Abstract
PU.1 is a key transcription factor for hematopoiesis and plays important roles in various hematological malignancies. To clarify the molecular function of PU.1, we initially tried to identify bona fide target genes regulated by PU.1. Dual microarrays were employed for this study to compare PU.1-knockdown K562 cells (K562PU.1KD) stably expressing PU.1 short inhibitory RNAs versus control cells and PU.1-overexpressing K562 cells (K562PU.1OE) versus control cells. In these analyses, we found that several genes, including metallothionein (MT)-1 isoforms (MT-1G and MT-1A) and vimentin (VIM), were markedly induced while Jun dimerization protein (JDP) 2 was suppressed in K562PU.1KD cells. Furthermore, the mRNA expressions of the MT-1 and VIM genes were inversely correlated and the mRNA expression of JDP2 was positively correlated with PU.1 mRNA expression in 43 primary acute myeloid leukemia specimens (MT-1G: R = −0.50, p < 0.001; MT-1A: R = −0.58, p < 0.0005; VIM: R = −0.39, p < 0.01; and JDP2: R = 0.30, p < 0.05). Next, we analyzed the regulation of the MT-1 and VIM genes. We observed increased associations of acetylated histones H3 and H4 with the promoters of these genes in K562PU.1KD cells. Sequence analyses of the regions ∼1 kb upstream from the transcription start sites of these genes revealed numerous CpG sites, which are potential targets for DNA methylation. Chromatin immunoprecipitation assays revealed that methyl CpG-binding protein 2 (MeCP2) and PU.1 bound to the CpG-rich regions in the MT-1 and VIM promoters. Bisulfite sequencing analyses of the PU.1-bound regions of these promoters revealed that the proportions of methylated CpG sites were tightly related to the PU.1 expression levels.
Keywords: DNA Methylation, Histone Modification, Transcription Factors, Transcription Repressor, Transcription Target Genes
Introduction
PU.1 is a member of the Ezb transformation-specific sequence family of transcription factors and is expressed in granulocytic, monocytic, and B-lymphoid cells (1). PU.1 expression levels increase during the differentiation of granulocytes (1). PU.1-deficient mice exhibit defects in the development of neutrophils, macrophages, and B cells (2). Therefore, PU.1 is indispensable for myelomonocytic differentiation during normal hematopoiesis. Mice carrying hypomorphic PU.1 alleles that reduce PU.1 expression to 20% of its normal levels were reported to develop acute myeloid leukemia (AML)2 (3). Moreover, down-regulation of PU.1 was reported to play a role in the pathogenesis of multiple myeloma (4) and is related to a poor prognosis in myelodysplastic syndrome (5). Therefore, characterizing the function and identifying the target genes of PU.1 are important for understanding the molecular biology of hematopoiesis and oncogenesis.
Recently, we cloned cell lines expressing reduced levels of PU.1 by stable transfection of PU.1 short inhibitory RNAs into human myeloid leukemia K562 cells (K562PU.1KD cells). By comparing the gene expressions between control short inhibitory RNA-transfected cells and K562PU.1KD cells, we found that annexin 1 (ANXA1) is a target gene of PU.1 in leukemia cells (6). To gain more knowledge about the downstream pathway of PU.1, we initially performed microarray analyses to compare PU.1-overexpressing K562 cells (K562PU.1OE cells) (6) versus control cells in the present study. From the combined analysis of these microarrays, we found that metallothionein (MT)-1 isoforms (MT-1G and MT-1A) and vimentin (VIM) were markedly induced while Jun dimerization protein (JDP) 2 was significantly suppressed, in K562PU.1KD cells.
MT proteins comprise a group of low molecular weight cysteine-rich intracellular proteins encoded by a family of genes containing several isoforms in humans (7). In humans, the MT genes are located on chromosome 16 in a cluster and can be activated by a variety of stimuli, including metal ions, oxidative stress, cytokines, glucocorticoids, and growth factors (7). The expression and induction of these proteins have been associated with protection against DNA damage, oxidative stress, and apoptosis (7). A number of studies have shown that increased MT expression is closely associated with tumor grade and proliferative activity in solid tumors (7). In addition, the MT-1 promoter was reported to be epigenetically regulated by histone deacetylase (HDAC) and DNA methyltransferase (Dnmt) in a synergistic manner (8). It has also been reported that PU.1 binds to methyl CpG-binding protein 2 (MeCP2), which recruits the corepressor mSin3A and forms a complex with HDAC1 (9, 10). The same research group recently reported that PU.1 forms a complex with Dnmt 3a and Dnmt 3b, and clearly demonstrated that CpG sites in the p16INK4A promoter are methylated by overexpression of PU.1 (11).
VIM is a cytoskeletal protein that belongs to the family of intermediate filaments (12). The steady-state levels of VIM mRNA are growth-regulated in many cells (13, 14). The mRNA levels are generally low in G0 cells, increase to a peak in the mid-G1 phase, and decrease to the prestimulation levels by the time the cells reach S phase (15). VIM mRNA levels are inducible by platelet-derived growth factor and serum (16). Consequently, VIM is of interest as a member of a group of growth-regulated genes as well as for its structural roles in the cytoskeleton. In addition, like the MT-1 genes, the VIM gene is regulated by modification of histones (17) as well as by methylation in certain types of cancer (18).
In the present study, we reveal that PU.1 directly binds and epigenetically suppresses the MT-1 and VIM promoters through HDAC and Dnmt activities. Reduction of PU.1 expression leads to aberrant induction of MT-1s and VIM through suppression of HDAC and Dnmt enzyme activities. The present study reveals a novel molecular pathway of PU.1 through the identification of MT-1s and VIM as PU.1 target genes.
EXPERIMENTAL PROCEDURES
Cell Culture of PU.1-knockdown and PU.1-overexpressing Cells
K562PU.1KD and K562PU.1OE cells were previously established in our laboratory (6) and employed for this study. K562PU.1KD cells were maintained in RPMI (Invitrogen) containing 10% heat-inactivated fetal bovine serum and 1 μg/ml puromycin. K562PU.1OE cells were grown in RPMI containing 10% heat-inactivated fetal bovine serum and 400 μg/ml neomycin. K562 cells were grown in RPMI containing 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cells were cultured in 5% CO2 at 37 °C in a humidified atmosphere.
