Abstract
The exocyst is an evolutionarily conserved octameric complex involved in polarized exocytosis from yeast to humans. The Sec3 subunit of the exocyst acts as a spatial landmark for exocytosis through its ability to bind phospholipids and small GTPases. The structure of the N-terminal domain of Sec3 (Sec3N) was determined ab initio and defines a new subclass of pleckstrin homology (PH) domains along with a new family of proteins carrying this domain. Respectively, N- and C-terminal to the PH domain Sec3N presents an additional α-helix and two β-strands that mediate dimerization through domain swapping. The structure identifies residues responsible for phospholipid binding, which when mutated in cells impair the localization of exocyst components at the plasma membrane and lead to defects in exocytosis. Through its ability to bind the small GTPase Cdc42 and phospholipids, the PH domain of Sec3 functions as a coincidence detector at the plasma membrane.
Keywords: Cdc42, Exocytosis, Membrane Trafficking, Phosphatidylinositol, Plasma Membrane, Secretion, Sec3, The Exocyst
Introduction
The exocyst is an evolutionarily conserved octameric protein complex composed of subunits Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84. This complex was first identified by genetic and biochemical methods in the budding yeast Saccharomyces cerevisiae (1, 2). A homologous complex was subsequently discovered in mammalian cells (3). The exocyst mediates initial tethering of post-Golgi secretory vesicles to the plasma membrane, a step that precedes SNARE7-driven membrane fusion (4, 5). The exocyst is regulated by numerous cellular factors, and in particular small GTPases, which are primarily responsible for the spatiotemporal control of exocytosis (6).
Recent studies have provided insights into the molecular architecture and function of tethering proteins. Crystal structures of nearly full-length Exo70 (7–9) and large fragments of Sec6 (10), Sec15 (11), and Exo84 (7) have been determined. Despite the lack of sequence similarity, these structures all reveal a similar fold, consisting of elongated tandem repeats of helical bundles, which are predicted to pack against one another during assembly of the exocyst complex (4). The recently determined structure of the yeast Dsl1p complex implicated in Golgi-to-endoplasmic reticulum transport provided the first glance into an assembled tethering complex consisting of helical bundles similar to those of exocyst subunits (12). The structure suggested a similar architecture, and possibly a common origin, among multisubunit tethering complexes. Structures have also been determined of the RalA-binding domains of the mammalian exocyst subunits Sec5 (13) and Exo84 (14), which display immunoglobulin-like and pleckstrin homology (PH) folds, respectively. However, these two domains are missing in the yeast complex and are not considered part of the conserved core of the exocyst (4).
Studies in yeast suggest that subunit Sec3 plays a pivotal role in exocyst function and vesicle tethering. Sec3 localizes, together with Exo70, to the growing end of the daughter cell (known as the “bud tip”). Although the localization of other exocyst components relies on the actin cables that serve as tracks for motor-driven vesicle transport to the daughter cell, the polarized localization of Sec3 is independent of actin assembly (15–17). Genetic analyses and live cell imaging have shown that the N-terminal 320-amino acid region of Sec3 is important for localization of the exocyst to the plasma membrane and exocytosis (18). This region has also been implicated in the binding of small Rho-family GTPases and phosphatidylinositol 4,5-bisphosphate (19–22).
As an important step toward understanding exocyst-mediated vesicle tethering, we identified the precise domain of Sec3 involved in plasma membrane and Cdc42 binding, determined its crystal structure, and studied its function in cells. We show that the core of this domain, consisting of residues 71–241 and referred to as Sec3N, defines a new subclass of PH domains. Respectively, N- and C-terminal to the PH domain, Sec3N contains an α-helix and two β-strands that mediate dimerization of the PH domain through domain swapping. A phosphate ion and the C terminus of a neighboring molecule in the crystal bind in the predicted phospholipid-binding pockets at the distal ends of the dimer, mimicking protein-phospholipid interactions. Mutations of positively charged residues identified by these interactions impair the polarized localization of exocyst components at the daughter cell membrane and lead to growth and secretion defects. Based on this structure, a new family of proteins was identified containing Sec3-like PH domains, including amisyn, a protein implicated in the regulation of SNARE complex assembly.
EXPERIMENTAL PROCEDURES
Protein Preparation
The cDNA encoding for yeast Sec3N (residues 71–241) was amplified by PCR and cloned into the BamHI site of vector pGEX4T-1 (Amersham Biosciences). This vector comprises a glutathione S-transferase affinity purification tag and a thrombin-cleavage site. BL21(DE3) competent cells (Invitrogen) were transformed with this construct and grown in LB medium at 37 °C to A600 = 0.6. Expression was induced by addition of 1 mm isopropyl-β-d-thiogalactopyranoside and carried out overnight at 20 °C. Cells were harvested by centrifugation, resuspended in phosphate-buffered saline (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4 (pH 7.4)), and lysed using a Microfluidizer apparatus (MicroFluidics Corp.). Affinity purification on a glutathione-Sepharose column (Amersham Biosciences) was done according to the manufacturer's protocol. Sec3N-glutathione S-transferase was eluted with 50 mm Tris-HCl (pH 7.5), 10 mm glutathione and dialyzed against 20 mm Tris-HCl (pH 7.5), 20 mm NaCl, 1 mm dithiothreitol. Cleavage of the glutathione S-transferase tag was done during dialysis with addition of thrombin at a molar ration of 1:1000 (Hematologic Technologies Inc.). Further purification was carried out on a Mono S ion-exchange column (Amersham Biosciences) in 20 mm Tris-HCl, (pH 7.5), 1 mm dithiothreitol, and 1 m NaCl gradient. Selenomethionine-substituted Sec3N was obtained by a similar procedure, growing cells in SelenoMet medium (Athena Enzyme Systems), supplemented with 70 mg/ml selenomethionine (Acros Organics).
Crystallization
Sec3N was dialyzed against 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 5 mm dithiothreitol and concentrated to 10 mg/ml using a Vivaspin centrifugal device (Sartorius Stedim Biotech). Crystals were obtained using the hanging drop, vapor-diffusion method. A typical setup consisted of a 1:1 (v/v) mixture of protein solution and a well solution containing 100 mm Bicine (pH 9.5), 14% polyethylene glycol monomethyl ether 5000, 1 mm dithiothreitol at 20 °C. Crystal quality was improved with addition of 10 mm of CdCl2 or BaCl2. Crystals were flash-frozen in liquid nitrogen using Paratone-N (Hampton Research) as cryoprotectant.
