Abstract
A disintegrin and metalloproteinase 10 (ADAM10) is a type I transmembrane glycoprotein responsible for the ectodomain shedding of a number of proteins implicated in the pathogenesis of diseases ranging from cancer to Alzheimer Disease. ADAM10 is synthesized in an inactive form, which is proteolytically activated during its forward transport along the secretory pathway and at the plasma membrane. Therefore, modulation of its trafficking could provide a mechanism to finely tune its shedding activity. Here we report the identification of an endoplasmic reticulum (ER) retention motif within the ADAM10 intracellular C-terminal tail. Sequential deletion/mutagenesis analyses showed that an arginine-rich (723RRR) sequence was responsible for the retention of ADAM10 in the ER and its inefficient surface trafficking. Mutating the second arginine to alanine was sufficient to allow ER exit and surface expression in both heterologous cells and hippocampal neurons. As synapse-associated protein 97 (SAP97) binds ADAM10 at its cytoplasmic tail and facilitates forward ADAM10 trafficking in neurons, we tested whether SAP97 could modulate ER export. However, neither expression nor Ser-39 phosphorylation of SAP97 in heterologous cells or hippocampal neurons were sufficient to allow the ER exit of ADAM10, suggesting that other signaling pathways or alternative binding partners are responsible for ADAM10 ER exit. Together, these results identify a novel mechanism regulating the intracellular trafficking and membrane delivery of ADAM10.
Keywords: Cell/Intracellular Processing, Cell/Trafficking, Protease/ADAM/ADAMts, Protein/Processing, Subcellular Organelles/Endoplasmic Reticulum, ADAM10
Introduction
A disintegrin and metalloproteinase 10 (ADAM10)2 belongs to a large family of membrane-anchored metalloproteases, which are known as the ADAM protein family. ADAMs mediate the proteolytic cleavage of transmembrane proteins in their juxtamembrane region, causing their shedding, i.e. the release of their extracellular domain in a soluble form. In addition, through the intracellularly retained stubs, ADAMs can initiate the activation of intracellular signaling cascades. Because of their metalloprotease, integrin binding, cell adhesion, and signaling functions, ADAMs are well positioned to coordinate cellular processes that are required for neural development, plasticity, and repair (1–3).
ADAM10 works as a sheddase for a large number of transmembrane proteins involved in a variety of biological functions, and has been implicated in the pathogenesis of diseases ranging from cancer to Alzheimer Disease (AD) (4, 5). Because of these links, much effort is currently directed toward developing tools which modulate ADAM10 activity and can be used to target these pathologies.
ADAM10 is a multidomain transmembrane glycoprotein which is expressed ubiquitously (6). It contains an N-terminal signal sequence followed by a prodomain, a metalloprotease domain, a disintegrin domain, a cysteine-rich region, an EGF-like repeat, a transmembrane domain and a SH3-binding cytoplasmic tail (7). ADAM10 is synthesized in an inactive form that carries a proprotein convertase (PC) recognition sequence between the prodomain and the catalytic domain. Both PC7 and furin can cleave ADAM10 at the predicted PC cleavage motif to yield a mature, active form (8). ADAM10 cleavage and maturation occur in the trans-Golgi network, in vesicles of the secretory pathway and at the cell surface (9, 10); indeed, it has been suggested that the active form of ADAM10 can exert its catalytic activity both along the secretory system and at the plasma membrane (7). Yet, despite the importance of forward secretory trafficking for ADAM10 activity, the mechanisms regulating its trafficking and localization are largely unknown.
Multiple signaling pathways could regulate ADAM10 trafficking and, consequently, modulate its activity in different cellular systems. For instance, calcium influx stimulates ADAM10 activity in fibroblasts, an effect that requires the intracellular C terminus (11). In neurons, the interaction between ADAM10 and synapse-associated protein-97 (SAP97), a protein involved in the dynamic trafficking of proteins to excitatory synapses, regulates the localization of ADAM10 at the postsynaptic membrane (12). However, before reaching the plasma membrane and inserting into synapses, newly synthesized ADAM10 must be transported through the endoplasmic reticulum (ER)-Golgi secretory pathway via a set of processes that remain poorly understood.
ER retention is a common mechanism used by many cell types to control the forward trafficking and surface expression of integral membrane proteins. Specific ER retention/retrieval signals and ER export signals have been identified in the intracellular domains of various channels and receptors, and these signals govern their surface density (13–15). ER exit has also been reported to be a rate-limiting step for the surface trafficking of ADAM12 and ADAM22, two members of the ADAMs family. This process is regulated by ER retention signals in their cytoplasmic tails, and binding to 14-3-3 proteins allows ER release and membrane targeting of ADAM22 by masking the retention signals (16–18). We have now investigated the trafficking and surface expression of ADAM10, and report the identification of a novel arginine-rich ER retention motif within its intracellular C terminus. Our results identify a novel mechanism regulating the intracellular trafficking and membrane delivery of ADAM10.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used: mouse anti-SAP97 (NeuroMab, Davis, CA), rabbit anti-calnexin (Stressgen Biotechnologies, Victoria, Canada), rabbit anti-calreticulin (Affinity Bioreagents, Golden, CO), rat anti-ADAM10 (R&D, Minneapolis, MN), chicken anti-GFP (Millipore, Billenica, MA), goat anti-Tac antibody (Sigma Aldrich, St. Louis, MO), rabbit anti-giantin (Covance, Princeton, NJ); mouse anti-Tac antibody (clone 7G7) was kindly provided by Dr. Bonifacino.
Cell Culture and Transfection
Primary neuronal cultures were prepared from E18-E19 rat hippocampi as described (19). Neurons were transfected using the calcium phosphate precipitation method at 10 days in vitro. COS7 cells were grown in DMEM supplemented with 10% bovine serum, 1 mm sodium pyruvate, and 50 units/ml penicillin/streptomycin. COS7 cells were transiently transfected with cDNA expression constructs using Superfect® Transfection Reagent (Qiagen, Hilden, Germany) and were grown for 24 h before fixation for immunocytochemistry or lysis for Western blot analysis.