RNA Preparation and Microarray Analysis
Total cellular RNA was isolated from parental, K562PU.1KD, and K562PU.1OE cells using an RNA Mini Purification Kit (Qiagen) according to the manufacturer's protocol. The samples were then subjected to a human 35K array analysis (Operon Biotechnologies, Huntsville, AL).
mRNA Expression Analyses
For RNA preparation for real-time PCR analyses, K562PU.1OE cells were seeded at a density of 2 × 105 cells/ml and treated with 5-aza-2′-deoxycytidine (5-azadC, Wako Pure Chemicals, Tokyo, Japan) at 0, 0.1, or 0.5 μm, sodium butyrate (SB, Sigma) at 0, 0.1, 0.5, or 1 mm or trichostatin A (TSA, Sigma) at 0, 0.1, 0.5, or 1 μm. The cells were harvested after 72 h for 5-azadC treatment and 12 h for SB and TSA treatments. cDNAs were prepared from the cells using reverse transcriptase (Superscript II, Invitrogen). Quantitative PCR was performed using the Quantitect SYBR Green PCR Reagent (Qiagen) according to the manufacturer's protocol and an Opticon Mini Real-time PCR Instrument (Bio-Rad, Hercules, CA) as previously described (19, 20). The sequences of the primers were: MT-1G: forward (5′-CTTCTCGCTTGGGAACTCTA-3′) and reverse (5′-AGGGGTCAAGATTGTAGCAAA-3′) (21); MT-1A: forward (5′-CTCGAAATGGACCCCAACT-3′) and reverse (5′-CAGGTTGTGCAGGTTGTTCTA-3′) (21); VIM: forward (5′-GGCTCAGATTCAGGGGAACAGC-3′) and reverse (5′-CAGGTTGTGCAGGTTGTTCTA-3′) (21); and JDP2: forward (5′-AAGAGGAGCGAAGGAAAAGG-3′) and reverse (5′-CTCTGCGTTCATGAGTTCCA-3′). Primers for PU.1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were employed as described previously (20). The thermal cycling conditions for PU.1 were: step 1, 95 °C for 15 min; step 2, 95 °C for 15 s; and step 3, 60 °C for 1 min. Steps 2–3 were repeated for 35 cycles. The conditions for the other genes were: step 1, 95 °C for 15 min; step 2, 95 °C for 30 s; step 3, 55 °C for 30 s; and step 4, 72 °C for 30 s. Steps 2–4 were repeated for 35 cycles. The copy number of each sample was calculated as previously described (22). Regarding patient samples, 43 specimens from AML patients in Tohoku University Hospital containing >60% blasts were subjected to density gradient centrifugation using Ficoll-Hypaque (Amersham Biosciences) and analyzed. The French-American-British classifications and numbers of the patients were as follows: AML-M0, 2; AML-M1, 9; AML-M2, 7; AML-M3, 12; AML-M4, 9; and AML-M5, 4. Informed consent was obtained from all patients. Ethical considerations according to the declaration of Helsinki were followed.
Plasmids
Human MT-1G, MT-1A, VIM, and ANXA1 promoters fused to a luciferase reporter gene were constructed. The constructs were first amplified with the following primers: MT-1G: forward (5′-AAGCCTCGAGGATTCACACCAAGCAGTGCC-3′) and reverse (5′-GACTAAGCTTCCCAAGCGAGAAGGGAAG-3′); MT-1A: forward (5′-TGCTCTCGAGTTTAAAGCAGGGTCAGCA-3′) and reverse (5′-CGGTAAGCTTGGAGATCCCAAGCAAGAA-3′); VIM: forward (5′- CAGGGGTACCCGGAGCCCGCTGAGACTTGA-3′) and reverse (5′-GCCCAAGCTTTGGGTGTGGGTGGTGGGAG-3′); and ANXA1: forward (5′-CAGGCTCGAGCAACAAGCTATATCTAGGAAC-3′) and reverse (5′-GCCCAAGCTTCTACCTTCTTGCAAAGAAC-3′). For the MT-1 and ANXA1 promoter constructs, the amplified DNA fragments were digested with XhoI and HindIII. The amplified VIM promoter fragments were digested with KpnI and HindIII. The digested fragments were inserted into the XhoI/HindIII and KpnI/ HindIII sites of the pGL3-Basic plasmid (Promega, Madison, WI), respectively. The DNA sequences were confirmed, and minor PCR errors were found in the VIM (−204 C/T mutated) and ANXA1 (−180 T/C mutated) promoters.
In Vitro Methylation and Luciferase Assay
The MT-1s, VIM, and ANXA1-PGL3 were in vitro-methylated by treatment with M. Sss1 methylase (New England Biolabs, County Road, MA) following the manufacturer's instructions. Aliquots (1 μg) of methylated or non-methylated promoter constructs together with Renilla vector PRL-TK (5 ng) were transfected into K562 cells using TransFectin (Bio-Rad), according to the manufacturer's instructions. The promoter activities were analyzed after 24 h using a dual reporter assay (Promega). The luciferase experiments were independently repeated six times, and their reproducibilities were confirmed.
ChIP Assay
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (23) with some modifications. Cells were cross-linked with 1% formaldehyde for 5 min at room temperature or 1 h at 4 °C, and the cross-linking reaction was stopped by incubation with 1.5 m glycine for 10 min at room temperature. The cells were then washed with ice-cold fluorescent-activated cell sorting solution (phosphate-buffered saline, 2% fetal bovine serum, 0.05% NaN3) and lysed on ice with lysis buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA pH 8.0, 1% SDS, Complete protease inhibitor mixture (Roche Applied Science)) for 20 min. The lysates were subjected to ultrasonic sonication and then centrifuged to remove cellular debris. The sonicated lysates were added to 9 volumes of ice-cold ChIP dilution buffer (50 mm Tris-HCl, pH 8.0, 167 mm NaCl, 1.1% Triton X-100, 0.11% sodium deoxycholate, Complete protease inhibitor mixture). For immunoprecipitation, the diluted lysates were incubated with 5 μg of rabbit polyclonal anti-acetyl-histone H3 (Upstate Biotechnology, Waltham, MA), rabbit antiserum anti-acetyl-histone H4 (Upstate Biotechnology), rabbit polyclonal anti-PU.1 (Santa Cruz Biotechnology), rabbit polyclonal anti-MeCP2 (Santa Cruz Biotechnology), or isotype control rabbit IgG (R&D Systems, Minneapolis, MN) antibodies for 2 h at 4 °C. Immunocomplexes were precipitated with 100 μl of Protein A-agarose beads (Roche Applied Science) for 16 h at 4 °C. After washing, the immunoprecipitates were eluted by incubation with ChIP direct elution buffer (10 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm EDTA, pH 8.0, 0.5% SDS). Proteins were removed by treatment with 30 μg of proteinase K at 37 °C overnight, followed by incubation at 65 °C for 6 h to reverse the cross-links. The samples were then incubated with 4 μg of RNase A for 1 h at 37 °C. The DNA was extracted with phenol-chloroform and precipitated with ethanol. After resuspension, the DNA was subjected to 35 cycles of PCR amplification. The ChIP products were quantified using various primer sets (supplemental Table 1) and real-time PCR as described above.