Data Collection, Structure Determination, and Analysis
X-ray datasets were collected from selenomethionine-substituted crystals (Table 1) using beamline 17-BM at the Industrial Macromolecular Crystallography Association-Collaborative Access Team facility of the Advance Photon Source (Argonne, IL). Data indexation and scaling were carried out with the program HKL2000 (HKL Research Inc.). The structure was determined using the multiwavelength anomalous dispersion method at 2.8-Å resolution. The positions of four selenium atoms (one per Sec3N monomer in the asymmetric unit) were found using the program SnB (23). The positions of the selenium atoms were refined, and phases were calculated with the program Phenix (24). The electron density map was improved by 4-fold symmetry averaging and density modification with the CCP4 program DM (25). Most of the structure was built into the averaged map using the graphics program Coot (26). Phases were extended to 2.0-Å resolution, and subsequent model building and refinement iterations were performed with the programs Coot and Phenix. Illustrations were prepared with the program PyMOL (DeLano Scientific LLC.). The solvent-accessible area buried at the dimer interface, defined as the locus of the center of a solvent probe of radius 1.4 Å as it rolls over the van der Waals surface of the protein, was calculated with the CCP4 program AREAIMOL.
TABLE 1.
Crystallographic data, phasing, and refinement statistics
Values in parentheses correspond to the highest resolution shell.
| Native | Selenium peak | Selenium inflection | Selenium remote | |
|---|---|---|---|---|
| Wavelength, Å | 1.0 | 0.9796 | 0.9798 | 0.9740 |
| Space group | P1 | P1 | ||
| Unit cell a/b/c, Å | 40.7/68.5/68.0 | 40.6/67.2/67.6 | ||
| Unit cell α/β/γ, ° | 89.9/82.2/71.8 | 85.5/82.0/73.5 | ||
| Resolution, Å | 2.00-50 (2.00-2.07) | 2.85-50 (2.85-2.95) | ||
| Completeness, % | 93.6 (62.7) | 96.3 (77.9) | 95.8 (73.5) | 95.9 (74.5) |
| Multiplicity | 10.1 (3.8) | 2.0 (1.9) | 2.0 (1.9) | 2.0 (1.9) |
| Rmerge,a % | 6.9 (52.0) | 3.4 (11.0) | 3.4 (11.7) | 3.5 (14.0) |
| I/σ | 24.6 (1.5) | 24.0 (6.2) | 23.8 (6.0) | 22.5 (4.9) |
| Rfactor,b % | 20.2 | |||
| Rfree,c % | 24.8 | |||
| Root mean square bonds, Å | 0.025 | |||
| Root mean square angles, ° | 1.251 | |||
| B-factor protein, Å2 | 60.25 | |||
| B-factor solvent, Å2 | 58.46 | |||
| PDB code | 3HIE |
a Rmerge = Σhkl(I − 〈I〉)/ΣI, where I and 〈I〉 are the observed and mean intensities of all observations of reflection hkl, including its symmetry-related equivalents.
b Rfactor = Σhkl‖Fobs| − |Fcalc‖/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors of reflection hkl.
c Rfree and Rfactor were calculated for a randomly selected subset of the reflections (5%) that were omitted during refinement.
Plasmids and Yeast Strains
Sec3 mutations were generated with the QuikChange site-directed mutagenesis kit (Agilent Technologies) using a CEN-LEU2 plasmid (pG1273) containing the full-length Sec3 gene as a template. To generate exo70 sec3 double mutants, yeast cells bearing the sec3ΔN mutation, exo70::HIS3 deletion, and carrying an EXO70 balancer plasmid (CEN-URA3) and plasmids with exo70–45 or exo70-47 were transformed with various sec3 mutant variants. The transformants were tested for growth upon losing the EXO70 balancer on 5-fluoroorotic acid (5-FOA) plates as described previously (18).
Bgl2 Secretion Assay
Wild-type, exo70-47, and exo70-47 sec3-303 cells were grown to early log phase at 25 °C. sec10-2 mutant cells were grown at 25 °C and moved for 1 h to 37 °C. NaF and NaN3 were added to a concentration of 10 mm. An amount of cells equivalent to 10 A600/ml was collected and washed three times with washing buffer (20 mm Tris-HCl, pH 7.5, 10 mm NaN3, and 10 mm NaF). Cells were resuspended in 300 μl of spheroplast solution (50 mm Tris-HCl, pH 7.5, 1.4 m sorbitol, 10 mm NaF, 10 mm NaN3, 30 mm 2-mercaptoethanol, and 0.2 mg/ml zymolyase), and incubated for 30 min at 37 °C. The resulting spheroplasts were pelleted at 2000 rpm, and supernatants (external pools) were collected. After washing, spheroplasts were dissolved in lysis buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm MgCl2, 0.5% Triton X-100, and protease inhibitor) on ice for 10 min. The samples were centrifuged at 500 × g for 5 min, and the supernatants (internal pools) were collected. Fractions from both pools were analyzed by Western blot using a rabbit anti-Bgl2 polyclonal antibody.
Microscopy
Chromosomal GFP tagging of exocyst components was performed as described (15). Cells were grown to early log phase (A600 0.6) in synthetic complete media and fixed in methanol/acetone before microscopy. Images were collected using a Leica DM IRB microscope equipped with a 100× oil immersion objective and a high resolution charge-coupled device camera (ORCA-ER, Hamamatsu Photonics). The fluorescence distribution in cell images was analyzed with the program ImageJ (rsb.info.nih.gov/ij). A straight line was drawn across the bud and into the mother cell. Plot Profile ImageJ was used to determine the gray value of each point along the line, a measure of the fluorescence intensity.
RESULTS
Sec3N Folds into a PH Domain
We had previously mapped the GTPase and membrane-binding activities of Sec3 to an N-terminal fragment comprising residues 1–320 (18, 19). Sequence analysis suggested that Sec3-(1–320) contained N- and C-terminal disordered, non-conserved regions. The fragment 71–250 appeared to form an independent globular domain, conserved across species and lacking the predicted disordered regions. Our genetic analysis also indicated that deletion of the first 70 amino acids of Sec3 did not affect its function in cells (supplemental Fig. S1). Attempts to crystallize Sec3-(71–250) yielded needle-like crystals of poor quality that could not be improved. Analysis of protein samples from old crystallization setups showed partial protein degradation, a condition known to affect crystallization. A stable fragment of slightly smaller mass (residues 71–241, referred to here as Sec3N) was identified using a combination of mass spectrometry and N-terminal sequencing. This fragment was subcloned and yielded excellent quality crystals, diffracting the x-rays to ∼2.0-Å resolution (Table 1 and “Experimental Procedures”). Importantly, Sec3N binds Cdc42 (supplemental Fig. S2) and, as shown below, accounts for the plasma membrane-binding activity of Sec3.
