Abstract
Prion-infected cells accumulate a heterogeneous population of aberrantly folded PrP conformers, including the disease-causing isoform (PrPSc). We found that specific chemicals can modulate the levels of various PrP conformers in cultured cells. Positively charged polyamidoamines (dendrimers) eliminated protease-resistant (r) PrPSc from prion-infected cells and induced the formation of insoluble, protease-sensitive PrP aggregates (designated PrPA). Larger, positively charged polyamidoamines more efficaciously induced the formation of PrPA and cleared rPrPSc, whereas negatively charged polyamidoamines neither induced PrPA nor cleared rPrPSc. Although the biochemical properties of PrPA were shown to be similar to protease-sensitive (s) PrPSc, bioassays of PrPA indicated that it is not infectious. Our studies argue that PrPA represents an aggregated PrP species that is off-pathway relative to the formation of rPrPSc. It remains to be established whether the formation of PrPA inhibits the formation of rPrPSc by sequestering PrPC in the form of benign, insoluble aggregates.
Keywords: Diseases/Aging, Diseases/Alzheimer's Disease, Diseases/Amyloid, Diseased/Prions, Polyamines, Prions, Protein/Conformation
Introduction
The prion hypothesis proposes that proteins can act as infectious, self-replicating agents (1). In prion diseases, an aberrantly folded conformer of the prion protein (PrP)2 propagates by catalyzing a post-translational conversion reaction, utilizing cellular PrP (PrPC) as substrate (2). This conversion reaction transforms endogenous, α-helix-rich PrPC to β-sheet-rich, disease-causing conformations (PrPSc) (3, 4). Traditionally, PrPSc has been distinguished from PrPC by its resistance to proteolysis (5–7). However, recent studies have shown that PrPSc encompasses a continuum of oligomeric states, some of which are protease sensitive (8–14). In prion-infected cells, multiple conformers of PrPSc with distinct biochemical properties coexist at steady state (9). It is not clear whether these conformers represent intermediates along a single misfolding pathway or distinct, thermodynamically stable, self-propagating, conformationally distinct strains. We sought to probe the spectrum of misfolded PrP conformers that accumulate in cultured, uninfected neuroblastoma (N2a) and prion-infected neuroblastoma (ScN2a) cells and evaluate the effect of known “antiprion” compounds on their distribution. We report that positively charged polyamidoamines clear protease-resistant PrPSc and induce the formation of a novel, non-infectious PrP aggregate.
EXPERIMENTAL PROCEDURES
Reagents
The following chemicals were obtained through Sigma-Aldrich: PAMAM generations 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 6.5, and 7; quinacrine; methyl-β-cyclodextran; Congo Red; phenylmethylsulfonyl fluoride (PMSF); sodium deoxycholate; bovine serum albumin; sodium phosphotungstic acid (PTA); dimethyl-sulfoxide (DMSO); sodium butyrate; and Nonidet P-40. Gleevec was purchased from Toronto Research Chemicals. Bis-acridine was prepared as preciously described (15). Minimal essential medium (MEM) with Earle's salts; Dulbecco's modified Eagle's medium (DMEM) high glucose 1× with 4.5 g/liter d-glucose and l-glutamine and without sodium pyruvate; cell dissociation buffer; fetal bovine serum (FBS); Geneticin (50 mg/ml); penicillin-streptomycin (10,000 units/ml and 10,000 μg/ml, respectively); and GlutaMAX were purchased from Invitrogen. Sybr GreenER qPCR supermix; qPCR oligonucleotides; dithiothreitol (0.5 m 10×); 4× loading buffer; and proteinase K (PK) were purchased from Invitrogen. 2,2′- azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) (ABTS) was obtained from KPL. Complete protease inhibitor (PI) mixture tablets were from Roche Diagnostics. Western blotting detection reagents 1 and 2 were from GE Healthcare. Anti-PrP antibodies Fab D18 (16), Fab EST123 (17), HRP-conjugated Fab D13 (18), and HRP-conjugated Fab 3F4 (19) were prepared as previously described.
Fresh media were prepared with MEM, 10% FBS, 1× GlutaMAX, and 1× penicillin-streptomycin. Lysis buffer was prepared as 10 mm Tris-HCl, pH 8, 15 mm NaCl, 0.5% Nonidet P-40, and 0.48% sodium deoxycholate in ddH2O.
The following compound stock solutions were prepared and used for all curing experiments: 10 mg/ml PAMAMs in PBS, 2 mm quinacrine in PBS, 0.5 mm bis-acridine in DMSO, 0.9 m methyl-β-cyclodextran in DMSO, 6 mm Gleevec in DMSO, 1 mg/ml Fab D18 in PBS, 6 mm Congo Red in DMSO, 10% (w/v) PTA, and 1 m sodium butyrate in PBS.
Cell Culture
The N2a cell line was purchased from American Tissue Culture Collection and stably transfected with pSPOX.neo vector expressing full-length mouse PrP under the control of the HCMV promoter with DOTAP liposomal transfection reagent (Roche) according to the manufacturer's protocol. Stably transfected lines were cloned by serial dilution, and 15 individual clones were assayed for PrP expression level by Western immunoblotting. The N2a-cl3 clone expressed the highest levels of PrP, 6× greater than that expressed in previous N2a lines. N2a-cl3 was infected with the Rocky Mountain Laboratory (RML) strain of mouse-adapted scrapie prions as previously described (20).
Noninfected N2a and N2a-cl3 as well as prion-infected ScN2a and ScN2a-cl3 cells were maintained at 37 °C in 10 ml of MEM supplemented with 10% FBS, 1% penicillin-streptomycin, and 1% GlutaMAX. N2a-cl3 and ScN2a-cl3 lines were also maintained at 37 °C in 10 ml of MEM supplemented with 10% FBS, 1% GlutaMAX, 1% penicillin-streptomycin, and 600 μg/ml Geneticin for selection of the transgene. GT1 and ScGT1 cells were maintained at 37 °C in 10 ml of DMEM supplemented with 10% FBS, 1% penicillin-streptomycin, and 1% GlutaMAX. Medium for non-dividing cell experiments was supplemented with 10 mm sodium butyrate. Treatment experiments with antiprion compounds lasting fewer than 14 days were performed in medium lacking both penicillin-streptomycin and Geneticin. The medium was refreshed every 2 days for dividing cells and daily for non-dividing cells. Typically, cells were propagated in 100-mm plates and allowed to grow to 95% confluence before dissociation with 1 ml of enzyme-free cell dissociation buffer. Cells were then replated at 10% confluence for further propagation.
