Abstract
Phogrin is a transmembrane protein expressed in cells with stimulus-coupled peptide hormone secretion, including pancreatic beta cells, in which it is localized to the membrane of insulin-containing dense-core vesicles. By sequence, phogrin is a member of the family of receptor-like protein-tyrosine phosphatases, but it contains substitutions in conserved catalytic sequences, and no significant enzymatic activity for phogrin has ever been reported. We report here that phogrin is able to dephosphorylate specific inositol phospholipids, including phosphatidylinositol (PI) 3-phosphate and PI 4,5-diphosphate but not PI 3,4,5-trisphosphate. The phosphatidylinositol phosphatase (PIPase) activity of phogrin was measurable but low when evaluated by the ability of a catalytic domain fusion protein to hydrolyze soluble short-chain phosphatidylinositol phospholipids. Unlike most PIPases, which are cytoplasmic proteins that associate with membranes, mature phogrin is a transmembrane protein. When the transmembrane form of phogrin was overexpressed in mammalian cells, it reduced plasma membrane phosphatidylinositol 4,5-disphosphate levels in a dose-dependent manner. When purified and assayed in vitro, the transmembrane form had a specific activity of 142 mol/min/mol, 75-fold more active than the catalytic domain fusion protein and comparable with the specific activities of the other PIPases. The PIPase activity of phogrin depended on the catalytic site cysteine and correlated with effects on glucose-stimulated insulin secretion. We propose that phogrin functions as a phosphatidylinositol phosphatase that contributes to maintaining subcellular differences in levels of PIP that are important for regulating stimulus-coupled exocytosis of insulin.
Keywords: Cell/Exocytosis, Cell/Secretion, Lipid/ Inositol Phospholipid, Lipid/Phospholipid/Turnover, Membrane/Enzymes, Receptors/Phosphatases, Signal Transduction/Inositol Phosphates, Subcellular Organelles/Vesicles
Introduction
IA-2 (also known as ICA512 and PTPRN) and IA-2β (also known as phogrin, NE-6, ICAAR, and PTPRN2) are closely related proteins that were first identified as targets of autoantibodies in Type I diabetics (1–3). The rat IA-2β ortholog is usually referred to as phogrin, which is how we will refer to it in this report because we have used a rat pancreatic beta cell line and rat transgenes. Phogrin is expressed in neuroendocrine cell types that exhibit stimulus-coupled secretion, including pancreatic beta cells and neurons but not in cells of the exocrine pancreas or in cells that lack a regulated secretion pathway (4–8). In pancreatic beta cells, phogrin is localized to the insulin-containing dense-core vesicles (DCVs)2 (4, 6, 8–11), suggesting that it plays some role in insulin secretion. Studies of mice with disruption of both IA-2 and IA-2β/phogrin genes have shown alterations in insulin or neuropeptide hormone secretion (12, 13), but expression of these genes in many neurosecretory cell types makes it difficult to clearly identify direct effects on pancreatic beta cells in vivo. Disruption of the IA-2β/phogrin gene (14) or both phogrin and IA-2 genes (12, 13) reduced glucose-stimulated insulin release from isolated islets, and overexpression of IA-2β/phogrin in the mouse beta cell line MIN6 almost eliminated glucose-stimulated insulin secretion (15). However, Torii et al. (16) reported the overexpression and knockdown of phogrin affected cell proliferation but not insulin secretion from Ins-1 or MIN6 cells. As discussed by Hu et al. (17), there is not yet a clear explanation for these differences in results.
By amino acid sequence similarity and domain structure, phogrin is related to receptor-like protein-tyrosine phosphatases (PTPases) with a single catalytic domain (3, 8, 18), but phogrin has been reported to lack PTPase activity (18–20). The consensus sequence for the catalytic site of an active PTPase is (I/V)HCXAGXXR(S/T)G. In phogrin, this sequence is VHCSDGAGRSG. Experimental mutation of Asp-936 to canonical Ala yields measurable PTPase activity (18, 21), and PTPase activity is increased further by mutation of amino acids in two other sequence motifs that are conserved in active PTPases but not in phogrin (22).
In the absence of detectable ability to dephosphorylate phosphoproteins or p-nitrophenyl phosphate (pNPP), it has been suggested that phogrin and the closely related DCV protein IA-2 may function as a dominant negatives, e.g. to bind to phosphotyrosine and block access by active PTPases (18), or may function by interacting with other proteins (23, 24). An alternative possibility is that phogrin is active against substrates other than phosphotyrosine. We report here that phogrin is able to dephosphorylate specific phosphatidylinositol phospholipids (PIPs), including PI(3)P and PI(4,5)P2 but not PI(3,4,5)P3. Unlike most PIPases, which are cytoplasmic proteins that associate with membranes, phogrin is a transmembrane protein, and full PIPase activity in vitro depends on the presence of the transmembrane domain. When overexpressed in cells, phogrin is able to reduce plasma membrane PI(4,5)P2 levels in a dose-dependent manner that depends on the catalytic site cysteine and correlates with inhibition of glucose-stimulated insulin secretion. Many secretory vesicle-associated proteins have domains that bind PI(4,5)P2, and PI(4,5)P2 has been implicated in the organization and function of secretory and endocytic vesicles. Domains that bind PI(3)P and PI(4,5)P2 serve to localize specific proteins to the correct subcellular compartment and/or to alter the activity of the bound proteins during exocytosis and endocytosis (25–39). We propose that phogrin functions as a phosphatidylinositol phosphatase and that it plays a role in regulating DCV and/or plasma membrane PIP content to facilitate stimulus-coupled exocytosis of insulin.