Sec3N had no detectable similarity to any known structure in the Protein Data Bank. Therefore, the structure was determined ab initio by multiple anomalous dispersion from crystals of selenomethionine-substituted protein. Sec3N has a single methionine residue in 171 amino acids, resulting in a weak anomalous signal. However, the crystal contained four Sec3N molecules in the asymmetric unit, and the implementation of 4-fold non-crystallographic symmetry averaging improved significantly the quality of the electron density map. The structure was built and refined to 2.0-Å resolution (Table 1).
During model building, the core domain of the Sec3N monomer was recognized as a PH domain. The PH domain belongs to a structural superfamily that includes the phosphotyrosine-binding, Ena/VASP homology, and Ran-binding domains (27). These domains lack sequence similarity, but all share a core structure of 100–120 amino acids consisting of a seven-stranded, semi-open, antiparallel β-barrel (strands β1 to β7), capped at one end by a C-terminal α-helix (α1). At the other end of the barrel three inter-strand variable loops (VL1, VL2, and VL3) form the canonical phosphoinositide-binding pocket. Sec3N shares this core structure, although it lacks statistically significant sequence identity with classic PH domains. For instance, the PH domain of Sec3 and that of the prototypical PH-domain-containing protein phospholipase Cδ (28) (PDB code: 1mai) superimpose with r.m.s.d. of 2.15 Å for 87 equivalent Cα positions, whereas the two proteins share only 7% sequence identity (Fig. 1A). The PH fold appears to be conserved among Sec3 sequences from different species (supplemental Fig. S3), including human Sec3, which shares 16.4% sequence identity (37.4% sequence similarity) with yeast Sec3 in this region (Fig. 1B). Importantly, amino acids predicted to be involved in phospholipid binding are well conserved (discussed below).
FIGURE 1.
Sec3N folds into a PH domain. A, two perpendicular views of a superimposition of the structure of the Sec3N monomer with that of the PH domain of phospholipase Cδ (28) (PDB code: 1MAI). Phospholipase Cδ (PLCD1) is colored blue and Sec3N is colored green (helices), yellow (β-strands in front), red (β-strands in back), and cyan (variable loops 1–3). Compared with PLCD1, Sec3N contains an additional α-helix at the N terminus (α0) and two β-strands at the C terminus (magenta). The two structures superimpose with an r.m.s.d. of 2.15 Å for 87 equivalent Cα positions. B, sequence alignment of yeast Sec3N with the equivalent region of human Sec3, and the Sec3N-related proteins amisyn and maize roothairless (or rth1). PLCD1 was also aligned based on a structure superimposition. The percentage identity and similarity are indicated in the bottom right corner. Secondary structure assignment is shown above the alignment and colored according to A. Residues predicted to be important for phosphoinositide binding are highlighted (red contour). Uniprot accession codes: SEC3_YEAST (P33332), SEC3_HUMAN (Q9NV70), AMISYN_HUMAN (Q8NFX7), rth1_MAIZE (Q5YLM3), and PLCD1_RAT (P10688).
In addition to Sec3-related sequences from different species, a BLAST search identified proteins previously unsuspected to contain PH domains (examples are given in Fig. 1 and supplemental Fig. S3). Most of these proteins are still uncharacterized. However, an interesting example is the protein amisyn (also known as syntaxin-binding protein 6 or STXBP6), a 210-amino acid polypeptide implicated in the regulation of SNARE complex assembly and exocytosis through its C-terminal coiled-coil, a vesicle-SNARE-homology domain (29, 30). Although its characterization is still limited, amisyn has been proposed to form part of a vesicle-docking complex and is known to partially co-sediment with membranes (30). Based on its relationship with Sec3N (Fig. 1B), we now predict that the association of amisyn with cellular membranes results from the presence of an N-terminal PH domain (residues 1–153).
Dimerization of Sec3N
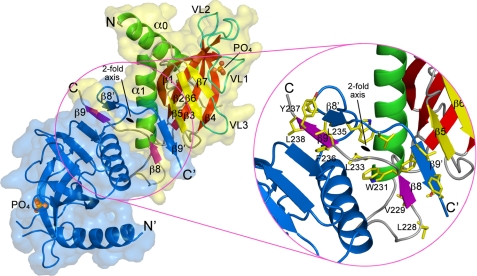
The Sec3N fragment crystallized here is 50–70 amino acids longer than the canonical PH domain, and the structure includes an additional α-helix at the N terminus (α0) and two β-strands at the C terminus (β8 and β9). To our surprise, the four Sec3N molecules in the asymmetric unit of the crystal are organized into two independent antiparallel dimers (Fig. 2). Dimerization involves domain swapping of strands β8 and β9 of each monomer, which overlap in “crossed arms” arrangement (supplemental movie S1). The dimer interface is extensive, burying 4074 Å2 (2037 Å2 per monomer) of solvent-accessible surface area (“Experimental Procedures”). This value is much larger than the average crystal packing contact (570 Å2) or protein-protein complex interface (1910 Å2), and even greater than the average homodimer interface (3880 Å2) (31–33). The dimerization interface is hydrophobic in character and involves residues from strands β8 and β9 and helix α1 (Tyr-217, Ile-218, Phe-221, Val-229, Trp-231, Phe-236, and Leu-238). Additional stabilization of the dimer interface results from incorporation of strands β8 and β9 from two different monomers into extended antiparallel β-sheets with strands β5, β6, and β7 of each of the PH domains (Fig. 2).
FIGURE 2.
Sec3N forms an antiparallel dimer of PH domains. The figure shows a ribbon diagram and surface representations of the Sec3 dimer. One of the molecules of the dimer is labeled and colored according to Fig. 1, while the other is colored blue. The phosphate ions that bind in the predicted phosphoinositide-binding pockets at the distal ends of the dimer are also shown. Note that dimerization results from domain swapping of strands β8 and β9. An enlarged view shows some of the hydrophobic amino acids involved in interactions at the dimer interface.