To collect cell lysates, cells were rinsed with PBS (3 × 10 ml) and lysed with 1 ml of cold lysis buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate). Lysates were centrifuged for 3 min at 10,000 × g to remove cell debris, and the total protein concentration was measured in the supernatant using the bicinchoninic acid assay (BCA, Pierce). Aliquots containing 500 μg of total protein were titrated by adding lysis buffer to achieve a final protein concentration of 1 mg/ml and then stored at −20 °C until further use.
PK Digestion
Unless otherwise stated, 500 μg of lysate aliquots (at 1 mg/ml) were digested with 10 μl of 1 mg/ml PK for 1 h at 37 °C. The enzyme/protein ratio by weight was 1:50. PK activity was quenched by adding 10 μl of 100 mm PMSF to achieve a final concentration of 2 mm.
PTA Precipitation
Unless otherwise stated, 500 μg of lysate aliquots (at 1 mg/ml) were supplemented with 0.75% PTA (final concentration from 10% stock solution in water, pH 7), 1% Sarkosyl (SA, from a 30% stock solution), and a PI mixture (Roche Diagnostics). The lysates were incubated for 3 h at 37 °C with shaking at 350 rpm and centrifuged at room temperature for 30 min at 16,000 × g. The pellets were washed with 0.75% PTA and PI-supplemented lysis buffer, then centrifuged again. The pellets were resuspended in 500 μl of lysis buffer by vigorous vortexing and stored at −20 °C until further use.
SA Precipitation
Unless otherwise stated, 500-μg lysate aliquots (at 1 mg/ml) were supplemented with 1% Sarkosyl (from a 30% stock solution) and a PI mixture (Roche Diagnostics). The lysates were centrifuged at 4 °C for 1 h at 100,000 × g. The pellets were resuspended in 500 μl of lysis buffer by vigorous vortexing and stored at −20 °C until further use.
Western Immunoblotting
Normalized aliquots of untreated, PK-digested, PTA-pelleted, and SA-pelleted extracts were prepared as described above. A volume of 30 μl of normalized lysate was added to 10 μl of 4× loading buffer and 1× dithiothreitol (50 mm). Samples were boiled for 5 min and run on an Invitrogen NuPage 4–12% Bis-Tris gel for Western blot analysis using either D13 or EST123 antibodies.
Analysis and Quantification of PrP Isoforms in Cell Lysates
Capture ELISA plates (96-well, Greiner) were coated with Fab D18 as described previously (21). Normalized aliquots of untreated, PK-digested, PTA-pelleted, or SA-pelleted extracts were prepared as described in the supporting online material. An aliquot of lysate was diluted with 8 m GdnHCl (Pierce) in 3:1 ratio and heated to 85 °C for 15 min. Upon cooling to room temperature, 60 μl of the denatured lysates were added to D18-coated ELISA plates containing 240 μl of 1% BSA in PBS per well. After incubation at 4 °C for 16 h, the plate was washed three times with TBST (10 mm TrisHCl, pH 8.0; 150 mm NaCl; 0.5% Tween-20). To each well of the plate, 100 μl of HRP-conjugated D13 (1:1000 dilution from 1 mg/ml stock) was added and incubated for 1 h at 37 °C. The plate was washed seven times with TBST and then 100 μl of ABTS was added per well and developed for 15–20 min before reading. The absorbance at 405 nm was determined by using a SpectraMax Plus microplate reader running SoftMaxPro. All readings were converted to PrP concentration by comparison to a standard concentration ladder composed of varying concentrations of purified, recombinant, full-length mouse PrP. Before plotting, all concentration measurements were normalized with respect to a negative reference sample.
Gel Filtration
Superdex 200 beads (Sigma) were used in 1 × 30-cm columns at room temperature using an Akta FPLC apparatus (Amersham Biosciences). Columns were loaded with 200 μl of lysates at 3 mg/ml and run at a rate of 0.5 ml/min with PBS (pH 7) and 1% Sarkosyl. Fractions of 0.5 ml were collected. For PK digestion, PTA centrifugation, SA centrifugation, and Western immunoblotting, 500 μl fractions were used.
GdnHCl Denaturation Curves
N2a-cl3 cells were grown to confluency in 10-cm tissue culture plates (Corning) in 10 ml of MEM (Invitrogen MEM, 10% FBS, 1× GlutaMAX, 1× penicillin-streptomycin, Geneticin), and lysed with 1 ml of lysis buffer. Total protein concentration was measured by BCA, and cell lysates were normalized to 1 μg/μl in lysis buffer. For insoluble PrPA, 150 μg of total protein was obtained from N2a-cl3 cells treated with 10 μg/ml PAMAM-G4 for 48 h. For rPrPSc, 150 μg of total protein from ScN2a-cl3 cells was digested with 3 μg of PK (1 μg/μl stock concentration) for 1 h at 37 °C; to stop the digestion, 3 μl of PMSF (100 mm in ethanol stock concentration) were added. Both N2a-cl3 and ScN2a-cl3 samples were precipitated with PTA; pellets were resuspended in lysis buffer with increasing concentrations of GdnHCl in a final volume of 480 μl and incubated for 1 h at room temperature. Then, 60 μl of each sample was loaded per well in triplicate for capture ELISA as described above.
mRNA Quantification using qRT-PCR
Total RNA was extracted from cells using the Qiagen RNeasy kit. Total RNA was quantified using a nanodrop spectrophotometer (Thermo Scientific). Total RNA (1.5 μg) was used for cDNA synthesis using a first-strand cDNA synthesis kit with oligo(dT) (23VN) primers for selection of messenger poly(A) tail RNA transcripts (New England Biolabs, Ipswich, MA). cDNA product was diluted 1:20 in DNase/RNase-free water (Invitrogen), and 4.2 μl was used for Sybr Green qPCR reaction. To the cDNA, 5 μl of Sybr GreenER qPCR master mix (Invitrogen) was added and 0.4 μl each of 0.5 μm 5′ and 3′ oligonucleotides (Invitrogen). Oligonucleotides passed primer efficiency quality tests. The sequences for the ActB primers were: 5′-gatcattgctcctcctgagc and 3′-ctcatcgtactcctgcttgc. The Prnp primers were 5′-cgagaccgatgtgaagatga and 3′-atcccacgatcaggaagatg. Total volume for the qRT-PCR reaction was 10 μl in a 384-well plate. Amplifications and readings were done in a 7900 HT Applied Biosystems instrument. Preliminary data analysis was performed using the SDS software, and subsequent comparative CT analysis was done using the average cycle threshold data from the SDS data.
