Abstract
The level of monoubiquitinated proliferating cell nuclear antigen (PCNA) is closely linked with DNA damage bypass to protect cells from a high level of mutagenesis. However, it remains unclear how the level of monoubiquitinated PCNA is regulated. Here, we demonstrate that human ELG1 protein, which comprises an alternative replication factor C (RFC) complex and plays an important role in preserving genomic stability, as an interacting partner for the USP1 (ubiquitin-specific protease 1)-UAF1 (USP1-associated factor 1) complex, a deubiquitinating enzyme complex for PCNA and FANCD2. ELG1 protein interacts with PCNAs that are localized at stalled replication forks. ELG1 knockdown specifically resulted in an increase in the level of PCNA monoubiquitination without affecting the level of FANCD2 ubiquitination. It is a novel function of ELG1 distinct from its role as an alternative RFC complex because knockdowns of any other RFC subunits or other alternative RFCs did not affect PCNA monoubiquitination. Lastly, we identified a highly conserved N-terminal domain in ELG1 that was responsible for the USP1-UAF1 interaction as well as the activity to down-regulate PCNA monoubiquitination. Taken together, ELG1 specifically directs USP1-UAF1 complex for PCNA deubiquitination.
Keywords: Deubiquitination, DNA Damage, DNA Repair, DNA Replication, Ubiquitination
Introduction
Proliferating cell nuclear antigen (PCNA)4 functions in DNA replication, repair, and recombination as a sliding clamp for various DNA replication polymerases and a scaffold for many DNA repair and recombination enzymes (1). Various post-translational modifications of PCNA, including ubiquitination, sumoylation, or phosphorylation, determine its specific functions (2, 3). PCNA ubiquitination directs different branches of the postreplication repair (PRR) pathway (4). When a DNA replication fork stalls at damaged DNA lesions, the ubiquitin conjugation enzyme, RAD6, and the ubiquitin ligase, RAD18, monoubiquitinate lysine 164 of PCNA (5). Monoubiquitinated PCNA recruits translesion synthesis (TLS) polymerases, which can insert a base opposite the damaged lesion to bypass the damaged DNA. Alternatively, PCNA can be polyubiquitinated to promote DNA damage bypass by a homologous recombination mechanism (6–8).
The process and consequence of PCNA deubiquitination after DNA damage bypass are not clearly understood. Recently, USP1 (ubiquitin-specific protease 1) was identified as a deubiquitinating enzyme for PCNA after DNA damage bypass (9, 10). USP1 reduces the accumulation of monoubiquitinated PCNA during normal cell division, which prevents mutagenesis by error-prone TLS polymerases. In response to damage that stalls DNA replication, USP1 is degraded, and consequently monoubiquitinated PCNA accumulates (9). However, the observation that levels of monoubiquitinated PCNA remain high, even in the presence of persistent levels of USP1 (11, 12) suggests that there might be additional mechanisms to regulate USP1 activity. The identification of UAF1 (USP1-associated factor 1) indicates that interacting proteins may regulate the activity of USP1 in vivo (13).
The loading and unloading of PCNA on DNA is executed by the replication factor C (RFC) complex in an ATP-dependent manner. The RFC complex is composed of five subunits, RFC1, -2, -3, -4, and -5 (14). RFC-dependent PCNA loading plays essential roles in PCNA-related processes during DNA replication and repair (15). Recently, alternative RFC complexes containing the RFC2 to -5 core complex and a substitute ATPase have been identified in eukaryotes (16–20). In human, three alternative RFC complexes exist: CTF18-RFC, RAD17-RFC, and ELG1-RFC. Although the canonical or alternative RFC complexes may function as clamp loaders, there is no evidence relating them to the processing of PCNA monoubiquitination.
Recently, we demonstrated that human ELG1 protein is required to suppress genomic instability (20), similar to the yeast homologue Elg1p (21–24). In the present study, we identified human ELG1 as an interacting protein of the USP1-UAF1 complex. Monoubiquitinated PCNA accumulated in the chromatin following knockdown of either ELG1 or USP1. The down-regulation of PCNA monoubiquitination by ELG1 was mediated through an N-terminal 17-amino acid sequence of ELG1, which directly interacts with UAF1. Taken together, these data indicate that ELG1 plays an important role in PCNA deubiquitination in cooperation with the USP1-UAF1 complex.
EXPERIMENTAL PROCEDURES
Cell Culture, Reagents, and Antibodies
Human embryonic kidney 293T (HEK293T) cells and the immortalized normal retinal pigment epithelial (RPE) cells were maintained in Dulbecco's modified Eagle's medium and Dulbecco's modified Eagle's medium/F-12 medium respectively, containing 10% fetal bovine serum (Hyclone), 100 units/ml penicillin G, and 100 μg/ml streptomycin. RPE cells stably expressing CFP-ELG1 protein were cultured as described previously (20). Methyl methane sulfate (MMS) was purchased from Sigma. The following antibodies were used: anti-PCNA (PC10), anti-FANCD2, anti-ubiquitin (P4D1), and anti-RFC4 antibodies (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)); anti-USP1 antibody (Bethyl Laboratories); anti-FLAG, anti-Myc, anti-RFC1, and anti-tubulin antibodies (Abcam); anti-histone H3 and anti-histone H4 antibodies (Upstate); and anti-HA (HA-7) antibody (Sigma). The antibody against human ELG1 was raised in rabbits against N-terminal 1–297 amino acid fragments of the protein.