The dimerization of several PH domains has been observed in crystal structures, but never confirmed in solution (34–38). Similarly, analysis by three different methods, including analytical size-exclusion chromatography, analytical ultracentrifugation, and multiangle light scattering (supplemental Fig. S4), suggests that Sec3N is a monomer in solution, at lower concentration. However, we cannot totally exclude the possibility that the region C-terminal to the PH domain contributes to dimerization. Indeed, the fact that there are two nearly identical (crystallographically independent) dimers in the asymmetric unit (r.m.s.d. of 0.85 Å between equivalent Cα atoms) suggests that dimerization is not a serendipitous crystallization artifact. Moreover, C-terminal to the PH domain Sec3 presents a region of predicted coiled-coil structure, which is also conserved in the human sequence (39). Finally, recruitment at the plasma membrane may increase the local concentration of Sec3 and facilitate dimerization, a possibility that has been proposed for other PH domains (34–36).
Lipid-binding Pocket
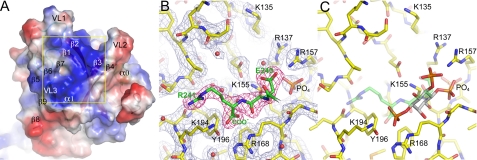
By analogy with other PH domains, the predicted phospholipid-binding site in Sec3 consists of a large, positively charged pocket, surrounded by VL1–3 (Fig. 3A). Two such pockets are symmetrically disposed at the distal ends of the Sec3N dimer (Fig. 2 and supplemental movie S1). Despite multiple attempts, Sec3N could not be co-crystallized with bound phosphoinositides. Furthermore, crystals of the unliganded protein soaked with various phosphoinositides dissolved immediately, suggesting that phosphoinositides bound Sec3N in the crystals, but their binding was incompatible with crystal packing contacts.
FIGURE 3.
Phosphoinositide-binding pocket. A, electrostatic surface representation of Sec3N near the predicted phosphoinositide-binding pocket surrounded by VL1–3. Note that the pocket is positively charged (blue color) due to the presence of various basic amino acids. B, electron density map (2Fo − Fc, contoured at 1.5σ) in the area corresponding to the yellow square in A. The C terminus of a symmetry-related molecule binds in the phosphoinositide-binding pocket (green backbone and red electron density map). A phosphate ion, the C terminus, and the carboxylic group of Glu-240 from the symmetry-related molecule are likely to mimic protein-phosphoinositide interactions. C, using these three negative charges as reference, inositol 1,4,5-trisphosphate can be modeled in the pocket. Although it is unlikely that phosphoinositides bind precisely in this manner, this model identifies some of the amino acids most likely to participate in protein-phosphoinositide interactions.
It has been previously shown that the locations of inorganic sulfate and phosphate ions in phosphoinositide-free structures of PH domains, such as those of Grp1 and Dapp1, coincide with the locations of the phosphate groups of phosphoinositides in the structures of the complexes (40). In this regard, it is very informative that one of the Sec3N dimers contains a phosphate ion bound in each of the phospholipid-binding pockets at the distal ends of the dimer (Fig. 2). The phosphates, which are distinguishable from solvent atoms at 2.0-Å resolution, present B-factors of ∼40 Å2, comparable to that of the neighboring residues, and were probably incorporated during purification, as phosphate was present in the purification buffer. What is more, the C terminus and the carboxylic group of Glu-240 from a neighboring molecule in the crystal also bind in the phospholipid-binding pocket near the phosphate ion (Fig. 3B). Although only one of the Sec3N dimers shows this arrangement, nearly identical interactions occur at the two distal ends of this dimer. Because the molecules of the dimer are crystallographically independent, this corresponds to two independent observations of the same set of interactions. The presence of these crystal contacts probably explains why the crystals could not withstand phosphoinositide soaking. More importantly, these contacts probably mimic protein-phosphoinositide interactions. We thus attempted to model inositol 1,4,5-trisphosphate in the phospholipid-binding pocket, so as to position the three phosphate groups of the phosphoinositide on top of the phosphate ion, the C terminus and the carboxylic group of Glu-240 from the neighboring molecule (Fig. 3C). Although the resulting model is unlikely to reflect the actual binding orientation or phosphoinositide specificity of the Sec3N pocket, it points to the residues most likely to be involved in protein-phosphoinositide interactions (Arg-137, Lys-155, Arg-157, Arg-168, Lys-194, and possibly Lys-135).
Structural studies have identified the determinants of PH domain-phosphoinositide interaction and specificity (28, 35, 40–43). Classic PH domains present a basic signature motif, KXm(R/K)XRXn(Y/N), with the first lysine residue located near the C terminus of strand β1, the (R/K)XR sequence near the N terminus of strand β2, and a tyrosine (or asparagine) residue in strand β3. The interactions of this motif are thought to mediate indiscriminate phosphoinositide binding but not specificity. Specificity appears to depend on additional interactions with non-conserved residues in the VLs, which for this reason are also known as the specificity determining regions.
Sec3 shares some of these features with classic PH domains but also displays important differences, defining a new subclass of PH domains. Thus, the position corresponding to the lysine residue in strand β1 is occupied by glutamic acid in Sec3, and in all the members of its PH domain subclass, except human Sec3, amisyn, and RTH1 (Fig. 1 and supplemental Fig. S3). This substitution of a deeply buried residue of the signature sequence is likely to affect the orientation of the bound phosphoinositide. Note that the overall orientation of bound phosphoinositides varies significantly among PH domains (40). A compensatory mutation occurs in Sec3, and all the members of its PH domain subclass, which present a lysine residue in strand β3 (Lys-155 in Sec3), corresponding to the location of the canonical tyrosine (or asparagine) residue of classic PH domains. The amino group of lysine 155 in Sec3 (Fig. 3) occupies a position very similar to that of the amino group of the conserved lysine residue in strand β1 of classic PH domains. Despite these important differences Sec3 shares with classic PH domains the (R/K)XR sequence in strand β2 (Lys-135, Lys-136, and Arg-137 in Sec3). This sequence corresponds to the basic patch (Lys-134 to Arg-137) previously identified by us as a critical determinant of Sec3 membrane binding and localization (18). In addition, other amino acids are likely to participate in lipid binding in Sec3 and could play a role in determining phosphoinositide specificity, including Arg-157, Arg-168, and Lys-194. These positions are highly conserved within the Sec3 PH domain subclass, but not among classic PH domains. It thus appears that the Sec3 subclass of PH domains has highly conserved basic amino acids, some of which are also present in classic PH domains, which are suitably positioned in the structure to allow for the specific binding of phosphoinositides. A more detailed knowledge of the specific lipid interactions will require the determination of phosphoinositide-bound structures of this subclass of PH domains. However, the information obtained here was invaluable in designing mutagenesis experiments to address the role of lipid binding in vivo.