Generation of TacADAM10 Chimera and Mutants of TacADAM10 and Full-length ADAM10
Chimeras of the extracellular domain of the human interleukin-2 receptor (Tac) with the intracellular C-terminal domain of mouse ADAM10 were generated by amplifying the ADAM10 C-terminal domain with the following set of primers: forward 5′-TGCCCAAGCTTCCGGATTTATCAAGATTTGCAGTG-3′ and reverse 5′-GCTCTAGATTAGCGTCGCATGTGTCCC-3′. Deletion mutants Tac737Δ, Tac734Δ, and Tac721Δ were generated by PCR amplification using the same forward primer and different reverse primers: 5′-GCTCTAGATTACCTCTGACGCGGGGGCTG-3′ for Tac737Δ, 5′-GCTCTAGATTACGGGGGCTGCTGAATGGG-3′ for Tac734Δ, 5′-GCTCTAGATTATAAAGTGCCTGGAAGTGGTTT-3′ for Tac721Δ. After digestion with HindIII and XbaI, PCR fragments were ligated into the linearized Tac pCDM8 expression vector. TacADAM10, full-length mouse ADAM10 (flADAM10) point mutations, and the stop codon resulting in the flADAM10 721Δ construct were introduced using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), following the manufacturer's instructions. All constructs were verified by sequencing.
Immunofluorescence
To evaluate surface and total staining, transfected COS7 cells or neurons were fixed with 4% paraformaldehyde, 4% sucrose in phosphate-buffered saline pH 7.4, and then incubated with either anti-Tac 7G7 antibody (for Tac constructs) or anti-ADAM10 antibody. To visualize surface expression, cells were then blocked with 4% normal serum, followed by a Cy3-conjugated secondary antibody. Afterward, cells were permeabilized with 0.1% Triton X-100 for 10 min and intracellular expression was determined by incubating cells with the appropriate antibody and labeling the total receptor fraction with a FITC-conjugated secondary antibody. For colocalization experiments, transfected COS7 cells were permeabilized with 0.2% saponin before incubation with anti-Tac 7G7 antibody and anti-calnexin/calreticulin (ER marker) or giantin antibodies (Golgi marker) followed by corresponding secondary antibodies. Wide-field fluorescence images were acquired in a Zeiss Axiovert 200M epifluorescence microscope with a Zeiss ×40 or 25 objective and a CoolSnap CCD camera. Images were analyzed using Metamorph Imaging software (Molecular Devices, Sunnyvale, CA).
Data Analysis
For quantification of surface and total expression intensities, images were acquired using the same settings and exposure times. The average intensity of surface fluorescence staining (Cy3, red) was determined after cell tracing and normalized to the total intensity (FITC, green) to correct for differences in expression. Surface ratios were obtained by dividing the background subtracted Cy3 and FITC fluorescence intensities. Statistical differences were analyzed by ANOVA followed by the Bonferroni test.
Analysis of Protein Glycosylation
Aliquots of transfected COS7 lysates were treated with or without 2 units of N-glycosidase F for 2 h at 37 °C in the buffer recommended by the supplier (Roche Diagnostics, Manheim, Germany). Endoglycosidase H (EndoH; 5 milliunits, Roche Diagnostics) was added to aliquots of transfected COS7 proteins and incubated in 40 mm sodium acetate, pH 5.4 for 17 h at 37 °C. Denaturing buffer was added to digested samples, which were loaded on SDS-PAGE and probed by immunoblotting using goat anti-Tac antibody. A control digestion with no enzyme added demonstrated that samples did not undergo spontaneous degradation during incubations.
RESULTS
A TacADAM10 Chimera Is Not Expressed at the Cell Surface
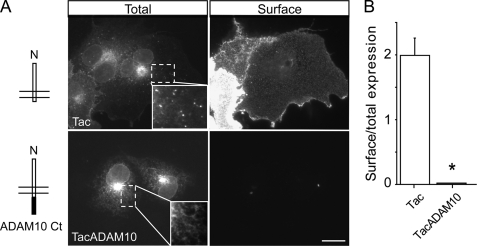
Previous studies suggested that ADAM10 traffics inefficiently to the plasma membrane in recombinant systems and is retained in intracellular compartments, with only a small fraction reaching the cell surface (7). Because we had previously reported that the C terminus of ADAM10 is responsible for its intracellular trafficking (12), we constructed chimeras of the ADAM10 C-terminal tail (695–749 amino acids of mouse ADAM10) with the surface reporter protein Tac (human interleukin-2 receptor α-subunit) (20). Tac is normally transported to the plasma membrane, where it accumulates at steady state and has been used extensively as a tool to define signals involved in secretory and endocytic membrane trafficking by attaching candidate sequences to its C terminus (15, 21, 22). Either Tac alone or TacADAM10 were transiently transfected into COS7 cells, and their surface expression was evaluated using an immunofluorescence-based antibody uptake assay. Consistent with previous reports, Tac displayed strong plasma membrane expression (Fig. 1A). Remarkably, addition of the C-terminal tail of ADAM10 completely prevented surface localization, despite intense intracellular labeling (Fig. 1, A and B, Tac surface/total expression = 1.99 ± 0.27; TacADAM10 surface/total expression = 0.02 ± 0.003; p < 0.001). The intracellular staining patterns of Tac and TacADAM10 also differed; whereas Tac was present in intracellular vesicles, intracellular TacADAM10 exhibited a reticular appearance (see insets in Fig. 1A) and accumulated in a perinuclear compartment.
FIGURE 1.
The intracellular C-terminal tail of ADAM10 limits its surface expression. A, left, schematic cartoons of the reporter Tac, a type I transmembrane protein, and the TacADAM10 chimera carrying the ADAM10 C terminus. Right, after transfection into COS7 cells, surface expression of Tac and TacADAM10 was visualized using antibody uptake assays and intracellular localization was visualized after permeabilization, as described under “Experimental Procedures.” Note that Tac displayed strong surface expression, whereas TacADAM10 was not detectable on the cell surface. Insets, intracellular TacADAM10 accumulates in a perinuclear compartment and displays a reticular pattern, whereas Tac is also found in vesicular structures. Scale bar, 20 μm. B, quantitative analysis of surface/total expression ratios. The average intensity of surface fluorescence staining was measured after cell tracing and normalized to the total intensity to correct for differences in expression. In this and all subsequent figures, data represent mean ± S.E. (*, p < 0.001; n = 6 cells per condition).
