Abstract
The myosin cross-reactive antigen (MCRA) protein family is highly conserved among different bacterial species ranging from Gram-positive to Gram-negative bacteria. Besides their ubiquitous occurrence, knowledge about the biochemical and physiological function of MCRA proteins is scarce. Here, we show that MCRA protein from Streptococcus pyogenes M49 is a FAD enzyme, which acts as hydratase on (9Z)- and (12Z)-double bonds of C-16, C-18 non-esterified fatty acids. Products are 10-hydroxy and 10,13-dihydroxy fatty acids. Kinetic analysis suggests that FAD rather stabilizes the active conformation of the enzyme and is not directly involved in catalysis. Analysis of S. pyogenes M49 grown in the presence of either oleic or linoleic acid showed that 10-hydroxy and 10,13-dihydroxy derivatives were the only products. No further metabolism of these hydroxy fatty acids was detected. Deletion of the hydratase gene caused a 2-fold decrease in minimum inhibitory concentration against oleic acid but increased survival of the mutant strain in whole blood. Adherence and internalization properties to human keratinocytes were reduced in comparison with the wild type. Based on these results, we conclude that the previously identified MCRA protein can be classified as a FAD-containing double bond hydratase, within the carbon-oxygen lyase family, that plays a role in virulence of at least S. pyogenes M49.
Keywords: Enzymes/Flavin, Enzymes/Lipid, Gene/Knockout, Lipid, Lipid/Fatty Acid, Lipid/Oxidation, Organisms/Bacteria
Introduction
In 1994, the first member of the MCRA5 protein family was identified in Streptococcus pyogenes as the result of a screening for antigens recognized by acute rheumatic fever sera. Its amino acid sequence did not exhibit significant similarity to any streptococcal protein with a known function but was conserved among pathogenic groups A, C, and G of Streptococci (1).
A BLAST search with the MCRA protein sequence reveals more than 148 conserved sequences across different Gram-positive and Gram-negative bacteria (supplemental Fig. S1). MCRA genes are not a part of any bacterial operon. Despite such conservation level only for two members of the family so far, biochemical features have been assigned: MCRA from Lactobacillus reuteri PYR8 was suggested to be a (9Z,11E)-conjugated linoleic acid (CLA)-forming isomerase (2), and only recently hydratase activity was shown for MCRA of Pseudomonas sp. strain 3266 (3).
To analyze biochemical properties and physiological functions of MCRA and their more ubiquitous activity as fatty acid hydratase, we have chosen S. pyogenes M49 as a representative of the group A streptococci (GAS). GAS species exclusively colonize humans and cause a wide range of primary infections of the skin, throat, and other mucosal surfaces, including pharyngitis and impetigo (4), and hence have a vast medical importance. We show that S. pyogenes MCRA is a FAD-containing hydratase that adds water to (9Z)- and (12Z)-double bonds of C-16 and C-18 fatty acids. Using a gene deletion strain, we show that the hydroxylated fatty acids are not further metabolized. Importantly, the mutant strain showed alteration in virulence properties, such as blood survival and adherence and internalization to human keratinocytes.
EXPERIMENTAL PROCEDURES
Materials
Chemicals were obtained from Sigma and Carl Roth & Co. (Karlsruhe, Germany). Agarose was from Biozym Scientific GmbH (Hess. Oldendorf, Germany). All fatty acids were purchased from Sigma or Cayman Chemical (Ann Arbor, MI). Acetonitrile was from Fisher. Restriction enzymes were provided by MBI Fermentas (St. Leon-Roth, Germany).
Bacterial Strains and Culture Conditions
S. pyogenes (GAS) serotype M49 strain 591 is a skin isolate originally obtained from R. Lutticken (Aachen, Germany). The Escherichia coli strains DH5α, XL Blue and BL21 Star were purchased from Invitrogen and served as a host for the plasmids. E. coli strains DH5α and XL Blue BL21 were used for DNA cloning purposes, and BL21 Star was used for the protein overproduction. The GAS wild type strain and mutant derivates were cultured in Todd-Hewitt broth or on Todd-Hewitt broth-agar (Oxoid-Unipath, Wesel, Germany), both supplemented with 0.5% yeast extract (THY). GAS mutants harboring recombinant pFW11 plasmid (5) were maintained in medium containing 100 μg/ml spectinomycin except when being used for functional analyses. GAS strains were grown as standing cultures at a temperature of 37 °C under a 5% CO2-20% O2 atmosphere unless otherwise indicated. All E. coli cultures were grown at 37 °C with agitation at 180 rpm except when used for expression of recombinant proteins.
Nucleic Acid Techniques
Chromosomal and plasmid DNA preparations, genetic manipulations, and other conventional DNA techniques, including electroporation of GAS and E. coli strains, were done as described previously (6).
Construction of Recombinant Vectors and S. pyogenes M49 Strains
To perform the deletion knock-out of sph, the upstream and downstream fragments of the gene (GenBankTM accession number ACI60731.1) were PCR-amplified using the following primer pairs: upstream fragment (5′-TTTAGTGTCGACCGTCCAGGAGATC-3′ (forward primer) and 5′-TTCAAGGATCCCTTGAATAGTCAAGCAAAG-3′ (reverse primer) containing SalI and BamHI recognition sites, respectively) and the downstream fragment (5′-CATGGCATGCAGTAGTTAAAGCTGATAAG-3′ (forward primer) and 5′-CTGCAGCCCGGGACCAAAAACCA-3′ (reverse primer) containing SphI and XmaI recognition sites, respectively). The PCRs were performed with the PhusionTM hot start high fidelity DNA polymerase system (Finnzymes, Espoo, Finland) according to the manufacturer's protocol. For expression in E. coli, the sph open reading frame was amplified with the primers 5′-AAAGCTAGCATGTATTATACTAGTGGTAATTACG-3′ (forward) and 5′-AAAGCGGCCGCTTACATAAGATTAGCATCTTTGAGC-3′ (reverse), containing NheI and NotI recognition sites, respectively, and cloned into the pET24a and pET28a expression vectors (Novagen), respectively, yielding the plasmid pET24a-SPH (non-tagged version) and pET28a-SPH (N-His-tagged version).
Reverse Transcription-PCR
To analyze sph expression during the exponential growth phase of wild type and the mutant, total mRNA was isolated by the FastRNA® Pro Blue Kit (MP Biomedicals) according to the manufacturer's protocol. RNA concentration and the quality were measured spectrophotometrically by Picodrop (Picodrop). Reverse transcription reaction was performed by the SuperScript® III first strand synthesis system (Invitrogen) according to the manufacturer's protocol; genomic DNA contamination was controlled by PCR with the upstream region primers, which were used for generation of the deletion knock-out (see above); and gyrA was used as the internal control (forward primer, 5′-CGACTTGTCTGAACGCCAAA-3′; reverse primer, 5′-TTATCACGTTCCAAACCAGTCAA-3′). The PCR reactions were performed with the PhusionTM hot start high fidelity DNA polymerase system (Finnzymes) according to the manufacturer's protocol.