DNA Constructs and Small Interfering RNAs (siRNAs)
Plasmids expressing full-length wild type ELG1 (p3XFLAG-ELG1) protein were described previously (20). The N-terminal deletion mutants of ELG1 were generated by PCR using specific primers with p3XFLAG-ELG1 as a template and subsequent cloning into the p3XFLAG-CMV10 mammalian expression vector. Site-directed mutagenesis was performed to generate a UAF1 interaction mutant of ELG1 using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions with p3XFLAG-ELG1 as a template. The DNA sequences of all constructs were verified by DNA sequencing. Plasmid expressing Myc-USP1 was as previously described (9). Plasmid expressing Myc-UAF1-FLAG was a gift from Dr. J. Jung (University of Southern California).
ON-TARGETplus NON-targeting pool (catalog number D-001810) and ON-TARGETplus SMART pool siRNAs for ELG1 (catalog number L-004738), RFC1 (catalog number L-009290), RFC4 (catalog number L-008691), CTF18 (catalog number L-013915), and RAD17 (catalog number L-003294) were purchased from Dharmacon. To target the 3′-untranslated region (UTR) of the ELG1 gene, siRNAs with the following sense and antisense sequences were purchased from Dharmacon and used: 1, 5′-GGA AGG UAG AGU UCA UUA AUU-3′ (sense) and 5′-UUC CUU CCA UCU CAA GUA AUU-3′ (antisense); 2, 5′- GUA UAU UUC UCG AUG UAC A-3′ (sense) and 5′-UGU ACA UCG AGA AAU AUA CUU-3′ (antisense).
Transfections and RNA Interference
Transfections of plasmid DNA, single siRNA synthetic duplexes, or SMART pool siRNAs (50–100 nm) were carried out using Lipofectamine2000 (Invitrogen) according to the manufacturer's instructions, and cells were further incubated for 48 h prior to harvesting. For RNA interference, transfection was performed two times with an interval of 24 h. In some experiments, the wild type or mutants of ELG1 were introduced into cells in which the endogenous ELG1 was forcefully reduced by knockdown of ELG1 with siRNA targeting the 3′-UTR of the ELG1 gene. In these experiments, plasmids expressing the wild type or mutant ELG1 were transfected 12 h after the first knockdown of ELG1.
MMS Treatment and Confocal Microscopy
RPE cells stably expressing CFP-ELG1 protein were plated in LabTek chamber slides (Nunc) 1 day prior to treatment of MMS. Cells were treated with 0.01% MMS for 1 h, washed with PBS three times, and then incubated for 12 h in growth medium. Cells were fixed with 50% (v/v) methanol at −20 °C for 15 min and 50% (v/v) acetone at room temperature for 1 min before staining with an anti-PCNA antibody. Fluorescence-conjugated anti-rabbit IgG antibodies (Invitrogen/Molecular Probes) were used as a secondary antibody, followed by washing with phosphate-buffered saline with 0.01% Tween 20. Confocal images were collected with a Zeiss LSM 510 NLO Meta system mounted on a Zeiss Axiovert 200M microscope with an oil immersion Plan-Apochromat ×63/1.4 differential interference contrast objective lens.
GST Pull-down Assay
GST fusion proteins were expressed in Escherichia coli strain BL21 (DE3)-RIL (Stratagene) and purified using glutathione-Sepharose beads (GE Healthcare). For the in vitro binding assay, cells were lysed with buffer X (100 mm Tris-HCl (pH 8.5), 250 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 100 μm NaVO4, 50 mm NaF, 2 mg/ml bovine serum albumin, and protease inhibitors). Extracts were incubated with 20 μl of glutathione-Sepharose beads (Amersham Biosciences) preincubated with 10 μg of purified GST fusion proteins. After incubation for 1 h at 4 °C, the beads were washed with buffer X, and the eluted proteins were used for immunoblot analysis and Coomassie Blue staining.
Immunoprecipitation and Immunoblot Analysis
The Triton X-100-insoluble fraction from the Triton X-100-soluble fraction was isolated with the methods described previously (7) with slight modifications. In brief, harvested cells were resuspended in buffer A (100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 10 mm Pipes (pH 6.8), 1 mm EGTA, 0.2% Triton X-100, 100 μm NaVO4, 50 mm NaF, and protease inhibitors (Roche Applied Science)) and incubated for 5 min on ice with gentle inverting. The supernatants were recovered as the “soluble fraction” after centrifugation. Followed by washing with the same buffer, the pellet was resuspended either in buffer X (100 mm Tris-HCl (pH 8.5), 250 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 100 μm NaVO4, 50 mm NaF, 2 mg/ml bovine serum albumin, and protease inhibitors) for immunoprecipitation or in buffer B (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.1% SDS, 100 μm NaVO4, 50 mm NaF, and protease inhibitors) for immunoblotting. Followed by a 10-min incubation on ice, the samples were sonicated and then incubated for another 10 min on ice before centrifugation to isolate the “chromatin-bound fraction.”