Confocal Microscopy
N2a-cl3 cells were plated on a poly-lysine D-coated coverslip (Fisher), then treated with 10 ng/ml of PAMAM-G7 for 24 h. Cells were briefly rinsed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. Cells were rinsed with wash buffer (0.2% BSA/Ca-free and Mg-free PBS) for 30 min, permeabilized with 0.3% Triton X-100 in PBS for 5 min at room temperature, and then rinsed with wash buffer for 15 min. Cells were treated with 3 m GdnSCN in 10 mm Tris-HCl, pH 8.0, for 8 min, then blocked with 10% normal goat serum (NGS) for 20 min. Cells were then incubated with D18 antibody (5 μg/ml) in 10% NGS overnight at 4 °C. Samples were rinsed with wash buffer for 15 min and incubated with a 1:200 dilution of Texas Red-conjugated AffiniPure goat anti-human IgG (H+L) (Jackson Immunoresearch) for 1 h at room temperature. Coverslips were then rinsed with wash buffer for 15 min, and with water briefly, air-dried, and then mounted on Superfrost plus microscope slides (Electron Microscopy Sciences, Hatfield, PA). Slides were coverslipped with Vectashield (Vector Laboratories), sealed with nail polish, and visualized under a Zeiss LSM510 confocal microscope.
Mouse Bioassays
To prepare whole cell homogenate samples for infectivity bioassays, 10-cm diameter plates of PAMAM-treated or untreated confluent cells were washed once with 5 ml of PBS, scraped into 1 ml of PBS, then homogenized by repeated extrusion through a 26-gauge needle. Cell homogenate (100 μl) was added to 900 μl of diluent, consisting of 5% BSA in PBS. A volume of 30 μl of this preparation was intracerebrally inoculated into Tg(MoPrP)4053 mice that express MoPrP-A at 8× levels compared to wild-type mice. To prepare PTA-precipitated pellets for infectivity bioassays, 250 μg of total protein from cell lysate was either left undigested or digested with PK as described, then precipitated with PTA as described. The PTA pellet was resuspended in 100 μl of PBS and added to 900 μl of diluent (5% BSA in PBS); 30 μl of this preparation was intracerebrally inoculated into Tg(MoPrP)4053 mice.
RESULTS
Quantification of Protease-sensitive, Misfolded PrP Conformers in Prion-infected N2a Cells Overexpressing PrP (ScN2a-cl3)
We created and cloned a new transgenic N2a cell line, denoted N2a-cl3, that expresses PrP at 6-fold greater levels compared with wild-type (wt) N2a cells (supplemental Fig. S1). N2a-cl3 cells are highly susceptible to infection with RML prions. Importantly, prion-infected N2a-cl3 (ScN2a-cl3) cells form rPrPSc at levels that are within the same order of magnitude as those found in the brains of RML-infected, wild-type mice (supplemental Fig. S1).
We characterized the PrPSc population in ScN2a-cl3 cells based on two biochemical properties: 1) resistance to proteolysis by PK, and 2) insolubility in PTA. These two properties have been previously attributed to prion preparations from infected cells and tissues, and both properties are absent in endogenous PrPC (8, 11). We found that in ScN2a-cl3, the amount of PrP that is PTA insoluble is greater than the amount that is resistant to PK digestion (Fig. 1A). The discrepancy between the amounts of PK-resistant and PTA-insoluble PrP is not an artifact of limited overdigestion of the protease-resistant population. Our data show that the PK digestion of the PrP pool is distinctly biphasic, with a protease-sensitive population that is rapidly digested and a protease-resistant population that is at steady-state within the time frames (60 min) and protease concentrations (1:50 PK/protein) used in our experiments (supplemental Fig. S2). These observations indicate that at least two distinct PrPSc populations in ScN2a-cl3 cells precipitate in the presence of PTA: one is protease-sensitive (sPrPSc, ∼30%) and another is protease-resistant (rPrPSc, ∼70%). sPrPSc may represent a metabolic precursor to rPrPSc, a transiently populated degradation product, or a distinct self-propagating strain that is off-pathway relative to the formation of rPrPSc.
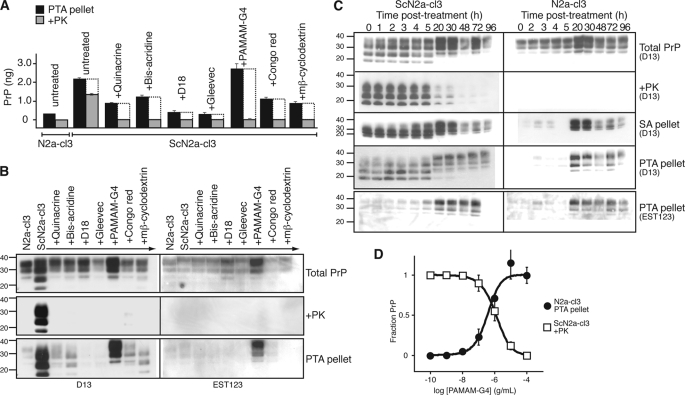
FIGURE 1.
Induction of PrPA by PAMAM-G4. A and B, various antiprion compounds were added to ScN2a-cl3 cells at their EC100 concentration (supplemental Table S1) for 5 days. Lysates were PK-digested or PTA-precipitated, as indicated; equivalent volumes were analyzed by ELISA (A) and Western immunoblotting (B) for PrP content. Differences between the PTA-insoluble PrPSc (black bars) and PK-resistant PrPSc (gray bars) represent the protease-sensitive, misfolded PrP population (white bars). C, kinetics of PrPA accumulation in ScN2a-cl3 and N2a-cl3 cells. Division-arrested ScN2a-cl3 and N2a-cl3 cells were treated with 10 μg/ml PAMAM-G4; extracts were collected at 0, 1, 2, 3, 4 5, 20, 30, 48, 72, and 96 h post-treatment. At 60 h, fresh media and PAMAM-G4 were added to the cells. Extracts were untreated (Total PrP), PK digested (+PK), pelleted by PTA (PTA pellet), or pelleted in the presence of SA (SA pellet). D, dose-response curves for the induction of PrPA in N2a-cl3 cells and clearance of rPrPSc in ScN2a-cl3 cells by PAMAM-G4. Levels of insoluble PrP in N2a-cl3 cells following PTA precipitation were measured by ELISA, then normalized to the signal obtained with samples treated with 100 μg/ml PAMAM. Levels of rPrPSc in ScN2a-cl3 following PTA precipitation and PK digestion were also measured by ELISA, then normalized to the signal obtained from samples treated with 0 μg/ml PAMAM. For ELISA measurements, antibodies D18 and D13 were used for capture and detection, respectively. Western blots were probed with D13 or EST123 (supplemental Fig. S6). Error bars indicate the S.D. of three replicate experiments. For the Western blots, the apparent molecular masses based on the migration of protein standards are shown in kilodaltons.