Overproduction of the Recombinant Protein
E. coli BL21 Star strain was transformed with pET24a-SPH or pET28a-SPH, and the bacteria was cultivated in 2× YT medium (30 g of Tryptone, 15 g of yeast extract, 5 g of NaCl per 1000 ml of H2O) supplied with 50 μg/ml kanamycin at 37 °C until A600 ∼0.6–0.8. Then isopropyl 1-thio-β-d-galactopyranoside was added to 0.1 mm, and the cells were shifted to 16 °C and harvested by centrifugation (10 min at 9100 × g) after an 18-h induction time.
Protein Purification
The non-tagged protein was purified as described below, and all steps were done at 4 °C. Cells were lysed by adding 0.1 mg/ml lysozyme and 1 μg/ml DNase I in 0.1 m Tris, pH 7.5, 0.05 m NaCl, 10 mm MgCl2 and centrifuged for 20 min at 70,000 × g. The supernatant was loaded on pre-equilibrated Source 30Q resin (25 ml, XK 16/20 column; GE Healthcare) at a flow rate of 2 ml/min using an ÄKTA-Prime-System (GE Healthcare). The column was washed with 3 column volumes of buffer A (0.1 m Tris, pH 7.5, 50 mm NaCl), followed by washing with 3 column volumes of buffer B (0.1 m MES, pH 6.0) at a flow rate of 3 ml/min, and the protein was eluted by linear NaCl gradient from 50 mm to 0.6 m using buffer C (0.1 m MES, pH 6.0, 1 m NaCl) at a flow rate of 2 ml/min, within 20 min. The eluted protein peak was collected and concentrated by ultrafiltration on Vivaspin 20 (50,000 molecular weight cut-off) (Sartorius Stedim Biotech GmbH, Goettingen, Germany). For gel filtration, the concentrated protein extract was loaded on a Superdex S200 26/60 pg column (GE Healthcare) equilibrated with buffer A. Elution was performed at a flow rate of 1 ml/min. Fractions corresponding to the SPH homodimers were combined and concentrated as above. In case a buffer exchange was required, the enzyme was loaded on a desalting 26/10 column (GE Healthcare) equilibrated with an appropriate buffer. Protein concentration was estimated spectrophotometrically using calculated ϵ280 = 104.335 m−1 cm−1. The N-His-tagged version was purified as follows. Cell lysis and the supernatant preparation were performed as above. The obtained supernatant was loaded on a HisTrap column (GE Healthcare), the column was washed by 10 volumes of buffer A, and the protein was eluted with buffer A containing 0.5 m imidazole.
Spectroscopic Assays and HPLC-MS/MS Cofactor Identification
For initial characterization, UV-visible spectra of purified proteins in the range 300–800 nm were recorded with the single-beam absorption spectrometer (Ultrospec 2100 pro, GE Healthcare). Reference spectra of FAD and FMN were recorded in the same way, using FAD and FMN concentrations of 10–40 μm in buffer A. To identify a flavin cofactor, an aliquot of 50 μm SPH in water was heated at 95 °C for 10 min, precipitated protein was removed by centrifugation at 20,000 × g for 10 min, and the supernatant was diluted 2-fold with acetonitrile and used for MS analysis. 20 μm FAD was used as a reference in the same solvent system. The analysis of constituents was performed using an Agilent 1100 HPLC system (Agilent, Waldbronn, Germany) coupled to an Applied Biosystems 3200 hybrid triple quadrupole/linear ion trap mass spectrometer (MDS Sciex, Toronto, Canada). Nanoelectrospray analysis was achieved using a chip ion source (TriVersa NanoMate, Advion BioSciences (Ithaca, NY)). Reversed-phase HPLC separation was performed on an EC 250/2 Nucleodure 100-5 C18ec column (250 × 2.1 mm, 5-μm particle size; Macherey and Nagel (Düren, Germany)) with a methanol, 5 mm NH4OAc, pH 6.0 (20:80, v/v) solvent system and a flow rate of 200 μl/min. For stable nanoelectrospray analysis, 100 μl min−1 2-propanol/acetonitrile/water/formic acid (70:20:10:0.1, v/v/v/v) delivered by a 515 HPLC pump (Waters, Milford, MA) were added just after the column via a mixing tee valve. By using another postcolumn splitter, 637 nl min−1 of the eluent were directed to the nanoelectrospray analysis chip. Ionization voltage was set to −1.7 kV. FMN and FAD were ionized in a positive mode and determined by product ion scanning. The collision energy, with nitrogen in the collision cell, was 50 V. Declustering potential was 80 V. Entrance potential was 11 V. The mass analyzers were adjusted to a resolution of 0.7 atomic mass units full width at half-height. The ion source temperature was 40 °C, and the curtain gas was set at 10 (given in arbitrary units).
Kinetic Parameters and Analysis of FAD Function
N-His-tagged SPH was purified as described above following buffer exchange in buffer A by the desalting column. The obtained amount of enzyme was split into two equal parts and dialyzed against buffer A with and without a 5-fold excess of FAD at 4 °C overnight. The next day, the protein preparations were centrifuged and concentrated by ultrafiltration on Vivaspin 20 (50,000 molecular weight cut-off), and the concentration was measured by a Bradford protein assay (7). The enzyme was diluted to a concentration of 10 mg/ml, frozen in liquid nitrogen, and stored at −70 °C until used. Kinetic measurements were performed in 200 μl of 50 mm MES, NaOH (pH 6.1), containing 50 mm NaCl, 2% ethanol and 10% glycerol with 32, 64, 128, or 180 μm of oleic or linoleic acid. The reactions were initiated by the addition of S. pyogenes M49 hydratase (SPH) to a final concentration of 500 nm and incubated at 37 °C for different times (0.5–15 min). The reactions were stopped by the addition of 1 ml of chloroform/methanol (2:1, v/v), and fatty acids were extracted by 10 min of immediate intensive vortexing in the presence of heptadecanoic acid as an internal standard. The organic phase was taken and dried under a stream of liquid nitrogen. The pellets were dissolved in 200 μl of methanol containing 0.1 m (trimethylsilyl)diazomethane and agitated at room temperature for 10 min. Methyl esters were diluted in acetonitrile and analyzed by the Agilent GC 6890 system coupled with a flame ionization detector, where the column and the parameters were identical to those described above. The Km and Vmax values were estimated by nonlinear fitting tools of OriginPro version 7.5 software.