Synthetic Growth Defects of sec3 Mutants with exo70-47
The exocyst subunits Exo70 and Sec3 display some functional redundancy, as mutants of either protein are individually viable, but lethal when combined (18, 44, 45). For this reason, mutant sec3ΔN (deletion of residues 1–320) lacks any detectable exocytosis defect but leads to severe exocytosis and growth defects, and even lethality, when combined with exo70 mutations. Therefore, to understand the functions of positively charged residues identified by the crystal structure to form part of the phosphoinositide-binding pocket, we studied sec3 mutants in an exo70 mutant background. Among the exo70 mutations previously studied by us, the point mutation Arg-595 → Ala in allele exo70-47 disrupts phosphoinositide binding of Exo70 and causes a synthetic growth defect with sec3ΔN (44). Therefore, we generated various combinations of mutants of Sec3 targeting residues Lys-134, Lys-135, Lys-136, Arg-137, Lys-155, Arg-157, and Arg-168 in an exo70-47 mutant background (Table 2).
TABLE 2.
Characterization of the sec3-exo70-47 double mutants
| Mutant alleles | Mutation sites | Viabilitya |
|---|---|---|
| SEC3 | N/A | + + + + |
| sec3-202 | Lys-134, Lys-135, Lys-136, Arg-137 | + |
| sec3-300 | Arg-137 | + + + |
| sec3-301 | Lys-135 | + + + |
| sec3-302 | Lys-134, Lys-136 | + + + + |
| sec3-303 | Lys-135, Arg-137 | + + |
| sec3-304 | Lys-155, Arg-157 | + + + |
| sec3-305 | Arg-168 | + + + |
| sec3-306 | Lys-135, Arg-137, Lys-155, Arg-157 | − |
| sec3-307 | Lys-135, Arg-137, Arg-168 | − |
| sec3-308 | Lys-155, Arg-157, Arg-168 | + + |
| sec3-309 | Arg-137, Lys-155 | + + + |
| sec3-310 | Arg-137, Lys-155, Arg-168 | + + + |
| sec3-311 | Arg-137, Arg-157 | + + + |
| sec3-312 | Arg-137, Arg-157, Arg-168 | + + + |
a Cell viability is indicated by symbols ranging from normal growth (+ + + +) to lethality (−).
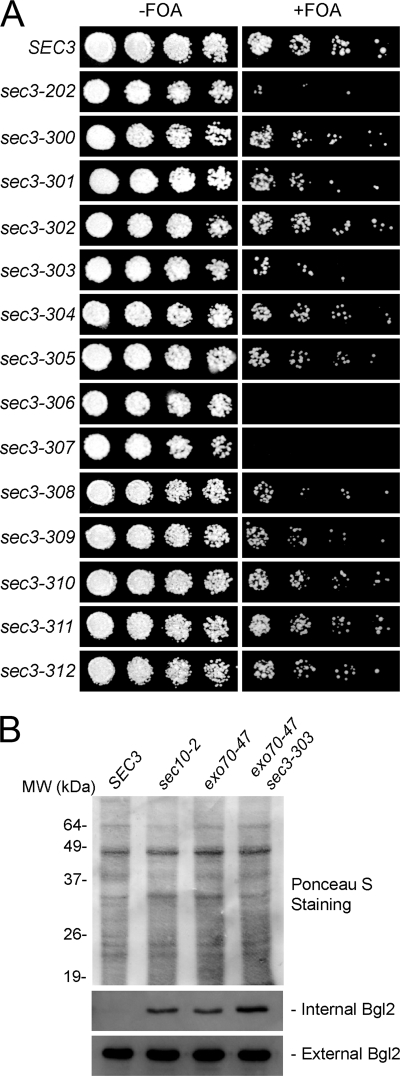
Compared with the single mutant exo70-47, the double mutant exo70-47 sec3-303, containing Sec3 mutations Lys-135 → Glu and Arg-137 → Glu, showed reduced growth on 5-FOA plates in which the wild-type exo70 balancer was removed (Fig. 4A). Moreover, single mutations of Sec3 residues Lys-135 or Arg-137 (alleles sec3-301 and sec3-300, respectively) also led to growth defects in the exo70-47 mutant background. In contrast, the double mutant exo70-47 sec3-302, containing Sec3 mutations Lys-134 → Glu and Arg-136 → Glu displayed normal growth. This is all consistent with the crystal structure in which the side chains of residues Lys-135 and Arg-137 are oriented toward the phosphoinositide-binding pocket (Fig. 3), whereas those of Lys-134 and Arg-136 are oriented in the opposite direction. Alleles sec3-306 (carrying mutations Lys-135 → Glu, Arg-137 → Glu, Lys-155 → Glu, and Arg-157 → Glu) and sec3-307 (mutations Lys-135 → Glu, Arg-137 → Glu, and Arg-168 → Glu) both resulted in lethality in the exo70-47 background, which is also consistent with the structure that predicts an important role for these residues in lipid binding (Fig. 3).
FIGURE 4.
The exo70-47 sec3-303 double mutant shows growth and secretion defects. A, various sec3 mutant alleles were introduced into the exo70-47 mutant strain, in which the chromosomal copy of EXO70 was deleted and replaced by the HIS3 gene. The exo70-47 mutant was expressed under the EXO70 promoter in a CEN TRP1 plasmid, and the cells were supplemented with an EXO70 balancer on a CEN URA3 plasmid. The cells were grown at 25 °C on 5-FOA-containing medium, which triggers the elimination of the EXO70 balancer plasmids and subsequent generation of the exo70-47 sec3 double mutants (right panel). Controls were grown on medium without 5-FOA (left panel). sec3-303 and several other mutants showed different degrees of synthetic growth defects with exo70-47. sec3-306 and sec3-307 were synthetically lethal with exo70-47. B, EXO70, exo70-47, and exo70-47 sec3-303 cells were grown to early log phase. The sec10-2 mutant cells were grown to early log phase and then shifted to 37 °C for 1 h. The internal and external pools of Bgl2 were analyzed by Western blot using an anti-Bgl2 antibody. The exo70-47 and sec10-2 mutants showed defects in Bgl2 secretion. The sec3-303 exo70-47 double mutant was more defective in Bgl2 secretion than the exo70-47 single mutant. Ponceau S staining is shown to indicate equal protein loading.
Taken together, the results suggest that residues Lys-135, Arg-137, Lys-155, Arg-157, and Arg-168, which are solvent-exposed and clustered together in a basic pocket in the crystal structure (Fig. 3), play an important role in Sec3 function in yeast cells, most likely through their involvement in phospholipid binding.