The Effect of Antiprion Compounds on the Relative Amounts of Protease-sensitive, Misfolded PrP Conformers
Having quantified relative levels of sPrPSc and rPrPSc conformers in ScN2a-cl3 cells, we investigated whether known antiprion compounds could alter their relative amounts. Numerous compounds have been identified that induce the clearance of rPrPSc in prion-infected cultured cells (for review, see Ref. 22). The effect of such compounds on protease-sensitive, misfolded PrP conformers has not been previously investigated, but it has been assumed that these antiprion compounds lead to the complete clearance of all PrP aggregates. We asked if some of these compounds might act by shifting the equilibrium of misfolded PrP conformers from protease-resistant to protease-sensitive aggregates. We analyzed the effects of 7 different antiprion compounds (Fig. 1, A and B and supplemental Table S1) that were previously reported to reduce PrPSc levels in ScN2a cells (15, 23–30). As expected, all compounds cleared ScN2a-cl3 cells of rPrPSc. However, their abilities to clear protease-sensitive, misfolded PrP conformers (measured by the difference between the levels of PTA-precipitated and PK-resistant PrP populations after treatment) were more limited. For many of these compounds, substantial levels of protease-sensitive, misfolded PrP conformers persisted after 5 days of treatment using concentrations that fully cleared rPrPSc. Notably, we observed that the addition of Generation 4 polyamidoamine (PAMAM-G4) caused a dramatic increase in the concentration of protease-sensitive, PTA-precipitable PrP conformations (Fig. 1, A and B).
The Induction of PrPA by PAMAM-G4
Western blots (Fig. 1B) indicate that the protease-sensitive, PTA-precipitable PrP population resulting from PAMAM-G4 treatment contains glycoforms of full-length PrP, with the diglycosylated band migrating at an apparent molecular mass of 33–35 kDa. In contrast, sPrPSc and rPrPSc in ScN2a-cl3 cells are composed primarily of N-terminally truncated PrP with a diglycosylated band of 27–30 kDa (Fig. 1B). To distinguish PAMAM-induced, PTA-precipitable PrP from sPrPSc, we refer to it as PrPA. Given the aforementioned difference in apparent molecular mass, we conclude that in ScN2a-cl3 cells, PAMAM-G4 treatment does not directly convert PrPSc to PrPA, but rather causes the de novo formation of PrPA from PrPC.
We monitored the accumulation of misfolded PrP conformers over the course of 4 days after the addition of PAMAM-G4 (Fig. 1C). During log-phase growth, the process of cell division leads to a transient decrease in rPrPSc levels even in the absence of antiprion compounds (21). To measure the kinetics of PrPA induction by PAMAM-G4 alone, cell growth was halted by the addition of sodium butyrate. Comparison of a confluent, stationary ScN2a-cl3 culture to division-arrested, sodium butyrate-treated culture indicated that sodium butyrate alone does not induce the clearance of rPrPSc (supplemental Fig. S3). We observed that after ∼20 h, PAMAM induced a marked increase in the total PrP populations in both infected and uninfected cells (Fig. 1C). The increase in PrP levels did not result from increased transcription of Prnp mRNA (supplemental Fig. S4) but coincided with the accumulation of PrPA that was insoluble in both PTA and the non-denaturing detergent sodium lauryl sarcosine (Sarkosyl). PrPA could be induced in both N2a-cl3 and ScN2a-cl3 cells and to a lesser extent in N2a and hypothalamic GT1 cells that do not overexpress PrP (supplemental Fig. S5). The PAMAM-G4 concentration required to induce PrPA in N2a-cl3 cells was similar to that needed to clear rPrPSc in infected ScN2a-cl3 cells (Fig. 1D).
Biochemical Characterization of PrPA
In contrast to rPrPSc, PrPA exhibited a broad size distribution as observed by size-exclusion chromatography (SEC, Fig. 2, A and B). Although most PrPA eluted within or near the void volume, we found PTA-insoluble PrP that eluted in later fractions, suggestive of low-order PrP oligomers. Similar to rPrPSc, PrPA has an altered conformation with a buried epitope for at least one of two anti-PrP antibodies: PrPA was undetectable by an ELISA that used D13 and D18 as capture and detection antibodies, respectively, prior to GdnHCl-induced unfolding (Fig. 2C). Upon treatment with increasing concentrations of GdnHCl, PrPA appeared to be almost as stable as rPrPSc (Fig. 2D).
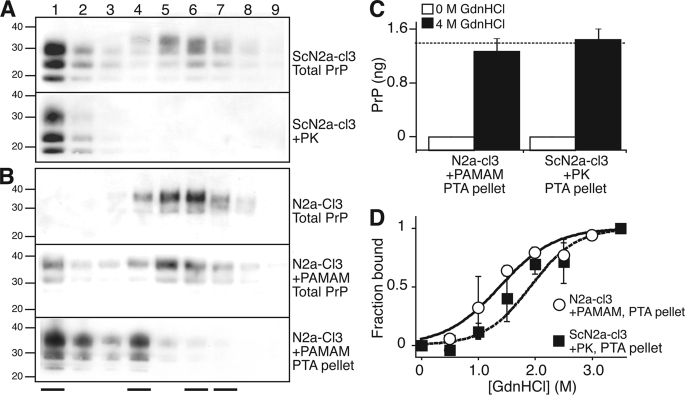
FIGURE 2.
Biochemical characterization of PrPA induced by PAMAM-G4. A and B, size distribution of PrP conformers in ScN2a-cl3 (A) and N2a-cl3 (B) cells. Lysates (600 μg of total protein) were fractionated on a Superdex 200 size-exclusion column. Each fraction (numbered lanes) was either left untreated (Total PrP), digested with PK (+PK), or pelleted in the presence of PTA (+PTA). Western blot analysis was performed on each fraction using D13 Ab. Western blots from untreated N2a-cl3 cells digested with PK or pelleted with PTA did not produce any signal and therefore are not shown. The fraction numbers begin immediately after the elution of the void volume. At the bottom of the panel, the four solid lines indicate, from left to right, the elution peaks for catalase (200 kDa), bovine serum albumin (66 kDa), ovalbumin (43 kDa), and ribonuclease A (13.7 kDa). C, binding of native (non-denatured) PrPA and rPrPSc to anti-PrP antibodies. Lysates containing PrPA were obtained by treating N2a-cl3 cells with 10 μg/ml PAMAM for 20 h; lysates containing rPrPSc were obtained by digesting untreated ScN2a-cl3 extracts with PK. Both lysates were PTA-precipitated, then resuspended in equivalent volumes of lysis buffer in the presence of 0 m or 4 m GdnHCl and incubated for 1 h at 25 °C. For samples originally incubated with 4.0 m GdnHCl, the GdnHCl concentration was adjusted to 0.4 m by addition of lysis buffer. The levels of epitope-exposed PrP were measured by ELISA using D18 and D13 as the capture and detection antibodies, respectively. The dashed line indicates the PrPC level detected in untreated N2a-cl3 lysates containing the same amount of total protein as the four experimental samples. D, stability of PrPA and rPrPSc to GdnHCl denaturation. Lysates were obtained and treated as described in panel C, but employing a range of GdnHCl concentrations (from 0–3.5 m, in 0.5 m increments). All measurements were normalized to the signal obtained from lysates incubated with 4.0 m GdnHCl to calculate the fraction bound. The error bars indicate the S.D. of three replicate measurements.