Stopped-flow kinetic analysis was performed in degassed buffer A by mixing equal volumes of SPH and linoleic acid solutions to a final concentration of 5 μm enzyme and 180 μm and 1.8 mm substrate, respectively. The formation of reduced FAD species was monitored by using a stopped-flow spectrophotometer (SX20, Applied Photophysics).
GC-MS Analysis of Lipids
To determine the substrate specificity, 50 μg of a substrate were mixed with 1 ml of buffer A and 10 μg of purified SPH and incubated for 1–24 h at 37 °C. For analysis of in vivo fatty acid metabolism, S. pyogenes cells at A600 = 2.5 were used. Substrates were either oleic acid or U-13C-labeled or non-labeled linoleic acid. Fatty acids were extracted by a chloroform/methanol (2:1) mixture, dissolved in methanol, and converted to methyl esters with (trimethylsilyl)diazomethane, hydroxyl groups were modified with N,O-bis-(trimethylsilyl)-trifluoroacetamide. Esterified fatty acids were transesterified by adding sodium methoxide in methanol/toluene (2:1, v/v). GC-MS analysis was carried out using an Agilent 5973 network mass-selective detector connected to an Agilent 6890 gas chromatograph equipped with a capillary DB-23 column (30 m × 0.25 mm; 0.25-μm coating thickness; Agilent, Waldbronn, Germany). Helium was used as carrier gas (1 ml/min). The temperature gradient was 150 °C for 1 min, 150–160 °C at 10 K/min, 160–200 °C at 7 K/min, 200–250 °C at 25 K/min, and 250 °C for 7 min. Electron energy of 70 eV, ion source temperature of 230 °C, and temperature of 260 °C for the transfer line were used (8).
Blood Survival Assay
Overnight cultures of the wild type and mutant strains were inoculated into fresh THY medium and grown to exponential growth phase. Bacteria was harvested by centrifugation, set to an optical density at 600 nm of 0.25, and further diluted 1:10,000 in PBS. Twenty microliters of each suspension were incubated with 480 μl of serum or heparinized blood for 3 h at 37 °C with agitation at 800 rpm. After this incubation, the remaining CFU were determined by plating and related to the serum inoculum, which was set to 100%. The CFU contained in this final suspension were determined by plating of serial dilutions on THY agar plates.
Eukaryotic Cell Adherence and Internalization Assays
Assessment of eukaryotic cell adherence and internalization was determined by an antibiotic protection assay. Briefly, for the antibiotic protection assay, the early stationary phase bacteria were suspended in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and were added to HaCaT cells (a human skin keratinocyte cell line) grown overnight to confluence, establishing a multiplicity of infection of 25. Previous studies indicated this multiplicity of infection to be in the linear range of such experiments. After 2 h, the cells were washed with phosphate-buffered saline. One-half of the cells were lysed with distilled water, and the number of bacteria contained in the lysate was assessed by viable counts. The other half of the eukaryotic cells were exposed to culture medium supplemented with penicillin and streptomycin for another 2 h. Then these cells were washed and lysed, and the bacterial numbers were counted as above.
MIC Determination
MICs for cerulenin and oleic and linoleic acid were determined by a series of double broth microdilutions according to the protocols of Andrews (9), followed by measurement of optical density after the overnight incubation.
Protein Sequences and Phylogenetic Analysis
Members of the MCRA gene family were identified using a reciprocal BLAST search. MCRA of S. pyogenes M49 591 (ZP_00366513.1) was used as an initial query sequence for a BLASTP (10) search with conditional compositional score matrix adjustment against the non-redundant protein data base (11). Obtained sequences were used for a BLASTP search of S. pyogenes M49 591 proteins, and only sequences that returned MCRA as a best hit were used for further analysis. Sequences obtained after the previous step were aligned using MUSCLE (12), and sequences with less than 55% of alignment extension were excluded from the data set. A draft phylogeny for 243 protein sequences was reconstructed using the neighbor-joining method (13), as implemented in MEGA4 (14). 148 sequences covering widest range of taxa were selected for in depth phylogenetic analysis. The JTT + GAMMA (15, 16) model was determined as the model of the best fit according to AIC (17) by the PROTTEST tool (18) with default parameters. Neighbor-joining phylogeny was reconstructed with the use of MEGA4 implementing the previously identified evolutionary model with 1000 bootstrap runs. The final tree was rooted using the ancestral node of all ascomycete sequences as an outgroup. Sequences from streptococcal, lactobacilli, and staphylococcal species were selected for detailed evolutionary analysis. Sequences identical at the amino acid level were excluded from the analysis. Protein sequences were aligned using MUSCLE, and gaps were inserted into DNA sequences using the PAL2NAL tool (19). Best fit evolutionary models for DNA were identified using the MODELTEST (20) program. The phylogenetic tree was reconstructed for DNA sequences using RAxML software (21) implementing the previously identified evolutionary model. The tree was rooted in order to maximize a number of monophyletic groups. DNA sequence data were analyzed with the CODELM program of the PAML4 software package (22).
RESULTS
SPH Protein Sequence Analysis
In order to identify all known homologues of MCRA protein from S. pyogenes M49 591, we conducted a BLAST search using the GenBankTM data base. 243 gene sequences from organisms of different domains of life were identified. The majority of these gene sequences are from Gram-positive and negative bacteria or from ascomycetes. A phylogenetic analysis of 148 sequences covering the widest range of taxa revealed that the MCRA protein family consists of three large clusters (supplemental Fig. S1). The first cluster includes all of the analyzed ascomycete genes. The rest of the sequences form two additional clusters. Based on high sequence similarity (more than 30% identity for the most divergent sequences), one may assume similar biological function for all of the family members. Disagreement of the gene tree topology with the accepted evolutionary relationship of considered organisms suggests evolution of the family through active horizontal gene transfer within at least the bacterial domain.
Evolutionary analysis of MCRA genes from streptococcal, lactobacilli, and staphylococcal species suggested that proteins evolved under strong negative selection because of the low non-synonymous/synonymous substitution rate over the branches of the gene tree (Kn/Ks rarely exceeds 1; supplemental Fig. S2, left). Alignment of the predicted FAD binding motif revealed a deviation from the canonical FAD binding motif GXGXXGX17–19E of the Rossmann fold for lactobacilli and staphylococcal proteins (supplemental Fig. S2, right). The third glycine residue of the motif is substituted by serine in staphylococcus (GXGXXSX15S and GXGXXSX21E) or by asparagine in lactobacillus (GXGXXN(A/S)X15(D/K/E) and GXGXXGX21E(G/S/A). The latter corresponds to members of the glutathione reductase family (23).
