotection (reviewed in Ref. 2). Homo- and heterotypic cis interactions of APP family members have been detected (3). APLP1 could also form trans interactions, suggesting a specific role of APLP1 in cell adhesion. Importantly, double knock-out studies in mice demonstrated a crucial role for APLP2 in survival because APP−/−/APLP2−/− and APLP1−/−/APLP2−/− mice died within the first week after birth, whereas APP−/−/APLP1−/− mice survived (4). Recently, the mechanism for the postnatal lethality in the APP−/−/APLP2−/− knock-out mice was correlated to hyperinsulinemia and hypoglycemia, suggesting that APP family proteins are essential modulators of glucose and insulin pathways (5). This is interesting because a relationship between diabetes mellitus and AD has been suggested, and disturbed insulin levels and signaling in AD brains have been observed (6). In addition, insulin and the related insulin-like growth factor-1 (IGF-1) have even been shown to affect APP processing (7–8).
Stimulation of α-secretase activity constitutes an important therapeutical target for AD, because this will decrease the production of Aβ. Several α-secretase candidates exist, and they are all members of the ADAM (a disintegrin and metalloproteinase) family. The two most likely candidates are ADAM10 and tumor necrosis factor-α converting enzyme (TACE, also known as ADAM17) (9–10). ADAM10 was established as an α-secretase candidate, as it was able to cleave a synthetic peptide spanning the α-secretase cleavage site (10). In addition, overexpression of ADAM10 in HEK293 cells significantly increased the levels of sAPPα. TACE has also been shown to cleave an α-site-spanning peptide (9). TACE gene silencing was further shown to completely block phorbol ester-induced sAPPα secretion. Furthermore, APP and TACE co-transfection in HEK293 cells increased basal sAPPα secretion in a dose-dependent manner in relation to TACE cDNA expression (11).
In addition to overlapping functions with APP, APLPs have also been demonstrated to be processed in a similar way. This includes α-, β-, and γ-like processing (12–13), which can be influenced by homo- and heterophilic interactions (14–15). Like APP, APLP2 processing has been demonstrated to be induced by phorbol ester (16). Furthermore, overexpression of both ADAM10 and TACE has been shown to increase the secretion of secreted APLP2 (sAPLP2) (17). Little is known about the processing of APLP1. However, α-secretase processing of APLP1 was demonstrated in APLP-1 transfected human neuroblastoma SH-SY5Y cells, as the production of an APLP1-derived p3-like fragment was strongly reduced by the ADAM inhibitors batimastat and tumor necrosis factor-α protease inhibitor-2 (12). The sAPLP1 secretion was also increased by treatment of cells with phorbol ester.
Although the APP family proteins can be processed in a similar way, there seem to be differences in how the induced processing of the three APP family members is regulated. In our previous study, insulin and IGF-1 were shown to increase ectodomain shedding of endogenously expressed APP, APLP1, and APLP2 in SH-SY5Y cells (7). In addition, it was demonstrated that there are different signaling pathways involved in the processing of the different paralogues. The IGF-1-induced secretion of sAPPα, concomitant with decreased production of Aβ, was dependent on phosphatidylinositol 3-kinase (PI3-K) activation. Stimulation of sAPLP1 secretion involved both MAPK and PI3-K signaling pathways. In contrast, APLP2 shedding was independent of both PI3-K and MAPK signaling pathways (7).
In this study, we wanted to clarify the differences between regulated processing of the APP family, with a focus on APP and APLP2. We used cells with endogenous expression of both the APP family members and the two main α-secretases. To investigate the role of protein kinase C (PKC) in ectodomain shedding, we used bisindolylmaleimide XI as a selective PKC inhibitor (18–19). Furthermore, to determine whether the differences in the regulated processing of the APP family members were due to cleavage by different α-secretases, we used an ADAM10 inhibitor (GI254023X) and siRNA targeted against TACE. Finally, [32P]phosphate labeling of SHSY-5Y cells demonstrated that IGF-1 induces PKC-dependent phosphorylation of TACE. Our results shed light on the mechanism behind IGF-1-induced processing of APP and APLP2 and clearly demonstrate that the stimulated processing of the APP family members is dependent on different signaling pathways and different processing enzymes.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment
SH-SY5Y human neuroblastoma cells (American Type Culture Collection) were routinely maintained as described previously (20). Cells were seeded at a density of 25,000 cells/cm2 in Nunc 60-mm dishes. The medium was changed to serum-free culture medium and grown for 6 days as described previously (7) in the presence or absence of 1 μm all-trans retinoic acid (RA) (Sigma-Aldrich). After washing for 30 min with 4 ml of serum-free culture medium devoid of insulin, cells were treated with 10 nm IGF-1 (Sigma-Aldrich) or 1 μm all-trans RA for 18 h in 2 ml of serum- and insulin-free culture medium. For inhibition studies, 5 μm curcumin, 5 μm bisindolylmaleimide XI (Sigma-Aldrich), a specific PKC inhibitor, 10 μm LY 292002 (Merck Biosciences), a specific PI3-K inhibitor, and 5 μm GI254023X (a kind gift from Dr. Andreas Ludwig), an ADAM10 inhibitor, were added to the cells during the washing step and the subsequent treatment.
siRNA Knockdown
For TACE knock-down, the cells were transfected 48 h prior to IGF-1 treatment with 100 nm SMARTpool siRNA (Dharmacon; NM_003183, referred to as sequence 1), or alternatively 5, 10, or 50 nm Silencer-validated siRNA (Invitrogen; siRNA s13718 and s13720, referred to as sequence 2 and sequence 3, respectively) using liposome-based transfection agents (Lipofectamine from Invitrogen or HiPerfect from Qiagen), according to the manufacturer's protocol. siGENOME nontargeting siRNA (Dharmacon) was used as a negative control. All cell culture reagents were purchased from Invitrogen unless otherwise indicated.