Induction of PrPA and Clearance of PrPSc by Different Generation PAMAMs
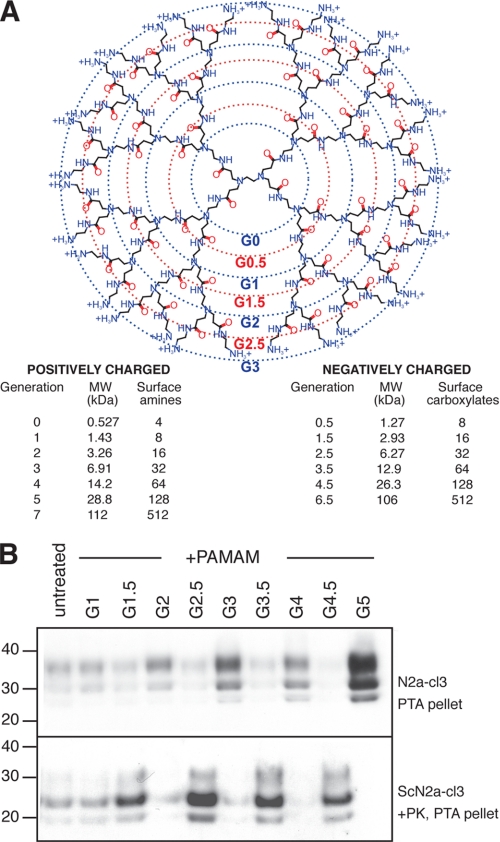
PAMAMs are branched, monodisperse polymers (also known as starburst dendrimers) consisting of a core ethylenediamine molecule and “spokes” of amidoamines radiating out in a quasi-spherical structure (Fig. 3A) (31). Successive polymerization reactions create larger (higher generation) PAMAMs with increasing numbers of free amino or carboxylate groups on the surface. PAMAMs have been used as antiviral drugs, tissue repair scaffolds, targeted carriers of chemotherapeutics and optical oxygen sensors. They are used extensively as transfection reagents and carriers of nucleic acids. In fact, the antiprion properties of PAMAMs were discovered by the observation that PAMAM-based transfection of prion-infected cell lines led to prion clearance (23). We investigated the ability of different generation PAMAMs to induce PrPA and clear rPrPSc (23, 24). We found that positively charged PAMAMs (G1, 2, 3, 4, 5) promote the formation of PrPA and induce the clearance of rPrPSc, whereas negatively charged PAMAMs (G1.5, 2.5, 3.5, 4.5) have the inverse effect (Fig. 3). These two diametrical properties are more pronounced for larger (higher generation) PAMAMs.
FIGURE 3.
The effect of different generation PAMAMs on PrPA and rPrPSc levels. A, structure and chemical properties of different generation PAMAMs. B, different generation PAMAMs were added to N2a-cl3 and ScN2a-cl3 cells at a concentration of 10 μg/ml for 20 h. N2a-cl3 lysates were precipitated with PTA; ScN2a-cl3 lysates were treated with PK prior to PTA precipitation. Western immunoblots were probed with D13 Ab. Apparent molecular masses based on the migration of protein standards are shown in kilodaltons.
Cellular Localization of PrPA
We utilized confocal microscopy to visualize the localization of PAMAM-induced PrPA in cells. Following the addition of PAMAM-G7, N2a-cl3 cells were fixed, permeabilized, and exposed to GdnHCl to expose all PrP epitopes. Cells were stained with D13 antibody, and images were collected by confocal microscopy (Fig. 4). When PAMAM-G7 was added to N2a-cl3 cells, we observed dramatic intracellular accumulation of PrPA. With the addition of PAMAM G5.5, PrPA induction was not observed (supplemental Fig. S7). The observation that both fixation and GdnHCl treatment were required to expose PrPA precluded co-staining with live cell organelle markers. However, although we cannot exclude endoplasmic reticulum localization, the punctate staining pattern was consistent with accumulation in the endocytic pathway and lysosomes, where PAMAM is known to accumulate (23).
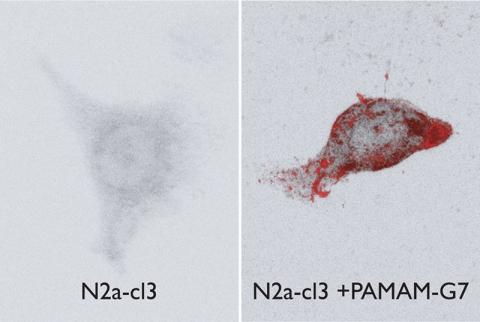
FIGURE 4.
Cellular localization of PrPA. N2a-cl3 cells were plated on a poly-lysine D-coated coverslip and treated with 10 ng/ml of PAMAM-G7 for 24 h (right panel). Untreated N2a-cl3 cells were used as controls (left panel). Cells were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100 for 5 min and treated with 3 m GdnSCN for 8 min prior to blocking with 10% normal goat serum. Cells were stained with D18 and Texas Red-conjugated secondary antibodies, then visualized with a Zeiss LSM510 confocal microscope. The total PrP signal in the presence of PAMAM is severalfold higher than in the absence of PAMAM (because of the accumulation of a large amount of PrPA). At the exposure time required to obtain an unsaturated image for the +PAMAM cells, PrP in the PAMAM cells is below the detection level.
Formation of PrPA in Cell-free Extracts
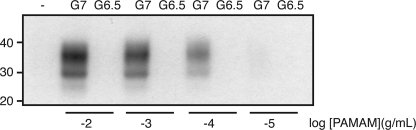
We next attempted induce the formation of PrPA in cell-free extracts. N2a-cl3 cells were lysed by the addition of detergents. Different concentrations of PAMAM-G7 and PAMAM-G6.5 were added to the extract and incubated for 1 h at room temperature. PTA precipitation and Western blots were conducted as described above. The results indicate that PAMAM-G7, but not PAMAM-G6.5, caused the formation of PrPA in a dose-dependent manner in cell-free extracts (Fig. 5).
FIGURE 5.
Induction of PrPA in cell-free extracts. N2a-cl3 lysates were incubated with different concentrations of PAMAM-G7 and PAMAM-G6.5, as indicated, for 1 h at room temperature. Samples were pelleted with PTA, then probed with D13 Ab in Western blots. Apparent molecular masses based on the migration of protein standards are shown in kilodaltons.