TacADAM10 Is Retained in the Endoplasmic Reticulum
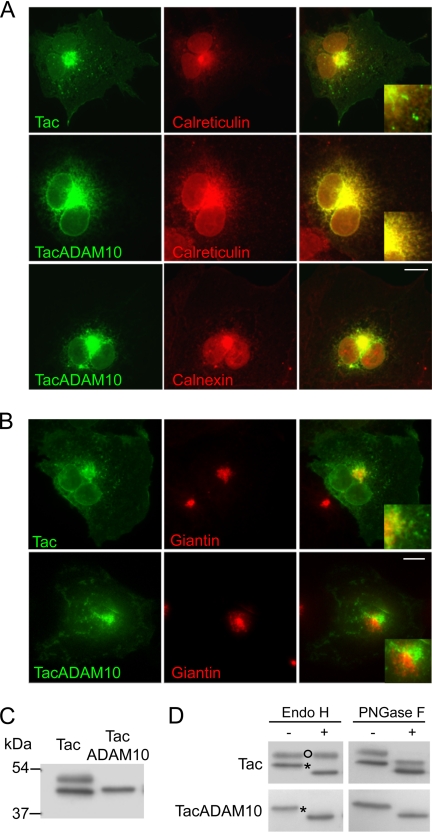
To identify the intracellular compartments where TacADAM10 was retained, we analyzed the subcellular localization of Tac and TacADAM10 using fluorescence microscopy. Tac was present along the secretory pathway, as showed by its partial colocalization with the ER marker calreticulin (Fig. 2A, top) and the Golgi-resident protein giantin (Fig. 2B, top), and its localization in vesicular structures. In contrast, TacADAM10 largely colocalized with two different ER markers, calreticulin and calnexin (Fig. 2A), and was excluded from Golgi structures (Fig. 2B, bottom). Further evidence of ER retention of TacADAM10 was obtained by Western blot analysis. Whereas a single band was present in homogenates of COS7 cells transfected with TacADAM10, two different bands appeared in cells transfected with Tac (Fig. 2C). The result is consistent with previous observations (15) that Tac is expressed as both mature high and immature low molecular weight species, and suggests that TacADAM10 is present only as an ER-retained immature species.
FIGURE 2.
The C-terminal tail of ADAM10 contains an endoplasmic reticulum retention signal. A, COS7 cells transfected with Tac or TacADAM10 were permeabilized and stained with antibodies against Tac (green) and either calreticulin or calnexin (red). Overlays (yellow) show extensive colocalization of TacADAM10 with both calreticulin and calnexin. Tac colocalizes only partially with calreticulin and is present in small intracellular vesicles. Scale bar, 20 μm. B, transfected COS7 cells were permeabilized and costained with antibodies against Tac (green) and the Golgi marker giantin (red). Image overlays show partial colocalization between Tac and giantin, whereas TacADAM10 is excluded from Golgi apparatus. Scale bar, 20 μm. C, Western blot analysis of Tac and TacADAM10. Two different bands were detected in lysates of COS7 cells transfected with Tac, while TacADAM10 appeared as a single band. D, TacADAM10 remained sensitive to EndoH. Note that Tac is normally present as both a mature, high molecular weight, EndoH-resistant species, and an immature EndoH-sensitive species that migrates at a lower molecular weight. After incubation with PNGase F, both Tac and TacADAM10 exhibited increased electrophoretic mobility, indicating PNGase F-sensitivity. Open circles indicates EndoH-resistant bands, which are transported through the Golgi; asterisks indicate EndoH-sensitive species, which are retained in the ER.
This was confirmed by deglycosylation experiments where we compared the glycosylation state of Tac and TacADAM10 by assessing their sensitivity to two different glycosidases: endoglycosidase H (EndoH), an enzyme that hydrolyzes the high mannose N-glycans present on immature secretory proteins in the ER (23) but not complex forms of N-linked oligosaccharide, and N-glycosidase F (PNGase F), an enzyme that cleaves all N-linked oligosaccharides (24). Because conversion of high mannose glycans into complex oligosaccharides occurs in the Golgi, resistance to EndoH indicates that a glycoprotein has reached this compartment. On the contrary, EndoH sensitivity is considered an indicator of immaturity. Treatment with EndoH demonstrated that the single TacADAM10 band was EndoH-sensitive and confirmed that it was retained in the ER as an immature protein (Fig. 2D, lower panels). In contrast, Tac appeared as both a mature, high molecular weight, EndoH-resistant species and a immature EndoH-sensitive species that migrated at a lower molecular weight (Fig. 2D, top panels). All Tac and TacADAM10 species exhibited increased electrophoretic mobility after PNGase F treatment, demonstrating PNGase F sensitivity (Fig. 2D). This combination of biochemical and immunocytochemical experiments provides strong evidence of selective ER retention mediated by the C-terminal domain of ADAM10.
An Amino Acid Stretch in ADAM10 Tail Is Responsible for ER Retention
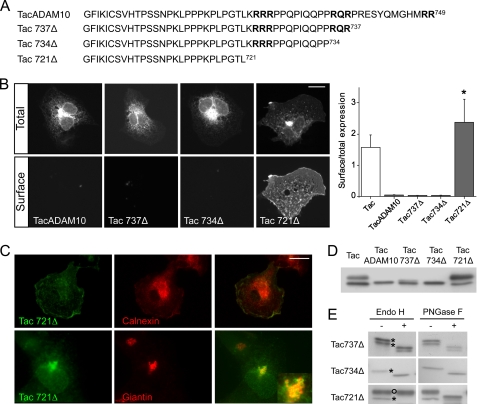
Next, we analyzed the amino acid sequence of the ADAM10 cytoplasmic tail to search for putative ER retention motifs. Based upon homology with known ER retention/retrieval consensus sequences in KATP channels and the NMDA receptor subunit NR1 (13–15), we noted three arginine-based motifs within the C-terminal tail that could be responsible for ER retention (for review see Refs. 25, 26 and Fig. 3A). Then we made a series of C-terminal truncations in TacADAM10 to evaluate the role of the motifs 723RRR, 734RQR, and 748RR in ER retention and surface expression. Deleting the last 12 amino acids of ADAM10 (Tac737Δ) did not allow surface expression. Similar results were obtained with the Tac734Δ mutant (Fig. 3B), indicating that the 748RR and 734RQR sequences do not play a major role in ER exit. In contrast, removal of the last 28 amino acids yielded robust surface staining of the Tac721Δ mutant when compared with TacADAM10 (TacADAM10 surface/total expression = 0.04 ± 0.001; Tac721Δ surface/total expression = 2.38 ± 0.74, p < 0.01, Fig. 3B).
FIGURE 3.