For immunoprecipitation, proteins were precleared by preincubation with protein G-Sepharose beads (GE Healthcare) and incubated with specific antibodies. The complex recognized by antibodies was precipitated with protein G-Sepharose beads. Proteins were separated on NuPAGE® Novex 4–12% BisTris gel (Invitrogen) and transferred to polyvinylidene difluoride membranes (Bio-Rad) for immunoblotting with antibodies following the manufacturer's recommendations. The band intensity was determined by using ImageJ software (version 1.41).
Homologous Recombination (HR) Assay
To determine double strand break (DSB)-induced HR frequency, we used a U2OS cell line carrying a DR-green fluorescent protein reporter in genome (20, 25). A DSB was introduced in the reporter in genome by expressing I-SceI endonuclease, and HR frequency was determined by comparing green fluorescent protein-positive cells upon DSB with no DSB.
SubF Mutation Analysis
SubF assay was performed with the methods described previously (26) with slight modifications. In brief, after irradiation with 0 or 500 J/m2 UV in a 10-μl droplet, 2 μg of shuttle plasmid pSP189 was introduced into HEK293T cells by using Lipofectamine2000 (Invitrogen). After incubation at 37 °C for 48 h, plasmid DNA was recovered by using a conventional plasmid isolation method. Isolated plasmid DNA was treated with DpnI and RNase I and introduced into MBM7070 E. coli strains, that carry an amber mutation in the lacZ gene, by electroporation. Transformants were plated on an LB plate with ampicillin, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal), and isopropyl-β-d-thiogalactoside. Mutation frequencies were determined as the number of white colonies over total colonies.
RESULTS
ELG1 Interacts with the USP1-UAF1 Complex and PCNA
To understand the regulatory mechanisms for USP1 activity, USP1-interacting proteins were purified from HeLa cells stably expressing epitope-tagged USP1 as previously described to identify UAF1 (13). From this purification, we identified human ELG1 as a protein interacting with USP1 (Fig. 1A). To check whether ELG1 is present in the USP1-UAF1 complex in vivo, we performed co-immunoprecipitations. Endogenous ELG1 was immunoprecipitated together with endogenous USP1 (Fig. 1B). Furthermore, UAF1 and USP1 were immunoprecipitated by FLAG-ELG1 (Fig. 1C) (data not shown). In addition, we observed that FLAG-ELG1 interacted with RFC4 (Fig. 1C), consistent with data previously reported in yeast (24). Therefore, ELG1 interacts with the deubiquitinating enzyme, USP1, and its associated factor, UAF1.
FIGURE 1.
ELG1 interacts with USP1-UAF1 complex. A, the USP1 complex was purified from HeLa nuclear extracts and stained by Coomassie blue stain. Proteins indicated were identified by mass spectrometry. B, nuclear extracts from HeLa cells were immunoprecipitated (IP) with anti-USP1-antibody or IgG control antibody. The immunoprecipitated proteins were identified with the antibodies indicated. C, HEK293T cells were transected with FLAG-ELG1 expression vectors or control vectors (Ctrl). Total extracts from the transfected cells were IP after 48 h with anti-FLAG-antibody. The immunoprecipitated proteins were identified with the antibodies indicated.
Because ELG1 interacts with the USP1-UAF1 complex and the yeast Elg1p protein interacts with PCNA (24), we examined whether human ELG1 could also interact with human PCNA. GST-fused PCNA (GST-PCNA) co-precipitated FLAG-ELG1 (Fig. 2A), whereas GST protein alone did not. In addition, FLAG-ELG1 co-immunoprecipitated PCNA in vivo (Fig. 2B). Previously, we demonstrated that ELG1 protein formed damage-induced nuclear foci at stalled DNA replication forks (20). The interaction between ELG1 and PCNA (Fig. 2, A and B) as well as the foci formation of PCNA at DNA replication forks (27) suggested that ELG1 may interact with PCNA localized at stalled DNA replication forks. To test this hypothesis, we monitored ELG1 and PCNA foci formation in RPE cells after 0.01% MMS treatment. Consistent with this hypothesis, ∼70% of PCNA foci co-localized with ELG1 foci (Fig. 2C). Therefore, ELG1 interacts with both the USP1-UAF1 complex and its target, PCNA.
FIGURE 2.