EXPERIMENTAL PROCEDURES
Cell Culture
INS-1 rat insulinoma cells were provided by the Diabetes Endocrinology Research Center Tissue Core at the University of Washington, Seattle, WA and used at passages 56–75. Unless otherwise noted, cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, antibiotics, l-glutamine, sodium pyruvate, and 50 μm β-mercaptoethanol. HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics.
Expression and Purification of Glutathione S-Transferase (GST) Fusion Proteins
Full-length rat phogrin cDNA was a generous gift of Dr. L. Fitzgerald (University of Maryland at Baltimore). The cytoplasmic domain, from amino acid Arg-629, the first amino acid after the transmembrane domain, was cloned into pGEX-4T-3 bacterial expression vector (Pharmacia Corp.) to generate GST-phogrinCat. As a positive control for an active PTPase, the cytoplasmic domain of PTPβ from Arg-1621 to the C terminus, was cloned into pGEX-4T-3 to generate GST-PTPβCat. As a positive control for an active PIP 5-phosphatase, the 5-phosphatase domain of synaptojanin-2 (40) (a generous gift of Dr. Marc Symons, Albert Einstein College of Medicine) was cloned into pGEX-4T-3 to generate GST-SJ-2PDCat. Mutations in phogrin-myc and the GST-phogrinCat constructs were created with a commercially available site-directed mutagenesis kit (QuikChange XL, Stratagene, La Jolla, CA) using protocols provided by the manufacturer. Preparation and purification of the GST fusion proteins is described in the online supplemental Material. GST-PTEN was purchased from Echelon Biosciences, Inc. (Salt Lake City, UT).
Preparation of Rabbit Polyclonal Anti-phogrin Catalytic Domain Antibody
Rabbit polyclonal antiserum was prepared (Alpha Diagnostic International, San Antonio, TX) against GST-phogrinCat by immunizing two male New Zealand White rabbits with multiple subcutaneous injections of 125 μg of GST-phogrinCat fusion protein emulsified in incomplete Freund's adjuvant. Injections were performed using a standard 63-day protocol. Preimmune serum was collected from both rabbits. Serum from immunized rabbits was affinity-purified through a column containing Sepharose coupled to the GST-phogrinCat antigen and tested for immunoreactivity in multiple formats including immunohistochemistry of fixed rat pancreatic islets and by Western blot analysis against purified GST-phogrinCat fusion protein (positive control for immunogen), cell lysates from INS-1 cells (positive control for endogenous phogrin), untransfected HEK293 cells (negative control), or HEK293 cells transfected with phogrin-myc (positive control).
Retroviral Vector Construction and Infection
Full-length rat phogrin cDNA (18) was cloned into pcDNA3.1(+)C myc-His (Invitrogen). The myc-His tagged rat phogrin was excised and cloned into the retroviral expression vector pBM-IRES-Puro (generous gift of Dr. E. Raines, University of Washington). High titer virus preparations were obtained using Phoenix amphotropic packaging cell line (Orbigen, San Diego, CA) as previously described (41). For infection of INS-1 and HEK293 cells, 6 × 104 cells were plated into 6-well plates 48 h before infection and incubated with 3 ml of virus stock for 18 h in the presence of 5 μg/ml Polybrene. After retroviral infection, cells were cultured for an additional 18 h in normal growth media. Phogrin-myc-overexpressing cells were enriched by puromycin selection (2 μg/ml for 36 h).
Assay for pNPP Hydrolysis
The reaction mixture containing 25 mm sodium acetate or 20 mm Tris, pH 5.5, 10 mm dithiothreitol, 1 μg of GST fusion protein, and 10 mm pNPP (Sigma) in a 50-μl final volume was incubated at 30 °C for 30 min. The reaction was terminated by the addition of 200 μl of 0.2 n NaOH. The amount of dephosphorylated reaction product, p-nitrophenol, was calculated from absorbance at 405 nm using the extinction coefficient of 1.8 × 104 m−1cm−1. The activity rates were corrected for non-enzymatic hydrolysis of pNPP in reactions without enzyme.
Assays of PIPase Activity
The malachite green assay for phosphate release was performed in a 50-μl reaction mixture containing 100 mm Tris HCl, pH 8.0, 10 mm dithiothreitol, water-soluble diC8-PIPs (Echelon Biosciences), and fusion protein. Samples were incubated for 30 min at 37 °C, and phosphate release was determined using a malachite green assay (Biomol Green Reagent, Biomol Research Laboratories, Inc., Plymouth Meeting, PA) in accordance with the manufacturer's instructions. The absorbance at 630 nm was measured in a SpectraMax plate reader (Molecular Devices, Sunnyvale, CA). A standard curve for inorganic phosphate was generated for each assay.
Cleavage of fluorescent BODIPY-FL di-C6 phosphoinositides including PI, PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3 (Echelon Biosciences) was evaluated as described by Taylor and Dixon (42). The fluorescent PIPs were diluted in distilled H2O to a final concentration of 1 μg/μl. Each phosphatase reaction contained 1.5 μg of substrate and 23.5 μl of reaction buffer (50 mm ammonium acetate, pH 7.0, 5 mm dithiothreitol). The phosphatase reactions were initiated by the addition of enzyme and incubated at 37 °C for the duration indicated. The reactions were terminated by the addition of 100 μl of acetone and spun at 17,000 × g for 1 min to pellet precipitated protein. The supernatant was then dried in a SpeedVac concentrator at medium heat and resuspended in 20 μl of CHCl3/2-propanol/glacial acetic acid (5:5:2) and spotted onto a silica gel thin layer chromatography plate (Fisher) activated before use by immersing in a 1.2% solution of potassium oxalate, air drying, and baking at 65 °C. The spotted plate was allowed to dry for 10 min and then developed in a solvent system consisting of CHCl3/MeOH/acetone/glacial acetic acid/distilled H2O (70:50:20:20:20). When the solvent was 2–3 cm from the top, the plate was removed and allowed to dry. The TLC plate was then placed with silica gel side up on a standard DNA gel UV illuminator, and the fluorescent phosphoinositide bands were visualized under UV light. Band intensity was quantified using ImageQuant software.