Western Blot Assay
Conditioned medium was collected and concentrated as described previously (21). Cells were harvested and analyzed by Western blot assay as described previously (21). Primary antibody concentrations were as follows: 1:2,000 for 6E10 (directed against Aβ1–17) and 1:4,000 for CT11 (directed against the C-terminal 11 amino acids of APLP1), 1:3,000 for 42464 (directed against amino acids 499–557 of APLP1; cf. 22) and 1:5,000 for DTII (directed against full-length APLP2), 1:1,000 for CT-TACE (directed against the C-terminal 18 amino acids of TACE), and subsequently 1:5,000 for horseradish peroxidase-coupled anti-mouse IgG, anti-rabbit IgG, and protein A. Western blot analysis of SH-SY5Y cell lysate using antibody 6E10 shows four major bands corresponding to APP751/770 N+O-glycosylated (139 kDa), APP695 N+O-glycosylated (120 kDa), APP751/770 N-glycosylated (113 kDa), and APP695 N-glycosylated (102 kDa). Western blot analysis of conditioned medium from SH-SY5Y cells using antibody 6E10 shows two major bands corresponding to the secreted sAPPα fragments of the isoforms APP695 and APP751/770, respectively. Antibody 42464 recognizes two bands of ∼80 and 90 kDa corresponding to immature and N-glycosylated APLP1, respectively. sAPLP1 is detected as one band of ∼80 kDa using antibody 42464. Antibody DTII recognizes bands of ∼110 kDa corresponding to APLP2707 and APLP2763. The secreted APLP2 is also recognized as two bands. DTII also recognizes chondroitin sulfate glucosaminoglycan modified APLP2/sAPLP2 (150–250 kDa), but this form was not quantified here. Western blot analysis of SH-SY5Y cell lysate using antibody CT-TACE shows major bands of 120 and 90 kDa corresponding to the pro-form and active form of TACE, respectively. The intensity in the region of the Western blots containing the specific bands (marked with a vertical line in Figs. 1–4) were quantified by densitometry using Image Gauge (version 3.46; Fujifilm), and the background was subtracted. Only bands that were not observed in the absence of primary antibody were quantified. The secreted levels of the APP family proteins were normalized to the corresponding full-length protein levels. All Western blotting reagents were from GE Healthcare or Bio-Rad Laboratories, except for CT11 and DTII (Calbiochem), 6E10 (Signet Laboratories), CT-TACE (Nordic Biosite), and protein A (Sigma-Aldrich).
FIGURE 1.
A PI3-K-specific inhibitor reduces the RA-induced secretion of sAPPα, whereas the sAPLP2 levels are not significantly changed. Relative abundance of sAPPα (A) and sAPLP2 (B) in culture medium, normalized to the relative abundance of full-length APP or APLP2 in cell lysates from SH-SY5Y cells treated with 1 μm RA for 6 days in the absence or presence of 10 μm LY 292002 (LY) for the last 18 h. Data represent mean ± S.E., n = 4–7. **, p < 0.01 significantly different from cells treated with RA. Representative Western blot analyses of secreted and full-length APP and APLP2 are shown below the graphs. Untreated control (C) cells are included for comparison. The vertical lines indicate the quantified bands.
FIGURE 2.
Curcumin and a PKC-specific inhibitor block IGF-1- and RA-induced secretion of sAPLP2. A, relative abundance of sAPPα, sAPLP2, and sAPLP1 in culture medium, normalized to the relative abundance of full-length APP, APLP2, or APLP1 in cell lysates from SH-SY5Y cells treated with 10 nm IGF-1 for 18 h in the absence or presence of 5 μm curcumin or 5 μm bisindolylmaleimide XI (BIM11) for the last 18 h. B, representative Western blot analyses of secreted and full-length APP, APLP2, and APLP1 after IGF-1 treatment. C, relative abundance of APPα and sAPLP2 in culture medium, normalized to the relative abundance of full-length APP or APLP2 in cell lysates from SH-SY5Y cells treated with 1 μm RA for 6 days in the absence or presence of 5 μm curcumin or 5 μm bisindolylmaleimide XI for the last 18 h. D, representative Western blot analyses of secreted and full-length APP and APLP2 after RA treatment. Data represent mean ± S.E., n = 4–7. *, p < 0.05 and **, p < 0.01 significantly different from cells treated with IGF-1 or RA. Untreated control (C) cells are included for comparison. The vertical lines indicate the quantified bands.
FIGURE 3.