ELG1 interacts with PCNA. A, purified GST or GST-PCNA proteins were mixed with protein extracts containing FLAG-ELG1 and pulled down for immunoblot analysis. B, HEK293T cells were transected with FLAG-ELG1 expression vectors. Immunoprecipitations were carried out on nuclear extracts using an anti-PCNA antibody, and the eluate was then subjected to immunoblot analysis. C, ELG1 and PCNA co-localize in response to DNA damage. RPE cells stably expressing CFP-ELG1 protein were treated with 0.01% MMS for 1 h and incubated in fresh medium for another 12 h. Cells were then fixed, permeabilized, and stained with anti-PCNA antibody for confocal microscopy analysis. The co-localization was shown by an overlay view of the blue and red channels in the same field. DIC, differential interference contrast image. Bar, 10 (top) and 20 μm (bottom).
ELG1 Knockdown Increases the Level of Monoubiquitinated PCNA in Chromatin
In response to DNA damage and stalled DNA replication, PCNA is monoubiquitinated (7). The interaction of ELG1 with PCNA and the USP1-UAF1 complex (Figs. 1 and 2) suggested that ELG1 might affect the level of PCNA monoubiquitination. To test this hypothesis, we reduced the expression of ELG1 by siRNA knockdown and determined the level of PCNA ubiquitination in chromatin. Six siRNAs targeting different regions of the ELG1 mRNA caused an increase in PCNA monoubiquitination (Fig. 3, A and B). The enhanced PCNA monoubiquitination, caused by siRNA targeting 3′-UTR of ELG1 mRNA, was rescued by ectopic expression of an ELG1 that was resistant to knockdown (Fig. 3C), indicating that the effect was specific. Similarly, PCNA monoubiquitination is increased by knockdown of USP1 or UAF1 (9). The USP1 protein level was not significantly affected by ELG1 knockdown (Fig. 3A), suggesting that ELG1 does not regulate PCNA ubiquitination by modulating the expression or stability of the USP1 protein. Additionally, there was no difference in the cell cycle profile between control and ELG1-silenced cells (Fig. 3D), excluding the possibility that the increased PCNA ubiquitination results indirectly from the accumulation of S-phase cells by ELG1 knockdown.
FIGURE 3.
ELG1 knockdown increases the level of monoubiquitinated PCNA in chromatin. A, HEK293T cells were transfected with various siRNAs (si-ELG1) targeting the coding region of the ELG1 mRNA or 3′-untranslated region (UTR) as indicated. Histone H4 protein was used as a loading control. PCNA-Ub, monoubiquitinated PCNA. B, nuclear extracts were prepared 48 h after transfection of HEK293T cells with siRNA targeting ELG1 or control siRNA and then subjected to immunoprecipitation with anti-PCNA antibody. Monoubiquitinated PCNA was identified with anti-ubiquitin (P4D1) antibody. An arrowhead indicates monoubiquitinated PCNA. C, the increase in PCNA monoubiquitination following ELG1 knockdown was not an off-target effect. HEK293T cells were cotransfected with the combination of siRNA targeting the 3′-UTR of ELG1 (#2 siRNA in A) and a FLAG-ELG1 expression vector as indicated. D, ELG1 knockdown does not affect cell cycle profile. HEK293T cells were transfected with siRNA targeting ELG1 or control siRNA twice with an interval of 24 h. Cells were harvested after 48 h from the second transfection, and their cell cycle profiles were determined by flow cytometry analysis. The x and y axes represent DNA content as measured by 7-amino-actinomycin D (7-AAD) staining and cells undergoing DNA replication as measured by bromodeoxyuridine incorporation, respectively. Each rectangle encompasses cells in G1, S, and G2/M phases of the cell cycle clockwise from the lower left. The numbers indicate the percentage of cells in the rectangle out of total cells. E, an increase in PCNA monoubiquitination was not observed following canonical RFC subunit (RFC1 or RFC4) knockdown. F, knockdown of other alternative RFC large subunits (CTF18 or RAD17) did not cause any significant change in PCNA monoubiquitination. The chromatin-bound fractions were prepared 72 h after transfection of HEK293T cells with the indicated siRNAs or a plasmid expressing FLAG-tagged ELG1 and then subjected to immunoblot analyses. The #3 siRNA in A that targets the coding region of ELG1 was used for ELG1 knockdown in B, E, and F.
In general, RFC complexes play essential roles in PCNA-related processes during DNA replication and repair (15). We checked whether other RFC complexes affect the level of PCNA ubiquitination. Unlike ELG1, when the expression of either RFC1 or RFC4 was reduced by siRNA knockdown, there was no increase in PCNA ubiquitination (Fig. 3E). Human cells have two alternative RFC complexes, CTF18-RFC (16, 18) and RAD17-RFC (19). The level of PCNA ubiquitination was not affected by reducing the expression of CTF18 or RAD17 (Fig. 3F). In conclusion, the regulation of the level of PCNA ubiquitination exclusively depends on ELG1 but not on other RFC complexes. Furthermore, because RFC4 knockdown did not increase PCNA ubiquitination, it appears that the function of ELG1 in regulating PCNA ubiquitination is independent of its role as an alternative RFC complex.