Protein Purification
In order to characterize the MRCA protein from S. pyogenes M49, we purified the non-tagged version after heterologous expression in E. coli Bl21 Star cells. The initial anion exchange chromatography yielded protein having a light yellow color (supplemental Fig. S3A, lines 1 and 2). With the following size exclusion chromatography, the protein was found in two fractions. The main fraction represented the homodimer of 134 kDa, and the second fraction represented the monomer of 67 kDa. Both fractions contained active protein, but in the fractions harboring the homodimer, only traces of other proteins were found and were used for further analysis (supplemental Fig. S3A, line 3). However, during this step, yellow color was lost. Therefore, N-His-tagged SPH was affinity-purified in one step (supplemental Fig. 3B). It eluted as intense yellow colored solution similar to the non-tagged enzyme after the anion exchange chromatography. However, in both colored and non-colored enzyme preparations, hydratase activity was detectable.
Spectral Properties and Cofactor Identification
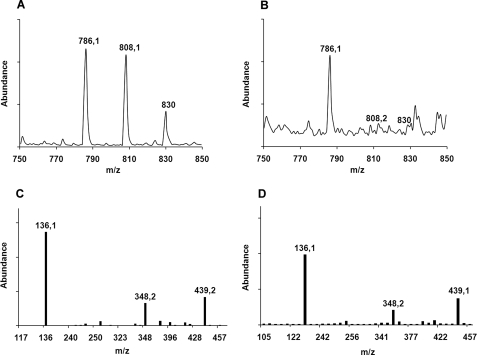
Concentrated affinity-purified SPH protein had yellow color, and the UV-visible spectrum of SPH showed maxima around 370 and 450 nm characteristic for flavin cofactors as well as small additional peaks at 420 and 480 nm, respectively (supplemental Fig. S4, solid line). To test for the presence of flavin cofactor in SPH, the protein was heat-denatured, and the spectrum of the protein-free supernatant was recorded (supplemental Fig. S4, thin solid line). In this case, the spectrum contained only two maxima at 447 and 373 nm, respectively, and closely resembled the absorption patterns of free FAD and FMN, confirming that SPH contained a non-covalently bound flavin cofactor. The cofactor was identified by mass spectrometry, and the spectra of the FAD standard and of the supernatant are shown in Fig. 1, A and B, respectively. The MS/MS spectrum of the parent ion M+ = 786.1 unambiguously identifies the cofactor as FAD, with three major signals (M+ = 136.1, M+ = 348.2, and M+ = 439.2) found both in the FAD standard and in the sample (Fig. 1, C and D).
FIGURE 1.
Identification of FAD in SPH. A and B, mass spectra of FAD standard and supernatant of SPH sample after protein precipitation, respectively. C and D, MS/MS spectra of molecular ion (M+ = 786.1) corresponding to A and B, respectively.
Products of SPH
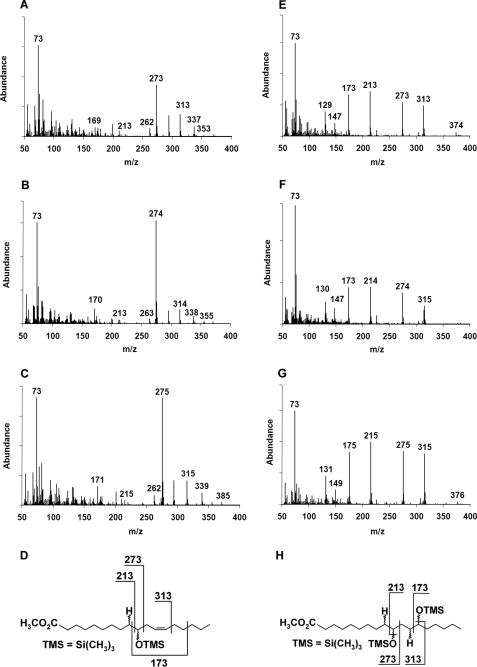
In activity assays with purified SPH protein and linoleic acid as a substrate, (12Z)-10-hydroxy-12-octadecenoic acid (10-HOE) was detected by GC-MS as the main reaction product (Fig. 2, A and D). The ion M+ = 273 is the predominant species in the spectrum and represents a C-10 fragment in which the 10-hydroxy group is modified by MS/MS. When an additional (12Z)-double bond is present in the substrate, it is also hydrated, yielding 10,13-dihydroxyoctadecanoic acid (10,13-DiHOA) (Fig. 2, E and H). In order to characterize the enzymatic preferences with respect to the double bond position and configuration and the polarity of the fatty acid headgroup, a range of different fatty acids and lipids were assayed (Table 1). SPH showed strict preference for the free carboxyl group and did not modify complex lipids. It also required the (9Z)-double bond, whereas the (9E)-, (11E)-, and (11Z)-double bonds were not hydrated. Any combination of cis and trans double bonds as well as double bonds in front of C-9 abolished SPH activity. The chain length of the substrate was limited to 16–18 carbons.
FIGURE 2.
Production of hydroxylated fatty acids by purified SPH. Linoleic acid was used as a substrate, and reaction products were analyzed by GC-MS. Mass spectra of 10-HOE after reaction in H2O (A), in 2H2O (B), and in H218O (C) show that one water molecule is added to the Δ9 double bond. The mass of C-10 fragment (m/z = 273) is changing due to the isotopic labeling either with 2H at position C-9 (+1 atomic mass unit) or with 18O at position C-10 (+2 atomic mass units). The structure of 10-HOE and its fragmentation pattern are shown in D. Mass spectra of 10,13-DiHOA obtained after reaction of SPH with linoleic acid in H2O (E), in 2H2O (F), and in H218O (G) show that two water molecules are incorporated into the substrate linoleic acid across the Δ9 and Δ12 double bonds. The structure of 10,13-DiHOA and its fragmentation pattern are shown in H.
TABLE 1.