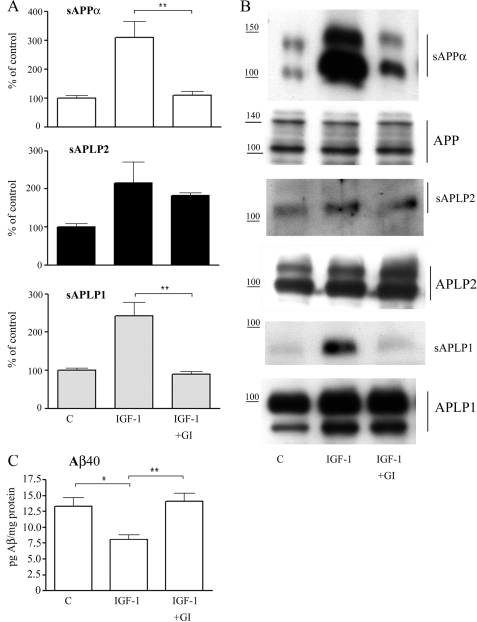
The ADAM10 inhibitor blocks IGF-1-induced secretion of sAPPα and sAPLP1. A, relative abundance of sAPPα, sAPLP2, and sAPLP1 in culture medium, normalized to the relative abundance of full-length APP, APLP2, or APLP1 in cell lysates from SH-SY5Y cells treated with 10 nm IGF-1 for 18 h in the absence or presence of 5 μm GI254023X (GI) for the last 18 h. Data represent mean ± S.E., n = 4–7. **, p < 0.01 significantly different from cells treated with IGF-1. B, representative Western blot analyses of secreted and full-length APP, APLP2, and APLP1. C, Aβ40 concentrations in conditioned cell medium determined by ELISA. Data represent mean ± S.E., n = 4. *, p < 0.05 and **, p < 0.01 significantly different from cells treated with IGF-1. Untreated control (C) cells are included for comparison. The vertical lines indicate the quantified bands.
FIGURE 4.
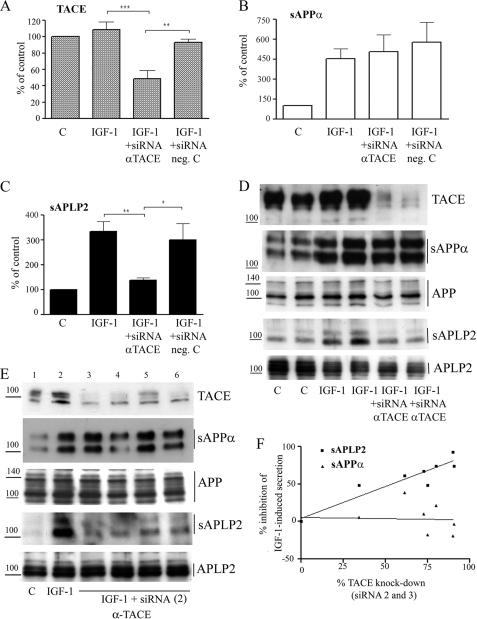
TACE knockdown blocks IGF-1-induced ectodomain shedding of APLP2. Relative abundance of TACE in cell lysates (A) and of sAPPα (B) and sAPLP2 (C) in culture, normalized to the relative abundance of full-length APP or APLP2 in cell lysates from SH-SY5Y cells treated with 10 nm IGF-1 for 18 h in the absence or presence of siRNA targeted against TACE (sequence 1). Data represent mean ± S.E., n = 4–7. **, p < 0.01 and ***, p < 0.001 significantly different from cells treated with IGF-1. D, representative Western blot analyses from SH-SY5Y cells treated with siRNA sequence 1. E, representative Western blots using siRNA sequence 2. Wells 3 and 4 correspond to 5 and 10 nm siRNA transfected with Lipofectamine. Wells 5 and 6 correspond to 10 and 50 nm siRNA transfected with HiPerfect. F, relative inhibition of IGF-1-induced sAPPα and sAPLP2 levels plotted against the silencing effect on TACE expression levels, by siRNA sequences 2 (shown in E) and 3. The siRNA sequences (5, 10, or 50 nm) were transfected using Lipofectamine or HiPerfect. Correlation coefficients (r) were 0.07 for sAPP and 0.95 for sAPLP2. Untreated control (C) cells are included for comparison. The vertical lines indicate the quantified bands.
ELISA
The levels of Aβ40 in conditioned cell medium were analyzed using a high sensitivity sandwich enzyme-linked immunosorbent assay (ELISA) according to the manufacturer (Invitrogen). The Aβ40 concentrations were normalized to the amount of cells in each culture (as determined by protein content in the cell lysate).
[32P]Phosphate Labeling
SH-SY5Y cells were seeded at a density of 100,000 cells/cm2 in a 6-well plate (Costar) and grown overnight. The next day, the cells were washed three times in 2 ml of phosphate-free Dulbecco's modified Eagle's medium (Invitrogen) and incubated for 1 h in phosphate-free Dulbecco's modified Eagle's medium. Next, the cells were incubated with 250 μCi/ml [32P]orthophosphate (Perkin and Elmer), in the presence or absence of 5 μm bisindolylmaleimid XI, in 1 ml of phosphate-free Dulbecco's modified Eagle's medium for 2 h. Some cells were then treated with 10 nm IGF-1 for 2 h. The radioactive media was removed and discarded, and the cells were washed twice in 2 ml of ice-cold phosphate-buffered saline and lysed in 37 μl radioimmune precipitation assay buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris, 1 mm EDTA; 0,25% Na deoxycholate, 1 mm NaF, and 1 mm Na3VO4). 50 μg of pre-cleared cell lysate was immunoprecipitated with 3 μg of TACE antibody (C15/sc-6414; Santa Cruz Biotechnology), previously rotated with 60 μl of protein G-Sepharose at 4 °C for 1 h. The solution was rotated for 1 h at 4 °C and washed with phosphate-buffered saline; the immunoprecipitated proteins were then resolved by SDS-PAGE, and the radioactive bands were detected by exposing the gel to a PhosphorImager screen. The intensity of the 120-kDa band, corresponding to uncleaved phosphorylated TACE, was quantified using Image Gauge (version 3.46, Fujifilm), and the background was subtracted.