ELG1 Is Required for USP1-mediated PCNA Deubiquitination
The increase in PCNA monoubiquitination following ELG1 knockdown (Fig. 3) and the physical interactions among ELG1, PCNA, and USP1-UAF1 complex (Figs. 1 and 2) raised the possibility that ELG1 down-regulates PCNA monoubiquitination by regulating USP1 activity or recruiting USP1 to monoubiquitinated PCNA. To explore this hypothesis, we ectopically overexpressed ELG1, USP1, or both proteins in HEK293T cells and investigated the level of PCNA monoubiquitination (Fig. 4A). USP1 overexpression suppressed PCNA monoubiquitination induced by 20J/m2 UV irradiation, as reported previously (9). When ELG1 was overexpressed, the level of monoubiquitinated PCNA was significantly reduced. Overexpression of both ELG1 and USP1 reduced the level of PCNA monoubiquitination further than the overexpression of a single protein (Fig. 4A). There was a strong increase in USP1 protein levels when both ELG1 and USP1 were co-expressed (compare lane 3 and lane 7), which appears to be due to an overexpression effect because endogenous USP1 levels were not affected by ELG1 knockdown (Fig. 3A). The suppression of PCNA monoubiquitination by USP1 overexpression was prevented when ELG1 expression was reduced by siRNA knockdown (Fig. 4B), indicating that ELG1 is required for the down-regulation of PCNA monoubiquitination by USP1. USP1 also deubiquitinates FANCD2, which is a major regulatory protein in the Fanconi anemia pathway (13). In contrast to USP1 knockdown, FANCD2 monoubiquitination was not affected by ELG1 knockdown (Fig. 4C). Taken together, these data suggest that ELG1 and USP1 cooperate to down-regulate PCNA monoubiquitination and that ELG1 specifically down-regulates PCNA monoubiquitination without affecting FANCD2 monoubiquitination.
FIGURE 4.
ELG1 facilitates USP1-mediated PCNA deubiquitination. A, overexpression of ELG1 and USP1 decreased UV-induced PCNA monoubiquitination (PCNA-Ub). HEK293T cells were transected with FLAG-ELG1 or Myc-USP1 expression vectors. Cells were irradiated with 20 J/m2 UV after 48 h and incubated further for 6 h. The soluble (S) and chromatin-bound (B) fractions were isolated and subjected to immunoblot analysis. Histone H3 and tubulin proteins were used as loading controls for chromatin-bound and soluble fractions, respectively. B, the increase in PCNA monoubiquitination following USP1 knockdown depends on ELG1. HEK293T cells were cotransfected with different combinations of control (Ctrl) or ELG1 siRNA with a Myc-USP1 expression vector as indicated. The chromatin-bound fractions were analyzed by immunoblotting. C, ELG1 knockdown did not affect FANCD2 monoubiquitination. The chromatin-bound fractions were prepared 72 h after transfection of HEK293T cells with the indicated siRNAs and then subjected to immunoblot analyses. D, the I-SceI-induced HR assay was performed as described previously (20) after knockdown of USP1 or ELG1. BLM depletion was used as a positive control. The inset shows the structure of the recombination substrate DR-green fluorescent protein and the location of the I-SceI recognition site. Error bars, S.D. of triplicate experiments. E, SupF mutation frequencies for UV-irradiated plasmids are indicated for control siRNA and ELG1 siRNA-treated cells. Error bars, S.D. of triplicate experiments. The number in the graph indicates probability (p value) calculated by Student's t tests.
Previously, we demonstrated that ELG1 is involved in DSB-induced HR. ELG1 knockdown reduced the HR frequency in the I-SceI DSB-induced HR assay (20). Furthermore, a deficiency of USP1 is also known to result in decreased HR frequency in the same assay (28). To further address the cooperation of ELG1 and USP1, we examined the effects of simultaneous knockdown of ELG1 and USP1 on I-SceI DSB-induced HR. Neither additive nor synergistic effects were observed, compared with the single knockdowns (Fig. 4D), suggesting that two proteins function in the same DNA repair pathway.
Accumulation of monoubiquitinated PCNA in the cells by USP1 knockdown leads to a slight increase of mutation frequency, probably due to more frequent use of TLS polymerase to bypass DNA damage (9). Similarly, there was significant increase of mutation frequency by ELG1 knockdown (Fig. 4E), further strengthening the role of ELG1 in USP1-mediated down-regulation of PCNA monoubiquitination.
UAF1-interacting Region of ELG1 Is Important for the Down-regulation of PCNA Monoubiquitination
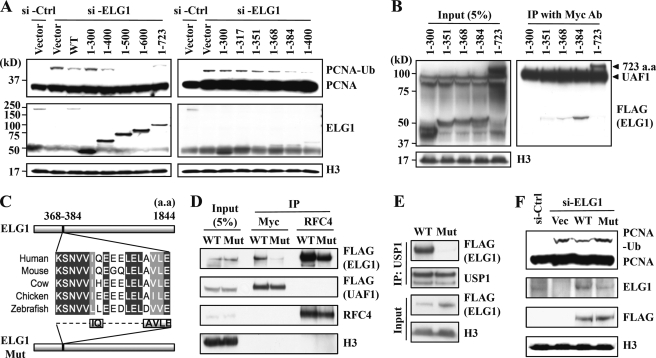
Last, to better understand the interaction between ELG1 and the USP1-UAF1 complex, we determined the region of ELG1 responsible for the down-regulation of PCNA monoubiquitination. The level of PCNA monoubiquitination was monitored in cells in which endogenous ELG1 was silenced and various N-terminal expressing mutants of ELG1 were introduced. First, we located the activity of the down-regulation of PCNA monoubiquitination to the amino acids between positions 300 and 400 of the ELG1 protein (Fig. 5A, left). We further defined this activity to amino acids between 368 and 384 of the ELG1 protein (Fig. 5A, right).