The Double Mutant exo70-47 sec3-303 Is Defective in Bgl2 Secretion
We next asked whether the double mutant exo70-47 sec3-303 displays exocytosis defects. This mutant was selected, because it showed growth defects, but was still viable (Fig. 4A), thus allowing us to perform secretion assays and fluorescence microscopy analyses. We examined the secretion of the cell wall modification enzyme endo-β1,3-glucanase (Bgl2), a widely used marker of polarized secretion at the daughter cell membrane. Wild-type cells did not accumulate Bgl2 (Fig. 4B), whereas cells expressing the single mutant allele exo70-47 showed moderate accumulation of Bgl2. In contrast, cells containing the double mutant exo70-47 sec3-303 displayed pronounced intracellular accumulation of Bgl2. As a positive control, we confirmed that mutant sec10-2, a temperature-sensitive allele of the exocyst subunit Sec10, accumulated Bgl2 in cells at the restrictive temperature of 37 °C (Fig. 4B).
Localization of Exocyst Components in the Double Mutant exo70-47 sec3-303
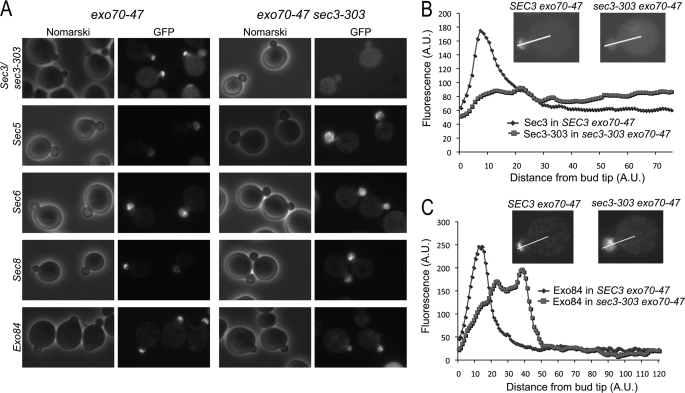
To examine the localization of exocyst components in mutant cells, we tagged subunits Sec5, Sec6, Sec8, Exo84, and wild-type and mutant Sec3 (sec3-303) with GFP by chromosomal integration in cells bearing the single exo70-47 mutation or double exo70-47 sec3-303 mutations. Cells were then analyzed using fluorescence microscopy. In the exo70-47 single mutant, GFP-tagged Sec3, Sec5, Sec8, or Exo84 localized normally to the daughter cell plasma membrane, as characterized by their crescent-shaped distribution at the bud tip (Fig. 5A). In the double mutant exo70-47 sec3-303, GFP-tagged Sec5, Sec8, and Exo84 remained polarized to the daughter cell, but were diffused throughout the bud, and not specifically associated with the bud tip membrane. On the other hand, the sec3-303-GFP mutant was diffused throughout the cell in the exo70-47 mutant background. Fig. 5 (B and C) shows representative plots of the fluorescence intensity along a line drawn through the middle of the bud and into the cell body in single and double mutants (expressing either Sec3-GFP, sec3-303-GFP, or Exo84-GFP). In exo70-47 cells, Sec3-GFP and Exo84-GFP showed localized staining at the bud tip, as indicated by a sharp peak of fluorescence coinciding with the location of the plasma membrane. In contrast, in sec3-303 exo70-47 cells, sec3-303-GFP was diffused throughout the cell, and there was no fluorescence peak associated with the plasma membrane (Fig. 5B). Exo84-GFP was polarized in sec3-303 exo70-47 cells, but the fluorescence distribution was diffused (Fig. 5C).
FIGURE 5.
Localization of exocyst components in exo70-47 and exo70-47 sec3-303 mutant cells. A, the exocyst components in the exo70-47 single mutant and the exo70-47 sec3-303 double mutant cells were GFP-tagged by chromosomal integration. GFP-tagged Sec3, Sec5, Sec8, and Exo84 were localized to the bud tip in the single mutants. In the double mutants, sec3-303-GFP was diffused throughout the cell, while the rest of the GFP-tagged exocyst proteins were localized to the entire bud instead of the bud tip. GFP-Sec6 was diffused in the entire bud in both single and double mutants. B and C, representative histograms depicting the fluorescence intensity along the bud and into the cell body (along the white line) in single (exo70-47) and double (exo70-47 sec3-303) mutants expressing Sec3-GFP or sec3-303-GFP (B) or Exo84-GFP (C) (see insets).
These results are consistent with the model of exocyst targeting proposed by Novick and colleagues (16), wherein most exocyst components associate with secretory vesicles traveling toward the bud tip along actin cables, whereas Sec3 and Exo70 bind to the bud tip membrane and act as landmarks for vesicle tethering. Thus, in the exo70-47 single mutant, Sec3 can still localize to the bud tip membrane, and the other exocyst components that travel to the bud are targeted to the membrane presumably though their interaction with Sec3. In the exo70-47 sec3-303 double mutant, the sec3-303 mutant protein is unable to bind directly to the bud tip membrane and is diffused throughout the cell. On the other hand, the other exocyst components are still able to enter the daughter cell by traveling with secretory vesicles along actin cables, but are no longer tethered to the bud tip membrane when Exo70 and Sec3 are both defective. Interestingly, GFP-Sec6 was distributed throughout the entire bud in both single and double Exo70 and Sec3 mutants, suggesting a unique targeting mechanism for this exocyst subunit. Taken together, our results demonstrate that mutations of residues Lys-135 and Arg-137 in the phosphoinositide-binding pocket of Sec3 lead to localization defects of the exocyst complex, which in turn results in impaired secretion and growth defects. According to the results described above, it could be expected that further disrupting the phosphoinositide-binding pocket of Sec3, for instance by introducing mutants sec3-306 and sec3-307, would lead to even more severe targeting and secretion defects, ultimately causing cell death (as shown in Fig. 4).
DISCUSSION
To understand the function and regulation of the exocyst during vesicle tethering it is important to know how the exocyst is targeted to the plasma membrane. Recent work has shown that Sec3 and Exo70 interact with phospholipids and small GTPases and play key roles in recruiting the exocyst to specific domains of the plasma membrane for exocytosis (16, 18–20, 44, 46). Mapping of the Rho3 GTPase and membrane-binding sites on Exo70 was aided by knowledge of its crystal structure (7). In the case of Sec3, the interactions with phospholipids and the small GTPases Rho1 and Cdc42 had been mapped to the N-terminal ∼320 amino acids (18, 19), but in the absence of structural information these results could not be rationalized. Here, we determined the structure of the N-terminal domain of Sec3 (Sec3N). The structure defines a new subclass of PH domains and enabled us to identify key residues involved in phospholipid binding. Genetic analysis and cell imaging further demonstrated that membrane binding through these residues is important for polarized localization of the exocyst at the plasma membrane and exocytosis in yeast.