Substrate specificity of SPH
The products were identified by GC/MS analysis after chloroform/methanol (2:1) extractions of the samples. The reactions were performed as described under “Experimental Procedures.” Two repetitions for each experiment were conducted.
| Substrate | Product(s) | Retention time | Fragmentation pattern |
|---|---|---|---|
| min | |||
| 14:1Δ9Z | No products detected | ||
| 16:1Δ9Z | 10-Hydroxyhexadecanoate | 6.88 | 357(M-1+), 273, 187, 169 |
| 18:1Δ9Z | 10-Hydroxyoctadecanoate | 8.1 | 385(M-1+), 273, 215, 169 |
| 18:1Δ9E | No products detected | ||
| 18:2Δ9Z,Δ12Z | (12Z)-10-Hydroxy-12-octadecenoate | 8.33 | 383(M-1+), 273, 213, 173, 169 |
| 10,13-Dihydroxyoctadecanoate | 8.81 | 313, 273, 213, 173 | |
| 18:3Δ9Z,Δ12Z,Δ15Z | (12Z,15Z)-10-Hydroxy-12,15-octadecadienoate | 8.77 | 380(M-1+), 273, 241, 169 |
| 18:3Δ6Z,Δ9Z,Δ12Z | No products detected | ||
| 20:3Δ11Z,Δ14Z,Δ17Z | No products detected | ||
| 20:4Δ5ZΔ8ZΔ11Z,Δ14Z | No products detected | ||
| 18:2Δ9Z,Δ12Z methyl ester | No products detected | ||
| Linoleyl-CoA | No products detected | ||
| Dilinoleylphosphatidylcholine | No products detected | ||
| Trilinoleylglycerol | No products detected |
The origin of oxygen in the hydroxylated fatty acids was analyzed with isotopically labeled water (Fig. 2, B, C, F, and G). In the mass spectrum of 10-HOE obtained after incubation in 2H2O, the mass shift of +1 was evident for the C-10 fragment described above (Fig. 2, A versus B, M+ = 273 versus 274). Furthermore, after the reaction in H218O, the mass of the C-10 fragment increased by +2 atomic mass units, indicating the addition of 18O from water (Fig. 2C, M+ = 275). The structures of 10,13-DiHOA confirmed these findings (Fig. 2, E–G).
Kinetic Parameters and Function of FAD
Next, properties of the SPH holoenzyme (FAD-harboring form) and the apoenzyme were compared. Supplemental Fig. 5 shows their UV-visible spectra together with that of a FAD standard. From the apoenzyme spectrum, we conclude that this form may contain only traces of the cofactor. For the holoenzyme the Km values for 18:2 and 18:1 were 49 ± 3 and 63 ± 6 μm, respectively. kcat values were 101 ± 3 and 67 ± 5 min−1, respectively. Linoleic acid (18:2) was the preferred substrate under the given conditions: kcat/Km 2 × 106 versus 1 × 106 m−1 min−1, respectively. In contrast to the holoenzyme, the apoenzyme showed a 10–50-times lower activity, making it impossible to reach substrate saturating conditions and therefore to determine its kinetic parameters.
To investigate direct involvement of FAD in catalysis, stopped-flow experiments with the holoenzyme in the presence of linoleic acid were performed. No difference in the functional range of absorbance (300–600 nm) of FAD was detected within 10–5000-ms time range at two different concentrations of linoleic acid (supplemental Fig. S6, A and B). Notably, absorbance spectra did not change even after prolonged incubation for 1–10 min. Together with the fact that molecular oxygen is not involved in this hydration reaction and thus the role of FAD in this process cannot be attributed to a redox reaction, these data suggest a stabilizing function of FAD in SPH rather than its involvement in catalysis.
Generation of Δsph Deletion Strain of S. pyogenes M49
To generate a sph gene deletion mutation, a replacement type targeting vector pFW11 was used and ligated with the upstream and downstream regions of the sph gene. After restriction and sequencing analysis, the vector was transformed into S. pyogenes M49 cells, and clones were selected in the presence of spectinomycin. Genomic DNA from surviving clones was isolated, and the deletion of the sph gene was confirmed by PCR. A fragment of 1782 bp corresponding to the sph open reading frame was amplified from the wild type DNA but not from the mutant DNA (supplemental Fig. S7A). Reverse transcription-PCR of total mRNA from wild type and the mutants confirmed sph expression in the wild type and no expression in the mutants during the exponential phase of the growth (supplemental Fig. S7B).
Metabolism of Oleic and Linoleic Acid and Their Influence on the Growth of Wild Type and Δsph Mutant Strains
Oleic and linoleic acid are toxic to a variety of bacteria (24–26). Therefore, growth curves for wild type cells in the presence of different concentrations of these fatty acids were recorded. We found that a 10 μg/ml concentration of either fatty acid did not inhibit growth. Next, growth kinetics of the wild type and mutant strains in THY medium without or with the addition of oleic and linoleic acid and 10% human serum alone were analyzed. As shown in supplemental Fig. S8, A–C, the mutant had a slight delay of 1 h in growth but very similar responses to all additives compared with wild type. The addition of fatty acids or serum enhanced growth speed. Because all mutant strains behaved in a similar way, only one mutant strain was further analyzed.
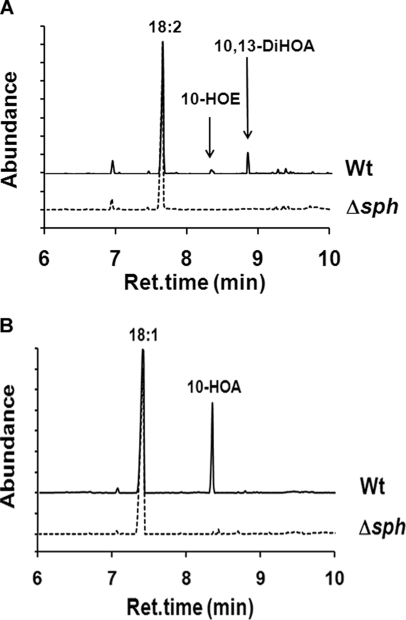
Wild type bacteria converted 18:2 to 10-HOE and 10,13-DiHOA and 18:1 to 10-hydroxyoctadecanoic acid (10-HOA), whereas the mutant strain did not metabolize the fatty acids to any of these compounds (Figs. 3, A and B). Incubations of either wild type or mutant strain with U-13C-labeled 18:2 did not reveal any further transformation to less saturated/unsaturated fatty acids. As a control, we measured free fatty acid consumption between control medium and medium containing the bacteria after the overnight incubation. As much as 43% of oleic and 37% of linoleic acid were utilized by wild type bacteria, and 16 and 19% were utilized by the mutant, respectively, and therefore the overall contribution of the hydratase activity of SPH may reach 50% of total free fatty acid utilization.
FIGURE 3.
Total ion chromatograms of GC/MS runs for identification of products formed from linoleic and oleic acid upon incubation with wild type and Δsph strains of S. pyogenes M49. A, products were formed from linoleic acid (18:2): 10-HOE and 10,13-DiHOA. None of these products were formed after incubation with the mutant. B, 10-HOA was formed from oleic acid (18:1) after incubation with the wild type (Wt), and 10-HOA was not detected after incubation with the mutant. For the experiments, 2.5 OD of the wild type and mutant cells were incubated with 50 μg of substrates in 200 μl of THY medium for 3 h at 37 °C, followed by extraction with chloroform/methanol (2:1). Representative data for one of two repetitions for the respective condition are presented.