FIGURE 5.
N terminus of ELG1 is important for the down-regulation of PCNA monoubiquitination and UAF1 interaction. A, amino acids 368–384 of ELG1 are important for the down-regulation of PCNA monoubiquitination (PCNA-Ub). HEK293T cells were co-transfected with the combination of siRNA targeting the 3′-UTR of ELG1 and vectors expressing various sizes of N terminus of ELG1 as indicated. The chromatin-bound fractions were analyzed to monitor PCNA ubiquitination. B, amino acids (a.a.) 368–384 of the ELG1 protein are important for its interaction with UAF1. HEK293T cells were transfected with a plasmids expressing different sizes of the FLAG-tagged N terminus of ELG1 and a plasmid expressing full-length Myc-UAF1-FLAG. Immunoprecipitations (IP) were carried out on nuclear extracts using an anti-Myc antibody. The immunoprecipitated proteins were analyzed by immunoblotting to detect the presence of the ELG1 protein. C, the sequence of the human ELG1 between amino acids 368 and 384 is evolutionarily conserved. The location and peptide sequences of amino acids 368–384 are shown in the diagram. The deleted amino acids (368–372 and 375–380) to generate a UAF1 interaction mutant of ELG1 (ELG1 Mut) are shown at the bottom as dotted lines. D, HEK293T cells were transected with vectors expressing wild type (WT) or deletion mutant (Mut) of ELG1 along with a Myc-UAF1-FLAG expression vector. Nuclear extracts were immunoprecipitated with anti-Myc or anti-RFC4 antibodies and analyzed by immunoblotting. E, HEK293T cells were transected with vectors expressing wild type or deletion mutant of ELG1. Nuclear extracts were immunoprecipitated with anti-USP1 antibody and analyzed by immunoblotting. F, HEK293T cells were cotransfected with the combination of ELG1 siRNA and vectors expressing wild type or deletion mutant of ELG1. The nuclear extracts were analyzed to monitor PCNA ubiquitination. The #2 siRNA in Fig. 3A that targets the 3′-UTR region of ELG1 was used for ELG1 knockdown in A and F.
ELG1 interacts with the USP1-UAF1 complex (Fig. 1) and is required for USP1-mediated down-regulation of PCNA monoubiquitination (Fig. 4B). Therefore, the domain of ELG1 responsible for the down-regulation of PCNA monoubiquitination would probably be a domain required for the interaction of ELG1 with the USP1-UAF1 complex. We examined the interaction between various deletion mutants of ELG1 and UAF1. Consistent with our hypothesis, two ELG1 mutant proteins expressing either the N-terminal 384 (positions 1–384) or 723 (positions 1–723) amino acids interacted strongly with UAF1 (Fig. 5B). In contrast, two ELG1 mutant proteins expressing the N-terminal 351 (positions 1–351) or 368 (positions 1–368) amino acids had significantly reduced interactions, and an ELG1 mutant protein having only the N-terminal 300 amino acids did not have any detectable interaction (Fig. 5B). The sequences of this region of ELG1 are highly conserved across several species (Fig. 5C). We generated a putative UAF1 interaction mutant of ELG1 by introducing partial deletions in the region (Fig. 5C). Although the mutant of ELG1 was still able to interact with RFC4, this deletion in ELG1 consequently caused defects in interaction with UAF1 and USP1 (Fig. 5, D and E). Furthermore, the UAF1 interaction mutant of ELG1 could not restore PCNA monoubiquitination that was increased by ELG1 depletion (Fig. 5F). Taken together, these data indicate that the down-regulation of PCNA monoubiquitination by ELG1 depends on its interaction with the USP1-UAF1 complex.
DISCUSSION
Precise regulation of PCNA ubiquitination is important for TLS pathway of postreplication repair as well as suppressing the mutagenic effects of TLS pathway in unperturbed cells (6, 7, 9). Although proteosomal degradation of USP1 was shown as a mechanism regulating PCNA monoubiquitination in DNA-damaged cells (9), several reports (11, 12) suggested the presence of additional mechanisms to regulate USP1 activity. Here we demonstrated that ELG1 was required for the down-regulation of PCNA monoubiquitination through its interaction with the USP1-UAF1 complex. Especially, the UAF1 interaction in the N-terminal region of ELG1 was important for the down-regulation of PCNA monoubiquitination by ELG1.