Ni-NTA Affinity Chromatography
Cells expressing phogrin-myc or phogrin(C934S)-myc were washed twice in 10 ml of Tris-buffered saline and lysed in 400 μl of buffer containing 1% Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 8.0, 20 mm 2-β-mercaptoethanol, 20 mm imidazole, and protease inhibitor mixture (Sigma). Lysates were spun at 10,000 rpm for 5 min at 4 °C. After incubation for 1 h at 4 °C with 40 μl of Ni-NTA magnetic agarose beads (Qiagen, Valencia CA), the beads were washed 4 times with 400 μl of cold washing buffer (20 mm Tris-HCl, 300 mm NaCl, and 20 mm imidazole, pH 8.0), and tagged protein was eluted with 40 μl of elution buffer (20 mm Tris-HCl, 300 mm NaCl, and 250 mm imidazole, pH 8.0).
Western Blot Analysis
Cells were rinsed in PBS and scraped into lysis buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl, pH 8.0) containing a protease inhibitor mixture to permit determination of protein concentrations (Pierce BCA assay kit). Alternatively, cells were lysed directly in SDS-PAGE sample buffer containing dithiothreitol. Protein samples were electrophoresed on 4–20% Tris-HEPES SDS gradient gels (Pierce) and electroblotted onto Protran nitrocellulose membranes (Schleicher and Schuell) using a semidry transfer apparatus. Membranes were blocked with Tris-buffered saline, 5% nonfat dried milk for 30 min, washed, and incubated with primary and secondary antibodies for 12 h and 30 min, respectively. The membranes were washed extensively with Tris-buffered saline, 0.1% Tween 20 after secondary antibody incubation and detected using the ECL Western blotting kit (Pierce) according to the manufacturer's suggested protocol.
TaqMan (Real-time Quantitative Reverse Transcription-PCR) Analysis
Total RNA from cultured cells was isolated using RNeasy (Qiagen). RNA was reverse-transcribed by standard methods using reverse transcriptase (Invitrogen). For TaqMan real-time PCR, phogrin and 18 S primers, TaqMan Gene Expression Assay probe, and primer sets were purchased from Applied Biosystems. An Applied Biosystems Prism 7500 Fast Real-time PCR System (Applied Biosystems, Foster City CA) was used with the default thermal cycling program (95 °C for 20 s followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s). Reactions were performed in triplicate and normalized to level of 18 S RNA transcript.
Immunofluorescence Microscopy
Cells were plated onto glass coverslips 2 days before analysis. For imaging, coverslips were set on ice, rinsed twice in ice-cold PBS, fixed in ice-cold 10% neutral buffered formalin solution and permeabilized with ice-cold methanol. All blocking, labeling, and washing steps were carried out in phosphate-buffered saline, 0.2% normal goat serum. Labeling was performed with the antibodies mouse monoclonal IgM anti-PI(4,5)P2 (Echelon Biosciences), mouse monoclonal anti-myc (9B11; Cell Signaling Technology, Inc.), mouse monoclonal anti-GST (Sigma), affinity-purified rabbit polyclonal anti-phogrin lumenal domain antisera (1:250) (generous gifts from Dr. John Hutton, University of Colorado, Barbara Davis Center for Childhood Diabetes, Aurora, CO) and visualized using either FITC-conjugated goat anti-rabbit (1:200), TRITC-labeled donkey anti-rabbit (1:200), FITC-conjugated goat anti-mouse IgM (1:200), or TRITC-labeled goat anti-mouse IgG (1:200) (Jackson ImmunoResearch Laboratories, West Grave PA). Images were acquired using a Zeiss LSM510 META based on an Axiovert 200 microscope. Zeiss LSM Image Analysis software was to quantitate signals by measuring pixel intensity for each fluor and comparing it to a control.
Phogrin Knockdown by siRNA
siRNA constructs (see the supplemental Material) were cloned into the mammalian expression vector pSUPER.retro.puro (OligoEngine, Seattle WA) according to the manufacturer's directions. To generate the retrovirus, plasmid DNA (10 μg) was transfected into the Phoenix amphotropic retrovirus packaging cell line (Orbigen, San Diego, CA) by calcium phosphate transfection. Medium containing virus was collected 48 h after transfection. INS-1 cells were infected and selected in 2 μg/ml puromycin (Sigma). Reduction of phogrin transcript level was evaluated by real-time PCR followed by Western blot evaluation of phogrin protein expression.
Insulin Determinations
Insulin secreted into incubation medium was determined using an enzyme-linked immunosorbent assay kit (Linco Research, St. Charles, MO), according to the manufacturer's instructions. To determine intracellular insulin content, cells were gently scraped with lysis buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris HCl, pH 8.0), and the insulin concentration in the cell lysate was determined as above and normalized to cell number determined from parallel wells.