Bisulfite DNA Sequencing Analysis
Genomic DNAs were prepared from PU.1 transgenic cells using a Blood & Cell Culture DNA Mini Kit (Qiagen) according to the manufacturer's protocol. The isolated genomic DNAs were subjected to bisulfite DNA treatment using an EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA) according to the manufacturer's protocol. The resulting DNAs were subjected to PCR amplification. Two primer sets were employed to analyze each gene as follows: MT-1A: forward (5′-GGGATAGGAGTAGGAGGTTGTGGTTGTATT-3′) and reverse (5′-ACACCTCTACTCCTAAAAACATCTATCCTATA-3′); MT-1A': forward (5′-GGAGGATTTGGATAAATGTG-3′) and reverse (5′-CTAAATACAACCACCTC-3′); MT-1G: forward (5′-TTTGGTAGATTTAGAAAGTGGAGTATAAGA-3′) and reverse (5′-CTCAAACCCAAAAACACTCTCTATAATATC-3′); MT-1G′: forward (5′-GGAGGATTTGGATAAATGGG-3′) and reverse (5′-CTAAATACAACCACAACCTC-3′); VIM: forward (5′-AGGATAYGGATTTGGTGGATATGGTTG-3′) and reverse (5′-CAACACCCCAAAATAAACCCAACTCAAACT-3′); and VIM′: forward (5′-GTTAAGGTAAGTTGATGGATAG-3′) and reverse (5′-CTAAACTCACCCTAAAATAC-3′. The amplified fragments were purified by 2% agarose gel electrophoresis, recovered using a Wizard SV Gel and PCR Clean-up System (Promega) and cloned into a pGEM-T Easy vector (Promega). The M13R primer was used for sequencing with a Big Dye Sequencing Kit (Applied Biosystems, Foster City, CA). The combination of bisulfite treatment and PCR amplification resulted in the conversion of unmethylated cytosine (C) residues to thymine (T) residues, whereas methylated C residues remained unconverted. The products were analyzed using an ABI Prism DNA Sequencer 3130 (Applied Biosystems) and changes from C to T were detected.
Statistical Analysis
Correlations between two continuous variables were calculated by the Spearman rank correlation test. For all analyses, the p values were two-tailed, and values of p < 0.05 were considered statistically significant.
RESULTS
MT-1s, VIM, and JDP2 Are Bona Fide Targets of PU.1
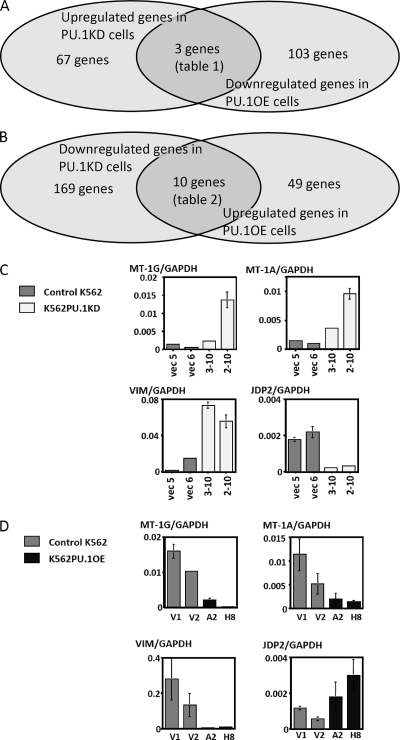
To identify the PU.1 target genes in leukemia cells, dual microarray analyses were carried out using Human 35-K arrays, in which 35,000 human genes were spotted. We employed K562PU.1KD cells (2-10 and 3-10) and K562PU.1OE cells (A2 and H8), which we recently established in our laboratory (6). In our previous study, the gene expression profiles were compared among K562PU.1KD 2-10 cells and their control vec6 cells. We identified ANXA1 as a target gene of PU.1 (6). In the present study, we performed further array analyses comparing K562PU.1OE cells (H8) and their control (V2) cells, and the data were analyzed in combination. In the comparison between K562PU.1KD and control cells, 70 genes were differentially expressed by ≥2-fold in PU.1KD cells, and 106 genes were differentially expressed by <0.5-fold in PU.1OE cells (Fig. 1A). Together with these genes, we found that three genes, including ANXA1 (6), were commonly affected, being induced by PU.1 down-regulation and suppressed by PU.1 up-regulation (Table 1). On the other hand, 179 genes were down-regulated in PU.1KD cells by ≥2-fold, and 59 genes were up-regulated by ≥2-fold in PU.1 OE cells (Fig. 1B). In these analyses, we identified 10 genes that were commonly affected by PU.1 (Table 2).
FIGURE 1.
MT-1s, VIM and JDP2 are targets of PU.1. A and B, results of the microarray analyses. C, expression levels of MT-1G, MT-1A, VIM, and JDP2 in K562PU.1KD cells evaluated by quantitative real-time PCR. Each gene transcript level was adjusted by the expression of GAPDH, and the relative levels are shown. D, mRNA expression levels of MT-1G, MT-1A, VIM, and JDP2 in K562PU.1OE cells evaluated by quantitative real-time PCR.
TABLE 1.
Commonly affected genes, induced by PU.1 down-regulation and suppressed by PU.1 up-regulation
| PU.1KD/control | PU.1OE/control | Gene |
|---|---|---|
| Fold intensity | Fold intensity | |
| 8.866838 | 0.324332 | Vimentin (VIM) |
| 5.078186 | 0.477423 | Metallothionein-1G (MT-1G) |
| 8.118739 | 0.191333 | Annexin Al (Annexinl) (ANXA1) |
TABLE 2.