Statistical Analysis
Statistical analysis was performed using analysis of variance followed by Tukey's multiple comparison test.
RESULTS
PKC Activation Is Required for IGF-1- and Retinoic Acid-induced Secretion of sAPLP2
Previous studies have determined that IGF-1-induced processing of the APP family proteins was dependent on different signaling pathways and that the dependence of PI3-K activation varied between the three proteins (7). IGF-1-induced processing of APP was completely blocked by PI3-K inhibition, whereas sAPLP2 secretion was unaffected. To further investigate PI3-K dependence, another known inducer of APP family processing, RA (21), was used. Western blot analysis of conditioned cell medium from human neuroblastoma SH-SY5Y cells showed an increase of both sAPPα and sAPLP2 levels in response to RA (Fig. 1, A and B), as shown previously (21). Note that APLP1 was excluded from the study because the level of secreted sAPLP1 in this experimental set-up is below detection limits. RA also induced an increase of full-length APP and APLP2 levels (Fig. 1, A and B), and therefore all secreted levels were normalized to the full-length levels. The RA-induced sAPPα secretion was significantly inhibited by the specific PI3-K inhibitor, LY294002 (∼52% reduction; Fig. 1A). On the contrary, the PI3-K inhibitor had no significant effect on RA-induced sAPLP2 levels (Fig. 1B).
To further investigate the signaling involved in IGF-1- and RA-induced processing of APLP2, we used a nonspecific and a specific PKC inhibitor, curcumin and bisindolylmaleimide XI, respectively. These inhibitors had a much stronger effect on the shedding of APLP2 compared with the other two mammalian APP family members. Western blot analysis demonstrated an ∼5.5-fold increase of sAPPα levels in culture medium (normalized to APP levels in the cell lysate) from SH-SY5Y cells in response to 10 nm IGF-1 (Fig. 2, A and B), as expected from previous studies (7). IGF-1 also induced an increase of both the sAPLP2 and sAPLP1 levels in the culture medium (Fig. 2, A and B), as observed previously (7). Curcumin or bisindolylmaleimide XI reduced the IGF-1-induced secretion of the ectodomains of all three APP family proteins, but to a various degree. The inhibitors only partly reduced the IGF-1-induced processing of APP (∼55% reduction), whereas the effect on the IGF-1-induced secretion of sAPLP2 was much more pronounced (∼97%; Fig. 2, A and B). As observed for the IGF-1-induced secretion, both curcumin and bisindolylmaleimide XI inhibited RA-induced secretion of sAPLP2 (by ∼69 and 96%, respectively; Fig. 2, C and D). However, neither curcumin nor Bisindolylmaleimide XI did significantly inhibit RA-induced secretion of sAPPα (Fig. 2, C and D). No significant effect of LY294002, curcumin or bisindolylmaleimide XI on full-length protein levels could be detected, indicating that the observed effects on the secreted fragments are only due to decreased proteolytic activity (Fig. 1 and Fig. 2, B and D).
ADAM10 Activity Is Required for IGF-1-induced Secretion of sAPPα
Because the IGF-1-induced processing of the three APP family proteins was dependent on different signaling pathways, we speculated that they may be cleaved by different processing enzymes. To investigate this, we used the ADAM10 inhibitor, GI254023X, which has a 100-fold higher potency to inhibit ADAM10 compared with TACE (23). Western blot analysis demonstrated that the ADAM10 inhibitor completely blocked the IGF-1-induced secretion of sAPPα and sAPLP1 (∼94 and 100% reduction, respectively; Fig. 3, A and B). On the contrary, the ADAM10 inhibitor did not have a significant effect on the IGF-1-induced secretion of sAPLP2 (∼20% nonsignificant reduction; Fig. 3, A and B). To further investigate the effect of the ADAM10 inhibitor on APP processing, the Aβ40 levels were analyzed (Fig. 3C). ELISA measurements of conditioned cell medium demonstrated that IGF-1 treatment induced an ∼40% decrease of secreted Aβ40, as shown previously (7). This effect was completely reversed by co-treatment with GI254023X (Fig. 3C).