RESULTS
Recombinant Phogrin Catalytic Domain Has Very Little Activity against Phosphotyrosine or pNPP
To evaluate the enzymatic activities of phogrin, we first expressed the cytoplasmic catalytic domain as a GST fusion protein (GST-phogrinCat) (Fig. 1B). As a negative control for enzymatic activity, we expressed a mutant version in which cysteine 934 in the “catalytic site” CX5R sequence was mutated to serine (GST-phogrinCatC934S). This cysteine plays a central role in the general catalytic mechanism proposed for the CX5R class of phosphatases, and eliminating it was expected to eliminate enzymatic activity against all substrates. As a positive control for PTPase activity, we expressed a GST fusion protein with the cytoplasmic catalytic domain of PTPβ, which is an active PTPase with the same general structure as phogrin, i.e. a transmembrane protein with a single catalytic domain, but with a canonical PTPase active site sequence (43). As a positive control for an established PIP 5-phosphatase, we expressed a GST fusion protein with the 5-phosphatase domain of synaptojanin 2 (GST-SJ2-PDCat) (37, 44–46). As a positive control for an established PIP 3-phosphatase, we used PTEN (47) as a GST fusion protein.
FIGURE 1.
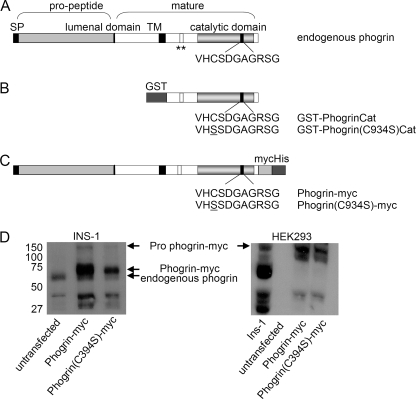
Structure of endogenous phogrin and fusion proteins. A, shown is a diagram of endogenous rat phogrin showing the major structural domains. Phogrin is synthesized as an ∼130-kDa transmembrane pro-form that includes a signal peptide (SP), lumenal domain, transmembrane domain (TM), and cytoplasmic catalytic domain. The N-terminal portion of the lumenal domain is cleaved during maturation such that the N-terminal portion (pro-peptide) is released into the lumen of the DCV. Mature phogrin is a 60/64-kDa transmembrane protein with a short lumenal domain and a PTPase-like cytoplasmic domain. Serine 680 and threonine 699 (location indicated by **) between TM and catalytic domains are sites of phosphorylation by protein kinase A (68, 69). B, the structure of GST-phogrinCat and GST-phogrin(C934S)Cat constructs expressed in bacteria is shown. C, shown is the structure of phogrin-myc and phogrin(C934S)-myc constructs expressed in mammalian cells. D, INS-1 and HEK293 cells were infected with retroviral constructs encoding phogrin-myc and phogrin(C934S)-myc and selected with puromycin. Cell lysates were evaluated by Western blot using a phogrin-specific rabbit antibody. In Ins-1 cells arrows indicate the ∼130-kDa pro-form and 73/77-kDa mature forms of phogrin-myc and the 60/64-kDa mature form of endogenous phogrin. The pro-form of endogenous phogrin is not present at a high enough level to be detected in this exposure. HEK293 cells do not express endogenous phogrin and phogrin-myc accumulates as unprocessed pro-form.
To evaluate the activity of GST-phogrinCat against phosphotyrosine, we used the malachite green assay to measure phosphate release from the commonly used tyrosine-phosphorylated peptide substrate RRLIEDAEpYAARG (RR-src). As previously reported (48), GST-PTPβCat was active against phosphorylated peptide RR-src, but GST-phogrinCat did not exhibit detectable activity (data not shown). We next evaluated the activity of GST-phogrinCat against pNPP, a small artificial substrate that is commonly used to measure general phosphatase activity. Table 1 shows that GST-PTPβCat showed strong activity against pNPP, GST-PTEN showed 100-fold lower activity, and GST-SJ2-PDCat and GST-phogrinCat showed barely detectable activity. These results are consistent with reports that phogrin has little or no detectable activity against phosphotyrosine or pNPP (18) and that, although pNPP is usually considered to be a substrate for all classes of phosphatase, even well documented PIPases like PTEN and synaptojanin are much less active against pNPP than are PTPases (44, 49).
TABLE 1.
Phosphatase activities of fusion proteins
The activity of fusion proteins against pNPP was evaluated by the rate of formation of the dephosphorylated product p-nitrophenol, determined by absorbance at 405 nm. The activity of fusion proteins against 200 μm diC8-PIP substrates was determined by a 30-min incubation at 37 °C. 1 mm Mg++ was included in assays using GST-SJ2-PD. Phosphate release was measured using the malachite green assay described under “Experimental Procedures.” Results are reported as the mean ± S.E. of specific activity expressed as mol of phosphate released/min/mol of recombinant protein. The dashes indicate activity less than detectable. NT indicates not tested.