Commonly affected genes, suppressed by PU.1 down-regulation and induced by PU.1 up-regulation
| PU.1KD/control | PU.1OE/control | Gene |
|---|---|---|
| Fold intensity | Fold intensity | |
| 0.461413 | 2.967024 | Jun dimerization protein (Homo sapiens). JDP2 |
| 0.191166 | 2.624883 | Ectoderm-neural cortex-1 protein (ENC-1) |
| 0.495526 | 2.313721 | Tryptophanyl-tRNA synthetase (EC 6.1.1.2) |
| 0.384764 | 2.31186 | Kynureninase (EC 3.7.1.3) (l-kynurenine hydrolase) |
| 0.479743 | 2.287885 | Tryptophanyl-tRNA synthetase (EC 6.1.1.2) |
| 0.424243 | 2.28255 | Kynureninase (EC 3.7.1.3) (l-kynurenine hydrolase) |
| 0.437378 | 2.247253 | Signal-regulatory protein α-1 |
| 0.174474 | 2.08398 | Neurotensin/neuromedin N precursor |
| 0.341625 | 2.073607 | Ras-related protein Rap-lb |
| 0.472486 | 2.007198 | 3′-Phosphoadenosine 5′-phosphosulfate synthetase 1 |
In the present study, we focused on the genes with fully characterized functions that are known to play roles in hematopoiesis and oncogenesis to clarify the roles of PU.1 in these processes. On these bases, we selected MT-1G (7), VIM (16), and JDP2 (24). Induction of VIM (Fig. 1C, lower left) and MT-1G (Fig. 1C, upper left) and suppression of JDP2 (Fig. 1C, lower right) were observed in both K562PU.1KD clones (2-10 and 3-10), suggesting that these genes are candidates for PU.1 target genes. Because MTs are encoded by a family of genes containing several isoforms in humans (7), we analyzed the expression changes of MT isoforms using this array. We found that the expression levels of all the functional MT isoforms expressed in humans (MT-1A, -B, -E, -F, -G, -H, and -X and MT-2) were increased by varying degrees in 2-10 cells (Table 3). Although the expression change was slightly lower than the experimental value (≥2-fold), we found that MT-1A was also significantly increased (1.922954) among these isoforms. These expression changes were confirmed by real-time PCR (Fig. 1C, upper right).
TABLE 3.
Expressions of metallothionein isoforms
| GenBankTM Acc. No. | 2-10 | vec6 | Relative exp. | |
|---|---|---|---|---|
| Signal intensity | Signal intensity | Fold intensity | ||
| NM_005946 | Metallothionein-IA (MT-1A) | 624.28571 | 324.64935 | 1.922954 |
| NM_005947 | Metallothionein-IB (MT-1B) | 1214.00793 | 1013.28993 | 1.198085 |
| NM_175617 | Metallothionein-IE (MT-1E) | 392.28306 | 221.43551 | 1.771545 |
| NM_005949 | Metallothionein-IF (MT-1F) | 760.57936 | 637.3582 | 1.193331 |
| NM_005950 | Metallothionein-IG (MT-1G) | 1634.8941 | 321.94452 | 5.078186 |
| NM_175622 | Metallothionein-IH (MT-1H) | 2237.67195 | 2063.2957 | 1.0845 |
| NM_005952 | Metallothionein-IX (MT-1X) | 1620.31481 | 1426.83900 | 1.135598 |
| NM_005953 | Metallothionein-IIA (MT-2A) | 826.55820 | 751.12693 | 1.100424 |
Next, we checked the expression levels of the MT-1, VIM, and JDP2 genes in PU.1OE cells. We found that MT-1G, MT-1A, and VIM were suppressed, whereas JDP2 was induced in proportion with the PU.1 expression (Fig. 1D), suggesting that these genes are bona fide targets of PU.1.
MT-1, VIM, JDP2, and PU.1 Transcript Levels in AML Patients
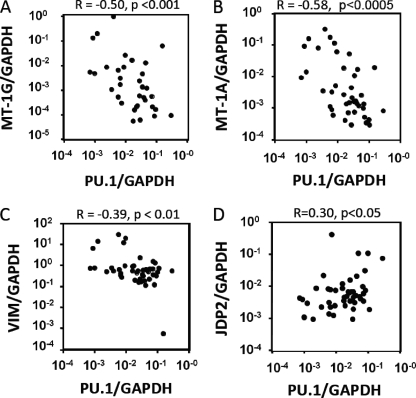
To address the potential relevance of our findings for human AML, we analyzed the gene expression levels in leukemic bone marrow cells from 43 individuals with AML. As expected, we detected negative correlations between the PU.1 mRNA expression level and the mRNA expression levels of the MT-1s and VIM in all 43 AML specimens (MT-1G: R = −0.50, p < 0.001, Fig. 2A; MT-1A: R = −0.58, p < 0.0005, Fig. 2B; VIM: R = −0.39, p < 0.01, Fig. 2C). In addition, there was a modest, but significant, positive correlation between JDP2 and PU.1 (R = 0.30, p < 0.05, Fig. 2D). These data suggest that the correlations of PU.1 with MT-1s, VIM, and JDP2 identified in the PU.1 transgenic cells play roles in human AML. Diminished PU.1 expression may contribute to the aberrant induction or suppression of these target genes in individuals with hematological malignancies.
FIGURE 2.
MT-1, VIM, JDP2, and PU.1 transcript levels in AML patients. A–D, correlations of PU.1 expression with the MT-1G (A), MT-1A (B), VIM (C), and JDP2 (D) expressions in 43 AML patients. Each dot represents the average target gene/GAPDH copy number ratio from two to three PCR amplifications. The correlation coefficient and p value are indicated at the top of each panel.
Changes in the Expressions of MT-1s, VIM, and ANXA1 Induced by the Dnmt Inhibitor 5-azadC and HDAC Inhibitors SB and TSA
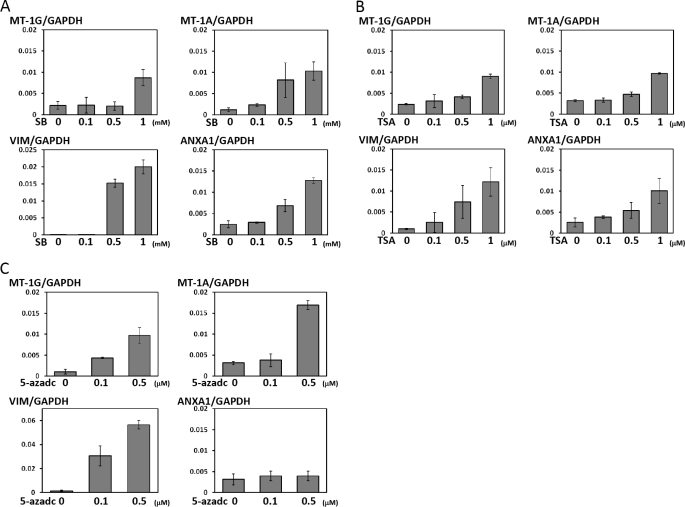
Next, we focused on how these induced genes are regulated by PU.1. Although PU.1 functions as a transcriptional activator (25), it also forms a complex with HDAC and Dnmt, and functions as a transcriptional repressor (9, 10, 26). In this study, we focused on the genes suppressed by PU.1 expression. We hypothesized that MT-1s and VIM may be epigenetically silenced by PU.1. In addition to these genes, we examined the regulation of the ANXA1 gene, because we recently identified an inverse correlation between its expression and PU.1 expression (6). To clarify the roles of the Dnmt and HDAC activities in the regulation of these genes, we treated K562PU.1OE cells with the Dnmt inhibitor 5-azadC and HDAC inhibitors SB and TSA and examined the changes in the gene expression levels. As shown in Fig. 3, MT-1 and VIM expressions were induced by the addition of these HDAC and Dnmt inhibitors (Fig. 3). In contrast, ANXA1 expression was induced by SB and TSA, but not by 5-azadC. These observations suggest that these genes are regulated, at least in part, by these epigenetic activities. The absence of ANXA1 gene induction by 5-azadC may indicate a difference in the regulation of this promoter.