An amino acid stretch within the ADAM10 cytoplasmic tail mediates ER retention. A, amino acid sequences of the ADAM10 C terminus deletion mutants fused to Tac. In bold, arginine-rich motifs located in the ADAM10 cytoplasmic tail. B, left, surface and total expression of TacADAM10 deletion mutants expressed in COS7 cells. Right, quantification of surface/total expression ratios. (*, p < 0.01 versus TacADAM10; n = 8–9 cells per condition). Scale bar, 20 μm. Note that only the Tac721Δ mutant was able to reach the cell surface. C, COS7 cells transfected with Tac721Δ were permeabilized and stained with antibodies against Tac (green) and either calnexin or giantin (red). Scale bar, 20 μm. Inset shows colocalization of Tac721Δ with giantin in a population of cells. D, comparative Western blot analysis of Tac, TacADAM10, and deletion mutants. Note that Tac and Tac721Δ deletion mutant were detected as two species. E, glycosylation analysis reveals that the Tac737Δ doublet and Tac734Δ band are EndoH-sensitive, indicating that they are retained in the ER. The Tac721Δ higher molecular weight species is EndoH-resistant but remains sensitive to PNGase F, indicating that it is able to leave the ER and enter the Golgi. Open circles, EndoH-resistant bands; asterisks, EndoH-sensitive bands.
We then assessed the intracellular localization of the deletion mutants by colocalization with calnexin. Tac737Δ and Tac734Δ mutants showed, as TacADAM10, strong perinuclear staining which colocalized with calnexin, suggesting that they were confined to the ER (supplemental Fig. S1). The surface-expressed Tac721Δ mutant displayed a more diffuse intracellular pattern and did not colocalize with calnexin (Fig. 3C, top). In a small percentage of cells, Tac721Δ accumulated intracellularly in giantin-positive structures (Fig. 3C, bottom), indicating that it was released from the ER and reached the Golgi apparatus. However, most cells exhibited a diffuse staining reflecting an efficient forward trafficking of this construct.
In Western blot analyses, the Tac721Δ mutant was present as both high and low molecular weight species, whereas TacADAM10 and Tac734Δ mutants appeared as single bands. The Tac737Δ mutant was expressed as a doublet (Fig. 3D). Both the single Tac734Δ band and the Tac737Δ doublet were, as TacADAM10, EndoH-sensitive (Fig. 3E), confirming that these constructs do not exit the ER and suggesting that the Tac737Δ doublet reflects multiple glycosylation sites rather than states of maturation. In contrast, the prominent higher molecular weight Tac721Δ band was EndoH-resistant, indicating transport to the late Golgi. As a forward trafficking index, we measured the optical densities of the mature and immature species, and compared the mature/immature ratio of the Tac721Δ mutant with that of Tac alone. The Tac721Δ mature/immature ratio was 2.26, higher than the ratio of 1 estimated for Tac. This result is consistent with a trend toward higher surface expression of the Tac721Δ mutant when compared with Tac (Fig. 3B) and indicates that removal of the distal part of the ADAM10 cytoplasmic tail, along with deleting the ER retention motif, could unmask forward trafficking signals. Taken together, these experiments demonstrate that a 12 amino acid fragment (721–734) in ADAM10 plays an inhibitory role on its trafficking to post-ER compartments and toward the cell surface.
The 723RRR Motif Is Responsible for ER Retention
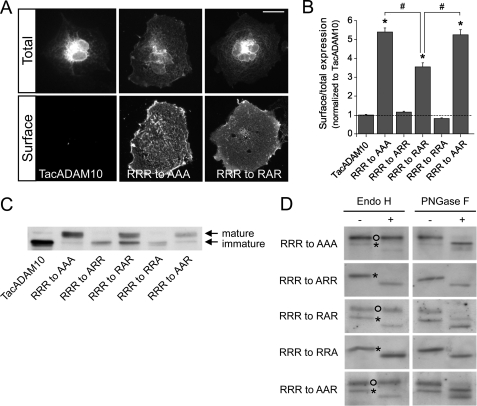
To map the motif responsible for ER retention, we carried out alanine substitutions in the arginine-rich putative ER retention sequence (723RRR) located within the 12 amino acid fragment and analyzed the behavior of the TacADAM10 mutants by immunofluorescence and deglycosylation experiments. Disruption of the RRR sequence by replacing all three arginine residues with alanine yielded strong surface expression (RRR to AAA = 5.53 ± 0.22-fold increase in the surface/total ratio compared with TacADAM10, p < 0.001, Fig. 4, A and B). Substituting the first or third arginines with alanine had no effect, but mutation of the second arginine was sufficient to allow surface expression (RRR to RAR = 3.64 ± 0.22-fold increase in the surface/total ratio compared with TacADAM10, p < 0.001, Fig. 4, A and B). Nevertheless, the effect on surface expression of this single substitution was significantly lower than that of the triple alanine mutant (p < 0.001, RRR to AAA versus RRR to RAR, Fig. 4B). To evaluate the importance of the sequence context, we generated double arginine mutants and observed a significantly increased surface expression of the RRR to AAR mutant when compared with the RRR to RAR mutant (p < 0.001, Fig. 4B). Indeed, the surface/total expression ratio of the RRR to AAR mutant was virtually identical to the triple mutant.
FIGURE 4.
Identification of an arginine-based ER retention signal and key residues required for ER retention. A, surface and total expression of TacADAM10 arginine mutants expressed in COS cells. B, surface/total expression ratios of TacADAM10 mutants were measured and normalized to TacADAM10 surface/total ratios (*, p < 0.001 versus TacADAM10; #, p < 0.001 RRR to AAA and RRR to AAR versus RRR to RAR; n = 14–27 cells per condition). Scale bar, 20 μm. C, Western blot analysis of Tac, TacADAM10, and point mutants. D, deglycosylation analysis reveals that RRR to ARR and RRR to RRA are sensitive to EndoH deglycosylation, indicating that they are retained in the ER. In contrast, higher molecular weight bands corresponding to RRR to AAA, RRR to AAR, and RRR to RAR mutants are resistant to Endo-H indicating that they leave the ER and enter the Golgi. Open circles, EndoH-resistant bands; asterisks, EndoH-sensitive bands.
Western blot and deglycosylation analyses revealed that the RRR to RAR mutant was present as both a mature, high molecular weight, EndoH-resistant species and a smaller, immature EndoH-sensitive species, as previously seen for Tac721Δ (Fig. 4, C and D). Notably, the RRR to AAA and RRR to AAR mutants were detected mainly as a high molecular weight mature species (Fig. 4C) not sensitive to EndoH treatment (Fig. 4D). Single RRR to ARR and RRR to RRA mutants were detected as a single EndoH-sensitive band, further confirming their ER retention (Fig. 4, C and D). Therefore, and despite a major role of the second arginine within the 723RRR motif in TacADAM10 ER retention, the amino acid context also influences TacADAM10 maturation and surface expression.