| GST-phogrinCat | GST-phogrin(C934S)Cat | GST-PTPβCat | GST-SJ-2PDCat | GST-PTEN | |
|---|---|---|---|---|---|
| pNPP | 0.28 ± 0.04 | 0.04 ± 0.03 | 1692 ± 30 | 0.07 ± 0.04 | 16 ± 2 |
| PI(3)P | 1.10 ± 0.07 | 0.18 ± 0.05 | – | – | 92 ± 8 |
| PI(4)P | 0.75 ± 0.08 | 0.11 ± 0.01 | – | – | 4.3 ± 0.7 |
| PI(5)P | 1.60 ± 0.04 | 0.25 ± 0.02 | – | 86 ± 8 | 17 ± 3 |
| PI(3,4)P2 | 0.19 ± 0.04 | 0.09 ± 0.00 | – | 0.08 | NT |
| PI(3,5)P2 | 0.31 ± 0.02 | 0.08 ± 0.01 | – | 0.09 | NT |
| PI(4,5)P2 | 1.90 ± 0.08 | 0.30 ± 0.01 | – | 101 ± 12 | 19 ± 2 |
| PI(3,4,5)P3 | 0.02 ± 0.01 | 0.02 ± 0.00 | – | 0.08 | 357 ± 17 |
Recombinant Phogrin Catalytic Domain Has PIPase Activity in Vitro
To quantitate the PIPase activity of phogrin, we used the colorometric malachite green assay to measure the release of phosphate from aqueous-soluble diC8-phosphatidylinositol phosphates (diC8-PIPs), in which the fatty acid chains are eight carbons long. Table 1 shows that when assayed with 200 μm diC8-PIPs, GST-phogrinCat was most active against PI(3)P, PI(5)P, and PI(4,5)P2 but that it was also active against other PIPs except PI(3,4,5)P3, the preferred substrate for PTEN. GST-phogrin(C934S)Cat did not have significant PIPase activity against any substrate, confirming that the activity of GST-phogrinCat is a function of the CX5R putative catalytic site. Consistent with this, the activity of GST-phogrinCat was not dependent on Mg++ (data not shown). This distinguishes it from the PI 5-phosphatase class of PIPases, including the 5-phosphatase domain of synaptojanin-2, which requires Mg++ and does not employ a CX5R catalytic site (44, 50). GST-PTPβCat, which is active as a PTPase, did not have detectable PIPase activity against any PIP. This indicates that the PIPase activity of GST-phogrinCat is not a general property of proteins with a CX5R-like catalytic site. To put the PIPase activity of GST-phogrinCat in perspective, Table 1 shows that the activity of GST-phogrinCat against 200 μm PI(4,5)P2 (1.9 mol/min/mol) is about 2% that of the positive-control 5-phosphatase GST-SJ2-PDCat (101 mol/min/mol) and about 0.5% that of the 3-phosphatase activity of GST-PTEN against PI(3,4,5)P3.
Full-length (Transmembrane) Phogrin Is Much More Active than GST-phogrinCat in Vitro
Synaptojanin and most other reported PIPases are cytosolic or membrane-associated proteins. Phogrin is a transmembrane protein. The transmembrane domain of phogrin may normally serve to orient the catalytic domain to membrane PIP substrates or may affect the conformation of the catalytic domain. We generated transgenes encoding full-length wild type (phogrin-myc) or C934S mutant phogrin (phogrin(C934S)-myc), tagged at the C terminus with a myc/His tag (Fig. 1C). We expressed the transgenes in both INS-1 and HEK293 cell lines. INS-1 is a rat insulinoma cell line that has been used by many investigators to study insulin secretion because it is functionally immortal and shows the secretory architecture and glucose sensitivity of a pancreatic beta cell. HEK293 is a kidney epithelial cell line that does not express endogenous phogrin or have a regulated secretory system.
In pancreatic beta cells, the endogenous phogrin is synthesized as an ∼130-kDa transmembrane glycoprotein pro-form, with the C terminus and catalytic domain in the cytoplasm and the N terminus in the lumen of the DCV (8, 10). This pro-form is cleaved in the post-trans Golgi/DCV compartment to release the pro-domain and generate the 60/64-kDa “mature” form with a short lumenal domain, transmembrane domain, and a PTPase-like cytoplasmic domain (8). When expressed in INS-1 cells and evaluated by a Western blot probed with rabbit antiserum against phogrin, which recognizes both endogenous phogrin and phogrin-myc, the phogrin-myc transgene protein was processed to an analogous mature 73/77-kDa form (Fig. 1D) with C-terminal myc/His tag. In four independent infections, the level of phogrin-myc expression was several times greater than that of endogenous phogrin and higher than the level of phogrin(C934S)-myc expression. When expressed in HEK293 cells, the phogrin-myc was not processed to a mature form, and wild type and mutant forms were expressed at comparable levels (Fig. 1D).
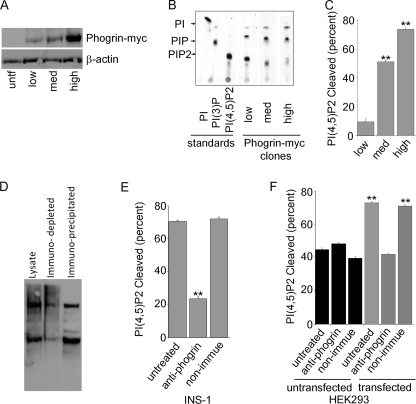
We initially evaluated the activity of phogrin-myc in detergent lysates. Because the cell lysates contained free phosphate that would interfere with an assay of phosphate release, we evaluated PIPase activity using fluorescently labeled PIP substrates and detected the appearance of products by changes in mobility on thin layer chromatography. To specifically determine the relationship between phogrin-myc expression level and the PI(4,5)P2 level, we isolated and evaluated three clones that expressed different levels of phogrin-myc, evaluated by Western blot in Fig. 2A and by quantitative immunofluoresence in supplemental Fig. S1, A and B. Fig. 2, B and C, show that detergent lysates of INS-1 clones expressing phogrin-myc cleaved fluor-PI(4,5)P2 and that the activity of extracts from the three clones paralleled phogrin-myc expression level. To confirm that the elevated PIPase activity in lysates of clones expressing phogrin-myc was due to phogrin and not to up-regulation of some other PIPase, we used anti-phogrin antiserum to immuno-deplete phogrin from the lysates (Fig. 2D). Treatment with anti-phogrin, but not with non-immune serum, reduced the PIPase activity of the detergent lysates of transfected INS-1 cells (Fig. 2E) and HEK293 cells (Fig. 2F). The remaining PIPase activity presumably represents the activity of other endogenous PIPases.
FIGURE 2.