FIGURE 3.
Changes in the expressions of MT-1s, VIM, and ANXA1 by treatment with HDAC and Dnmt inhibitors. A–C, K562PU.1OE cells were treated at indicated doses with SB (A) or TSA (B) for 12 h or 5-azadC for 72 h (C). The changes in the MT-1G, MT-1A, VIM, and ANXA1 expressions were evaluated by quantitative real-time PCR. Each gene transcript level was adjusted by the expression of GAPDH, and the relative levels are shown.
DNA Methylation Suppresses MT-1, VIM, and ANXA1 Promoter Activities in Vitro
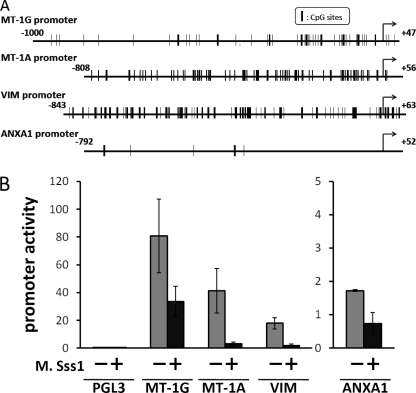
Next, we examined whether these promoter activities were affected by DNA methylation. The promoter region of the human MT-1G gene has been characterized and found to be GC-rich (70%) (27). This region has relatively high homology (67%) to the MT-1A promoter, suggesting that common regulatory mechanisms may underlie the regulations of these genes. The VIM promoter also has a high GC content (15, 17). Therefore, these promoters may be potential targets for DNA methylation. In contrast, CpG sites were relatively scarce in the ANXA1 promoter (Fig. 4A). Hence, we examined the regulation of these genes by DNA methylation. To analyze the effects of DNA methylation on the promoter activities, we generated and employed MT-1G (−1000 to +47), MT-1A (−808 to +56), VIM (−843 to +63), and ANXA1 (−792 to +52) promoter luciferase constructs. These constructs were treated with M. Sss1 methylase to determine whether these promoters are regulated by DNA methylation. As shown in Fig. 4B, the promoter activities were significantly suppressed by treatment with this enzyme. Although this was an in vitro assay, these findings indicate that the regions depicted in Fig. 4A confer promoter activity and are subject to DNA methylation.
FIGURE 4.
Suppression of the MT-1, VIM, and ANXA1 promoters by DNA methylation. A, schematic representations of the genomic structures of the 5′ regions of the MT-1G, MT-1A, VIM, and ANXA1 genes. The arrow indicates the transcription start site. The regions corresponding to −1000 to +47 for the MT-1G promoter, −808 to +56 for the MT-1A promoter, −843 to +63 for the VIM promoter, and −792 to +52 for the ANXA1 gene promoter were amplified and fused to a luciferase reporter gene for further analyses. B, the MT-1G, MT-1A, VIM, and ANXA1 luciferase reporter constructs and empty vector (PGL3) were transfected into K562 cells. The constructs were incubated with or without M. Sss1 methylase. The results are indicated as ratios of the luciferase activity to the Renilla luciferase activity. The data presented were calculated from six independent experiments.
Histone Modifications in the MT-1, VIM, and ANXA1 Promoters Induced by PU.1 Down-regulation
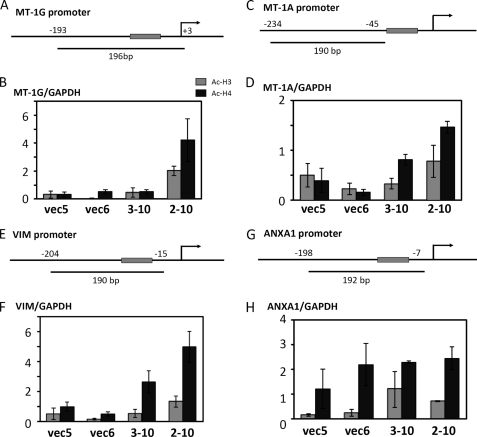
To examine whether histone modifications in the MT-1, VIM, and ANXA1 promoters are correlated with changes in PU.1 expression, quantitative ChIP assays were performed using polyclonal antibodies against acetylated H3 and acetylated H4. The indicated regions of the MT-1, VIM, and ANXA1 promoters were amplified by ChIP assays (Fig. 5, A, C, E, and G). In K562PU.1KD cells (2-10 and 3-10), the histone H4 acetylation levels were clearly increased in the MT-1 and VIM promoters, but not in the ANXA1 promoter. In addition, acetylated H3 showed a tendency to be increased in these promoters in K562PU.1KD cells (Fig. 5, B, D, F, and H). These findings suggest that the transcription levels of the MT-1, VIM, and ANXA1 genes are regulated by histone acetylation in their promoters. In addition, the histone modification of the ANXA1 gene promoter by PU.1 is different from that of the other three promoters.
FIGURE 5.
Analyses of acetylated histones by ChIP assays. A–H, the indicated regions of the MT-1G (A), MT-1A (C), VIM (E), and ANXA1 (G) promoters were amplified by ChIP assays. A TATA (-like) box is shown in the shaded square. ChIP assays were performed with the indicated K562PU.1KD cells and their control cells. Chromatin modification by histone H3 and H4 acetylations was analyzed. The results for the MT-1G (B), MT-1A (D), VIM (F), and ANXA1 (H) promoters are shown. Antibodies against acetylated histone H3 (Ac-H3) or acetylated histone H4 (Ac-H4) were used. The ChIP products were quantified by PCR. The y-axis shows the relative expression calculated from the copy numbers divided by the copy numbers of GAPDH. The results shown were calculated from three independent experiments.