TACE Is Required for IGF-induced Secretion of sAPLP2 but Not of sAPPα
Our results from the studies using PKC inhibitors and the ADAM10 inhibitor clearly demonstrate that APP and APLP2 strongly differ in how the IGF-1-induced processing of the proteins is mediated. To further analyze this, we wanted to determine the effect of inhibiting the other major α-secretase candidate, namely TACE. To reduce the expression levels of TACE, we transfected SH-SY5Y cells with siRNA directed against TACE. Nontargeting siRNA was used as a negative control. Western blot analysis of cell lysates demonstrated that the siRNA directed against TACE down-regulated the expression levels of TACE by ∼60% (Fig. 4, A and D). The conditioned cell medium was also analyzed by Western blot. Even though the TACE down-regulation was not complete, an ∼83% reduction of the IGF-1-induced secretion of sAPLP2 was observed (Fig. 4, C and D). On the contrary, TACE down-regulation had no significant effect on IGF-1-induced processing of APP (Fig. 4, B and D). Furthermore, we found a strong correlation between the degree of TACE down-regulation and the degree of inhibition of IGF-1-induced sAPLP2 secretion (correlation coefficient, r = 0.79). In some of the blots, a weak tendency to decreased levels of full-length proteins (not significant) was observed (Fig. 4D). The sAPPα secretion showed no significant correlation with the TACE levels (correlation coefficient r = 0.28). As a control, and to exclude off-target effects, two other siRNA sequences against TACE were used to knock down the expression and analyze the effect on APP and APLP2 processing (Fig. 4, E and F). Effects on sAPLP2, but not on sAPPα, secretion were strongly correlated to effects on TACE expression (Fig. 4, E and F; correlation coefficients, r = 0.95 for sAPLP2 and r = 0.07 for sAPPα). No effects induced by these two other siRNAs on full-length APP or APLP2 levels could be observed (Fig. 4E).
IGF-1 Induces Phosphorylation of TACE
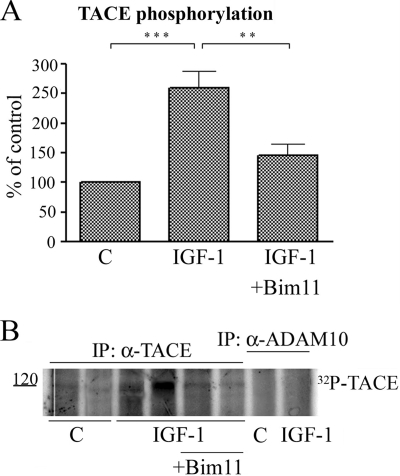
Together, our results show that IGF-1-induced shedding of APLP2 is dependent on both TACE and PKC. It has previously been shown that TACE activity is increased by phosphorylation (24–25). To further investigate the mechanism of IGF-1-induced APLP2 shedding, we analyzed whether IGF-1 in fact could induce phosphorylation of TACE. After [32P]phosphate labeling of SHSY-5Y cells and immunoprecipitation with TACE antibodies, a radioactive band corresponding to ∼120 kDa could be detected (Fig. 5, A and B), corresponding to phosphorylated uncleaved TACE (25–26). A significant 2.5-fold increase in TACE phosphorylation was observed after 2 h of IGF-1 treatment. The results suggest that the IGF-1-induced shedding of APLP2 is a result of increased TACE phosphorylation. Immunoprecipitation with anti-ADAM10 antibodies did not result in any specific radioactive bands on the gel, suggesting that ADAM10 is not phosphorylated under the conditions used in this study (Fig. 5B).
FIGURE 5.
IGF-1 induces phosphorylation of TACE in a PKC-dependent manner. A, relative abundance of phosphorylated TACE in cell lysate from SH-SY5Y cells treated with 10 nm IGF-1 for 18 h in the absence or presence of 5 μm bisindolylmaleimide XI (BIM11) or in the absence of IGF-1 (C). Data represent mean ± S.E., n = 3. **, p < 0.01 and **, p < 0.01 significantly different from cells treated with IGF-1. B, representative SDS-PAGE gel analyzed by autoradiography. IP, immunoprecipitation.
IGF-1-induced Phosphorylation of TACE Is Dependent on PKC
Next, we wanted to investigate the role of PKC in the IGF-1-induced phosphorylation of TACE. As shown in Fig. 5. the selective PKC inhibitor, bisindolylmaleimid XI, significantly decreased the IGF-1-induced phosphorylation of TACE (by ∼77%). Our results suggest that PKC, when activated by IGF-1, mediates phosphorylation and thereby activates TACE, which then preferentially cleaves APLP2, and not APP (Fig. 6).
FIGURE 6.
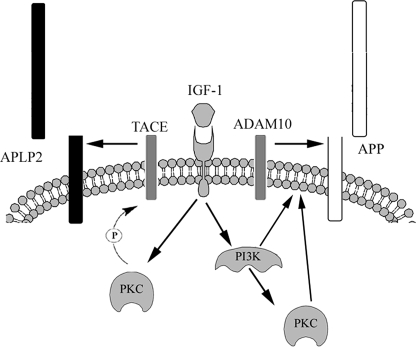
Schematic and simplified illustration of the signaling proposed to mediate IGF-1-induced processing of APP and APLP2.
DISCUSSION
In this study, we show that IGF-1-induced ectodomain shedding of APP and APLP2 are mediated by different α-secretases. ADAM10 is the enzyme responsible for IGF-1-induced processing of APP, whereas the other main α-secretase candidate, TACE, mediates the induced processing of APLP2 upon IGF-1 stimulation. We further demonstrate that the IGF-1-induced processing of the three APP family members is dependent on different signaling pathways. In addition, we demonstrate that TACE is phosphorylated in a PKC-dependent manner in response to IGF-1, thereby shedding light on the signaling behind IGF-1-induced processing of APLP2 (Fig. 6). α-Secretase constitutes an important therapeutical target, because increased α-secretase activity would reduce generation of Aβ. Furthermore, sAPPα, in contrast to the neurotoxic Aβ peptide, has been shown to be neuroprotective (27–28). Recently, BACE-dependent shedding of APP was proposed to generate an N-terminal fragment that could trigger degeneration via activation of death receptor 6 (29). This further supports a beneficial effect of increased α-secretase activity. There are several known stimulators of α-secretase, including insulin and IGF-1 (7–8). We have previously shown that IGF-1 induced a shift in the processing of APP, increasing the levels of sAPPα concomitant with decreased Aβ levels (7). In addition to stimulating α-secretase, insulin and IGF-1 may be even more beneficial for AD patients because impaired insulin and IGF-1 signaling has been observed as a part of AD pathogenesis. AD patients have been shown to have lower insulin and IGF-1 levels in the cerebrospinal fluid and higher plasma insulin levels than non-AD individuals (6). Furthermore, a reduced insulin receptor density and activity has been observed in AD brains. Increased serum IGF-1 levels have even been demonstrated to decrease Aβ levels in the brain of aging rats (30) and in AD transgenic mice (31).