We investigated whether PAMAM was directly incorporated into PrPA aggregates. PAMAM-G7 was immobilized by coupling to CNBr-activated Sepharose beads. The immobilized PAMAM was incubated for 1 h at room temperature with uninfected brain homogenates from FVB mice. As a control, uninfected brain homogenates were incubated with uncoupled beads. After the beads were removed by centrifugation, brain homogenates were left untreated or precipitated with PTA. Samples were analyzed by Western blots to detect PrP. PTA-precipitable PrP (PrPA) was found in brain homogenates incubated with the PAMAM-coupled beads, but not in samples with uncoupled beads (supplemental Fig. S8). Additionally, we boiled the beads in SDS, then performed Western immunoblotting with the resulting sample to detect bound PrP. We did not detect any PrP bound to immobilized PAMAM (data not shown). These results indicate that PAMAM does not form a stable complex with PrP and need not be directly incorporated into PrPA.
Self-propagation of PrPA in the Absence of PAMAM
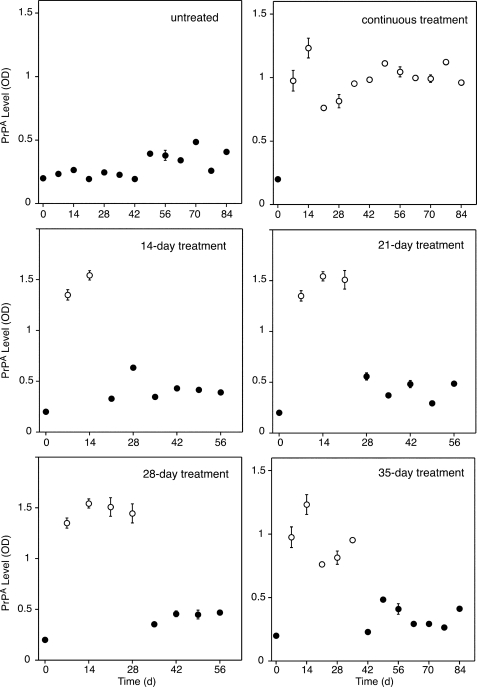
We aimed to determine whether PrPA could be stably propagated from mother to daughter cells in successive generations. N2a-cl3 cells were treated with PAMAM-G7 for 14, 21, 28, or 35 days, after which PAMAM-G7 was removed, and the treated cells were passaged for at least 28 days (split 1:10 every 7 days). Our results show that PrPA did not persist during this time course. Unlike PrPSc, PrPA does not stably self-propagate and is cleared by the cell upon the removal of PAMAM (Fig. 6). It remains to be determined whether persistently heritable PrPA can be formed in cells by longer term exposure to positively charged PAMAM under different media conditions.
FIGURE 6.
Propagation of PrPA in culture. N2a-cl3 cells were treated with 10 μg/ml of PAMAM-G7 for different durations, as indicated. After treatment, PAMAM-G7 was removed, and the cells were continuously passaged for up to 84 days. For continuous treatment, PAMAM-G7 was not removed from N2a-cl3 cells. Every 7th day, extracts were collected from confluent cells and centrifuged in the presence of PTA; the pellet was analyzed by ELISA using D18 and D13 Abs as the capture and detection antibodies, respectively. Open and closed circles indicate the presence and absence of PAMAM-G7, respectively.
Infectivity of PrPA
First, we attempted to induce the formation of de novo PrPA in N3a-cl3 cells by adding cell extracts of PAMAM-G4-treated cells. In these cell culture studies, we failed to demonstrate measurable infectivity of PrPA (data not shown). Second, we tried to measure the infectivity of PrPA by mouse bioassay (Table 1). N2a-cl3 and ScN2a-cl3 cells were treated with PAMAM-G7 or PAMAM-G4, and the cell extracts were inoculated intracerebrally into Tg4053 mice overexpressing MoPrP. Some inocula were PTA-purified and/or PK-digested prior to infection. Homogenates of untreated ScN2a-cl3 cells caused disease in Tg4053 mice in ∼68 days. PAMAM-induced PrPA aggregates failed to produce neurological disease, arguing that PrPA is neither pathogenic nor infectious in mice.
TABLE 1.
Infectivity of PrPA in Tg4053 mouse bioassays
| Inoculum |
n/n0b | Incubation time | |||
|---|---|---|---|---|---|
| Cells | Treatment | Purification | PKa | ||
| days ± S.E. | |||||
| N2a-cl3 | None | PTA | – | 0/12 | 336c |
| N2a-cl3 | PAMAM-G4 | PTA | – | 0/7 | 336c |
| N2a-cl3 | PAMAM-G7 | PTA | – | 0/12 | 315c |
| N2a-cl3 | PAMAM-G7 | PTA | + | 0/11 | 287c |
| N2a-cl3 | PAMAM-G7 | Homogenate | – | 0/10 | 287c |
| ScN2a-cl3 | None | PTA | + | 12/12 | 74 ± 6 |
| ScN2a-cl3 | None | Homogenate | – | 12/12 | 68 ± 8 |
a −, undigested; +, digested with PK.
b n, number of mice showing clinical signs of disease; n0, number of inoculated mice.
c No animals showed clinical signs. Bioassays were terminated at these times after inoculation.
DISCUSSION
Taken together, our data argue that the accumulation of PrPA is directly correlated to the clearance of rPrPSc in ScN2a-cl3 cells exposed to positively charged PAMAMs (Figs. 1 and 3) and suggest that PrPA represents a PrP species that is off-pathway relative to the formation of rPrPSc. However, given that PrPA can be formed in uninfected cells that do not harbor PrPSc and from uninfected lysates, the disaggregation of PrPSc is not necessary for the formation of PrPA, which is composed of full-length PrP. In contrast, truncation of PrP is necessary to form PrP 27–30 (the protease-resistant core of the infectious prion particle), which aggregates into rod-shaped polymers that are insoluble in nonionic detergent. Our results indicate that truncation of PrP is not necessary to produce PrPA. In one interpretation of our data, PrPA may serve to sequester PrPC and prevent it from entering the unfolding pathway that leads to formation of rPrPSc. Because rPrPSc is dynamically catabolized in the cell with a half-life of ∼1 day (28, 32), a decrease in the rate of formation results in a rapid lowering of its steady-state levels.