Evaluation of the PIPase activity of phogrin-myc by thin layer chromatography. A mass culture of INS-1 cells infected with phogrin-myc vector was plated at low density, and 10 individual clones were picked and evaluated by Western blot. From these 10 clones, 3 clones expressing low, medium, or high levels of phogrin-myc were chosen for further evaluation and use. A, shown is a Western blot of cell extracts probed with anti-myc and with anti-β-actin as a loading control. untf, untransfected. B, BODIPY-FL di-C6 phosphoinositides were distinguished by thin layer chromatography as described under “Experimental Procedures.” The first three lanes show the chromatographic mobility of 1 μg of fluorescent PIP standards. The final three lanes show the products of 1 μg of PI(4,5)P2 incubated for 5 min at 37 °C with 5 μg of lysate protein from the INS-1 clones expressing low, medium, or high levels of phogrin-myc. C, shown is a densitometric quantitation of a chromatographic analysis performed as for panel B and plotted as the percentage of fluorescent PI(4,5)P2 cleaved at the end of the incubation period; data are the average ±S.E. of triplicate determinations. Data were analyzed by ImageQuant software. **, p < 0.01 compared with the low-expressing clone. D, lysates from the INS-1 clone expressing the highest level of phogrin-myc were incubated for 3 h with anti-phogrin serum or non-immune serum followed by 1 h of incubation with protein A and then centrifugation to pellet bound protein. A Western blot probed with anti-phogrin shows immunodepletion of mature and precursor forms of phogrin (endogenous phogrin and phogrin-myc) from the INS-1 transfected clone. E, the control and immunodepleted lysates were assayed for cleavage of fluorescent-PI(4,5)P2 as for panel B. **, p < 0.01 compared with untreated. F, lysates from control HEK293 cells or from HEK293 cells expressing phogrin-myc were immunodepleted with control or anti-phogrin antiserum as for panel C and evaluated for the ability to cleave fluorescent PI(4,5)P2. Anti-phogrin did not affect the basal PIPase activity in HEK293 lysates and eliminated all of the additional PIPase activity contributed by phogrin-myc expression. **, p < 0.01 compared with untreated untransfected.
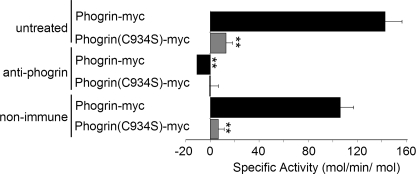
We used nickel affinity chromatography to purify phogrin-myc via the polyhistidine sequence in the Myc-His tag. Fig. 3 shows that phogrin-myc purified from Triton-solubilized transfected INS-1 cells had a specific activity of 142 mol/min/mol with 100 μm diC8-PI(4,5)P2 as determined in the malachite green assay for phosphate release. This is 75-fold higher than the activity measured for the GST-phogrinCat fusion protein and comparable with that of the GST synaptojanin fusion protein (Table 1). Purified phogrin(C934S)-myc had little activity, indicating that the activity of the affinity-purified preparation was not likely to be due to contaminating or co-precipitating endogenous PIPases. To directly evaluate the contribution of phogrin to the activity of the affinity-purified phogrin-myc, we incubated the preparation with rabbit antibody prepared against the phogrin catalytic domain. This reduced PIPase activity to undetectable levels (Fig. 3). Incubation with non-immune IgG had no significant effect on PIPase activity. These results indicate that full-length phogrin-myc has significant intrinsic PIPase activity, much higher than GST-phogrinCat and comparable with established PIPases.
FIGURE 3.
Affinity-purified phogrin-myc has PIPase activity. Triton X-100 lysates of INS-1 cells expressing phogrin-myc or phogrin(C934S)-myc were incubated with Ni-NTA magnetic- agarose beads to bind phogrin-myc via the C-terminal poly-His tag. The beads were washed, and the bound phogrin-myc eluted as described under “Experimental Procedures.” The amount of phogrin-myc in the eluate was determined by Western blot with rabbit anti-phogrin catalytic domain antibody using dilutions of purified GST-phogrinCat as standards. Aliquots containing 0.6 μg of phogrin-myc or phogrin(C934S)-myc were incubated with rabbit anti-phogrin IgG or control non-immune IgG then assayed for hydrolysis of 100 μm diC8-PI(4,5)P2. Phosphate release was measured using the malachite green assay and normalized to the amount of phogrin transgene present (mean ± S.E. of triplicate determinations). **, p < 0.01 compared with the activity of untreated phogrin-myc samples.
Expression of Phogrin-myc in Cells Reduces PI(4,5)P2 Levels
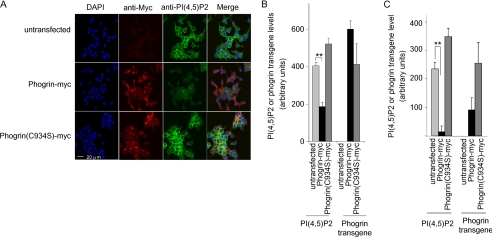
To evaluate the PIPase activity of full-length phogrin in a native membrane environment, we determined how expression of phogrin-myc affects cell PI(4,5)P2 levels. We initially detected PI(4,5)P2 using anti-PI(4,5)P2 monoclonal antibody 2C11, visualized with FITC-labeled second antibody and imaged by confocal microscopy. We confirmed these results using a recombinant fusion protein consisting of the PI(4,5)P2 binding domain of PLCδ1 fused to GST (51) and visualized by fluorescein-conjugated anti-GST (data not shown). In untransfected cells, PI(4,5)P2 was detected largely in the plasma membrane (Fig. 4A). Phogrin-myc and phogrin(C934S)-myc were localized to areas of the cytoplasm enriched in DCVs, as confirmed by co-immunostaining with antibodies against insulin (data not shown). Expression of phogrin-myc greatly reduced PI(4,5)P2 staining in the plasma membrane. As quantitated by image analysis, expression of phogrin-myc reduced PI(4,5)P2 levels by about 50% (Fig. 4B). Analysis of clones expressing different levels of phogrin-myc showed that PI(4,5)P2 levels are lowest in the clones that expressed the highest level of phogrin-myc (supplemental Fig. S1). Expression of catalytically inactive phogrin(C934S)-myc never decreased cell PI(4,5)P2 levels and showed a trend (not statistically significant) to increase PI(4,5)P2 levels. This indicates that the ability of wild type phogrin-Myc to decrease PI(4,5)P2 levels results from its intrinsic PIPase activity and not from some indirect effect, e.g. binding to other cell proteins.