Given the importance of α-site cleavage of APP, further insight about the identity, properties, and regulation of the enzyme(s) involved is of great interest. Previously, we demonstrated that IGF-1-induced secretion of sAPPα was totally blocked by a PI3-K inhibitor, whereas APLP2 processing was completely unaffected (7). In this study, we showed that IGF-1-induced ectodomain shedding of APLP2 was blocked by curcumin as well as by bisindolylmaleimide XI, a specific PKC inhibitor. The results from the present study together with our previous studies (7) suggest that IGF-1 stimulation activates a PKC-dependent pathway that is independent of both PI3-K and MAPK, possibly through receptor tyrosine kinase activation of phospholipase Cγ (cf. 32).
In this study, we also observed that the IGF-1-induced processing of APP was dependent on PKC, but not to the same extent as for APLP2. As PKC inhibition only partially blocked the IGF-1-induced secretion of sAPPα, one plausible explanation could be that PI3-K acts upstream of PKC. However, it should be noted that a PKCα specific inhibitor was reported to reduce IGF-1-induced PI3-K activation and IGF-1 receptor phosphorylation in a rat embryonic heart cell line (33). As for APP, IGF-1-induced processing of APLP1 was shown to involve activation of PKC- as well as PI3-K-dependent signaling pathways (7).
To further investigate the involvement of PKC in APLP2 processing, we used RA, another known stimulator of ectodomain shedding of the APP family (21), to analyze the signaling pathways involved. Our results show that the RA-induced, similar to the IGF-1-induced, ectodomain shedding of APLP2 was inhibited by curcumin as well as by the specific PKC inhibitor, but was independent of PI3-K activation. In contrast, neither curcumin nor bisindolylmaleimide XI did significantly reduce RA-induced secretion of sAPPα. Instead, we show that also the RA-induced secretion of sAPPα is dependent on PI3-K. In addition, we have observed previously that RA treatment of SH-SY5Y cells resulted in significantly increased levels of ADAM10 and TACE (34). The up-regulation of ADAM10 was dependent on PI3-K and independent on PKC. However, like APLP2 processing, TACE up-regulation was shown to be dependent on PKC and independent on PI3-K and MAPK. Together, these results indicate that IGF-1 induces activation of PI3-K and PKC, which induce increased ADAM10- and TACE-mediated processing of APP and APLP2, respectively (Fig. 6).
In addition to the APP family, insulin has also been demonstrated to induce processing of the anti-aging protein Klotho (35). This shedding was shown to be mediated by both ADAM10 and TACE. In our study, silencing of TACE only had an effect on the IGF-1-induced processing of APLP2. On the other hand, pharmacological inhibition of ADAM10 only affected APP processing during IGF-1 stimulation. Clearly, IGF-1 stimulation activates both ADAM10 and TACE. Overexpression and gene silencing studies have previously shown that both APP and APLP2 can be processed by ADAM10 and TACE (9–10, 17). In addition, phorbol ester stimulation is known to induce secretion of both sAPPα and sAPLP2 (16–17, 36–38). Together, this raises the question: why is APP preferentially cleaved by ADAM10 and APLP2 by TACE during IGF-1 stimulation?
Members of the ADAM family generally have broad substrate specificity, and the sequence of the cleavage site is usually not an important factor for substrate recognition (39–40). Instead, it seems like the membrane proximity and the length of stalk (i.e. the susceptible membrane-proximal region between the ectoplasmic globular domain and the transmembrane domain) are more important. It is possible that APP and APLP2 differ in these criteria, favoring them for processing by different ADAMs. Different subcellular localization of APP and APLP2 may also contribute to the difference in processing. Previous studies with ADAM10 and TACE have demonstrated that TACE activity is increased by phorbol ester stimulation, whereas ADAM10 activity is modulated by increased calcium influx (41–42). Furthermore, TACE was shown to colocalize to the plasma membrane with one of its other substrates, CD44, within 10 min after phorbol ester stimulation (43). Furthermore, Soond and colleagues (26) demonstrated that TACE phosphorylation induced translocation to the cell surface. Previous studies have also shown that TACE activity can be regulated by phosphorylation (24–26). Reddy et al. (25) demonstrated that TACE was phosphorylated in response to high glucose, resulting in increased shedding of tumor necrosis factor-α. The effect of high glucose was prevented by inhibition of PKCδ (25). Furthermore, mutation at TACE Thr735 was shown to abolish the carbachol-induced increase of TACE phosphorylation as well as the induced processing of the cellular prion protein (24). This study demonstrates a direct correlation between TACE phosphorylation and induced activity. Here, we demonstrate that TACE also is phosphorylated in response to IGF-1, in a PKC-dependent manner (Fig. 5). One can speculate that upon IGF-1 treatment, TACE is phosphorylated in a PKC-dependent manner, causing co-localization with APLP2, whereas ADAM10 co-localizes with APP.