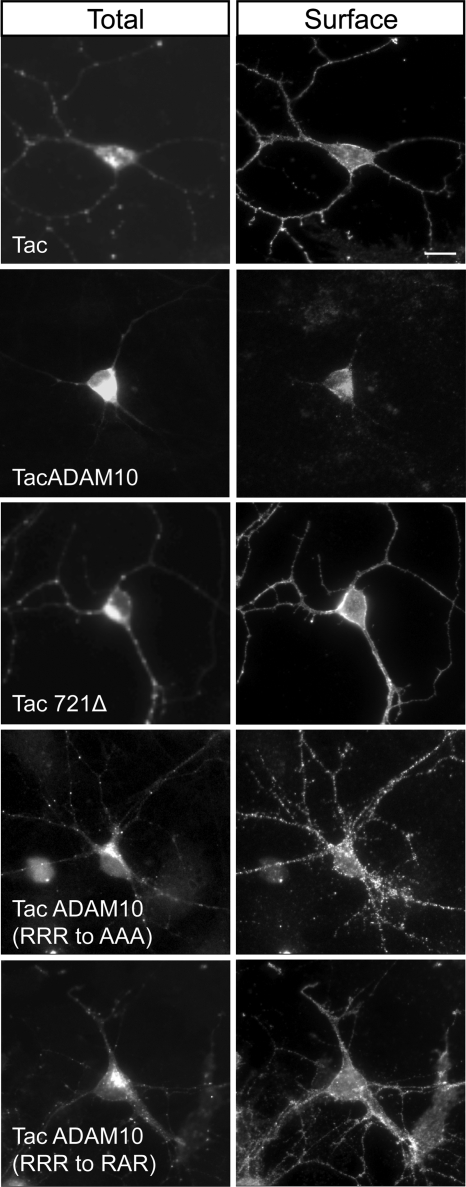
To rule out the possibility that ER retention of TacADAM10 reflects, rather than an endogenous mechanism to limit surface expression, a lack of trafficking factors in COS7 cells which would be available in a native environment for ADAM10 expression, we monitored the surface expression of TacADAM10 in neurons. Cultured hippocampal neurons were transfected with either Tac or TacADAM10 constructs, and their localization was analyzed by immunofluorescence. Whereas Tac was expressed at the surface and displayed punctate intracellular labeling along dendrites, TacADAM10 was mainly found intracellularly in the cell body (Fig. 5). Surface staining of TacADAM10 represented a very faint signal at the soma, matching our results in COS7 cells (Fig. 5). In contrast, Tac721Δ and TacADAM10 mutants lacking the retention motif (RRR to AAA and RRR to RAR) were located at the neuronal surface and in intracellular vesicles along dendrites (Fig. 5), suggesting that removal of the ER retention signal favors the dendritic targeting and surface expression of TacADAM10 in hippocampal neurons and indicating that ER retention is a mechanism controlling ADAM10 localization in neurons.
FIGURE 5.
Deletion/mutagenesis of the ER retention motif allows TacADAM10 surface expression in neurons. Primary hippocampal neurons were transfected with the indicated Tac constructs at DIV10, and surface/total staining analyzed 48 h later. Note that TacADAM10 was faintly expressed at the surface, and exclusively at the soma, whereas TacADAM10 constructs lacking the ER motif were targeted to the cell surface and displayed a punctate staining along dendrites. Scale bar, 20 μm.
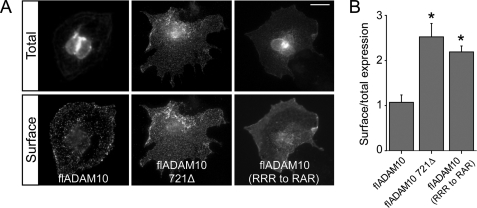
We then asked whether ER retention plays a role in the context of the full-length ADAM10 protein, as it is possible that folding or other factors permit ER release. Indeed, it has been previously reported that ADAM10 is localized in the Golgi, albeit it trafficks inefficiently to the surface (7). COS7 cells were transfected with full-length ADAM10 (flADAM10), a deletion mutant lacking the last 28 amino acids (flADAM10 721Δ) or a mutant lacking the second arginine of the ER retention motif, and their surface expression was analyzed by immunofluorescence. flADAM10 showed a strong perinuclear staining and was weakly expressed at the surface, whereas the mutants displayed a more diffuse intracellular pattern and stronger surface staining (Fig. 6A). Quantification revealed significant increases in surface/total expression ratios when the RRR motif was removed (Fig. 6B, p < 0.001 versus wild-type flADAM10).
FIGURE 6.
Mutations in the ER retention motif favor surface expression of the full-length ADAM10 protein. A, wild-type flADAM10 and deletion or point mutants lacking the ER retention motif were transfected into COS7 cells and surface/total labeling was analyzed. flADAM10 was faintly localized at the surface despite intense intracellular labeling. In contrast, all the mutants tested displayed strong surface staining. Scale bar, 20 μm. B, quantification of surface expression ratios (*, p < 0.001 versus flADAM10, n = 7–20 cells per condition).
Neither Expression nor Ser-39 Phosphorylation of SAP97 Allow ADAM10 ER Exit
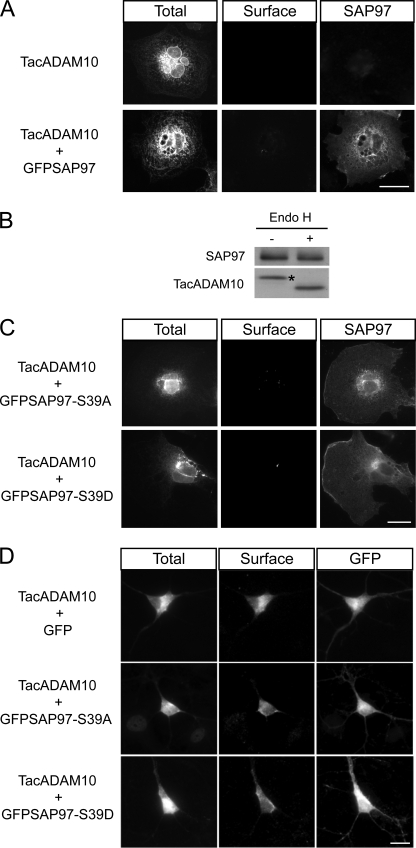
Recent studies showed that PDZ (PSD95/DLG/ZO-1) domain-containing scaffolding proteins that associate with receptors and channels early in the secretory pathway can facilitate or inhibit their ER to Golgi transport (14, 15, 27). ADAM10 binds the PDZ domain protein SAP97, interaction which facilitates its trafficking to postsynaptic compartments (12). To test whether SAP97 could favor ADAM10 ER exit, TacADAM10 and SAP97 were cotransfected in COS7 cells and surface expression was determined. However, SAP97 did not allow TacADAM10 surface expression (Fig. 7A, center panels). The result was confirmed by deglycosylation analysis, which showed that SAP97 coexpression did not modify TacADAM10 maturation, because only the EndoH-sensitive state was attained (Fig. 7B).
