Abstract
Sphingosine 1-phosphate (S1P), a potent sphingolipid mediator produced by sphingosine kinase isoenzymes (SphK1 and SphK2), regulates diverse cellular processes important for breast cancer progression acting in an autocrine and/or paracrine manner. Here we show that SphK1, but not SphK2, increased S1P export from MCF-7 cells. Whereas for both estradiol (E2) and epidermal growth factor-activated SphK1 and production of S1P, only E2 stimulated rapid release of S1P and dihydro-S1P from MCF-7 cells. E2-induced S1P and dihydro-S1P export required estrogen receptor-α, not GPR30, and was suppressed either by pharmacological inhibitors or gene silencing of ABCC1 (multidrug resistant protein 1) or ABCG2 (breast cancer resistance protein). Inhibiting these transporters also blocked E2-induced activation of ERK1/2, indicating that E2 activates ERK via downstream signaling of S1P. Taken together, our findings suggest that E2-induced export of S1P mediated by ABCC1 and ABCG2 transporters and consequent activation of S1P receptors may contribute to nongenomic signaling of E2 important for breast cancer pathophysiology.
Keywords: Receptors/Steroid/Thyroid, Signal Transduction/Sphingolipid, Transport/Multidrug, Lipid, Sphingolipid, ABC Transporters, Estradiol, Sphingosine Kinase, Sphingosine 1-Phosphate
Introduction
Sphingosine 1-phosphate (S1P)3 is a potent sphingolipid metabolite that regulates a number of biological processes critical for breast cancer such as cell growth and survival, movement and invasion, angiogenesis, and immunity (1–3). Most of the biological effects of S1P are mediated by five specific G protein-coupled receptors located on the cell surface (designated S1P1–5) (2, 4). S1P is produced in cells by two closely related sphingosine kinase (SphK) isoenzymes, SphK1 and SphK2, which have distinct localizations and functions (5). SphK1 has been implicated in breast cancer progression, whereas the role of SphK2 still remains unclear (1). Enforced expression of SphK1 in estrogen receptor (ER)-positive MCF-7 human breast cancer cells significantly increased cell growth in response to 17β-estradiol (E2) (6), enhanced resistance to anti-cancer drugs (7, 8), and produced larger xenograft tumors with higher microvessel density in an E2-dependent manner (9). SphK1 is overexpressed in human breast cancer and has been associated with tumor angiogenesis and resistance to radiation and chemotherapy (10, 11). Moreover, microarray analyses of 1,269 breast tumor samples revealed a worse outcome for patients with high SphK1 expression (12).
SphK1 activity is enhanced by growth factors important for breast cancer progression, such as epidermal growth factor (EGF) (7) and E2 (6). The physiological actions of E2 mainly result from regulation of transcription by estrogen receptors ER-α and ER-β (13). Because ERs are overexpressed in ∼70% of breast cancers, anti-estrogen hormonal therapies alone are as effective as any other chemotherapeutic drugs for breast cancer treatment (14). E2 also elicits a variety of rapid nongenomic events mediated by either ER located at the plasma membrane (15), or the seven-transmembrane receptor, GPR30 (16–18). Some of these rapid actions of E2 in breast cancer cells may result from stimulation of SphK1, and “inside-out” signaling of S1P leading to transactivation of EGF receptors (19). Yet, the mechanism by which S1P is released from breast cancers is unknown. How S1P generated inside cancer cells is exported to the outside is an important issue to resolve as it does not spontaneously traverse the lipid bilayer due to its polar head group. In this study, we show that SphK1, but not SphK2, is responsible for production of the S1P that is released from MCF-7 breast cancer cells. Moreover, E2, but not EGF, induces S1P release although both activate SphK1 and generate S1P inside the cell. Furthermore, ABCG2 and ABCC1 are involved in E2-mediated export of S1P raising the possibility that secretion of S1P by ABC transporters may be a contributing factor to the poor prognosis of patients overexpressing these drug-resistant proteins.
EXPERIMENTAL PROCEDURES
Reagents
S1P and MK571 were from Biomol Research Laboratory (Plymouth Meeting, PA). d-erythro-[3-3H]Sphingosine (23 Ci/mmol) and [γ-32P]ATP (3000 Ci/mmol) were from PerkinElmer Life Sciences. E2 and ICI 182,780 were from Sigma. Fumitremorgin C (FTC) was purchased from Alexis Biochemicals (San Diego, CA) and GPR30 from Calbiochem/EMD Biosciences (San Diego, CA). Anti-tubulin and anti-ERK2 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibodies against SphK1 and SphK2 were described previously (20, 21). Anti-lamin A/C, anti-ER-α, anti-ABCC1, anti-ABCG2, and anti-phospho-ERK1/2 (Thr202/Tyr204) antibodies were from Cell Signaling (Beverly, MA); anti-phospho-SphK1 (Ser225) was from Exalpha (Maynard, MA); anti-ABCC1 was from Alexis Biochemicals (San Diego, CA); and anti-ABCG2 was from Novus Biologicals (Littleton, CO). Secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA). The internal standard mixture for mass spectrometry containing C17-sphingosine, C17-sphinganine, C17-S1P, and C17-dihydro-S1P was obtained from Avanti Polar Lipids (Alabaster, AL).
Cell Culture and Transfection
MCF-7 and MDA-MB-231 human breast carcinoma cells were obtained from American Type Culture Collection (Manassas, VA). MCF-7 cells were grown in phenol red-free improved minimal essential medium (supplemented with 0.25% glucose and 10% fetal bovine serum as described (22)). For down-regulation with small interfering RNAs (siRNA), MCF-7 cells were transfected with ON-TARGET plus SMARTpool siRNAs targeted to SphK1, SphK2, ABCC1, or ABCG2 or control siRNA (Dharmacon, Lafayette, CO) according to the manufacturer's instructions.
Quantitative PCR
Total RNA was isolated with TRIzol reagent (Invitrogen). RNA was reverse transcribed with SuperScript II (Invitrogen). For real-time PCR, pre-mixed primer-probe sets were purchased from Applied Biosystems (Foster City, CA) and cDNA was amplified with the ABI Prism 7900HT as described (23).
Western Blotting
MCF-7 cells were lysed and proteins were detected by immunoblotting, as described previously (22).
SphK Assays
SphK1 and SphK2 activities were determined exactly as described previously (24). Activity is expressed as picomole of S1P formed/min/mg of protein.
Measurement of Cellular [3H]S1P and Its Release
S1P release was determined as previously described (25). Briefly cells were labeled for 10 min at 37 °C with [3H]sphingosine (1.5 μm, 0.4 μCi), washed with ice-cold medium, and incubated at 37 °C for the indicated times. Lipids were extracted from the medium by addition of chloroform, methanol, and 3 n NaOH (1:1:0.1, v/v/v), followed by vigorous mixing and phase separation. The basic aqueous phase contained S1P, devoid of sphingosine, whereas the majority of phospholipids were in the organic phase. Cells were washed with cold phosphate-buffered saline, scraped with methanol, sonicated on ice, chloroform added, and vortexed. 1 m NaCl and 3 n NaOH were added to make the final ratios of methanol, chloroform, 1 m NaCl, 3 m NaOH (1:1:1:0.1) (v/v/v/v). Radioactivity in 100-μl aliquots of aqueous and organic phases was determined by scintillation counting. Cellular [3H]S1P was expressed as picomole/million cells based on the specific activity of [3H]sphingosine. [3H]S1P in the medium was expressed as picomole/ml secreted by one million cells.