Even though members of the ADAM family favor certain substrates, there also seems to be a functional overlap between the enzymes. In a recent study by Le Gall et al. (42), it was demonstrated that calcium influx induced ADAM10-mediated processing of several TACE substrates in TACE−/− cells. However, acute treatment of wild type cells with a specific TACE inhibitor showed that TACE is the main processing enzyme of the substrate tested when both ADAM10 and TACE are present. We propose that although APP can be cleaved by TACE, ADAM10 is the main secretase cleaving APP in response to a stimulus like IGF-1 that activates both ADAM10 and TACE.
Today, it is not known what part of the protein that mediates the most important function of APP or the other family members. Increased knowledge of the processing of these proteins is vital for understanding their function and for the design of therapies aiming at blocking the APP processing enzymes resulting in Aβ production. In the present study, we show that the mechanisms underlying the stimulated processing of the APP family are different and thus it may be possible to selectively modulate APP processing. APP and APLP1 processing is dependent on PI3-K, whereas for APLP2 the PKC signaling pathway is the main mediator. Our results also indicate that ADAM10 is the main mediator of stimulated processing of APP, whereas APLP2 is cleaved by TACE.
This work was supported by the Swedish Research Council (to K. I.), Salén Stiftelsen (to K. I.), Magn. Bergvalls Stiftelse (to K. I.), Alzheimerfonden (to K. I. and L. A.), Längmanska kulturfonden (to K. I.), and the Deutsche Forschungsgemeinschaft (to G. M.).
- AD
- Alzheimer disease
- Aβ
- amyloid-β peptide
- siRNA
- small interfering RNA
- APP
- amyloid precursor protein
- APLP1
- APP-like protein 1
- ELISA
- enzyme-linked immunosorbent assay
- IGF-1
- insulin-like growth factor-1
- MAPK
- mitogen-activated protein kinase
- PI3-K
- phosphatidylinositol 3-kinase
- PKC
- protein kinase C
- RA
- retinoic acid
- sAPP
- secreted APP
- TACE
- tumor necrosis factor-α converting enzyme.
REFERENCES
- 1.Esler W. P., Wolfe M. S. (2001) Science 293, 1449–1454 [DOI] [PubMed] [Google Scholar]
- 2.Jacobsen K. T., Iverfeldt K. (2009) Cell Mol. Life Sci. 66, 2299–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaden D., Voigt P., Munter L. M., Bobowski K. D., Schaefer M., Multhaup G. (2009) J. Cell Sci. 122, 368–377 [DOI] [PubMed] [Google Scholar]
- 4.Heber S., Herms J., Gajic V., Hainfellner J., Aguzzi A., Rülicke T., von Kretzschmar H., von Koch C., Sisodia S., Tremml P., Lipp H. P., Wolfer D. P., Müller U. (2000) J. Neurosci. 20, 7951–7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Needham B. E., Wlodek M. E., Ciccotosto G. D., Fam B. C., Masters C. L., Proietto J., Andrikopoulos S., Cappai R. (2008) J. Pathol. 215, 155–163 [DOI] [PubMed] [Google Scholar]
- 6.Steen E., Terry B. M., Rivera E. J., Cannon J. L., Neely T. R., Tavares R., Xu X. J., Wands J. R., de la Monte S. M. (2005) J. Alzheimers. Dis. 7, 63–80 [DOI] [PubMed] [Google Scholar]
- 7.Adlerz L., Holback S., Multhaup G., Iverfeldt K. (2007) J. Biol. Chem. 282, 10203–10209 [DOI] [PubMed] [Google Scholar]
- 8.Solano D. C., Sironi M., Bonfini C., Solerte S. B., Govoni S., Racchi M. (2000) FASEB J. 14, 1015–1022 [DOI] [PubMed] [Google Scholar]
- 9.Buxbaum J. D., Liu K. N., Luo Y., Slack J. L., Stocking K. L., Peschon J. J., Johnson R. S., Castner B. J., Cerretti D. P., Black R. A. (1998) J. Biol. Chem. 273, 27765–27767 [DOI] [PubMed] [Google Scholar]
- 10.Lammich S., Kojro E., Postina R., Gilbert S., Pfeiffer R., Jasionowski M., Haass C., Fahrenholz F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3922–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slack B. E., Ma L. K., Seah C. C. (2001) Biochem. J. 357, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eggert S., Paliga K., Soba P., Evin G., Masters C. L., Weidemann A., Beyreuther K. (2004) J. Biol. Chem. 279, 18146–18156 [DOI] [PubMed] [Google Scholar]
- 13.Minogue A. M., Stubbs A. K., Frigerio C. S., Boland B., Fadeeva J. V., Tang J., Selkoe D. J., Walsh D. M. (2009) Brain Res. 1262, 89–99 [DOI] [PubMed] [Google Scholar]
- 14.Munter L. M., Voigt P., Harmeier A., Kaden D., Gottschalk K. E., Weise C., Pipkorn R., Schaefer M., Langosch D., Multhaup G. (2007) EMBO J. 26, 1702–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaden D., Munter L. M., Joshi M., Treiber C., Weise C., Bethge T., Voigt P., Schaefer M., Beyermann M., Reif B., Multhaup G. (2008) J. Biol. Chem. 283, 7271–7279 [DOI] [PubMed] [Google Scholar]