How does ELG1 specifically affect USP1 activity toward monoubiquitinated PCNA? One possibility is that the ELG1-RFC complex unloads monoubiquitinated PCNA from chromatin, thereby providing monoubiquitinated PCNA to USP1 as a substrate. However, several pieces of evidence argue against this model. Previous in vitro studies in yeast showed a dependence of PCNA unloading on the ATPase activities of RFC1 or CTF18 (29, 30). A mutant ELG1 expressing only the N terminus that lacks the ATPase domain could still reduce the level of PCNA monoubiquitination (Fig. 5A). In addition, the N-terminal mutant of ELG1 would probably also be defective in forming the functional RFC complex. Previous reports showed that the C-terminal regions of the yeast Elg1p protein and the large subunit of the human RFC complex are essential for complex formation with the RFC2 to -5 core subunits (31–33). These results indicate that intact alternative RFC complex formation is not necessary for ELG1 to down-regulate PCNA monoubiquitination. In accordance, our data demonstrate that the knockdown of RFC4 did not alter PCNA monoubiquitination (Fig. 3E). Collectively, it is unlikely that PCNA unloading by the ELG1-RFC complex from chromatin contributes to the down-regulation of PCNA monoubiquitination.
Another possibility is that ELG1 enhances or stabilizes the formation of the USP1-UAF1 complex. USP1 is degraded in a proteasome-dependent manner when DNA is damaged (9). However, the complex formation of USP1 with UAF1 stabilizes USP1 as well as stimulates the activity of USP1 (13). When we tested the possibility by checking the interaction between USP1 and UAF1 in ELG1 knockdown, we could not find any significant change in their interaction (data not shown), which excludes a possible role of ELG1 as a stabilizer for USP1-UAF1 interaction.
The USP1-UAF1 complex can deubiquitinate both PCNA and FANCD2 (9, 13). Unexpectedly, we found that ELG1 knockdown only affected PCNA ubiquitination and not FANCD2 ubiquitination (Fig. 4C). This result suggests that ELG1 promotes the selective recruitment of USP1-UAF1 to monoubiquitinated PCNA through its interaction with PCNA and the USP1-UAF1 complex. The recruitment of the USP1-UAF1 complex to monoubiquitinated PCNA through ELG1 interaction appears to be transient because the chromatin-bound USP1 level was not changed after ELG1 knockdown (Figs. 3A and 4C). Consistently, unlike with ELG1, we did not observe the formation of either USP1 or UAF1 foci in response to DNA damage (data not shown).
The absence of ELG1 led to the accumulation of monoubiquitinated PCNA (Fig. 3A). In a previous report (20), we showed that ELG1 knockdown increased spontaneous DNA damage. ELG1 may therefore have a physiological role in repairing spontaneous DNA damage with the USP1-UAF1 complex despite the fact that the level of ELG1 without DNA damage is relatively low (20). Several types of spontaneous DNA damage could be escaped to induce PCNA monoubiquitination in the absence of ELG1. Constant oxidative stress and DNA sequences that could stall DNA replication, such as repetitive sequences, could be sources for spontaneous DNA damage in the absence of ELG1. Spontaneous DNA damage could be produced by abnormal Okazaki fragment maturation in the lagging strand synthesis because yeast Elg1p interacts with Rad27p (a yeast homologue of human FEN1 flap endonuclease specific to remove RNA primers during lagging strand synthesis) (24).
Besides forming a deubiquitinating enzyme complex with USP1, UAF1 makes other complexes with USP12 and USP46, respectively (34). Maintaining the optimal ratio among these complexes in response to cellular needs may be important for each complex to efficiently execute its required functions. In summary, our results support a model in which ELG1 facilitates the formation and/or appropriate recruitment of the USP1-UAF1 complex when PCNA deubiquitination is required, which will reduce the likelihood of the mutagenic effects by the TLS pathway after DNA-damaged legions are bypassed.
Acknowledgments
We thank D. Bodine (NHGRI, National Institutes of Health (NIH)), M. Lichten (NCI, NIH), P. Liu (NHGRI, NIH), P. Meltzer (NHGRI, NIH), and Y. Shiloh (Tel Aviv University) for helpful discussions; J. Jung (University of Southern California) for a plasmid expressing UAF1; F. Candotti (NHGRI, NIH) and members of the Myung laboratory for comments on the manuscript; and J. Fekecs (NHGRI, NIH) for figure preparation. K. M. especially thanks E. Cho.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1-HL52725, RO1-DK43889, and PO1-CA092584 (to A. D.) and National Institutes of Health, National Human Genome Research Institute, Intramural Research Program Grant HG012003-07 (to K. M.). This work was also supported by Korea Research Foundation Grant KRF-2007-357-C00092 funded by the Korean Government (to K. Y. L.).
- PCNA
- proliferating cell nuclear antigen
- PPR
- postreplication repair
- TLS
- translesion synthesis
- RFC
- replication factor C
- HEK293T
- human embryonic kidney 293T
- MMS
- methyl methane sulfate
- siRNA
- small interfering RNA
- UTR
- untranslated region
- DSB
- double strand break
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GST
- glutathione S-transferase
- HR
- homologous recombination.