Measurement of Mass Levels of Released S1P and Dihydro-S1P by Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS)
MCF-7 cells (2.5 × 105/well) cultured for 24 h in 6-well plates were washed twice with 2 ml of phosphate-buffered saline, and treated as described in the figure legends. 1-ml aliquots of medium were centrifuged at 2000 × g for 10 min. 150 μl of phosphatase inhibitor mixture containing sodium orthovanadate (2 mm), sodium pyrophosphate (4 mm), and sodium fluoride (100 mm) were added to 850 μl of supernatant in 13 × 100-mm borosilicate tubes with Teflon-lined caps and mixed with CH3OH (5 ml) and CHCl3 (2.5 ml) and the internal standard mixture (500 pmol of each sphingolipid in 20 μl of ethanol). After sonication for 30 s, the single-phase mixture was incubated at 48 °C overnight. 1 m KOH in CH3OH (0.75 ml) was added followed by incubation in a shaking water bath for 2 h at 37 °C to hydrolyze glycerolipids. Extracts were neutralized with glacial acetic acid, 75% transferred to a new tube, and reduced to dryness with a SpeedVac. Dried residues were reconstituted in 0.3 ml of mobile phase for LC-ESI-MS/MS analysis and centrifuged at 16,000 × g for 30 s to remove particulates before transfer to autoinjector vials.
For LC-ESI-MS/MS analyses, a Shimadzu LC-20 AD binary pump system coupled to a SIL-20AC autoinjector and DGU20A3 degasser interfaced with an ABI 4000 QTrap mass spectrometer (Applied Biosystems) operating in a triple quadrupole mode was used. Q1 and Q3 were set to pass molecularly distinctive precursor and product ions (m/z 380.4 to 264.4 for S1P and m/z 382.4 to 266.4 for dihydro-S1P), using N2 to collisionally induce dissociations in Q2. Lipids were separated with a binary solvent system at a flow rate of 1.5 ml/min using a Supelco 2.1 × 50-mm Discovery C18 column (Sigma). Prior to injection of the sample, the column was equilibrated for 0.5 min with a solvent mixture of 80% solvent A (CH3OH/H2O/HCOOH, 58:41:1, v/v/v, containing 5 mm ammonium formate) and 20% solvent B (CH3OH/HCOOH, 99:1, v/v, containing 5 mm ammonium formate). After injection, the A/B ratio was maintained at 80:20 for 0.8 min, followed by a linear gradient to 100% B over 1.2 min, held at 100% B for 1.4 min, followed by a 0.3-min gradient return to 80:20 A/B. The column was re-equilibrated with 80:20 A/B for 0.5 min before each injection. S1P and dihydro-S1P in the medium was expressed as picomole/ml secreted by one million cells.
ERK1/2 Activation
MCF-7 cells were cultured in 6-well plates in phenol red-free improved minimal essential/Ham's F-12 medium (1:1) containing 10% fetal bovine serum. The following day cells were washed once in phosphate-buffered saline and then cultured in fresh phenol red-free, serum-free medium with glucose for 3 days as described (26) to minimize basal levels of ERK1/2 phosphorylation.
Statistical Analysis
Experiments were repeated at least three times with consistent results. Statistical differences between groups were determined with unpaired Student's t test. p < 0.05 was considered significant.
RESULTS
SphK1, but Not SphK2, Is Involved in Export of S1P by MCF-7 Cells
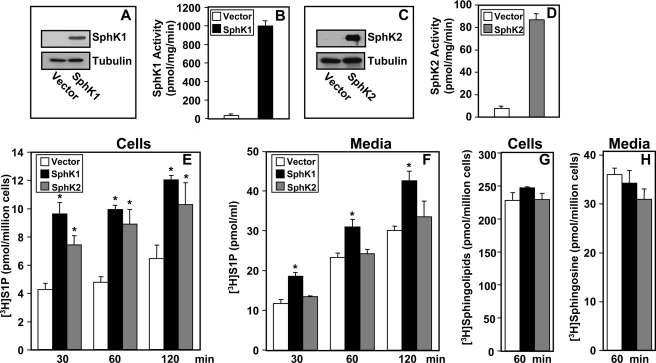
In ER positive MCF-7 breast cancer cells, E2 stimulates production of S1P that has been implicated in tumorigenesis by activation of its receptors present on the same or neighboring cells (6, 7, 9, 19, 20, 27). However, it is not clear how breast cancer cells secrete S1P or which of the two sphingosine kinase isozymes expressed are involved. To address these questions, we utilized a recently developed differential extraction assay to examine [3H]S1P export from MCF-7 cells overexpressing SphK1 or SphK2 (25). In agreement with previous studies (7, 20), transient transfection of SphK1 or SphK2 caused large increases in isoenzyme-specific enzymatic activity (Fig. 1, B and D), accompanied by protein expression with the expected molecular masses (Fig. 1, A and C). Expression of both SphK1 and SphK2 increased intracellular [3H]S1P formation in MCF-7 cells pre-labeled with [3H]sphingosine (Fig. 1E). However, there was only a significant increase in export of [3H]S1P from MCF-7 cells expressing SphK1, but not from those expressing SphK2 (Fig. 1F). SphK1 or SphK2 expression did not affect 3H-labeled sphingolipids in the cells (Fig. 1G) or uptake of sphingosine from the medium (Fig. 1H).
FIGURE 1.
Expression of SphK1 but not SphK2 increases S1P secretion. MCF-7 cells were transfected with vector, V5-SphK1, or V5-SphK2. A and C, equal amounts of cell lysate proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-V5 antibody. Blots were then stripped and re-probed with anti-tubulin antibody to demonstrate equal loading. B and D, SphK1 and SphK2 activities in lysates were determined with isoenzyme-specific assays. E and F, vector (open bars), V5-SphK1 (black bars), or V5-SphK2 (gray bars)-transfected MCF-7 cells were incubated for 10 min with [3H]sphingosine (1.5 μm, 0.45 μCi) in serum-free medium, washed extensively, fresh medium was added, and labeled lipids were extracted differentially at the indicated times from medium (F and H) and cells (E and G) into aqueous phases containing [3H]S1P (E and F) and organic phases (G and H) and quantified by scintillation counting as described under “Experimental Procedures.” Data are the mean ± S.D. of duplicate determinations. Cellular [3H]S1P is expressed as picomole/million cells and [3H]S1P in the medium is expressed as picomole/ml secreted by one million cells. *, p < 0.05 compared with vector. Similar results were obtained in two additional experiments.