FIGURE 7.
SAP97 does not influence ADAM10 ER exit. A, COS7 cells were cotransfected with constructs encoding TacADAM10 and GFP-tagged SAP97, and stained for TacADAM10 surface/total expression. Scale bar, 20 μm. B, deglycosylation analysis of TacADAM10 in SAP97-transfected COS7 cells shows a single EndoH-sensitive band, indicating that SAP97 does not affect ADAM10 maturation and ER exit. Asterisk indicates EndoH-sensitive band. C, COS7 cells were transfected with TacADAM10 and either GFP-SAP97-S39A or GFP-SAP97-S39D as indicated, and stained for TacADAM10 surface/total expression. Scale bar, 20 μm. D, primary hippocampal neurons were transfected with TacADAM10 and either GFP or GFP-SAP97-S39A or GFP-SAP97-S39D. Neither GFP-SAP97-S39A nor GFP-SAP97-S39D cotransfection affected TacADAM10 surface expression or intracellular colocalization. Scale bar, 20 μm.
Alternatively, the activation of specific signaling pathways might be required for ADAM10 transport along the secretory pathway. For instance, SAP97-Ser-39 phosphorylation by CaMKII favors the release of a SAP97/NR2A complex from the ER (28). In order to evaluate the putative role of SAP97 phosphorylation in modulating the ER release of ADAM10, we cotransfected COS7 cells with TacADAM10 and SAP97 mutants that prevent (GFPSAP97-S39A) or mimic (GFPSAP97-S39D) CaMKII-dependent phosphorylation. Neither SAP97-S39D nor SAP97-S39A cotransfection allowed TacADAM10 surface expression in COS7 cells (Fig. 7C).
Given that SAP97 can interact with several neuronal proteins which would be absent from COS7 cells, we evaluated whether phosphorylated SAP97 could mediate ADAM10 ER exit in neurons. Surface expression of TacADAM10 in hippocampal neurons cotransfected with TacADAM10 and either GFPSAP97-S39A or GFPSAP97-S39D was analyzed. As shown in Fig. 7D, neither SAP97-S39D nor SAP97-S39A allowed redistribution of TacADAM10 to dendrites or modified surface expression. These data suggest that other signaling pathways or alternative binding partners are responsible for ADAM10 ER exit.
DISCUSSION
Previous reports suggested that correct secretory trafficking and localization of ADAM10 at the plasma membrane are required for the shedding activity of the enzyme toward its substrates. Here we demonstrate that the C-terminal tail of ADAM10 carries an ER retention signal that limits ADAM10 ER exit and forward secretory trafficking. Our findings are in line with previous reports showing limited surface expression of ADAM10 in heterologous systems (7), and provide a functional explanation by demonstrating a role for ER retention on ADAM10 localization at the cell surface.
It was generally thought that once folded and assembled, proteins targeted to later compartments of the secretory pathway leave the ER by default, with no need for specific traffic signals (29). However, studies on the trafficking of oligomeric ion channels and receptors have revealed that, instead, complex interactions between ER retention/retrieval signals (13–15, 30, 31) and anterograde export signals located within intracellular domains work in concert to retain or favor the ER exit of fully folded and assembled proteins (26, 32, 33). These mechanisms are crucial to prevent ER export of malfunctioning proteins, and to maintain adequate surface expression levels of certain plasma membrane proteins. The process is regulated via interactions with chaperones or scaffolds, such as PDZ-domain containing proteins or Homer, that affect ER to Golgi transport of the associated protein, and/or by post-translational modifications such as phosphorylation (14, 15, 27, 34, 35).
The mechanisms regulating ER retention/exit of KATP channels (13), GABAB receptors (30), NR1 subunit of NMDA receptor (14, 15) and potassium channels (26) have been extensively studied. Although less known, ER retention motifs also regulate the membrane transport of other transmembrane proteins, including members of the ADAMs family such as ADAM12 and ADAM22 (17, 18). In this study, we focused on ADAM10. We provide the first description of an ER quality control mechanism that regulates its delivery to the plasma membrane, and reveal the presence of an ER retention signal within the C-terminal intracellular domain of ADAM10, which controls its maturation and surface expression.
To restrict the search for putative trafficking signals to the ADAM10 cytoplasmic tail and minimize effects of other domains, we chose to isolate the contribution of the C-terminal domain by using chimeras with the Tac protein, approach, which has been used extensively to identify ER retention signals (36, 37). Immunofluorescence analyses showed that TacADAM10 was not able to reach the cell surface, and demonstrated that the C-terminal domain of ADAM10 is sufficient to prohibit the surface expression of TacADAM10 in heterologous cells. Consistent with its retention in the ER and failure to reach the Golgi, TacADAM10 remained sensitive to EndoH, enzyme that preferentially hydrolyzes the high mannose N-glycans present on immature ER-resident proteins.
An analysis of the amino acid sequence of the C terminus of ADAM10 led to the identification of three putative arginine-based ER retention motifs. Arginine motifs (typically RR or RXR) are a class of cytosolic ER-sorting signals that, in contrast to the better known C-terminal di-lysine signals, do not need to be exposed at the distal terminus of a membrane protein (38). Imaging and biochemical assays of a battery of constructs created by sequential truncation or mutagenesis of the ADAM10 cytoplasmic tail demonstrated that one arginine stretch, 723RRR, was responsible for ER retention. Within this motif, the second arginine was sufficient to mediate ER retention of TacADAM10. One of the adjacent arginines was important as well, because imaging analysis showed that the surface expression of the RRR to AAR mutant was greater than the RRR to RAR mutant, and indeed matched surface ratios of the triple RRR to AAA mutant. Additionally, our quantitative data showing higher surface expression of the Tac721Δ mutant versus Tac suggest that the C terminus of ADAM10 might contain additional forward trafficking signals which are unmasked by removal of discrete ER retention motifs, and point toward a possible second mechanism for controlling ADAM10 targeting to the cell surface.