FIGURE 4.
Phogrin-myc expression decreases PI(4,5)P2 levels in cells. Transfected or untransfected INS-1 cells (panels A and B) or HIK293 cells (panel C) were cultured in medium with 11.1 mm glucose, fixed with cold formalin, permeabilized with cold methanol, and incubated with mouse monoclonal anti-myc IgG visualized with TRITC-conjugated goat anti-mouse IgG (red) and with mouse monoclonal IgM anti-PI(4,5)P2 visualized with FITC-conjugated goat anti-mouse IgM (green). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). A, representative confocal images of INS-1 cells are shown. Expression of phogrin-myc, but not phogrin(C934S)-myc, substantially decreased the levels of PI(4,5)42. B, to quantitate the levels of transgene and PI(4,5)P2 in INS-1 cells, images were evaluated by Zeiss LSM Image Analysis software as described under “Experimental Procedures.” At least 3 low power fields (no fewer than 20 cells/field) were evaluated for each condition, and fluorescence intensity was normalized to the numbers of cells per field based on numbers of nuclei stained with 4′,6-diamidino-2-phenylindole. The data are presented as the means ± S.D. of triplicate determinations. Comparable results were obtained in five experiments. **, p < 0.01 compared with untransfected. C, HEK293 cells expressing phogrin-myc or phogrin(C934S)-myc were evaluated as for panels A and B. The data are presented as the means ± S.D. of five fields/condition. Comparable results were obtained in three experiments. **, p < 0.01 compared with untransfected.
In INS-1 cells, most phogrin-myc is in DCV membranes, and most of the PI(4,5)P2 is in the plasma membrane. The HEK293 line is derived from human embryonic kidney and does not contain DCVs. We predicted that when expressed in HEK293 cells, more phogrin-myc transgene would be localized to the plasma membrane and result in more efficient reduction of plasma membrane PI(4,5)P2 levels. Fig. 4C shows that expression of phogrin-myc eliminated 90% of the PI(4,5)P2 signal in HEK293 cells, a greater reduction than seen in INS-1 cells. Expression of phogrinC936S-myc increased PI(4,5)P2 levels slightly, although the increase did not reach statistical significance.
Reducing Levels of Endogenous Phogrin Increases Levels of PI(4,5)P2
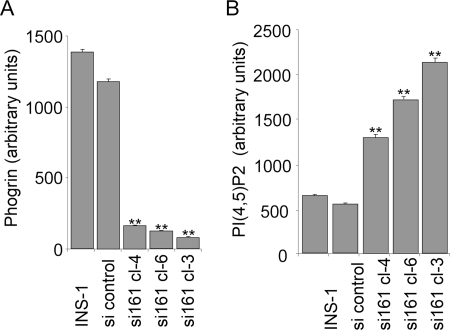
To evaluate the contribution of endogenous phogrin to regulation of PI(4,5)P2 levels in INS-1 cells, we reduced expression of endogenous phogrin using siRNA. We infected INS-1 cells with retroviral vectors expressing siRNA constructs targeting three sequences within the phogrin transcript (see “Experimental Procedures”). We evaluated efficacy by measuring phogrin transcript levels by quantitative PCR and phogrin protein levels by Western blot using a rabbit antibody against the lumenal domain (supplemental Fig. S2). From the sequence that produced the greatest decrease in bulk transfected population, we selected three clones for further analysis by confocal microscopy. Fig. 5 shows that clone 3, in which phogrin expression was reduced by 94%, showed a 3-fold increase in PI(4,5)P2 level. The other clones showed a smaller decrease in phogrin level and a smaller increase in PI(4,5)P2 levels, although the dose-response trend did not reach statistical significance.
FIGURE 5.
Reducing endogenous phogrin levels by SI-knockdown increases PI(4,5)P2 levels in INS-1 cells. Anti-phogrin construct si161 or a control sequence was transfected into INS-1 cells, and three clones were picked for evaluation by immunostaining, confocal microscopy, and quantitation of expression levels by image analysis as described for Fig. 4. A, endogenous phogrin was detected using rabbit antiserum against the lumenal domain of mature phogrin and visualized with TRITC-labeled donkey anti-rabbit IgG. B, PI(4,5)P2 was detected using mouse monoclonal IgM anti-PI(4,5)P2 visualized with FITC-conjugated goat anti-mouse IgM. The data are presented as the means ± S.D. of triplicate determinations. Comparable results were obtained in three experiments. **, p < 0.01 compared with untransfected INS-1.