PU.1 Directly Binds to the MT-1 and VIM Promoters in Concert with MeCP2
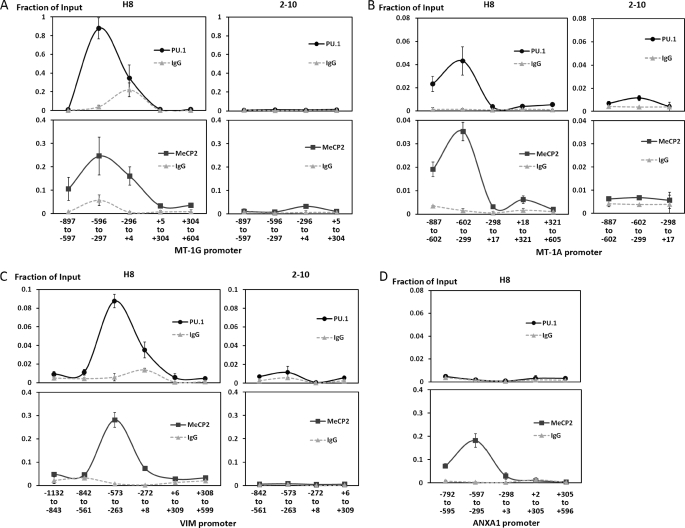
PU.1 forms a complex with MeCP2, which recruits mSin3A-HDAC and Dnmt (10). MeCP2 is known to induce transcriptional repression of methylated CpG promoters by recruiting Dnmts and HDACs (28). Therefore, the recruitments of PU.1 and MeCP2 to these promoters were examined. ChIP assays using K562PU.1OE cells (H8) revealed that PU.1 primarily bound to the −596 to −297 region of the MT-1G promoter (Fig. 6A, upper left panel). MeCP2 was observed to overlap with the PU.1 occupancy (Fig. 6A, lower left panel). In the MT-1A and VIM promoters, PU.1 and MeCP2 were also observed to bind to overlapping regions (Fig. 6, B and C, left). In contrast, using K562PU.1KD cells (2-10), the recruitment of PU.1, as well as MeCP2, to these promoters was obviously diminished by the down-regulation of PU.1. Additionally, we were unable to detect any recruitment of PU.1 to the ANXA1 promoter (Fig. 6D, upper panel), although MeCP2 primarily bound to the −597 to −295 region of this promoter (Fig. 6D, lower panel). Taken together, these findings demonstrate that PU.1 directly binds to the MT-1 and VIM promoters in concert with MeCP2 and epigenetically represses the expressions of these genes.
FIGURE 6.
Analyses of PU.1 and MeCP2 recruitments to the MT-1, VIM, and ANXA1 promoters by ChIP assays. A–D, ChIP assays of K562PU.1OE (H8) cells (A–D), and K562PU.1KD (2-10) cells (A–C) were performed with antibodies against PU.1 and MeCP2 or the IgG isotype control. The ChIP products were quantified by PCR using primers flanking the MT-1G (A), MT-1A (B), VIM (C), and ANXA1 (D) promoter regions as indicated. The y-axis shows the relative expression calculated from the copy numbers divided by the input copy numbers. The results shown are the means of three independent experiments.
Proportions of Methylated CpG Sites in the MT-1 and VIM Promoters Are Tightly Correlated with the PU.1 Expression Levels
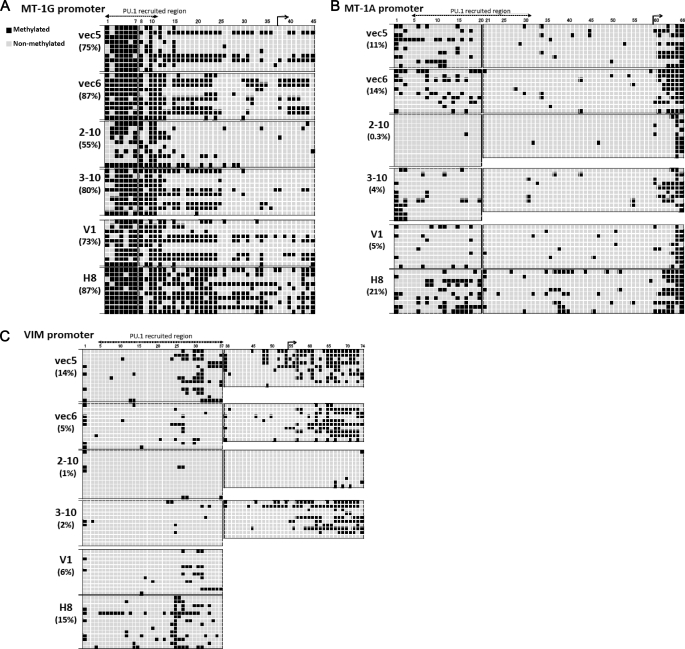
We observed that MT-1s and VIM were direct target genes of PU.1. Therefore, we determined the methylation statuses of the PU.1- and MeCP2-recruited regions in these promoters by bisulfite DNA sequencing analyses. As shown in Fig. 7A, the proportion of methylated CpG islands in the PU.1-recruited region in the MT-1G promoter was significantly reduced in K562PU.1KD cells, most prominently in 2-10 cells (∼55%), compared with control cells (75–87%). In addition, the methylated CpG sites in K562PU.1OE cells were modestly increased (87%) compared with their control cells (73%) (Fig. 7A). The methylation status of the MT-1A promoter was also correlated with PU.1 expression, ranging from 0.3% (2-10 cells) to 14% (vec6 cells) and from 21% (H8 cells) to 5% (V1 cells) (Fig. 7B). Furthermore, the proportion of methylated CpG islands in the VIM promoter was also correlated with the expression of PU.1, ranging from ∼1% (2-10 cells) to 14% (vec5 cells) and from 15% (H8 cells) to 6% (V1 cells) (Fig. 7C). These findings suggest that MT-1s and VIM are regulated by promoter methylation associated with PU.1 expression. Taken together, these findings demonstrate that PU.1 directly binds to the MT-1 and VIM promoters in concert with MeCP2 and epigenetically represses the expression of these genes in leukemia cells.
FIGURE 7.
The proportions of methylated CpG sites in the MT-1 and VIM promoters are tightly correlated with the PU.1 expression levels. A–C, results of bisulfite DNA sequencing analyses of the MT-1G (−637 to +170, 45 CpG sites) (A), MT-1A (−678 to +175, 66 CpG sites) (B), and VIM (−610 to +164, 74 CpG sites) (C) promoters. Genomic DNA extracts from the indicated cells were treated with sodium bisulfite and amplified with specific primer sets. The PCR products were cloned, and individual clones were randomly selected for DNA sequencing. The clone numbers are shown in the left column, and the CpG sites are numbered across the top. An arrow indicates the location of the transcription start site in each gene. Filled squares represent methylated CpGs, and open squares represent unmethylated CpGs. The percentages of methylated CpGs in the PU.1-recruited regions are shown in parentheses.