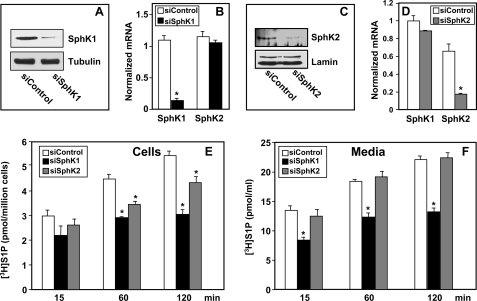
To investigate the involvement of endogenous SphK1 and SphK2, the expression of each isoform in MCF-7 cells was down-regulated. Endogenous SphK1 and SphK2 mRNA levels were reduced by 90 and 75%, respectively, by transfection with siRNA targeted to SphK1 or SphK2, compared with scrambled siRNA (Fig. 2, B and D). The specificity of these siRNAs was demonstrated by their inability to inhibit expression of the other isoform of SphK (Fig. 2, B and D). Reduced mRNA levels of SphK1 and SphK2 were accompanied by markedly reduced protein expression as determined by Western blotting with isotype-specific antibodies (Fig. 2, A and C). Although reduction of expression of either SphK1 or SphK2 significantly decreased intracellular [3H]S1P formation (Fig. 2E), only down-regulation of SphK1, not SphK2, significantly decreased export of S1P compared with control siRNA transfectants (Fig. 2, E and F).
FIGURE 2.
Down-regulation of SphK1 but not SphK2 decreases S1P secretion. MCF-7 cells were transfected with control siRNA or siRNA targeted to SphK1 or SphK2, as indicated. Equal amounts of lysate or nuclear proteins were immunoblotted with anti-SphK1 (A) or anti-SphK2 (C). Blots were stripped and reprobed with anti-tubulin (A) or lamin (C) to confirm equal loading. B and D, RNA was isolated from duplicate cultures and SphK1 and SphK2 mRNA determined by quantitative real time PCR and normalized to levels of glyceraldehyde-3-phosphate dehydrogenase mRNA. E and F, siControl (open bars), siSphK1 (black bars), or siSphK2 (gray bars) transfected MCF-7 cells were incubated for 10 min with [3H]sphingosine (1.5 μm, 0.45 μCi) in serum-free medium, washed extensively, fresh medium was added, and [3H]S1P in cells (E) or secreted into the medium (F) was measured at the indicated times. Data are the mean ± S.D. of duplicate determinations. Similar results were obtained in two additional experiments. *, p < 0.05 compared with siControl.
E2, but Not EGF, Induces Export of S1P
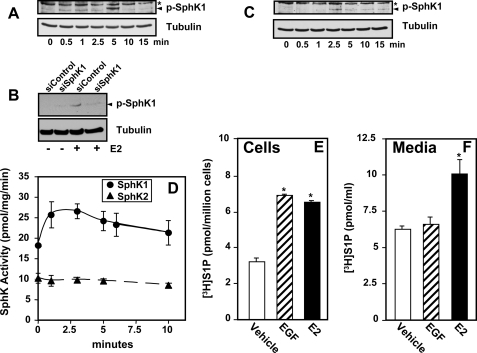
E2 and EGF have been shown to stimulate SphK1 activity in MCF-7 cells leading to increased intracellular S1P (7, 19, 20, 27). In agreement, both E2 and EGF rapidly activated SphK1 as determined by enhanced phosphorylation on Ser225 (Fig. 3, A–C), an event important for its activation (28). As expected, down-regulation of SphK1 eliminated the phosphorylated SphK1 immunoreactive band (Fig. 3B). Consistent with a previous study (19), E2 stimulated the enzymatic activity of SphK1 determined with an isoenzyme-specific assay and had almost no effect on SphK2 activity (Fig. 3D). E2 and EGF increased formation of intracellular [3H]S1P in MCF-7 cells to similar extents within 5 min (Fig. 3E). Interestingly, however, only E2 significantly increased export of S1P (Fig. 3F).
FIGURE 3.
Effect of E2 and EGF on SphK1 and S1P formation and secretion. A, MCF-7 cells were stimulated with E2 for the indicated times. B, MCF-7 cells were transfected with control siRNA or siRNA targeted to SphK1 and stimulated without or with E2. C, MCF-7 cells were stimulated with EGF for the indicated times. A–C, cell lysates were prepared, and equal amounts of protein were separated by SDS-PAGE and analyzed by immunoblotting with anti-phospho-SphK1 antibody. Blots were stripped and re-probed with anti-tubulin to ensure equal loading and transfer. The asterisks indicate nonspecific immunostained bands and the arrowheads indicate SphK1. D, MCF-7 cells were stimulated without or with E2 for the indicated times. Cells were lysed and SphK1 activity (circles) was measured with sphingosine added as Triton X-100 mixed micelles and SphK2 activity (triangles) with sphingosine added as a bovine serum albumin complex in the presence of 1 m KCl. *, p < 0.05 for SphK1 activity at all time points compared with unstimulated (t = 0). E and F, MCF-7 cells were serum starved overnight, prelabled with [3H]sphingosine (1.5 μm, 0.45 μCi), and washed extensively, fresh medium was added and then stimulated without (vehicle) or with EGF (50 ng/ml) or E2 (10 nm) for 5 min, and the [3H]S1P in cells (E) or secreted into the medium (F) determined. Data are expressed as mean ± S.D. Similar results were obtained in two independent experiments. *, p < 0.05.
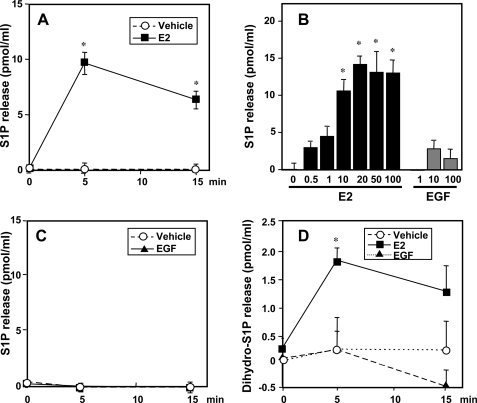
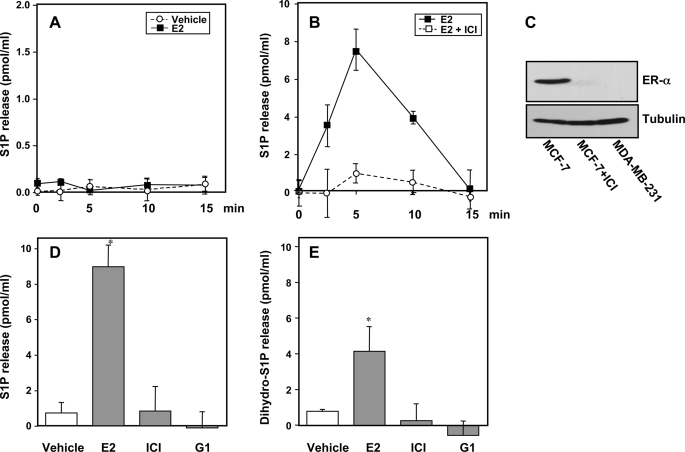
It was important to confirm these results by determination of mass levels of S1P produced from endogenous pools of sphingosine and released into the medium. Analysis of S1P released from cells presents a challenge due to the low amounts relative to the large volume of medium. For this purpose, we developed a sensitive and quantitative mass spectrometric method capable of measuring femtomole amounts of S1P. E2 induced a significant increase in S1P released from MCF cells, peaking within 5 min (Fig. 4A), and was dose-dependent, reaching a maximum at a concentration of 10–20 nm (Fig. 4B). This increase in S1P as well as the levels of S1P present in the medium at the beginning of the experiment were not due to the presence of fragments of cell membranes as centrifugation of the medium at 100,000 × g for 1 h did not alter these values.