The PH domain is best known for its ability to bind phospholipids. However, it is becoming increasingly clear that only a subset of PH domains share this property (47), whereas a growing number of PH domains have been implicated in protein-protein interactions (27, 48). The interactions of various PH domains with protein targets have been visualized crystallographically, including complexes with small GTPases (14, 49–51) (PDB codes: 1rrp, 1zc3, 2fju, and 2w2x), and heterotrimeric G proteins subunits (56, 57) (PDB codes: 1omw and 2rgn). These structures suggest that the interactions of the PH domain with protein targets are highly variable and difficult to predict, because they can involve virtually any interface of the PH fold. This is in sharp contrast with the interactions of the PH domain with phosphoinositides, which are constrained to the pocket formed by loops VL1–3.
Some PH domains share both properties, by binding phosphoinositides and specific protein targets, in particular small GTPases. In such cases, the PH domain is thought to function as a coincidence detector (48, 52). It is proposed that such PH domains may bind a particular type of lipid and a specific membrane-associated protein target, so as to recruit a PH-containing protein (or complex) selectively to membranes that contain both the lipid and the protein target. Such a mechanism, providing both added affinity and increased selectivity for certain membrane loci, has been implicated in specific Golgi targeting of certain PH domains (53, 54). As shown here and in previous work (18, 19), Sec3 binds lipid membranes and GTPases, and both activities reside within the Sec3N fragment studied here. We have now shown that a PH domain mediates both these interactions. Thus, Sec3, and probably other members of its structural subclass, join the growing number of PH domain-containing proteins that function as coincidence detectors at the membrane.
Yeast Sec3 is a large protein of 1336 amino acids. The region C-terminal to the PH domain is predicted to be predominantly helical in structure. Residues 320–470 within this region are predicted with high probability to form a coiled-coil structure. Fold recognition programs further suggest that the 727-amino acid region starting from residue 610 to the C terminus of the protein consists of a series of long helices separated by short loops. This is the signature pattern of helical bundles, characteristic of other subunits of the exocyst (4, 10, 39). Indeed, the crystal structures of four other subunits of the exocyst show a related fold consisting of consecutively stacked helical bundles (7–11). Each bundle typically contains three helices. The third helix of each bundle is usually longer and contributes its C-terminal half to the next bundle, helping to stabilize bundle-bundle interactions, a fold somewhat related to that of the spectrin repeat (55). The human Sec3 sequence is relatively shorter (894 amino acids) and displays low overall sequence identity (∼12%) with yeast Sec3 but appears to share a similar domain organization, including an N-terminal PH domain (residues 1–140), followed by a predicted coiled-coil domain (residues 155–255) and a series of C-terminal helical bundles (residues 258–894). As we have shown here, residues implicated in phospholipid binding in yeast Sec3N are also well conserved in the human sequence, as well as in other eukaryotes. This fact was previously underappreciated, as the N terminus of Sec3 shows relatively low sequence similarity across species. Our preliminary results indicate that the N terminus of human Sec3 also binds phospholipids8; further cell biological studies are being carried out to investigate the function of this domain in mammalian cells.
The structure of Sec3N also identifies a number of proteins that carries this new subclass of PH domains. One of these proteins is amisyn, implicated in binding to the t-SNARE protein syntaxin in neuronal cells and known to partially co-sediment with membranes (30). Future studies will examine whether the PH domain of amisyn binds phospholipids and whether this interaction is implicated in the recruitment of amisyn to the plasma membrane and regulation of SNARE function during vesicle docking and fusion.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM073791 (to R. D.) and R01-GM64690 (to W. G.). Use of the Industrial Macromolecular Crystallography Association-Collaborative Access Team (IMCA-CAT) beamline 17-BM was supported by the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. The Advanced Photon Source was supported by Department of Energy Contract W-31-109-Eng-38.
The atomic coordinates and structure factors (code 3HIE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Movie S1.
K. Baek, A. Knödler, S. H. Lee, X. Zhang, K. Orlando, J. Zhang, T. J. Foskett, W. Guo, and R. Dominguez, unpublished observation.
- SNARE
- soluble NSF attachment protein receptors
- PH
- pleckstrin homology
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- GFP
- green fluorescent protein
- Bgl2
- endo-β1,3-glucanase
- 5-FOA
- 5-fluoroorotic acid.
REFERENCES
- 1.Novick P., Field C., Schekman R. (1980) Cell 21, 205–215 [DOI] [PubMed] [Google Scholar]
- 2.TerBush D. R., Novick P. (1995) J. Cell Biol. 130, 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu S. C., Ting A. E., Hazuka C. D., Davanger S., Kenny J. W., Kee Y., Scheller R. H. (1996) Neuron 17, 1209–1219 [DOI] [PubMed] [Google Scholar]
- 4.Munson M., Novick P. (2006) Nat. Struct. Mol. Biol. 13, 577–581 [DOI] [PubMed] [Google Scholar]
- 5.He B., Guo W. (2009) Curr. Opin. Cell Biol. 21, 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu S. C., TerBush D., Abraham M., Guo W. (2004) Int. Rev. Cytol. 233, 243–265 [DOI] [PubMed] [Google Scholar]
- 7.Dong G., Hutagalung A. H., Fu C., Novick P., Reinisch K. M. (2005) Nat. Struct. Mol. Biol. 12, 1094–1100 [DOI] [PubMed] [Google Scholar]
- 8.Hamburger Z. A., Hamburger A. E., West A. P., Jr., Weis W. I. (2006) J. Mol. Biol. 356, 9–21 [DOI] [PubMed] [Google Scholar]