DISCUSSION
MT was reported to be a potential negative regulator of apoptosis (29). Strong linear negative correlations were found between the basal MT levels and etoposide-induced apoptosis in the human tumor cell lines PLC/PRF5/5, H460, and HepG2 (29). These findings suggest that MT may play roles in carcinogenesis and drug resistance, in at least a portion of cancer cells. Compared with analyses of solid tumors, few studies have analyzed the roles of MT in hematological malignancies. Sauerbrey et al. (30) investigated the expression levels of the resistance-related proteins Pgp, GST-pi, topoisomerase-II, and MT in leukemic cells from 19 children with newly diagnosed acute non-lymphoblastic leukemia. Although the number of patients was small, they found that MT was expressed in leukemic cells isolated from 68% of these newly diagnosed acute non-lymphoblastic leukemia cases. Patients who developed relapses showed a poorer prognosis and frequently expressed more than two resistance-related proteins, including MT, compared with patients who remained in remission (30). In children with acute leukemia at diagnosis and during treatment, in vivo expression of MT by tumor cells constituted a cellular protective mechanism that prevented chemotherapy-induced apoptosis (30). Of note, it was recently reported that, in 115 diffuse large B-cell lymphoma patients, MT-1 labeling of >20% of the lymphoma cells was associated with a significantly poorer 5-year survival rate, independent of age, stage, or international prognostic index (31). We speculate that down-regulation of PU.1, which leads to overexpression of MT-1s, may play a role in the drug resistance of AML cells. Indeed, a previous study showed that MT can inhibit anthracycline-induced mitochondrial cytochrome c release and caspase-3 activation (32). Together with the present findings, further analyses of the expression levels of MT-1s in hematological malignancies are desired. Studies to clarify the resistance mechanisms of K562PU.1KD cells toward anticancer drugs are underway in our laboratory.
Caspase-induced cleavage of the cytoskeletal network has been demonstrated and, in particular, VIM is degraded in response to various inducers of apoptosis (33). In Jurkat cells transfected with full-length VIM, apoptosis was partly suppressed by photodynamic treatment with the silicon phthalocyanine Pc4 (33). Therefore, in addition to functions in growth regulation (13, 14), induced expression of VIM, as well as MT, may function in anti-apoptosis mechanisms in leukemia cells. Consequently, it is possible that up-regulated levels of MT and/or VIM mRNAs may be useful for identifying particular subgroups of leukemia patients who could benefit from more intensive therapies.
In follicular lymphoma, strong PU.1 expression, together with CD20 and CD75 expressions, showed significant associations with longer progression-free survival and overall survival (34). We recently reported that, in 24 AML cases, PU.1 expression was inversely correlated with Flt3 expression (20), whereas strong expression of Flt3 was an unfavorable prognostic factor for overall survival in 91 AML cases without Flt3-ITD mutations (35). Together with the present findings, decreased PU.1 expression may represent a poor prognostic marker for AML or other types of leukemia and hematopoietic malignancies.
JDP2 was identified as a c-Jun-interacting protein (36). JDP2 also interacts with other Jun family transcription factors, JunB and JunD (36). In addition, JDP2 inhibits cell transformation and acts as a tumor suppressor (24). In analyses of samples from primary AML patients, Steidl et al. (37) demonstrated a significant positive correlation between PU.1 expression and JunB expression, but not c-Jun expression. Because we detected significant correlations between PU.1 and JDP2 in our AML specimens, and the role of c-Jun in the pathogenesis of AML is well documented (37), we speculate that JDP2 may serve as a cofactor for c-Jun function in the pathogenesis of PU.1 knockdown-induced AML. Taken together, the suppression of the tumor suppressor JDP2 with c-Jun and/or JunB mediated by PU.1 down-regulation may play a role in leukemogenesis. The current report may complement previous studies (3, 37) regarding the roles of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells.
We have clearly demonstrated that PU.1 binds to the MT-1 and VIM promoters in concert with MeCP2. Although MeCP2 was also observed to bind to a region of the ANXA1 promoter, we found that the proximal region of this promoter was not affected by PU.1. In addition, we found that JDP2 expression was positively correlated with PU.1. Intriguingly, based on a Transcriptional Factor Search (available on-line), there are three putative AP-1-related JDP2-binding sites in the −950 to −430 region in the ANXA1 promoter. JDP2 is a transcriptional repressor that recruits HDAC complexes and leads to changes in the histone acetylation statuses of the promoter regions of its target genes (38). Although the methylation activity of JDP2 has not yet been reported, we speculate that the down-regulation of JDP2, or another transcriptional repressor related to PU.1, may be responsible for the induction of the ANXA1 gene.
In summary, we have identified MT-1s and VIM as direct target genes for PU.1 suppression in leukemia cells. Although aberrant DNA methylation and acetylation of histones are believed to be important for the pathogenesis of leukemia, this study and others (39, 40) have indicated that insufficient recruitments of these epigenetic factors also play roles in leukemia pathogenesis. This notion may represent an essential insight toward understanding of the mechanisms of hematopoietic malignancies.
Supplementary Material
Acknowledgment
We thank all the staff and students in the Division of Hematology for help with this study.
This work was supported in part by grants from Nakayama Foundation for Human Science, the Foundation from the Kitasato University School of Allied Health Sciences (Grants-in-Aid for Research Project, 2007-1003, 2008-1001, and 2009-1001), and the Foundation from the Kitasato University Graduate School of Medical Sciences Research Project.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- AML
- acute myeloid leukemia
- MT