FIGURE 4.
E2 but not EGF stimulates release of S1P and dihydro-S1P from MCF-7 cells. MCF-7 cells were stimulated with vehicle (open circles) or 20 nm E2 (filled squares) for the indicated times (A and C), for 5 min with the indicated concentrations of E2 or EGF (B), or with 10 ng/ml of EGF (filled triangles) for the indicated times (C and D). S1P (A–C) and dihydro-S1P (D) released into the medium were measured by LC-ESI-MS/MS. Data are expressed as picomole of phosphorylated sphingoid base/ml and are mean ± S.D. Levels of S1P and dihydro-S1P in the cells at zero time (7.4 ± 1 and 2.8 ± 0.2 pmol/ml, respectively) were subtracted from all values. *, p < 0.01 compared with vehicle. Similar results were obtained in three independent experiments.
In agreement with the [3H]S1P secretion results (Fig. 3F), stimulation with EGF at any concentration examined did not significantly increase export of S1P, throughout a time course of 15 min (Fig. 4C). Interestingly, LC-ESI-MS/MS analysis revealed that export of dihydro-S1P was also enhanced by treatment with E2 but not by EGF (Fig. 4D).
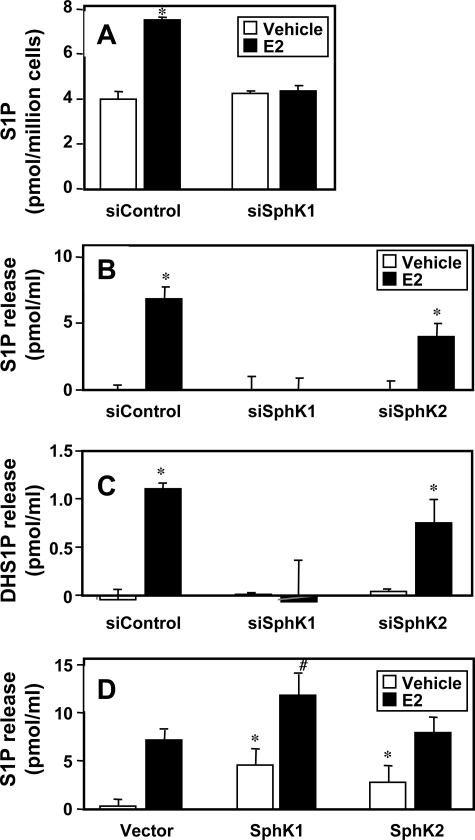
Previous studies showed that transfection with catalytically inactive SphK1 suppressed secretion of S1P after E2 stimulation (19), suggesting that SphK1 activation is necessary for increased S1P production and release. In agreement, E2 increased intracellular levels of S1P that was dependent on the expression of SphK1 (Fig. 5A). Depletion of SphK1 also abolished the elevated release of S1P (Fig. 5B) and dihydro-S1P (Fig. 5C) in response to E2. In contrast, down-regulation of SphK2 did not eliminate the E2-stimulated release of S1P (Fig. 5B) and dihydro-S1P (Fig. 5C). Moreover, although ectopic expression of SphK1 or SphK2 increased release of S1P into the medium, only expression of SphK1 but not SphK2 further enhanced the E2-stimulated release of S1P (Fig. 5D). These results suggest that SphK1 is mainly responsible for formation of S1P and dihydro-S1P secreted in response to E2.
FIGURE 5.
SphK1 is important for E2-mediated release of S1P and dihydro-S1P from MCF-7 cells. A, MCF-7 cells were transfected with siControl or siSphK1 and treated with vehicle (open bars) or 10 nm E2 (black bars) for 5 min. Lipids were extracted from 106 cells and S1P was determined by LC-ESI-MS/MS. *, p < 0.01 compared with vehicle. B, MCF-7 cells were transfected with siControl, siSphK1, or siSphK2 and treated with vehicle (open bars) or 10 nm E2 (black bars) for 5 min. S1P (B) and dihydro-S1P (DHS1P) (C) released into the medium was determined by LC-ESI-MS/MS. Levels at zero time were subtracted and data expressed as mean ± S.D. *, p < 0.01 compared with vehicle. Similar results were obtained in two additional experiments. D, MCF-7 cells were transfected with vector, SphK1, or SphK2 and treated with vehicle (open bars) or with 10 nm E2 (black bars) for 5 min. S1P released into the medium was determined by LC-ESI-MS/MS. Levels at zero time were subtracted and data expressed as mean ± S.D. *, p < 0.05 compared with unstimulated vector; #, p < 0.05 compared with E2-stimulated vector. Similar results were obtained in two additional experiments.
ER-α, but Not GPR30, Is Required for E2-mediated Rapid Release of S1P
Nongenomic responses to E2 are mediated by either ER located at the plasma membrane (15, 29) or by a G protein-coupled receptor, GPR30 (17, 26). To clarify whether ER-α and/or GPR30 are responsible for E2-mediated S1P export, we first utilized MDA-MB-231 breast cancer cells that lack both ER-α and GPR30 (18, 30). In agreement with previous studies (6), E2 did not activate SphK1 in MDA-MB-231 cells as determined by Western blotting with anti-phospho-Ser225 (data not shown). In contrast to MCF-7 cells, which express both ER-α and GPR30, E2 did not stimulate export of S1P from MDA-MB-231 cells (Fig. 6A). Moreover, when ER-α was down-regulated in MCF-7 cells by prolonged treatment with the pure estrogen antagonist ICI 182,780 (Fig. 6C), the ability of E2 to stimulate S1P export was completely abolished (Fig. 6B). Unlike E2, neither ICI 182,780, which is also a GPR30 agonist (17), nor G1, a selective GPR30 agonist (31), significantly increased S1P (Fig. 6D) or dihydro-S1P (Fig. 6E) export from MCF-7 cells determined by LC-ESI-MS/MS. Hence, only ER-α is necessary for E2-induced S1P export and not GPR30.
FIGURE 6.