Several possible mechanisms may explain how positively charged PAMAMs influence the off-pathway aggregation of PrP (33). First, PAMAMs and different PrP conformers may interact directly. PrP has an isoelectric point of ∼9 and exists as a positively charged protein at neutral pH. The positive charge is further increased in the low pH environment of the endocytic pathway where aggregated PrP conformers are thought to accumulate (34). Positively charged PAMAMs localize to lysosomes, where they are thought to interact directly with endocytosed, aggregated PrP (23, 24). This localization may create an unfavorable environment for the native PrPC molecule and thermodynamically favor certain aggregation states. Ordered amyloidic structures of prions are theorized to have β-helical conformations, with positively charged residues facing away from the core of the growing fiber (35). Thus, positively charged PAMAMs may disfavor both PrPC and β-helical PrPSc conformations in favor of amorphous aggregates that reduce the solvent exposure of positively charged side chains. Alternatively, the formation of PrPA may be a result of gross cellular disruptions caused by PAMAM. For example, PAMAM has been shown to alter the pH and cause osmotic swelling of endosomes (36). We doubt this latter explanation, as PAMAM appears to be relatively innocuous at concentrations utilized in this study. N2a-cl3 cells can tolerate concentrations of PAMAMs (up to 100 μg/mL), demonstrating its low cellular toxicity. Additionally, we have not observed the induction of morphological and cellular changes by PAMAM.
The different roles of protease-sensitive and protease-resistant misfolded PrP conformers in prion disease remain to be elucidated. Protease-resistant PrPSc has been associated with the vast majority of human prion diseases described to date (37). Protease-sensitive, misfolded PrP conformers have been shown to accumulate at substantial levels in diseased tissues and cells (11). These protease-sensitive conformers feature prominently in Gerstmann-Straüssler-Scheinker (GSS) disease (38) and in protease-sensitive prionopathies (39). Additionally, the accumulation of protease-sensitive misfolded PrP conformers was found in a Tg mouse model of GSS disease (10, 14, 40, 41) as well as in synthetic prion strains induced by the polymerization of recombinant PrP into amyloids (42, 43). PrPA and protease-sensitive, misfolded PrP conformers that accumulate in Tg mouse models and human patients both show PTA and detergent insolubility, retain protease sensitivity, have buried D13/D18 epitopes, and contain a heterogeneous population of aggregated, full-length PrP. However, our data suggest that PrPA is neither pathogenic nor transmissible in cell culture or mouse bioassays (Table 1). Our initial infectivity studies showed that intracerebral inoculation of dendrimer-treated, scrapie-infected cells did not transmit disease to Tg4053 mice overexpressing PrP (24).
The ability to induce non-pathogenic conformational changes in PrPC is unlikely to be unique to PAMAMs. Proteasome inhibitors have been reported to induce the accumulation of cytosolic PrP aggregates that appear to be self-propagating on a short time scale (44), although this phenomenon was only observed for a transgenic, truncated form of PrP. Suramin, a bis-hexasulfonated naphthylurea, has been shown to induce the aggregation of PrP in a post-ER/Golgi compartment, inhibiting PrP trafficking to the outer plasma membrane (45). A more recent study showed that the excess addition of epigallocatechin gallate (EGCG), the main polyphenol in green tea, induces a conformational transition in PrPC, rendering it insoluble in detergents (46). However, this alternative conformer could only be induced by overexpression of a sequence-tagged PrP. The structural relationship between PrPA and other non-native PrP conformations remains to be determined.
In summary, we have chemically induced the formation of non-infectious PrP aggregates in mammalian cells. PrPA has chemical properties that are distinct from disease-causing PrPSc and exists in culture as protease-sensitive aggregates of full-length PrP. More generally, our results demonstrate that chemicals can rapidly modulate the relative amounts of different PrP conformers present in cells. Our observations suggest that the chemical induction of alternative, possibly benign, off-pathway aggregates may be a viable strategy for halting the accumulation of malignant PrPSc conformers and other amyloid-forming proteins.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AG02132, AG10770, and AG021601 as well as by gifts from the G. Harold and Leila Y. Mathers Foundation, Hillblom Foundation, Fight for Mike Homer Program, and Mr. Robert Galvin.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S8.
- PrP
- prion protein
- PrPA
- PrP aggregated
- PrPC
- PrP cellular isoform
- PrPSc
- disease-causing PrP isoform
- rPrPSc
- protease-resistant PrPSc isoform
- sPrPSc
- protease-sensitive PrPSc isoform
- N2a
- neuroblastoma
- ScN2a
- prion-infected neuroblastoma
- PMSF
- phenylmethylsulfonyl fluoride
- BSA
- bovine serum albumin
- PTA
- sodium phosphotungstic acid
- FBS
- fetal bovine serum
- PK
- proteinase K
- ABTS
- 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid)
- RML
- Rocky Mountain Laboratory
- SA
- Sarkosyl
- PAMAM
- polyamidoamines
- HRP
- horseradish peroxidase
- PBS
- phosphate-buffered saline
- ELISA
- enzyme-linked immunosorbent assay
- PI
- protease inhibitor.
REFERENCES
- 1.Prusiner S. B. (1982) Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S. B. (2001) N. Engl. J. Med. 344, 1516–1526 [DOI] [PubMed] [Google Scholar]
- 3.Pan Y. T., Hori H., Saul R., Sanford B. A., Molyneux R. J., Elbein A. D. (1983) Biochemistry 22, 3975–3984 [DOI] [PubMed] [Google Scholar]
- 4.Basler K., Oesch B., Scott M., Westaway D., Wälchli M., Groth D. F., McKinley M. P., Prusiner S. B., Weissmann C. (1986) Cell 46, 417–428 [DOI] [PubMed] [Google Scholar]
- 5.Bolton D. C., McKinley M. P., Prusiner S. B. (1982) Science 218, 1309–1311 [DOI] [PubMed] [Google Scholar]
- 6.Prusiner S. B., McKinley M. P., Bowman K. A., Bolton D. C., Bendheim P. E., Groth D. F., Glenner G. G. (1983) Cell 35, 349–358 [DOI] [PubMed] [Google Scholar]
- 7.Meyer R. K., McKinley M. P., Bowman K. A., Braunfeld M. B., Barry R. A., Prusiner S. B. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 2310–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safar J., Wille H., Itri V., Groth D., Serban H., Torchia M., Cohen F. E., Prusiner S. B. (1998) Nat. Med. 4, 1157–1165 [DOI] [PubMed] [Google Scholar]