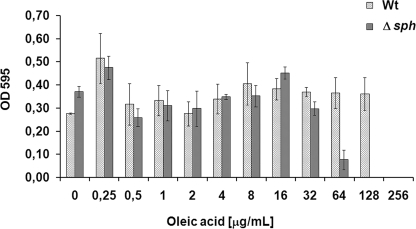
Bacteria detoxify unsaturated fatty acids by hydrogenating them to less unsaturated or saturated fatty acids (25, 27), and hydratase activity may be involved in this process. To test this, MICs of free oleic and linoleic acid for the wild type and the mutant strain were determined. MICs for 18:1 and 18:2 were 256 and 16 μg/ml for the wild type and 128 and 16 μg/ml for the mutant, respectively (Fig. 4). Hence, the mutant was 2 times more sensitive toward oleic acid than the wild type. No difference between the two strains in sensitivity to linoleic acid was observed.
FIGURE 4.
Minimal inhibitory concentration of oleic acid for wild type and Δsph strains of S. pyogenes M49. Optical density was measured after overnight growth in the presence of different concentrations of free oleic acid in THY medium. Data represent mean and S.D. of three independent experiments. Wt, wild type.
To further analyze a possible involvement of SPH in the bacterial fatty acid metabolism, we determined MIC for cerulenin. Cerulenin acts as an irreversible inhibitor of β-ketoacyl-acyl carrier protein synthases, which are crucial for fatty acid biosynthesis (28, 29). The experiments were carried out in the presence of oleic, linoleic, and palmitic acid with or without human serum albumin and with 10% human serum alone. An increase of the growth at similar levels was recorded for both strains in the presence of all additives (supplemental Fig. S9). The addition of either free fatty acids or fatty acids supplemented with human serum albumin (to increase availability of fatty acids to the bacterial cells) showed no phenotypic differences; nor did the addition of 10% human serum alone.
Analysis of Δsph Mutant Strain in Human Blood and HaCaT Cells
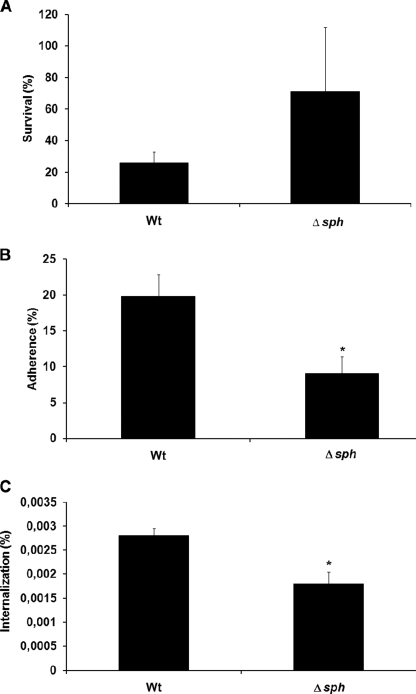
In order to characterize virulence-associated roles of SPH, the wild type and Δsph mutant strain were compared for survival in human blood and their capability to adhere to and internalize into HaCaT cells. The mutant strain survived 2 times better in human blood than the wild type (Fig. 5A). However, the mutant strain was significantly attenuated in its capacity to adhere to and internalize into HaCaT cells (Fig. 5, B and C).
FIGURE 5.
Blood survival and adherence to and internalization into HaCaT cells of wild type and Δsph strains of S. pyogenes M49. A, blood survival in heparinized human whole blood. The number of surviving CFU in whole human blood was represented as a percentage of CFU of the respective serum. Data represent means and S.D. of three independent experiments with three different blood samples. B and C, adherence and internalization, respectively, represented as a percentage from the CFU count of the original inoculum. Data represent means and S.D. of three independent experiments. The asterisks mark data with statistical significance (p < 0.05). Wt, wild type.
DISCUSSION
Because the first member of the MCRA protein family was identified in S. pyogenes (1), different functions have been discussed for them, ranging from their similarity to the major histocompatibility complex class II antigen to a catalytic activity as CLA forming double bond isomerase (2) or hydratase (3). A high degree of sequence similarity between the family members and a low level of non-synonymous substitutions suggests a negative selection and therefore a preservation of their yet unclear function. Sequence analysis of the proteins of the genera Streptococcus, Staphylococcus, and Lactobacillus revealed only a semiconserved FAD/dinucleotide binding motif similar to that of the glutathione reductase family (GXGXXGX17–19(E/D)) (supplemental Fig. S2). GXGXXG is part of a loop connecting the first β-strand and α-helix in the βαβαβ-Rossmann fold with the N-terminal end of the first α-helix pointing toward the pyrophosphate moiety for charge compensation. The role of the glycine residues is well understood. The first glycine allows for a tight turn of the main chain, which is important for positioning the second glycine. The second glycine permits close contact of the main chain to the pyrophosphate of FAD, and the third glycine allows close packing of the helix with the β-sheet (30, 31). According to our analysis, the most drastic changes in the canonical motif were observed for MCRA proteins from Staphylococcus and Lactobacillus species, where the third glycine is substituted either with a serine or asparagine residue, respectively (supplemental Fig. S2). The substitution of glycine with these larger residues may perturb proper orientation of the first α-helix with the β-sheet and therefore may significantly affect FAD binding. In fact, the MCRA proteins from lactobacilli have even lower affinity with FAD in comparison with SPH (data not shown). Noticeably, MCRA proteins have different numbers of amino acid residues between the third glycine and the conserved glutamate residue compared with the canonical motif, namely 15 or 21 residues instead of 17–19 for the glutathione reductase family. The glutamate/aspartate residues are involved in hydrogen bonding with the ribose 2′-hydroxyl of the adenosine moiety (30), and therefore the different positioning may also affect the strength of FAD binding. In accordance with this observation, heterologously expressed SPH appeared to be a yellow protein after the first step of purification by anion exchange chromatography but appeared colorless after the size exclusion chromatography or dialysis. Purification of His-tagged protein allowed us to obtain colored enzyme solution of higher purity, which was used for flavin cofactor identification and analysis of kinetic parameters (supplemental Fig. S3). Most likely, the purification of His-tagged SPH avoids multiple washing steps and therefore allows retention of a larger proportion of the cofactor. HPLC-MS/MS analysis unambiguously identified FAD as a non-covalent bound cofactor of SPH (Fig. 1).