ER-α is necessary for E2-mediated release of S1P and dihydro-S1P. A, MDA-MB-231 cells were stimulated with vehicle (open circles) or 10 nm E2 (filled squares) for the indicated times and S1P released into the medium was measured by LC-ESI-MS/MS. Levels of S1P in the medium of MDA-MB-231 cells at zero time (0.6 ± 0.1 pmol/ml) were subtracted from all values. B, MCF-7 cells were pretreated for 18 h without or with 10 μm ICI 182,780 and then stimulated with 10 nm E2 and S1P release was determined at the indicated times by mass spectrometry. Levels of S1P in the medium of MCF-7 cells at zero time (8.8 ± 0.7 pmol/ml) was subtracted and data expressed as mean ± S.D. C, equal amounts of proteins from lysates of duplicate cultures of MDA-MB-231 and MCF-7 cells treated as in A and B, respectively, were separated by SDS-PAGE and analyzed by immunoblotting with anti-ER-α antibody. Blots were stripped and reprobed with anti-tubulin to confirm equal loading. D and E, MCF-7 cells were treated with vehicle, 10 nm E2, 10 μm ICI 182,780, or 10 nm G1 for 5 min. S1P (D) and dihydro-S1P (E) released into the medium were determined by LC-ESI-MS/MS. Levels at zero time were subtracted and data are expressed as mean ± S.D. *, p < 0.01 compared with vehicle. Similar results were obtained in two additional experiments.
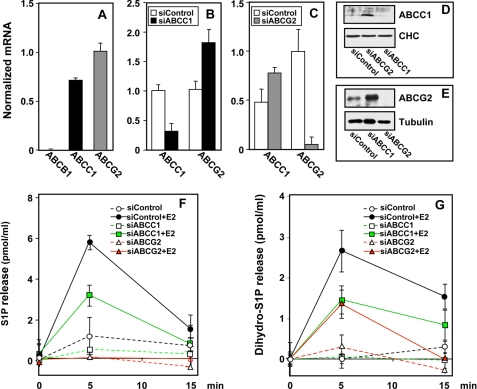
ABCC1 and ABCG2 Mediate E2-induced Export of S1P
Recent studies suggest that members of the ABC transporter family might be involved in release of S1P from several types of cells (32–35). Three ABC transporters, ABCB1 (also known as multidrug resistant gene 1 (MDR1)) or p-glycoprotein, ABCC1 (also known as multidrug resistance protein 1 (MRP1)), and ABCG2 (also known as breast cancer resistance protein), appear to account for 80–90% of multidrug-resistant human tumor cells (36). In agreement with previous reports (36, 37), MCF-7 cells have high expression levels of ABCG2, less ABCC1, and no ABCB1 (Fig. 7A). To examine the involvement of these transporters in S1P and dihydro-S1P export from MCF-7 cells, their expression was down-regulated by gene silencing. Transfection with siRNA targeted to ABCC1 or ABCG2 reduced endogenous ABCC1 and ABCG2 mRNA levels by more than 60 and 90%, respectively, compared with scrambled control siRNA (Fig. 7, B and C) and also reduced the corresponding protein expression (Fig. 7, D and E). Interestingly, down-regulation of ABCC1 increased ABCG2 mRNA with a concomitant increase in ABCG2 protein (Fig. 7, B and E), and conversely, decreasing ABCG2 mRNA resulted in increased ABCC1 mRNA and protein (Fig. 7, C and D). Although the magnitude of the response to E2 by cells transfected with control siRNA was similar to that of the naïve cells (Fig. 4A), down-regulation of ABCC1 significantly decreased E2-induced S1P release, and down-regulation of ABCG2 completely abrogated it (Fig. 7F). The greater susceptibility to depletion of ABCG2 might be due to its higher expression and greater down-regulation compared with the decrease in expression of ABCC1 (Fig. 7). Down-regulation of either ABCC1 or ABCG2 also decreased E2-induced dihydro-S1P release (Fig. 7G).
FIGURE 7.
Down-regulation of ABCC1 or ABCG2 decreases E2-mediated release of S1P or dihydro-S1P. A, RNA was isolated from MCF-7 cells and mRNA of ABCB1, ABCC1, and ABCG2 determined by quantitative real time reverse transcription-PCR and normalized to cyclophilin mRNA. B–E, MCF-7 cells were transfected with control siRNA (open bars) or siRNA targeted to ABCC1 (black bars) or ABCG2 (gray bars), as indicated. Expression of ABCC1 and ABCG2 was determined by quantitative real-time reverse transcription-PCR and normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA. D and E, equal amounts of cell lysate proteins from duplicate cultures were immunoblotted with anti-ABCC1 (D) or anti-ABCG2 (E). Blots were stripped and reprobed with anti-clathrin heavy chain (CHC) or anti-tubulin, as indicated, to confirm equal loading. F and G, MCF-7 cells were transfected with siControl, siABCC1, or siABCG2 and stimulated with vehicle or E2 (10 nm) for the indicated times. S1P (F) and dihydro-S1P (G) released into the medium was determined by LC-ESI-MS/MS. Levels of S1P and dihydro-S1P at zero time (7 ± 0.7 and 1.5 ± 0.3 pmol/ml, respectively) were subtracted and data expressed as mean ± S.D. All values at 5 min were statistically significant (p < 0.01) compared with siControl treated with E2 at 5 min. Similar results were obtained in two additional experiments.
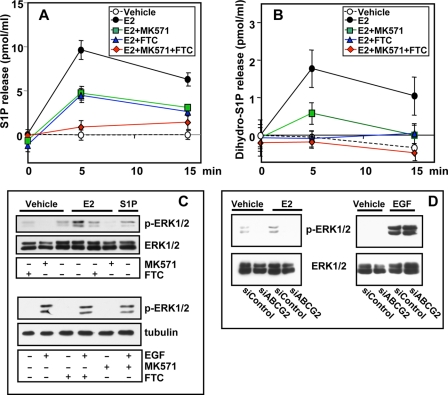
To further substantiate the involvement of ABCC1 and ABCG2 in S1P and dihydro-S1P export from MCF-7 cells, the effects of MK571 or FTC, pharmacological inhibitors of ABCC1 and ABCG2, respectively (32, 36, 38), were also examined. E2-stimulated release of S1P and dihydro-S1P from MCF-7 cells was significantly reduced by preincubation with MK571 or FTC (Fig. 8, A and B). Moreover, a combination of both inhibitors totally abrogated E2-induced release of S1P (Fig. 8, A and B).
FIGURE 8.
Pharmacological inhibitors of ABCC1 or ABCG2 decrease E2-mediated release of S1P and dihydro-S1P and ERK1/2 activation. MCF-7 cells were preincubated for 2 h without or with 20 μm MK571 or 20 μm FTC, or both, followed by stimulation with vehicle or E2 (10 nm) for the indicated times (A and B), for 10 min with 10 nm E2, or with 10 ng/ml EGF (C). S1P (A) and dihydro-S1P (B) released was determined by LC-ESI-MS/MS. Levels of S1P and dihydro-S1P in the cells at zero time (7 ± 0.6 and 2.9 ± 0.2 pmol/ml, respectively) were subtracted and data expressed as mean ± S.D. All values at 5 and 15 min were statistically significant (p < 0.05) compared with control cells treated with E2. Similar results were obtained in two additional experiments. C, equal amounts of proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-phospho-ERK1/2 antibody. D, MCF-7 cells were transfected with siControl or siABCG2 and stimulated with vehicle, E2 (20 nm), or EGF (10 ng/ml) for 10 min, and ERK1/2 activation was determined by immunoblotting with anti-phospho-ERK1/2 antibody. Blots were stripped and reprobed with anti-total ERK antibodies to confirm equal loading and transfer.