Importantly, deletion or mutations of the ER-retention motif identified in the context of TacADAM10 also affected the surface expression of the full-length ADAM10 protein, proving that this motif is relevant not only for the trafficking of chimeric TacADAM10 but regulates the trafficking of the wild-type species.
Despite the existence of ER retention mechanisms, a significant pool of ADAM10 reaches the surface in neurons (12). What are the mechanisms for unmasking? ER retention signals can be overcome via different mechanisms. ER retention motifs in ATP-sensitive potassium channels and GABA receptors are masked during subunit oligomerization, allowing forward trafficking of only correctly folded assemblies (13, 30). ER retention mediated by an RXR motif in the NMDA receptor subunit NR1 can be suppressed by PKC phosphorylation of serine residues adjacent to the retention signal (15). In addition to their organizational role, scaffold proteins that associate with channels and receptors early in the secretory pathway can facilitate/inhibit their ER to Golgi transport (14, 15). For instance, the PDZ-domain protein SAP97 is involved in the correct targeting and clustering of ionotropic glutamate receptors and potassium channels (39); interactions between SAP97 and the potassium channel Kv 1.4 begin in the ER, causing ER retention of both proteins (35). Interestingly, SAP97 also binds the cytoplasmic tail of ADAM10, and activation of NMDA receptors in hippocampal neurons promotes SAP97-mediated ADAM10 delivery to the postsynaptic compartment (12). However, and despite its reported roles on synaptic ADAM10 trafficking and channel ER export, expression of SAP97 was not sufficient for ER exit suggesting that ADAM10/SAP97 complexes are likely formed in later secretory compartments and that interaction with other proteins, yet to be identified, or activation of specific signaling pathways might be required for ADAM10 ER exit and forward transport. Because phosphorylation of SAP97 by CaMKII represents the driving force to release SAP97/NR2A complex from the ER (28), we considered a potential role of SAP97-Ser-39 phosphorylation in modulating ADAM10 release from the ER. However, TacADAM10 surface expression was not affected.
ER retention motifs play roles in permitting the forward secretory trafficking of only properly folded ion channels, and regulate the number of functional channels and receptors exposed at the cell surface (13–15). In this context, ADAM10 retention in early secretory compartments might constitute a biological checkpoint to control ADAM10 folding, but also a rate-limiting step for surface expression which would affect its enzymatic activity (7, 12). Given that PC cleavage occurs after ADAM10 leaves the ER, limiting the amount of ADAM10 which reaches the late secretory pathway might provide a means to keep the enzyme in an inactive, latent form by slowing down trafficking rates. Regulated exit might then enable processing and activation, and regulate the supply of active ADAM10 to the surface.
ADAM10 is crucial to many biological processes, and is thought to be involved in the pathogenesis of different diseases because various surface proteins undergo regulated cleavage by ADAM10. The list of substrates includes molecules involved in brain pathology, inflammation, and cancer. In the brain, ADAM10 mediates neuroprotective cleavage events such as prevention of amyloid β production; proteolytic cleavage of the amyloid precursor protein (APP) by ADAM10 precludes the formation of amyloidogenic peptides and favors the release of soluble APP fragments into the extracellular milieu. A moderate neuronal overexpression of ADAM10 in transgenic mouse models of AD reduces the formation of amyloid β peptides, prevents their deposition in plaques and alleviates cognitive deficits (40). Conversely, expression of a dominant-negative form of ADAM10 potentiates plaque formation in AD mouse models (41). In inflammatory settings, cytokine signaling is triggered by proteolytic release of soluble agonists and leukocyte recruitment is controlled by the cleavage of adhesion molecules. In tumors, ADAM10-mediated shedding events trigger proliferative signaling via activation of growth factors, including ErbB family members. As a result, specific and potent inhibitors of ADAM10 can be of use to suppress tumor progression in vivo (3).
Nevertheless, and despite the therapeutic promise of targeting pathways that increase ADAM10 activity to limit neurodegeneration, or suppress it to block inflammation or tumor growth, the feasibility of such strategies has yet to be carefully studied. A more detailed knowledge of the mechanisms regulating ADAM10 intracellular trafficking and activity will be essential to understand if this process is affected (and how) in disease, and to design specific tools to overcome such alterations. If so, manipulations of the cellular systems controlling ADAM10 trafficking could foster the development of novel therapeutic strategies which took advantage of the endogenous physiological processes that modulate ADAM10 enzymatic activity.
Supplementary Material
Acknowledgments
We thank A. Longhi, A. Zandueta, and E. Zianni for technical assistance and V. Marinaccio for excellent practical work. We appreciate the gift of mouse ADAM10 cDNA from Dr. Saftig (Biochemical Institute, Christian-Albrechts-University Kiel, Germany).
This work was supported by EC Contract LSHM-CT-2008-217902 (cPADs), CARIPLO 2008, MIUR Italy-Spain Integrated Action (to M. D. L.), Kemali Foundation fellowship (to E. M.), UTE project CIMA and Spanish Ministry of Education and Science grants (Spain-Italy Integrated Action, SAF2006-10025, CSD2008-00005) (to I. P.-O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ADAM
- a disintegrin and metalloproteinase
- AD
- Alzheimer Disease
- FITC
- fluorescein isothiocyanate
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- PC
- proprotein convertase.