- 9.Tzaban S., Friedlander G., Schonberger O., Horonchik L., Yedidia Y., Shaked G., Gabizon R., Taraboulos A. (2002) Biochemistry 41, 12868–12875 [DOI] [PubMed] [Google Scholar]
- 10.Tremblay P., Ball H. L., Kaneko K., Groth D., Hegde R. S., Cohen F. E., DeArmond S. J., Prusiner S. B., Safar J. G. (2004) J. Virol. 78, 2088–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safar J. G., Geschwind M. D., Deering C., Didorenko S., Sattavat M., Sanchez H., Serban A., Vey M., Baron H., Giles K., Miller B. L., DeArmond S. J., Prusiner S. B. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3501–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Legname G., Nguyen H. O., Peretz D., Cohen F. E., DeArmond S. J., Prusiner S. B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19105–19110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastrana M. A., Sajnani G., Onisko B., Castilla J., Morales R., Soto C., Requena J. R. (2006) Biochemistry 45, 15710–15717 [DOI] [PubMed] [Google Scholar]
- 14.Colby D. W., Zhang Q., Wang S., Groth D., Legname G., Riesner D., Prusiner S. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20914–20919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May B. C., Fafarman A. T., Hong S. B., Rogers M., Deady L. W., Prusiner S. B., Cohen F. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3416–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson R. A., Peretz D., Pinilla C., Ball H., Bastidas R. B., Rozenshteyn R., Houghten R. A., Prusiner S. B., Burton D. R. (1998) J. Virol. 72, 9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leclerc E., Peretz D., Ball H., Solforosi L., Legname G., Safar J., Serban A., Prusiner S. B., Burton D. R., Williamson R. A. (2003) J. Mol. Biol. 326, 475–483 [DOI] [PubMed] [Google Scholar]
- 18.Williamson T. L., Bruijn L. I., Zhu Q., Anderson K. L., Anderson S. D., Julien J. P., Cleveland D. W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9631–9636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., Diringer H. (1987) J. Virol. 61, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler D. A., Scott M. R., Bockman J. M., Borchelt D. R., Taraboulos A., Hsiao K. K., Kingsbury D. T., Prusiner S. B. (1988) J. Virol. 62, 1558–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghaemmaghami S., Phuan P. W., Perkins B., Ullman J., May B. C., Cohen F. E., Prusiner S. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17971–17976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trevitt C. R., Collinge J. (2006) Brain 129, 2241–2265 [DOI] [PubMed] [Google Scholar]
- 23.Supattapone S., Nguyen H. O., Cohen F. E., Prusiner S. B., Scott M. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14529–14534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Supattapone S., Wille H., Uyechi L., Safar J., Tremblay P., Szoka F. C., Cohen F. E., Prusiner S. B., Scott M. R. (2001) J. Virol. 75, 3453–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doh-Ura K., Iwaki T., Caughey B. (2000) J. Virol. 74, 4894–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korth C., May B. C., Cohen F. E., Prusiner S. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9836–9841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caughey B., Race R. E. (1992) J. Neurochem. 59, 768–771 [DOI] [PubMed] [Google Scholar]
- 28.Peretz D., Williamson R. A., Kaneko K., Vergara J., Leclerc E., Schmitt-Ulms G., Mehlhorn I. R., Legname G., Wormald M. R., Rudd P. M., Dwek R. A., Burton D. R., Prusiner S. B. (2001) Nature 412, 739–743 [DOI] [PubMed] [Google Scholar]
- 29.Ertmer A., Gilch S., Yun S. W., Flechsig E., Klebl B., Stein-Gerlach M., Klein M. A., Schätzl H. M. (2004) J. Biol. Chem. 279, 41918–41927 [DOI] [PubMed] [Google Scholar]
- 30.Prior M., Lehmann S., Sy M. S., Molloy B., McMahon H. E. (2007) J. Virol. 81, 11195–11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee I. S., Long J. R., Prusiner S. B., Safar J. G. (2005) J. Am. Chem. Soc. 127, 13802–13803 [DOI] [PubMed] [Google Scholar]
- 32.Safar J. G., DeArmond S. J., Kociuba K., Deering C., Didorenko S., Bouzamondo-Bernstein E., Prusiner S. B., Tremblay P. (2005) J. Gen. Virol. 86, 2913–2923 [DOI] [PubMed] [Google Scholar]
- 33.Baskakov I. V., Legname G., Baldwin M. A., Prusiner S. B., Cohen F. E. (2002) J. Biol. Chem. 277, 21140–21148 [DOI] [PubMed] [Google Scholar]
- 34.Borchelt D. R., Taraboulos A., Prusiner S. B. (1992) J. Biol. Chem. 267, 16188–16199 [PubMed] [Google Scholar]
- 35.Govaerts C., Wille H., Prusiner S. B., Cohen F. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonawane N. D., Szoka F. C., Jr., Verkman A. S. (2003) J. Biol. Chem. 278, 44826–44831 [DOI] [PubMed] [Google Scholar]
- 37.Gambetti P., Kong Q., Zou W., Parchi P., Chen S. G. (2003) Br. Med. Bull. 66, 213–239 [DOI] [PubMed] [Google Scholar]
- 38.Kong Q., Surewicz W. K., Petersen R. B., Zou W., Chen S. G., Gambetti P., Parchi P., Capellari S., Goldfarb L., Montagna P., Lugaresi E., Piccardo P., Ghetti B. (2004) in Prion Biology and Diseases (Prusiner S. B. ed), pp. 673–775, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39.Gambetti P., Dong Z., Yuan J., Xiao X., Zheng M., Alshekhlee A., Castellani R., Cohen M., Barria M. A., Gonzalez-Romero D., Belay E. D., Schonberger L. B., Marder K., Harris C., Burke J. R., Montine T., Wisniewski T., Dickson D. W., Soto C., Hulette C. M., Mastrianni J. A., Kong Q., Zou W. Q. (2008) Ann. Neurol. 63, 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsiao K. K., Scott M., Foster D., Groth D. F., DeArmond S. J., Prusiner S. B. (1990) Science 250, 1587–1590 [DOI] [PubMed] [Google Scholar]
- 41.Hegde R. S., Mastrianni J. A., Scott M. R., DeFea K. A., Tremblay P., Torchia M., DeArmond S. J., Prusiner S. B., Lingappa V. R. (1998) Science 279, 827–834 [DOI] [PubMed] [Google Scholar]
- 42.Legname G., Baskakov I. V., Nguyen H. O., Riesner D., Cohen F. E., DeArmond S. J., Prusiner S. B. (2004) Science 305, 673–676 [DOI] [PubMed] [Google Scholar]
- 43.Colby D. W., Giles K., Legname G., Wille H., Baskakov I. V., DeArmond S. J., Prusiner S. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20417–20422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma J., Lindquist S. (2002) Science 298, 1785–1788 [DOI] [PubMed] [Google Scholar]
- 45.Gilch S., Winklhofer K. F., Groschup M. H., Nunziante M., Lucassen R., Spielhaupter C., Muranyi W., Riesner D., Tatzelt J., Schätzl H. M. (2001) EMBO J. 20, 3957–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rambold A. S., Miesbauer M., Olschewski D., Seidel R., Riemer C., Smale L., Brumm L., Levy M., Gazit E., Oesterhelt D., Baier M., Becker C. F., Engelhard M., Winklhofer K. F., Tatzelt J. (2008) J. Neurochem. 107, 218–229 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.