E2-induced S1P Secreted via ABCC1 or ABCG2 Activates ERK1/2
One of the most well known nongenomic effects of E2 is ERK1/2 activation (reviewed in Refs. 15, 18, and 26). Previous studies have suggested that activation of SphK1 and production of S1P might be involved in E2-induced ERK1/2 activation (6, 19). If so, prevention of S1P release should negate this response to E2. In agreement with previous studies (6, 18, 19, 26, 39), treatment of MCF-7 cells with E2 rapidly activated ERK1/2, as demonstrated with phosphospecific antibodies, with maximal response obtained at 10 min, which returned to basal levels within 30 min (data not shown). Inhibition of either ABCC1 or ABCG2 using MK571 or FTC, respectively, drastically reduced activation of ERK1/2 induced by E2 (Fig. 8C), consistent with the ability of these inhibitors to reduce export of S1P (Fig. 8A) and dihydro-S1P (Fig. 8B). As expected, ERK1/2 activation by EGF, which does not enhance S1P secretion from MCF7 cells, was not inhibited by inhibition of ABCC1 or ABCG2 (Fig. 8C). Moreover, down-regulation of ABCG2 markedly reduced E2- but not EGF-mediated ERK1/2 activation (Fig. 8D).
DISCUSSION
Numerous studies have suggested that S1P is important in breast cancer, acting in an autocrine and/or paracrine manner (1, 11, 40). In this study, we have shown that SphK1, not SphK2, is necessary for release of S1P from MCF-7 ER-positive breast cancer cells. This is probably due to the distinct subcellular localization of the two kinase isoforms. SphK1 is primarily cytosolic and rapidly translocated to the plasma membrane upon stimulation, where produced S1P may be readily available to be transported outside the cells (41, 42). In contrast, SphK2 has been shown to be localized to the nucleus of MCF-7 cells, as determined by confocal microscopy and Western blotting (22, 43, 44). Surprisingly, although both E2 and EGF stimulated SphK1 in MCF-7 cells, only E2 rapidly stimulated export of S1P and dihydro-S1P. This suggests that activation of SphK1 and production of S1P at the plasma membrane is not sufficient for its release and indicates that active transport might also be involved. Although our initial efforts to examine secretion of S1P utilized labeled sphingosine, this method has the caveat that it requires treatment of cells with exogenous sphingosine and does not provide any information on secretion of endogenously produced S1P. However, consistent trends were obtained when secretion of mass levels of S1P was determined by a sensitive LC-ESI-MS/MS method that we developed. Moreover, we were able to demonstrate for the first time that MCF-7 cells also export dihydro-S1P, which is also a ligand for all of the S1P receptors. E2-stimulated release of both S1P and dihydro-S1P was dependent on SphK1.
Studies from several laboratories, including ours, have suggested the involvement of ABC transporters in S1P export from mast cells (32), platelets (33), endothelial cells (34), astrocytes (35), thyroid follicular cell (45), fibroblasts (46), and erythrocytes (47). Similarly, using pharmacological and molecular approaches, we demonstrated that ABCC1 and ABCG2 are involved in E2-mediated transport of S1P and dihydro-S1P out of MCF-7 cells. Yet in MCF7 cells, ABCG2 predominates, which might be due to its greater expression in these cells. It is still not completely understood whether these two transporters operate serially rather than in parallel and whether both are necessary or whether one is sufficient.
Overexpression of ABCC1 and ABCG2 appear to account for a large fraction of multidrug-resistant breast tumors (48, 49). It is tempting to speculate that enhanced expression of these ABC transporters is involved in multidrug resistance not only due to increased drug efflux from the cell, but also due to export of S1P and dihydro-S1P, because by binding to S1P receptors they can play significant roles in regulating angiogenesis, metastasis, and breast cancer progression. Hence, export of S1P by ABC transporters may be a contributing factor to the poor prognosis of patients who overexpress these multidrug-resistant proteins. Interestingly, MCF7 cells constitutively export relatively high levels of S1P into the culture medium compared with MDA-MB-231 cells that are not dependent on ABCC1 and ABCG2 transporter activity or expression and are not affected by decreased temperature nor did they significantly increase with time.
ABCC1 and ABCG2 transporters have been shown to play a major role in resistance of breast cancer to some important chemotherapy agents, such as doxorubicin (49). ABCG2 is often expressed in cancer stem cell populations and could help them survive cytotoxic or targeted therapies leading to tumor regrowth or relapse (38, 49). A fascinating connection is that S1P, which is secreted by ABCG2, has also been shown to be important for stem cell growth, survival, maintenance of pluripotentiality (50), and homing to sites of action (51). Of note, it was recently shown that expression of SphK1 is linked with resistance of MCF-7 cells to tamoxifen (52), suggesting that targeting the SphK1/S1P axis might be a useful approach to overcome anti-estrogen resistance in human breast cancer.
E2 stimulates mitogenesis of MCF-7 cells in the absence of any other growth factors. In the classical or genomic mechanism, E2 molecules diffuse into the cell and bind to the ER, which exists as two different forms, ER-α and ER-β. The binding of E2 to either of its receptors induces a conformational change, resulting in interactions with the transcriptional co-regulators and regulatory DNA sequences of target genes. The resulting changes in levels of the proteins they encode underlie many overall physiological responses following E2 exposure. However, there are rapid biochemical and physiological responses that occur within minutes after E2 administration that cannot be accounted for by changes in gene expression mediated by nuclear ERs. There is still controversy regarding the rapid nongenomic responses to E2 and the receptors that mediate these events. One school of thought is that plasma membrane-localized ER-α initiates E2-dependent rapid signaling cascades (29). On the other hand, it has been suggested that GPR30 is the membrane receptor that mediates some of the nongenomic signaling of E2 (16, 18, 26). Because these E2 receptors have different pharmacologies, it was possible to determine which was involved in E2-stimulated export of S1P and dihydro-S1P. Interestingly, neither ICI 182,780, a known antagonist of ER-α and an agonist for GPR30 (17), nor G1, a specific GPR30 agonist (31), had significant effects on S1P or dihydro-S1P export, in contrast to E2. In accordance, down-regulation of ER-α abolished E2-mediated release of S1P and dihydro-S1P. Importantly, suppression of export of S1P and dihydro-S1P by inhibitors of ABCC1 and ABCG2 markedly decreased activation of ERK1/2 in response to E2. Our results support the notion that activation of SphK1 by E2, export of S1P via ABC transporters, and consequent activation of S1P receptors (Fig. 9) may contribute to nongenomic signaling of E2 important for breast cancer pathophysiology. Furthermore, the S1P receptors may be the G protein-coupled receptors that mediate rapid nongenomic responses to E2 rather than GPR30, which has been shown to be present in endoplasmic reticulum, not the plasma membrane (17, 53). Our work, taken together with the criss-cross model for E2 signaling proposed by Sukocheva et al. (19), suggests that E2 stimulates SphK1, increasing production of S1P in the vicinity of the ABC transporters, ABCC1 and ABCG2, at the plasma membrane, which facilitate its export (Fig. 8). This S1P in turn binds to and activates S1P receptors, to stimulate ERK1/2 and increase EGFR transactivation in a matrix metalloprotease-dependent manner. Finally, the demonstration that ER-α expression is necessary for S1P and dihydro-S1P export from breast cancer cells may, at least in part, explain why ER-positive breast cancer shows reduced responses to modern combined chemotherapy compared with ER-negative tumors, as found in a randomized clinical trial involving more than 6500 advanced breast cancer patients (54).