- 9.Moore B. A., Robinson H. H., Xu Z. (2007) J. Mol. Biol. 371, 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivaram M. V., Furgason M. L., Brewer D. N., Munson M. (2006) Nat. Struct. Mol. Biol. 13, 555–556 [DOI] [PubMed] [Google Scholar]
- 11.Wu S., Mehta S. Q., Pichaud F., Bellen H. J., Quiocho F. A. (2005) Nat. Struct. Mol. Biol. 12, 879–885 [DOI] [PubMed] [Google Scholar]
- 12.Tripathi A., Ren Y., Jeffrey P. D., Hughson F. M. (2009) Nat. Struct. Mol. Biol. 16, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukai S., Matern H. T., Jagath J. R., Scheller R. H., Brunger A. T. (2003) EMBO J. 22, 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin R., Junutula J. R., Matern H. T., Ervin K. E., Scheller R. H., Brunger A. T. (2005) EMBO J. 24, 2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finger F. P., Hughes T. E., Novick P. (1998) Cell 92, 559–571 [DOI] [PubMed] [Google Scholar]
- 16.Boyd C., Hughes T., Pypaert M., Novick P. (2004) J. Cell Biol. 167, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X., Zajac A., Zhang J., Wang P., Li M., Murray J., TerBush D., Guo W. (2005) J. Biol. Chem. 280, 20356–20364 [DOI] [PubMed] [Google Scholar]
- 18.Zhang X., Orlando K., He B., Xi F., Zhang J., Zajac A., Guo W. (2008) J. Cell Biol. 180, 145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo W., Tamanoi F., Novick P. (2001) Nat. Cell Biol. 3, 353–360 [DOI] [PubMed] [Google Scholar]
- 20.Zhang X., Bi E., Novick P., Du L., Kozminski K. G., Lipschutz J. H., Guo W. (2001) J. Biol. Chem. 276, 46745–46750 [DOI] [PubMed] [Google Scholar]
- 21.Adamo J. E., Rossi G., Brennwald P. (1999) Mol. Biol. Cell 10, 4121–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson N. G., Guo L., Imai J., Toh-E. A., Matsui Y., Tamanoi F. (1999) Mol. Cell. Biol. 19, 3580–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weeks C. M., Miller R. (1999) J. Appl. Crystallogr. 32, 120–124 [Google Scholar]
- 24.Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Adams P. D. (2008) Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 25.Cowtan K., Main P. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 487–493 [DOI] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27.Lemmon M. A. (2004) Biochem. Soc. Trans. 32, 707–711 [DOI] [PubMed] [Google Scholar]
- 28.Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B. (1995) Cell 83, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 29.Constable J. R., Graham M. E., Morgan A., Burgoyne R. D. (2005) J. Biol. Chem. 280, 31615–31623 [DOI] [PubMed] [Google Scholar]
- 30.Scales S. J., Hesser B. A., Masuda E. S., Scheller R. H. (2002) J. Biol. Chem. 277, 28271–28279 [DOI] [PubMed] [Google Scholar]
- 31.Janin J., Rodier F. (1995) Proteins 23, 580–587 [DOI] [PubMed] [Google Scholar]
- 32.Lo Conte L., Chothia C., Janin J. (1999) J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 33.Bahadur R. P., Chakrabarti P., Rodier F., Janin J. (2004) J. Mol. Biol. 336, 943–955 [DOI] [PubMed] [Google Scholar]
- 34.Depetris R. S., Wu J., Hubbard S. R. (2009) Nat. Struct. Mol. Biol. 16, 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baraldi E., Djinovic Carugo K., Hyvönen M., Surdo P. L., Riley A. M., Potter B. V., O'Brien R., Ladbury J. E., Saraste M. (1999) Structure 7, 449–460 [DOI] [PubMed] [Google Scholar]
- 36.Murayama K., Kato-Murayama M., Mishima C., Akasaka R., Shirouzu M., Fukui Y., Yokoyama S. (2008) Biochem. Biophys. Res. Commun. 377, 23–28 [DOI] [PubMed] [Google Scholar]
- 37.Jackson S. G., Zhang Y., Bao X., Zhang K., Summerfield R., Haslam R. J., Junop M. S. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 324–330 [DOI] [PubMed] [Google Scholar]
- 38.Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B. (1994) Cell 79, 199–209 [DOI] [PubMed] [Google Scholar]
- 39.Croteau N. J., Furgason M. L., Devos D., Munson M. (2009) PloS one 4, e4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cronin T. C., DiNitto J. P., Czech M. P., Lambright D. G. (2004) EMBO J. 23, 3711–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lietzke S. E., Bose S., Cronin T., Klarlund J., Chawla A., Czech M. P., Lambright D. G. (2000) Mol. Cell 6, 385–394 [DOI] [PubMed] [Google Scholar]
- 42.Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., Lemmon M. A. (2000) Mol. Cell 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 43.DiNitto J. P., Cronin T. C., Lambright D. G. (2003) Sci. STKE 2003, re16. [DOI] [PubMed] [Google Scholar]
- 44.He B., Xi F., Zhang X., Zhang J., Guo W. (2007) EMBO J. 26, 4053–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutagalung A. H., Coleman J., Pypaert M., Novick P. J. (2009) Mol. Biol. Cell 20, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., Zuo X., Yue P., Guo W. (2007) Mol. Biol. Cell 18, 4483–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., Lemmon M. A. (2004) Mol. Cell 13, 677–688 [DOI] [PubMed] [Google Scholar]
- 48.Lemmon M. A. (2007) Biochem. Soc. Symp. 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vetter I. R., Nowak C., Nishimoto T., Kuhlmann J., Wittinghofer A. (1999) Nature 398, 39–46 [DOI] [PubMed] [Google Scholar]
- 50.Jezyk M. R., Snyder J. T., Gershberg S., Worthylake D. K., Harden T. K., Sondek J. (2006) Nat. Struct. Mol. Biol 13, 1135–1140 [DOI] [PubMed] [Google Scholar]
- 51.Bunney T. D., Opaleye O., Roe S. M., Vatter P., Baxendale R. W., Walliser C., Everett K. L., Josephs M. B., Christow C., Rodrigues-Lima F., Gierschik P., Pearl L. H., Katan M. (2009) Mol. Cell 34, 223–233 [DOI] [PubMed] [Google Scholar]
- 52.Lemmon M. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 53.Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. (2004) Nat. Cell Biol. 6, 393–404 [DOI] [PubMed] [Google Scholar]
- 54.Levine T. P., Munro S. (2002) Curr. Biol. 12, 695–704 [DOI] [PubMed] [Google Scholar]
- 55.Djinovic-Carugo K., Gautel M., Ylänne J., Young P. (2002) FEBS Lett. 513, 119–123 [DOI] [PubMed] [Google Scholar]
- 56.Lodowski D. T., Pitcher J. A., Capel W. D., Lefkowitz R. J., Tesmer J. J. (2003) Science 300, 1256–1262 [DOI] [PubMed] [Google Scholar]
- 57.Lutz S., Shankaranarayanan A., Coco C., Ridilla M., Nance M. R., Vettel C., Baltus D., Evelyn C. R., Neubig R. R., Wieland T., Tesmer J. J. (2007) Science 318, 1923–1927 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.