The first attempt to identify an enzymatic activity for MCRA protein was reported by Rosson et al. (2). Their study focused on the isolation of a (9Z,11E)-CLA-forming double bond isomerase from L. reuteri PYR8. In this case, the majority of the activity was found in the membrane fraction. The isolated isomerase protein had more than 70% identity and 85% similarity to the known MCRA from S. pyogenes, for which no enzymatic activity was previously reported. However, when the putative (9Z,11E)-CLA isomerase was heterologously expressed in E. coli and B. subtilis, no isomerase activity was detected. Instead, we were able to show that the MCRA protein from S. pyogenes, when it was heterologously expressed in E. coli, acts as a hydratase. The enzyme is specific for (9Z)- and (12Z)-double bonds of C-16 and C-18 fatty acids and catalyzes the formation of 10-hydroxy and 10,13-dihydroxy derivatives (Table 1 and Fig. 2), and no CLA-forming isomerase activity was detected (32). Analysis of substrate specificity showed that SPH strictly prefers free fatty acids over the esterified species (Table 1). The ability to add a water molecule to a double bond classifies SPH as a new member of the carbon-oxygen lyase family. In contrast to the well studied enoyl-CoA hydratases involved in fatty acid β-oxidation, which add a water molecule to the polarized α-double bond in CoA-thioester (33, 34), SPH hydrates a non-activated carbon-carbon double bond. Our experiments show that molecular oxygen is not involved in this hydration reaction; thus, the role of FAD in this process cannot be attributed to a redox reaction (Fig. 2). Additionally, FAD was not reduced during reaction of the holoenzyme with linoleic acid at least under our conditions (supplemental Fig. S6). Kinetic analysis of holo- and apo-SPH forms demonstrated that apo-SPH was at least 10–50 times less active than the holoenzyme, whereas linoleic acid was the preferred substrate. Together, these data provide good evidence of a structural role of FAD in SPH function.
There are several examples of flavoenzymes that require FAD to maintain a proper architecture of the active site but where the cofactor is not directly involved in catalysis (35–37). The direct role of FAD in the hydrogen transfer during (10E,12Z)-CLA formation by double bond isomerase from Propionibacterium acnes (PAI) was demonstrated previously (38, 39). SPH has similar substrate requirements as the PAI and (9Z,11E)-CLA isomerase of the cell envelope preparation of Butyrivibrio fibrisolvens (40). So far, PAI is the only CLA-forming double bond isomerase for which structural information is available. In contrast to SPH, PAI retained FAD during purification and crystallization, and the crystal structures of the free enzyme and its complex with (10E,12Z)-CLA provided insight into the substrate binding and reaction mechanism (38). However, the sequence alignment of PAI and SPH did not reveal any significant similarity between the two proteins except the short stretch of the FAD binding motif. With respect to an involvement of SPH in CLA formation, it is still possible that recombinant MCRA enzymes from CLA-producing species like lactobacilli require critical protein partners or a membrane environment in order to retain a double bond isomerase activity.
To investigate the possible role of the hydroxy fatty acids formed by SPH in the transformation of unsaturated fatty acids to saturated ones, we used feeding experiments with U-13C-labeled linoleic acid. Analysis of free and esterified fatty acid fraction by GC/MS from the wild type strain and the Δsph mutant showed conversion to the hydroxy derivatives by wild type cells but not by the mutant (Fig. 3). However, in neither the wild type strain nor in the mutant did we detect CLA formation. This observation argues against a link between the SPH-dependent hydration and CLA formation, at least under our experimental conditions. On the other hand, SPH seems to be involved in fatty acid metabolism by a yet unknown mechanism because in the mutant strain, consumption of the free fatty acids from the medium is reduced to about 50% of that of the wild type. However, it seems not to be involved in fatty acid homeostasis because MICs of cerulenin for wild type and the mutant in the presence of different fatty acids were similar (supplemental Fig. S9).
Unsaturated fatty acids are toxic for many bacteria (41) due to deteriorating effect on bacterial cellular membrane (42). They also inhibit enoyl-ACP reductase (FabI) and thus disrupt bacterial fatty acid synthesis (43). We hypothesize that hydration of unsaturated fatty acids may represent an additional detoxification mechanism in bacteria harboring MCRA enzymes. Such detoxification could be essential for bacterial colonization and survival in fatty acid-rich environments (e.g. skin and inflamed tissues). In fact, the mutant appeared to be 2-fold more sensitive to oleic acid than wild type, whereas no changes in sensitivity to linoleic acid was observed (Fig. 4). The later may be explained by the fact that linoleic acid is 10 times more toxic for S. pyogenes M49.
No growth inhibition was observed for the mutant in human serum, which can be explained by a much lower concentration (up to 5 orders of magnitude, 7.5 nm) of free oleic and linoleic acid in the serum (44, 45) than the measured MIC (58 μm for linoleic acid, 458 μm for oleic acid). This may explain as well why the virulence properties are independent of the fatty acid concentrations in the serum. Here, the mutant showed increased survival in blood with reduced adherence and internalization properties to HaCaT cells (Fig. 5, A–C). Therefore, the deletion of MCRA protein may lead to reduced opsonization of the bacterial cell surface, resulting in lower efficiency of antibody-dependent phagocytosis (46, 47). Whether the involvement of MCRA in pathogenicity, however, is connected to its enzymatic activity remains an open question at this stage.
In summary, our study shows for the first time a detailed characterization of biochemical and physiological properties for a member of the MCRA protein family. The SPH protein of S. pyogenes M49 was identified as fatty acid hydratase that harbors FAD as a cofactor that stabilizes its active conformation. It forms 10-hydroxy and 10,13-dihydroxy derivatives from C-16 and C-18 non-esterified fatty acids. Moreover, the deletion of SPH led to a 2-fold decrease in resistance to oleic acid, increased survival of the mutant strain in the whole blood, and reduced adhesion and internalization to human keratinocytes in comparison with the wild type.
Supplementary Material
Acknowledgments
We thank Prof. Kai Tittmann and Dr. Danilo Meyer for support with stopped-flow measurements and for constant discussions.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
- MCRA
- myosin cross-reactive antigen
- 10,13-DiHOA
- 10,13-dihydroxyoctadecanoic acid
- GAS
- group A streptococci
- GC
- gas chromatography
- 10-HOA
- 10-hydroxyoctadecanoic acid
- 10-HOE
- (12Z)-10-hydroxy-12-octadecenoic acid
- HPLC
- high pressure liquid chromatography
- MIC
- minimal inhibitory concentration
- MS
- mass spectrometry
- MS/MS
- tandem MS
- PAI
- P. acnes isomerase
- SPH
- S. pyogenes hydratase
- THY
- Todd-Hewitt broth supplemented with yeast extract
- CLA
- (9Z,11E)-conjugated linoleic acid
- MES
- 4-morpholineethanesulfonic acid
- CFU
- colony-forming unit(s).