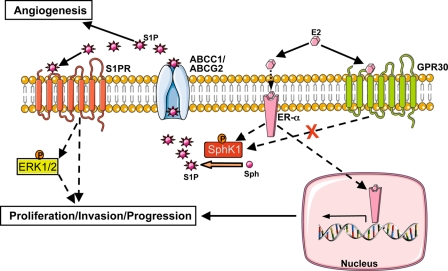
FIGURE 9.
Scheme highlighting the importance of ABC transporters and S1P/dihydro-S1P export from the cell in nongenomic effects of E2. See text for more details. For simplicity, many other known signaling pathways downstream of ER are not shown. Binding of E2 to ER-α and not GPR30 stimulates release of S1P (and dihydro-S1P, not shown here) via ABC transporters, ABCC1 and ABCG2. This S1P in turn binds to and activates S1P receptors to stimulate ERK1/2 leading to downstream signaling events important for breast cancer proliferation, progression, and invasion.
This work was supported, in whole or in part, by National Institutes of Health Grants R37GM043880 and R01CA61774 (to S. S.), Virginia Commonwealth University Grant BIRCWH K12HD055881, and Susan G. Komen for the Cure Career Catalyst Research Grant KG090510 (to K. T.).
- S1P
- sphingosine 1-phosphate
- EGF
- epidermal growth factor
- ER
- estrogen receptor
- ERK
- extracellular signal-regulated kinase
- E2
- 17β-estradiol
- FTC
- fumitremorgin C
- LC-ESI-MS/MS
- liquid chromatography electrospray ionization tandem mass spectrometry
- siRNA
- small interfering RNA
- SphK
- sphingosine kinase.
REFERENCES
- 1.Takabe K., Paugh S. W., Milstien S., Spiegel S. (2008) Pharmacol. Rev. 60, 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spiegel S., Milstien S. (2003) Nat. Rev. Mol. Cell Biol. 4, 397–407 [DOI] [PubMed] [Google Scholar]
- 3.Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 4.Saba J. D., Hla T. (2004) Circ. Res. 94, 724–734 [DOI] [PubMed] [Google Scholar]
- 5.Okada T., Ding G., Sonoda H., Kajimoto T., Haga Y., Khosrowbeygi A., Gao S., Miwa N., Jahangeer S., Nakamura S. (2005) J. Biol. Chem. 280, 36318–36325 [DOI] [PubMed] [Google Scholar]
- 6.Sukocheva O. A., Wang L., Albanese N., Pitson S. M., Vadas M. A., Xia P. (2003) Mol. Endocrinol. 17, 2002–2012 [DOI] [PubMed] [Google Scholar]
- 7.Sarkar S., Maceyka M., Hait N. C., Paugh S. W., Sankala H., Milstien S., Spiegel S. (2005) FEBS Lett. 579, 5313–5317 [DOI] [PubMed] [Google Scholar]
- 8.Taha T. A., Kitatani K., El-Alwani M., Bielawski J., Hannun Y. A., Obeid L. M. (2006) FASEB J. 20, 482–484 [DOI] [PubMed] [Google Scholar]
- 9.Nava V. E., Hobson J. P., Murthy S., Milstien S., Spiegel S. (2002) Exp. Cell Res. 281, 115–127 [DOI] [PubMed] [Google Scholar]
- 10.French K. J., Schrecengost R. S., Lee B. D., Zhuang Y., Smith S. N., Eberly J. L., Yun J. K., Smith C. D. (2003) Cancer Res. 63, 5962–5969 [PubMed] [Google Scholar]
- 11.Shida D., Takabe K., Kapitonov D., Milstien S., Spiegel S. (2008) Curr. Drug Targets 9, 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruckhäberle E., Rody A., Engels K., Gaetje R., von Minckwitz G., Schiffmann S., Grösch S., Geisslinger G., Holtrich U., Karn T., Kaufmann M. (2008) Breast Cancer Res. Treat. 112, 41–52 [DOI] [PubMed] [Google Scholar]
- 13.McKenna N. J., O'Malley B. W. (2002) Cell 108, 465–474 [DOI] [PubMed] [Google Scholar]
- 14.Thürlimann B., Keshaviah A., Coates A. S., Mouridsen H., Mauriac L., Forbes J. F., Paridaens R., Castiglione-Gertsch M., Gelber R. D., Rabaglio M., Smith I., Wardley A., Price K. N., Goldhirsch A. (2005) N. Engl. J. Med. 353, 2747–2757 [DOI] [PubMed] [Google Scholar]
- 15.Levin E. R. (2005) Mol. Endocrinol. 19, 1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas P., Pang Y., Filardo E. J., Dong J. (2005) Endocrinology 146, 624–632 [DOI] [PubMed] [Google Scholar]
- 17.Revankar C. M., Cimino D. F., Sklar L. A., Arterburn J. B., Prossnitz E. R. (2005) Science 307, 1625–1630 [DOI] [PubMed] [Google Scholar]
- 18.Filardo E., Quinn J., Pang Y., Graeber C., Shaw S., Dong J., Thomas P. (2007) Endocrinology 148, 3236–3245 [DOI] [PubMed] [Google Scholar]
- 19.Sukocheva O., Wadham C., Holmes A., Albanese N., Verrier E., Feng F., Bernal A., Derian C. K., Ullrich A., Vadas M. A., Xia P. (2006) J. Cell Biol. 173, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hait N. C., Sarkar S., Le Stunff H., Mikami A., Maceyka M., Milstien S., Spiegel S. (2005) J. Biol. Chem. 280, 29462–29469 [DOI] [PubMed] [Google Scholar]
- 21.Hait N. C., Bellamy A., Milstien S., Kordula T., Spiegel S. (2007) J. Biol. Chem. 282, 12058–12065 [DOI] [PubMed] [Google Scholar]
- 22.Sankala H. M., Hait N. C., Paugh S. W., Shida D., Lépine S., Elmore L. W., Dent P., Milstien S., Spiegel S. (2007) Cancer Res. 67, 10466–10474 [DOI] [PubMed] [Google Scholar]
- 23.Shida D., Fang X., Kordula T., Takabe K., Lépine S., Alvarez S. E., Milstien S., Spiegel S. (2008) Cancer Res. 68, 6569–6577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maceyka M., Sankala H., Hait N. C., Le Stunff H., Liu H., Toman R., Collier C., Zhang M., Satin L., Merrill A. H., Jr., Milstien S., Spiegel S. (2005) J. Biol. Chem. 280, 37118–37129 [DOI] [PubMed] [Google Scholar]