- metallothionein
- VIM
- vimentin
- ANXA1
- annexin 1
- JDP2
- Jun dimerization protein 2
- 5-azadC
- 5-aza-2′-deoxycytidine
- SB
- sodium butyrate
- TSA
- trichostatin A
- ChIP
- chromatin immunoprecipitation
- MeCP2
- methyl-CpG-binding protein 2
- HDAC
- histone deacetylase
- Dnmt
- DNA methyltransferase
- ANLL
- acute non-lymphoblastic leukemia
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Chen H. M., Zhang P., Voso M. T., Hohaus S., Gonzalez D. A., Glass C. K., Zhang D. E., Tenen D. G. (1995) Blood 85, 2918–2928 [PubMed] [Google Scholar]
- 2.Scott E. W., Simon M. C., Anastasi J., Singh H. (1994) Science 265, 1573–1577 [DOI] [PubMed] [Google Scholar]
- 3.Rosenbauer F., Wagner K., Kutok J. L., Iwasaki H., Le Beau M. M., Okuno Y., Akashi K., Fiering S., Tenen D. G. (2004) Nat. Genet. 36, 624–630 [DOI] [PubMed] [Google Scholar]
- 4.Pettersson M., Sundström C., Nilsson K., Larsson L. G. (1995) Blood 86, 2747–2753 [PubMed] [Google Scholar]
- 5.Huh H. J., Chae S. L., Lee M., Hong K. S., Mun Y. C., Seong C. M., Chung W. S., Huh J. W. (2009) Int. J. Lab. Hematol. 31, 344–351 [DOI] [PubMed] [Google Scholar]
- 6.Iseki Y., Imoto A., Okazaki T., Harigae H., Takahashi S. (2009) Leuk. Res. 33, 1658–1663 [DOI] [PubMed] [Google Scholar]
- 7.Cherian M. G., Jayasurya A., Bay B. H. (2003) Mutat. Res. 533, 201–209 [DOI] [PubMed] [Google Scholar]
- 8.Ghoshal K., Datta J., Majumder S., Bai S., Dong X., Parthun M., Jacob S. T. (2002) Mol. Cell. Biol. 22, 8302–8319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kihara-Negishi F., Yamamoto H., Suzuki M., Yamada T., Sakurai T., Tamura T., Oikawa T. (2001) Oncogene 20, 6039–6047 [DOI] [PubMed] [Google Scholar]
- 10.Suzuki M., Yamada T., Kihara-Negishi F., Sakurai T., Oikawa T. (2003) Oncogene 22, 8688–8698 [DOI] [PubMed] [Google Scholar]
- 11.Suzuki M., Yamada T., Kihara-Negishi F., Sakurai T., Hara E., Tenen D. G., Hozumi N., Oikawa T. (2006) Oncogene 25, 2477–2488 [DOI] [PubMed] [Google Scholar]
- 12.Lazarides E. (1982) Annu. Rev. Biochem. 51, 219–250 [DOI] [PubMed] [Google Scholar]
- 13.Ferrier A. F., Hirschhorn R. R. (1992) J. Cell. Biochem. 50, 245–254 [DOI] [PubMed] [Google Scholar]
- 14.Kaczmarek L., Calabretta B., Elfenbein I. B., Mercer W. E. (1987) Exp. Cell Res. 173, 70–79 [DOI] [PubMed] [Google Scholar]
- 15.Rittling S. R., Baserga R. (1987) Mol. Cell. Biol. 7, 3908–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari S., Battini R., Kaczmarek L., Rittling S., Calabretta B., de Riel J. K., Philiponis V., Wei J. F., Baserga R. (1986) Mol. Cell. Biol. 6, 3614–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y., Zhang X., Salmon M., Zehner Z. E. (2007) Genes Cells 12, 905–918 [DOI] [PubMed] [Google Scholar]
- 18.Zou H., Harrington J. J., Shire A. M., Rego R. L., Wang L., Campbell M. E., Oberg A. L., Ahlquist D. A. (2007) Cancer Epidemiol. Biomarkers Prev. 16, 2686–2696 [DOI] [PubMed] [Google Scholar]
- 19.Takahashi S., Harigae H., Kameoka J., Sasaki T., Kaku M. (2005) Br. J. Haematol. 130, 428–436 [DOI] [PubMed] [Google Scholar]
- 20.Inomata M., Takahashi S., Harigae H., Kameoka J., Kaku M., Sasaki T. (2006) Leuk. Res. 30, 659–664 [DOI] [PubMed] [Google Scholar]
- 21.Mididoddi S., McGuirt J. P., Sens M. A., Todd J. H., Sens D. A. (1996) Toxicol. Lett. 85, 17–27 [DOI] [PubMed] [Google Scholar]
- 22.Takahashi S., Harigae H., Ishii K. K., Inomata M., Fujiwara T., Yokoyama H., Ishizawa K., Kameoka J., Licht J. D., Sasaki T., Kaku M. (2005) Leuk. Res. 29, 893–899 [DOI] [PubMed] [Google Scholar]
- 23.Ye S. K., Agata Y., Lee H. C., Kurooka H., Kitamura T., Shimizu A., Honjo T., Ikuta K. (2001) Immunity 15, 813–823 [DOI] [PubMed] [Google Scholar]
- 24.Heinrich R., Livne E., Ben-Izhak O., Aronheim A. (2004) J. Biol. Chem. 279, 5708–5715 [DOI] [PubMed] [Google Scholar]
- 25.Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A. (1990) Cell 61, 113–124 [DOI] [PubMed] [Google Scholar]
- 26.Sugimoto T., Tomita A., Hiraga J., Shimada K., Kiyoi H., Kinoshita T., Naoe T. (2009) Biochem. Biophys. Res. Commun. 390, 48–53 [DOI] [PubMed] [Google Scholar]
- 27.Foster R., Jahroudi N., Varshney U., Gedamu L. (1988) J. Biol. Chem. 263, 11528–11535 [PubMed] [Google Scholar]
- 28.Hendrich B., Bird A. (1998) Mol. Cell. Biol. 18, 6538–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimoda R., Achanzar W. E., Qu W., Nagamine T., Takagi H., Mori M., Waalkes M. P. (2003) Toxicol. Sci. 73, 294–300 [DOI] [PubMed] [Google Scholar]
- 30.Sauerbrey A., Zintl F., Hermann J., Volm M. (1998) Anticancer Res. 18, 1231–1236 [PubMed] [Google Scholar]
- 31.Poulsen C. B., Borup R., Borregaard N., Nielsen F. C., Møller M. B., Ralfkiaer E. (2006) Blood 108, 3514–3519 [DOI] [PubMed] [Google Scholar]
- 32.Wang G. W., Klein J. B., Kang Y. J. (2001) J. Pharmacol. Exp. Ther. 298, 461–468 [PubMed] [Google Scholar]
- 33.Belichenko I., Morishima N., Separovic D. (2001) Arch. Biochem. Biophys. 390, 57–63 [DOI] [PubMed] [Google Scholar]
- 34.Torlakovic E. E., Bilalovic N., Golouh R., Zidar A., Angel S. (2006) J. Pathol. 209, 352–359 [DOI] [PubMed] [Google Scholar]
- 35.Ozeki K., Kiyoi H., Hirose Y., Iwai M., Ninomiya M., Kodera Y., Miyawaki S., Kuriyama K., Shimazaki C., Akiyama H., Nishimura M., Motoji T., Shinagawa K., Takeshita A., Ueda R., Ohno R., Emi N., Naoe T. (2004) Blood 103, 1901–1908 [DOI] [PubMed] [Google Scholar]
- 36.Aronheim A., Zandi E., Hennemann H., Elledge S. J., Karin M. (1997) Mol. Cell. Biol. 17, 3094–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steidl U., Rosenbauer F., Verhaak R. G., Gu X., Ebralidze A., Otu H. H., Klippel S., Steidl C., Bruns I., Costa D. B., Wagner K., Aivado M., Kobbe G., Valk P. J., Passegué E., Libermann T. A., Delwel R., Tenen D. G. (2006) Nat. Genet. 38, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 38.Jin C., Li H., Murata T., Sun K., Horikoshi M., Chiu R., Yokoyama K. K. (2002) Mol. Cell. Biol. 22, 4815–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan M., Burel S. A., Peterson L. F., Kanbe E., Iwasaki H., Boyapati A., Hines R., Akashi K., Zhang D. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17186–17191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi S., McConnell M. J., Harigae H., Kaku M., Sasaki T., Melnick A. M., Licht J. D. (2004) Blood 103, 4650–4658 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.