REFERENCES
- 1.Moldovan G. L., Pfander B., Jentsch S. (2007) Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 2.Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 3.Wang S. C., Nakajima Y., Yu Y. L., Xia W., Chen C. T., Yang C. C., McIntush E. W., Li L. Y., Hawke D. H., Kobayashi R., Hung M. C. (2006) Nat. Cell Biol. 8, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 4.Lee K. Y., Myung K. (2008) Mol. Cells 26, 5–11 [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. (2004) EMBO J. 23, 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I. (2005) Science 310, 1821–1824 [DOI] [PubMed] [Google Scholar]
- 7.Kannouche P. L., Wing J., Lehmann A. R. (2004) Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 8.Plosky B. S., Vidal A. E., Fernández de Henestrosa A. R., McLenigan M. P., McDonald J. P., Mead S., Woodgate R. (2006) EMBO J. 25, 2847–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang T. T., Nijman S. M., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., D'Andrea A. D. (2006) Nat. Cell Biol. 8, 339–347 [DOI] [PubMed] [Google Scholar]
- 10.Motegi A., Liaw H. J., Lee K. Y., Roest H. P., Maas A., Wu X., Moinova H., Markowitz S. D., Ding H., Hoeijmakers J. H., Myung K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niimi A., Brown S., Sabbioneda S., Kannouche P. L., Scott A., Yasui A., Green C. M., Lehmann A. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown S., Niimi A., Lehmann A. R. (2009) Cell Cycle 8, 689–692 [DOI] [PubMed] [Google Scholar]
- 13.Cohn M. A., Kowal P., Yang K., Haas W., Huang T. T., Gygi S. P., D'Andrea A. D. (2007) Mol. Cell 28, 786–797 [DOI] [PubMed] [Google Scholar]
- 14.Johnson A., O'Donnell M. (2005) Annu. Rev. Biochem. 74, 283–315 [DOI] [PubMed] [Google Scholar]
- 15.Majka J., Burgers P. M. (2004) Prog Nucleic Acids Res. Mol Biol 78, 227–260 [DOI] [PubMed] [Google Scholar]
- 16.Merkle C. J., Karnitz L. M., Henry-Sánchez J. T., Chen J. (2003) J. Biol. Chem. 278, 30051–30056 [DOI] [PubMed] [Google Scholar]
- 17.Banerjee S., Sikdar N., Myung K. (2007) Biochem. Biophys. Res. Commun. 362, 546–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bermudez V. P., Maniwa Y., Tappin I., Ozato K., Yokomori K., Hurwitz J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10237–10242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsey-Boltz L. A., Bermudez V. P., Hurwitz J., Sancar A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11236–11241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sikdar N., Banerjee S., Lee K. Y., Wincovitch S., Pak E., Nakanishi K., Jasin M., Dutra A., Myung K. (2009) Cell Cycle 8, 3199–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee S., Myung K. (2004) Eukaryot. Cell 3, 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellaoui M., Chang M., Ou J., Xu H., Boone C., Brown G. W. (2003) EMBO J. 22, 4304–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Aroya S., Koren A., Liefshitz B., Steinlauf R., Kupiec M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9906–9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanellis P., Agyei R., Durocher D. (2003) Curr. Biol. 13, 1583–1595 [DOI] [PubMed] [Google Scholar]
- 25.Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F. J., Livingston D. M. (2006) Mol. Cell 22, 719–729 [DOI] [PubMed] [Google Scholar]
- 26.Ishizaki K., Nishizawa K., Mimaki S., Aizawa S. (1996) Mutat. Res. 364, 43–49 [DOI] [PubMed] [Google Scholar]
- 27.Bravo R., Macdonald-Bravo H. (1987) J. Cell Biol. 105, 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J. M., Parmar K., Huang M., Weinstock D. M., Ruit C. A., Kutok J. L., D'Andrea A. D. (2009) Dev. Cell 16, 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibahara K., Stillman B. (1999) Cell 96, 575–585 [DOI] [PubMed] [Google Scholar]
- 30.Bylund G. O., Burgers P. M. (2005) Mol. Cell. Biol. 25, 5445–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidson M. B., Brown G. W. (2008) DNA Repair 7, 1221–1232 [DOI] [PubMed] [Google Scholar]
- 32.Uhlmann F., Cai J., Gibbs E., O'Donnell M., Hurwitz J. (1997) J. Biol. Chem. 272, 10058–10064 [DOI] [PubMed] [Google Scholar]
- 33.Uhlmann F., Gibbs E., Cai J., O'Donnell M., Hurwitz J. (1997) J. Biol. Chem. 272, 10065–10071 [DOI] [PubMed] [Google Scholar]
- 34.Cohn M. A., Kee Y., Haas W., Gygi S. P., D'Andrea A. D. (2009) J. Biol. Chem. 284, 5343–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]