- 25.Mitra P., Payne S. G., Milstien S., Spiegel S. (2007) Methods Enzymol. 434, 257–264 [DOI] [PubMed] [Google Scholar]
- 26.Filardo E. J., Quinn J. A., Bland K. I., Frackelton A. R., Jr. (2000) Mol. Endocrinol. 14, 1649–1660 [DOI] [PubMed] [Google Scholar]
- 27.Döll F., Pfeilschifter J., Huwiler A. (2007) Endocr. Relat. Cancer 14, 325–335 [DOI] [PubMed] [Google Scholar]
- 28.Pitson S. M., Moretti P. A., Zebol J. R., Lynn H. E., Xia P., Vadas M. A., Wattenberg B. W. (2003) EMBO J. 22, 5491–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedram A., Razandi M., Levin E. R. (2006) Mol. Endocrinol. 20, 1996–2009 [DOI] [PubMed] [Google Scholar]
- 30.Kleuser B., Malek D., Gust R., Pertz H. H., Potteck H. (2008) Mol. Pharmacol. 74, 1533–1543 [DOI] [PubMed] [Google Scholar]
- 31.Bologa C. G., Revankar C. M., Young S. M., Edwards B. S., Arterburn J. B., Kiselyov A. S., Parker M. A., Tkachenko S. E., Savchuck N. P., Sklar L. A., Oprea T. I., Prossnitz E. R. (2006) Nat. Chem. Biol. 2, 207–212 [DOI] [PubMed] [Google Scholar]
- 32.Mitra P., Oskeritzian C. A., Payne S. G., Beaven M. A., Milstien S., Spiegel S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16394–16399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi N., Nishi T., Hirata T., Kihara A., Sano T., Igarashi Y., Yamaguchi A. (2006) J. Lipid Res. 47, 614–621 [DOI] [PubMed] [Google Scholar]
- 34.Lee Y. M., Venkataraman K., Hwang S. I., Han D. K., Hla T. (2007) Prostaglandins Other Lipid Mediat. 84, 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato K., Malchinkhuu E., Horiuchi Y., Mogi C., Tomura H., Tosaka M., Yoshimoto Y., Kuwabara A., Okajima F. (2007) J. Neurochem. 103, 2610–2619 [DOI] [PubMed] [Google Scholar]
- 36.Dean M., Rzhetsky A., Allikmets R. (2001) Genome Res. 11, 1156–1166 [DOI] [PubMed] [Google Scholar]
- 37.Szakács G., Annereau J. P., Lababidi S., Shankavaram U., Arciello A., Bussey K. J., Reinhold W., Guo Y., Kruh G. D., Reimers M., Weinstein J. N., Gottesman M. M. (2004) Cancer Cell 6, 129–137 [DOI] [PubMed] [Google Scholar]
- 38.Robey R. W., To K. K., Polgar O., Dohse M., Fetsch P., Dean M., Bates S. E. (2009) Adv. Drug Deliv. Rev. 61, 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visram H., Greer P. A. (2006) Cancer Biol. Ther. 5, 1677–1682 [DOI] [PubMed] [Google Scholar]
- 40.Visentin B., Vekich J. A., Sibbald B. J., Cavalli A. L., Moreno K. M., Matteo R. G., Garland W. A., Lu Y., Yu S., Hall H. S., Kundra V., Mills G. B., Sabbadini R. A. (2006) Cancer Cell 9, 225–238 [DOI] [PubMed] [Google Scholar]
- 41.Johnson K. R., Becker K. P., Facchinetti M. M., Hannun Y. A., Obeid L. M. (2002) J. Biol. Chem. 277, 35257–35262 [DOI] [PubMed] [Google Scholar]
- 42.Pitson S. M., D'Andrea R. J., Vandeleur L., Moretti P. A., Xia P., Gamble J. R., Vadas M. A., Wattenberg B. W. (2000) Biochem. J. 350, 429–441 [PMC free article] [PubMed] [Google Scholar]
- 43.Igarashi N., Okada T., Hayashi S., Fujita T., Jahangeer S., Nakamura S. I. (2003) J. Biol. Chem. 278, 46832–46839 [DOI] [PubMed] [Google Scholar]
- 44.Hait N. C., Allegood J., Maceyka M., Strub G. M., Harikumar K. B., Singh S. K., Luo C., Marmorstein R., Kordula T., Milstien S., Spiegel S. (2009) Science 325, 1254–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergelin N., Blom T., Heikkilä J., Löf C., Alam C., Balthasar S., Slotte J. P., Hinkkanen A., Törnquist K. (2009) Endocrinology 150, 2055–2063 [DOI] [PubMed] [Google Scholar]
- 46.Nieuwenhuis B., Lüth A., Chun J., Huwiler A., Pfeilschifter J., Schäfer-Korting M., Kleuser B. (2009) J. Mol. Med. 87, 645–657 [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi N., Yamaguchi A., Nishi T. (2009) J. Biol. Chem. 284, 21192–21200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonard G. D., Fojo T., Bates S. E. (2003) Oncologist 8, 411–424 [DOI] [PubMed] [Google Scholar]
- 49.Dean M. (2009) J. Mammary Gland Biol. Neoplasia 14, 3–9 [DOI] [PubMed] [Google Scholar]
- 50.Pébay A., Wong R. C., Pitson S. M., Wolvetang E. J., Peh G. S., Filipczyk A., Koh K. L., Tellis I., Nguyen L. T., Pera M. F. (2005) Stem Cells 23, 1541–1548 [DOI] [PubMed] [Google Scholar]
- 51.Pébay A., Bonder C. S., Pitson S. M. (2007) Prostaglandins Other Lipid Mediat. 84, 83–97 [DOI] [PubMed] [Google Scholar]
- 52.Sukocheva O., Wang L., Verrier E., Vadas M. A., Xia P. (2009) Endocrinology 150, 4484–4492 [DOI] [PubMed] [Google Scholar]
- 53.Otto C., Rohde-Schulz B., Schwarz G., Fuchs I., Klewer M., Brittain D., Langer G., Bader B., Prelle K., Nubbemeyer R., Fritzemeier K. H. (2008) Endocrinology 149, 4846–4856 [DOI] [PubMed] [Google Scholar]
- 54.Berry D. A., Cirrincione C., Henderson I. C., Citron M. L., Budman D. R., Goldstein L. J., Martino S., Perez E. A., Muss H. B., Norton L., Hudis C., Winer E. P. (2006) J. Am. Med. Assoc. 295, 1658–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]