- 16.Xu K. P., Zoukhri D., Zieske J. D., Dartt D. A., Sergheraert C., Loing E., Yu F. S. (2001) Am. J. Physiol. Cell Physiol. 281, C603–C614 [DOI] [PubMed] [Google Scholar]
- 17.Endres K., Postina R., Schroeder A., Mueller U., Fahrenholz F. (2005) FEBS J. 272, 5808–5820 [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson S. E., Parker P. J., Nixon J. S. (1993) Biochem. J. 294, 335–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis P. D., Elliott L. H., Harris W., Hill C. H., Hurst S. A., Keech E., Kumar M. K., Lawton G., Nixon J. S., Wilkinson S. E. (1992) J. Med. Chem. 35, 994–1001 [DOI] [PubMed] [Google Scholar]
- 20.Beckman M., Iverfeldt K. (1997) Neurosci. Lett. 221, 73–76 [DOI] [PubMed] [Google Scholar]
- 21.Holback S., Adlerz L., Iverfeldt K. (2005) J. Neurochem. 95, 1059–1068 [DOI] [PubMed] [Google Scholar]
- 22.Paliga K., Peraus G., Kreger S., Dürrwang U., Hesse L., Multhaup G., Masters C. L., Beyreuther K., Weidemann A. (1997) Eur. J. Biochem. 250, 354–363 [DOI] [PubMed] [Google Scholar]
- 23.Hundhausen C., Misztela D., Berkhout T. A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., Kallen K. J., Rose-John S., Ludwig A. (2003) Blood 102, 1186–1195 [DOI] [PubMed] [Google Scholar]
- 24.Alfa Cissé M., Sunyach C., Slack B. E., Fisher A., Vincent B., Checler F. (2007) J. Neurosci. 27, 4083–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy A. B., Ramana K. V., Srivastava S., Bhatnagar A., Srivastava S. K. (2009) Endocrinology 150, 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soond S. M., Everson B., Riches D. W., Murphy G. (2005) J. Cell Sci. 118, 2371–2380 [DOI] [PubMed] [Google Scholar]
- 27.Araki W., Kitaguchi N., Tokushima Y., Ishii K., Aratake H., Shimohama S., Nakamura S., Kimura J. (1991) Biochem. Biophys. Res. Commun. 181, 265–271 [DOI] [PubMed] [Google Scholar]
- 28.Mattson M. P., Cheng B., Culwell A. R., Esch F. S., Lieberburg I., Rydel R. E. (1993) Neuron 10, 243–254 [DOI] [PubMed] [Google Scholar]
- 29.Nikolaev A., McLaughlin T., O'Leary D. D., Tessier-Lavigne M. (2009) Nature 457, 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Carro E., Trejo J. L., Gomez-Isla T., LeRoith D., Torres-Aleman I. (2002) Nat. Med. 8, 1390–1397 [DOI] [PubMed] [Google Scholar]
- 31.Carro E., Trejo J. L., Gerber A., Loetscher H., Torrado J., Metzger F., Torres-Aleman I. (2006) Neurobiol. Aging 27, 1250–1257 [DOI] [PubMed] [Google Scholar]
- 32.Hong F., Moon Ka, Kim S. S., Kim Y. S., Choi Y. K., Bae Y. S., Suh P. G., Ryu S. H., Choi E. J., Ha J., Kim S. S. (2001) Biochem. Biophys. Res. Commun. 282, 816–822 [DOI] [PubMed] [Google Scholar]
- 33.Maniar R., Pecherskaya A., Ila R., Solem M. (2005) Mol. Cell Biochem. 275, 15–24 [DOI] [PubMed] [Google Scholar]
- 34.Holback S., Adlerz L., Gatsinzi T., Jacobsen K. T., Iverfeldt K. (2008) Biochem. Biophys. Res. Commun. 365, 298–303 [DOI] [PubMed] [Google Scholar]
- 35.Chen C. D., Podvin S., Gillespie E., Leeman S. E., Abraham C. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19796–19801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slack B. E., Breu J., Muchnicki L., Wurtman R. J. (1997) Biochem. J. 327, 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caporaso G. L., Gandy S. E., Buxbaum J. D., Ramabhadran T. V., Greengard P. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3055–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buxbaum J. D., Koo E. H., Greengard P. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9195–9198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsakadze N. L., Sithu S. D., Sen U., English W. R., Murphy G., D'Souza S. E. (2006) J. Biol. Chem. 281, 3157–3164 [DOI] [PubMed] [Google Scholar]
- 40.Deuss M., Reiss K., Hartmann D. (2008) Curr. Alzheimer Res. 5, 187–201 [DOI] [PubMed] [Google Scholar]
- 41.Sahin U., Weskamp G., Kelly K., Zhou H. M., Higashiyama S., Peschon J., Hartmann D., Saftig P., Blobel C. P. (2004) J. Cell Biol. 164, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Gall S. M., Bobé P., Reiss K., Horiuchi K., Niu X. D., Lundell D., Gibb D. R., Conrad D., Saftig P., Blobel C. P. (2009) Mol. Biol. Cell 20, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagano O., Murakami D., Hartmann D., De Strooper B., Saftig P., Iwatsubo T., Nakajima M., Shinohara M., Saya H. (2004) J. Cell Biol. 165, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]