REFERENCES
- 1.Kil K. S., Cunningham M. W., Barnett L. A. (1994) Infect. Immun. 62, 2440–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosson R. A., Grund A. D., Deng M., Sanchez-Riera F. (June1, 2004) United States Patent US 6,743,609 B1
- 3.Bevers L. E., Pinkse M. W., Verhaert P. D., Hagen W. R. (2009) J. Bacteriol. 191, 5010–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham M. W. (2000) Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podbielski A., Spellerberg B., Woischnik M., Pohl B., Lütticken R. (1996) Gene 177, 137–147 [DOI] [PubMed] [Google Scholar]
- 6.Kreikemeyer B., Boyle M. D., Buttaro B. A., Heinemann M., Podbielski A. (2001) Mol. Microbiol. 39, 392–406 [DOI] [PubMed] [Google Scholar]
- 7.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 8.Liavonchanka A., Rudolph M. G., Tittmann K., Hamberg M., Feussner I. (2009) J. Biol. Chem. 284, 8005–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews J. M. (2001) J. Antimicrob. Chemother. 48, 5–16 [DOI] [PubMed] [Google Scholar]
- 10.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 11.Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W. (2009) Nucleic Acids Res. 37, D26–D31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saitou N., Nei M. (1987) Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 14.Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 15.Jones D. T., Taylor W. R., Thornton J. M. (1992) Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 16.Yang Z. H. (1993) Mol. Biol. Evol. 10, 1396–1401 [DOI] [PubMed] [Google Scholar]
- 17.Sugiura N. (1978) Commun. Statistics 7a, 13–26 [Google Scholar]
- 18.Abascal F., Zardoya R., Posada D. (2005) Bioinformatics 21, 2104–2105 [DOI] [PubMed] [Google Scholar]
- 19.Suyama M., Torrents D., Bork P. (2006) Nucleic Acids Res. 34, W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Posada D., Crandall K. A. (1998) Bioinformatics 14, 817–818 [DOI] [PubMed] [Google Scholar]
- 21.Stamatakis A. (2006) Bioinformatics 22, 2688–2690 [DOI] [PubMed] [Google Scholar]
- 22.Yang Z. H. (2007) Mol. Biol. Evol. 24, 1586–1591 [DOI] [PubMed] [Google Scholar]
- 23.Dym O., Eisenberg D. (2001) Protein Sci. 10, 1712–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kepler C. R., Tove S. B. (1967) J. Biol. Chem. 242, 5686–5692 [PubMed] [Google Scholar]
- 25.Maia M. R., Chaudhary L. C., Figueres L., Wallace R. J. (2007) Antonie Van Leeuwenhoek 91, 303–314 [DOI] [PubMed] [Google Scholar]
- 26.Stenz L., François P., Fischer A., Huyghe A., Tangomo M., Hernandez D., Cassat J., Linder P., Schrenzel J. (2008) FEMS Microbiol. Lett. 287, 149–155 [DOI] [PubMed] [Google Scholar]
- 27.Kepler C. R., Hirons K. P., McNeill J. J., Tove S. B. (1966) J. Biol. Chem. 241, 1350–1354 [PubMed] [Google Scholar]
- 28.Kauppinen S., Siggaard-Andersen M., von Wettstein-Knowles P. (1988) Carlsberg Res. Commun. 53, 357–370 [DOI] [PubMed] [Google Scholar]
- 29.Price A. C., Choi K. H., Heath R. J., Li Z., White S. W., Rock C. O. (2001) J. Biol. Chem. 276, 6551–6559 [DOI] [PubMed] [Google Scholar]
- 30.Wierenga R. K., Terpstra P., Hol W. G. (1986) J. Mol. Biol. 187, 101–107 [DOI] [PubMed] [Google Scholar]
- 31.Kleiger G., Eisenberg D. (2002) J. Mol. Biol. 323, 69–76 [DOI] [PubMed] [Google Scholar]
- 32.Feussner I., Hornung E., Liavonchanka A. (September10, 2008) International Patent WO2008 119735
- 33.Engel C. K., Kiema T. R., Hiltunen J. K., Wierenga R. K. (1998) J. Mol. Biol. 275, 847–859 [DOI] [PubMed] [Google Scholar]
- 34.Bahnson B. J., Anderson V. E., Petsko G. A. (2002) Biochemistry 41, 2621–2629 [DOI] [PubMed] [Google Scholar]
- 35.Cromartie T. H., Walsh C. T. (1976) J. Biol. Chem. 251, 329–333 [PubMed] [Google Scholar]
- 36.Vargo D., Pokora A., Wang S. W., Jorns M. S. (1981) J. Biol. Chem. 256, 6027–6033 [PubMed] [Google Scholar]
- 37.Duggleby R. G., Pang S. S. (2000) J. Biochem. Mol. Biol. 33, 1–36 [Google Scholar]
- 38.Liavonchanka A., Hornung E., Feussner I., Rudolph M. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2576–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liavonchanka A., Feussner I. (2008) ChemBioChem 9, 1867–1872 [DOI] [PubMed] [Google Scholar]
- 40.Kepler C. R., Tucker W. P., Tove S. B. (1970) J. Biol. Chem. 245, 3612–3620 [PubMed] [Google Scholar]
- 41.Raychowdhury M. K., Goswami R., Chakrabarti P. (1985) J. Appl. Bacteriol. 59, 183–188 [DOI] [PubMed] [Google Scholar]
- 42.Greenway D. L., Dyke K. G. (1979) J. Gen. Microbiol. 115, 233–245 [DOI] [PubMed] [Google Scholar]
- 43.Zheng C. J., Yoo J. S., Lee T. G., Cho H. Y., Kim Y. H., Kim W. G. (2005) FEBS Lett. 579, 5157–5162 [DOI] [PubMed] [Google Scholar]
- 44.Richieri G. V., Kleinfeld A. M. (1995) J. Lipid Res. 36, 229–240 [PubMed] [Google Scholar]
- 45.Kleinfeld A. M., Prothro D., Brown D. L., Davis R. C., Richieri G. V., DeMaria A. (1996) Am. J. Cardiol. 78, 1350–1354 [DOI] [PubMed] [Google Scholar]
- 46.Verbrugh H. A., Lee D. A., Elliott G. R., Keane W. F., Hoidal J. R., Peterson P. K. (1985) Immunology 54, 643–653 [PMC free article] [PubMed] [Google Scholar]
- 47.Valenti-Weigand P., Benkel P., Rohde M., Chhatwal G. S. (1996) Infect. Immun. 64, 2467–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.