REFERENCES
- 1.Blobel C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 2.Seals D. F., Courtneidge S. A. (2003) Genes Dev. 17, 7–30 [DOI] [PubMed] [Google Scholar]
- 3.Pruessmeyer J., Ludwig A. (2009) Semin Cell Dev. Biol. 20, 164–174 [DOI] [PubMed] [Google Scholar]
- 4.Kheradmand F., Werb Z. (2002) Bioessays 24, 8–12 [DOI] [PubMed] [Google Scholar]
- 5.Moss M. L., Bartsch J. W. (2004) Biochemistry 43, 7227–7235 [DOI] [PubMed] [Google Scholar]
- 6.Prinzen C., Muller U., Endres K., Fahrenholz F., Postina R. (2005) Faseb. J. 19, 522–1524 [DOI] [PubMed] [Google Scholar]
- 7.Lammich S., Kojro E., Postina R., Gilbert S., Pfeiffer R., Jasionowski M., Haass C., Fahrenholz F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3922–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anders A., Gilbert S., Garten W., Postina R., Fahrenholz F. (2001) Faseb. J. 15, 1837–1839 [DOI] [PubMed] [Google Scholar]
- 9.Schäfer W., Stroh A., Berghöfer S., Seiler J., Vey M., Kruse M. L., Kern H. F., Klenk H. D., Garten W. (1995) EMBO J. 14, 2424–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wouters S., Leruth M., Decroly E., Vandenbranden M., Creemers J. W., van de Loo J. W., Ruysschaert J. M., Courtoy P. J. (1998) Biochem. J. 336, 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horiuchi K., Le Gall S., Schulte M., Yamaguchi T., Reiss K., Murphy G., Toyama Y., Hartmann D., Saftig P., Blobel C. P. (2007) Mol. Biol. Cell 18, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcello E., Gardoni F., Mauceri D., Romorini S., Jeromin A., Epis R., Borroni B., Cattabeni F., Sala C., Padovani A., Di Luca M. (2007) J. Neurosci. 27, 1682–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. (1999) Neuron 22, 537–548 [DOI] [PubMed] [Google Scholar]
- 14.Standley S., Roche K. W., McCallum J., Sans N., Wenthold R. J. (2000) Neuron 28, 887–898 [DOI] [PubMed] [Google Scholar]
- 15.Scott D. B., Blanpied T. A., Swanson G. T., Zhang C., Ehlers M. D. (2001) J. Neurosci. 21, 3063–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hougaard S., Loechel F., Xu X., Tajima R., Albrechtsen R., Wewer U. M. (2000) Biochem. Biophys. Res. Commun. 275, 261–267 [DOI] [PubMed] [Google Scholar]
- 17.Cao Y., Kang Q., Zhao Z., Zolkiewska A. (2002) J. Biol. Chem. 277, 26403–26411 [DOI] [PubMed] [Google Scholar]
- 18.Gödde N. J., D'Abaco G. M., Paradiso L., Novak U. (2006) J. Cell Sci. 119, 3296–3305 [DOI] [PubMed] [Google Scholar]
- 19.Gardoni F., Bellone C., Viviani B., Marinovich M., Meli E., Pellegrini-Giampietro D. E., Cattabeni F., Di Luca M. (2002) Eur. J. Neurosci. 16, 777–786 [DOI] [PubMed] [Google Scholar]
- 20.Bonifacino J. S., Cosson P., Klausner R. D. (1990) Cell 63, 503–513 [DOI] [PubMed] [Google Scholar]
- 21.Jansen E. J., Holthuis J. C., McGrouther C., Burbach J. P., Martens G. J. (1998) J. Cell Sci. 111, 2999–3006 [DOI] [PubMed] [Google Scholar]
- 22.Ghosh R. N., Mallet W. G., Soe T. T., McGraw T. E., Maxfield F. R. (1998) J. Cell Biol. 142, 923–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trimble R. B., Maley F. (1984) Anal. Biochem. 141, 515–522 [DOI] [PubMed] [Google Scholar]
- 24.Tarentino A. L., Gómez C. M., Plummer T. H., Jr. (1985) Biochemistry 24, 4665–4671 [DOI] [PubMed] [Google Scholar]
- 25.Teasdale R. D., Jackson M. R. (1996) Annu. Rev. Cell Dev. Biol. 12, 27–54 [DOI] [PubMed] [Google Scholar]
- 26.Ma D., Jan L. Y. (2002) Curr. Opin. Neurobiol. 12, 287–292 [DOI] [PubMed] [Google Scholar]
- 27.Roche K. W., Tu J. C., Petralia R. S., Xiao B., Wenthold R. J., Worley P. F. (1999) J. Biol. Chem. 274, 25953–25957 [DOI] [PubMed] [Google Scholar]
- 28.Mauceri D., Gardoni F., Marcello E., Di Luca M. (2007) J. Neurochem. 100, 1032–1046 [DOI] [PubMed] [Google Scholar]
- 29.Wieland F. T., Gleason M. L., Serafini T. A., Rothman J. E. (1987) Cell 50, 289–300 [DOI] [PubMed] [Google Scholar]
- 30.Margeta-Mitrovic M., Jan Y. N., Jan L. Y. (2000) Neuron 27, 97–106 [DOI] [PubMed] [Google Scholar]
- 31.Xia H., Hornby Z. D., Malenka R. C. (2001) Neuropharmacology 41, 714–723 [DOI] [PubMed] [Google Scholar]
- 32.Stockklausner C., Ludwig J., Ruppersberg J. P., Klöcker N. (2001) FEBS Lett. 493, 129–133 [DOI] [PubMed] [Google Scholar]
- 33.Ma D., Zerangue N., Raab-Graham K., Fried S. R., Jan Y. N., Jan L. Y. (2002) Neuron 33, 715–729 [DOI] [PubMed] [Google Scholar]
- 34.Fernández-Larrea J., Merlos-Suárez A., Ureña J. M., Baselga J., Arribas J. (1999) Mol. Cell 3, 423–433 [DOI] [PubMed] [Google Scholar]
- 35.Tiffany A. M., Manganas L. N., Kim E., Hsueh Y. P., Sheng M., Trimmer J. S. (2000) J. Cell Biol. 148, 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Meskini R., Galano G. J., Marx R., Mains R. E., Eipper B. A. (2001) J. Biol. Chem. 276, 3384–3393 [DOI] [PubMed] [Google Scholar]
- 37.Tan P. K., Waites C., Liu Y., Krantz D. E., Edwards R. H. (1998) J. Biol. Chem. 273, 17351–17360 [DOI] [PubMed] [Google Scholar]
- 38.Michelsen K., Yuan H., Schwappach B. (2005) EMBO Rep. 6, 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garner C. C., Nash J., Huganir R. L. (2000) Trends Cell Biol. 10, 274–280 [DOI] [PubMed] [Google Scholar]
- 40.Postina R., Schroeder A., Dewachter I., Bohl J., Schmitt U., Kojro E., Prinzen C., Endres K., Hiemke C., Blessing M., Flamez P., Dequenne A., Godaux E., van Leuven F., Fahrenholz F. (2004) J. Clin. Invest. 113, 1456–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schroeder A., Fahrenholz F., Schmitt U. (2009) J. Alzheimers Dis. 16, 309–314 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.