The Ability of Phogrin to Affect Insulin Secretion Depends on Its PIPase Activity
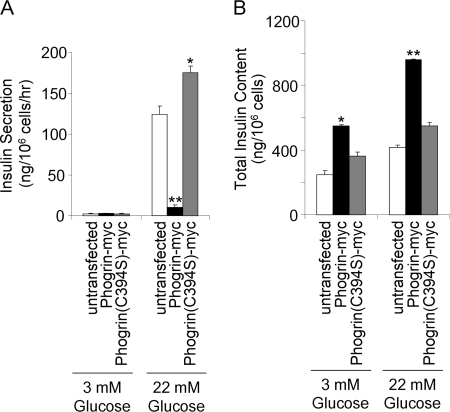
The localization of phogrin to the DCV in pancreatic beta cells suggests that it plays some role in insulin secretion. Fig. 6A shows that overexpression of phogrin-myc in INS-1 cells reduced glucose-stimulated insulin secretion during a 60-min assay period by 83%. Fig. 6B shows that the decrease in secretion did not result from a decrease in insulin content. In fact, insulin content was increased in cells expressing phogrin-myc, probably because insulin synthesis was not affected whereas secretion was inhibited. Time course studies showed that the phogrin-myc affected the second phase of glucose-stimulated insulin secretion with little effect on the first phase (supplemental Fig. S3), suggesting that the effect is on the rate of priming rather than on the initial response to secretion. Insulin secretion was not decreased by overexpression of phogrin(C934S)-myc, indicating that the effect of phogrin on insulin secretion depends on its enzymatic activity.
FIGURE 6.
Inhibition of glucose-stimulated insulin secretion by phogrin-myc depends on its PIPase activity. A, cultures of INS-1 cells were incubated for 1 h in RPMI without glucose then for 1 h in Krebs-Ringer bicarbonate-HEPES buffer with 3 or 22.2 mm glucose or 3 or 22.2 mm mannitol (not shown) as negative controls for possible osmotic effects (none were seen). Insulin released into the KRBH was determined by enzyme-linked immunosorbent assay and expressed as ng/106 cells/h. Expression of phogrin-myc, but not phogrin(C934S)-myc, reduced insulin secretion by about 90%. B, after collecting the supernatant, cells were washed and lysed, and insulin content was determined by enzyme-linked immunosorbent assay and normalized to cell number determined from parallel cultures. *, p < 0.05; **, p < 0.01 compared with untransfected.
DISCUSSION
The Cytoplasmic Domain of Phogrin Has PIPase Activity That Is Greatly Enhanced by Its Transmembrane Domain
Most well characterized PIPases are cytoplasmic or reversibly associated with membranes. When these cytoplasmic PIPases are assayed as recombinant proteins in vitro, they can display high specific activities against PIP substrates; e.g. 300–1000 mol/min/mol for hydrolysis of PI(3,4,5)P3 by PTEN (52, 53) and >4000 mol/min/mol for hydrolysis of PI(3)P by myotubularin (53). The activity that we measured for the phogrin catalytic domain GST fusion protein, ∼2 mol/min/mol against 200 μm diC8-PI(4,5)P2 (Table 1), was much lower than this, and comparable results may have discouraged other investigators from reporting or further investigating this activity. We reasoned that because endogenous phogrin is an integral membrane protein, an isolated catalytic domain-GST fusion protein may not display full enzymatic activity. This possibility is supported by evidence that the PIPase activities and biological activities of well characterized cytoplasmic PIPases are strongly dependent on membrane-interaction domains, including phospholipid binding pleckstrin homology and C2 domains (54–57). For example, the C2 domain of PTEN can bind phospholipid membranes in vitro and may serve to position the catalytic domain to cleave plasma membrane PIPs (55–57) or to bind PI(4,5)P2 as an allosteric activator (58). Mutations in the C2 domain of PTEN reduce its PIPase activity in vitro and its growth-suppressing activity in cells (57, 59). These membrane-interacting domains are almost always included in the assayed fusion proteins, but the most likely such domain in phogrin, the transmembrane domain, is absent from GST-phogrinCat. To begin to determine whether the transmembrane domain and/or other domains are important for full PIPase activity, we expressed a myc-His-tagged full-length phogrin or a catalytic site mutant as a negative control in INS-1 cells. Detergent extracts of phogrin-myc-transfected cells contained severalfold higher PIPase activity against PI(4,5)P2 than did extracts from untransfected cells (Fig. 2). This excess activity was proportional to the level of phogrin-myc expression (Fig. 2C) and was eliminated by immunodepletion by anti-phogrin antiserum (Fig. 2E). We partially purified phogrin-myc via binding of the myc-His tag to a nickel affinity column. The eluted phogrin-myc had a specific activity of 142 mol/min/mol, i.e. about 75-fold higher than the specific activity calculated for the GST-catalytic domain fusion protein. It is not likely that this PIPase activity reflects contaminating or co-precipitating cell PIPases, because parallel purification of the catalytic site mutant phogrin(C934S)-myc brought down much less PIPase activity (Fig. 3) and because the activity was inhibited by antibody against the catalytic domain of phogrin (Fig. 3).
A few other transmembrane PIPases have been identified. Like phogrin, their catalytic domains display relatively low PIPase activities when assayed as soluble catalytic domain fusion proteins despite evidence for biologically significant activity of the full-length protein in vivo. PTPRQ is a transmembrane PIPase that, like phogrin, is a single-pass transmembrane protein in the PTPase family that differs from canonical active PTPases at several sites in the catalytic domain (48). Best characterized of the transmembrane PIPases is a newly recognized group in which a CX5R PTPase-like domain is coupled to multiple putative transmembrane domains, similar to those present in voltage-dependent ion channels. The first member to be described in this way is Ci-VSP, a four-pass integral membrane protein from the tunicate Ciona intestinalis (52, 60, 61). A fusion protein of GST with the PIPase-like domain of Ci-VSP showed only modest in vitro activity (about 10–20 mol/min/mol) against PI(3,4,5)P3 (52, 60), but the full-length protein expressed in living cells was able to effectively lower plasma membrane PIP levels in response to membrane depolarization (61). Mammalian proteins have been identified that have the same domain structure as Ci-VSP, including TPIP (62), TPTE (62–64), and PTEN2 (65), but less is known about their PIPase activities.
Phogrin Is Active as a PIPase in Cells
The in vitro assay results with purified full-length phogrin indicated that phogrin has substantial PIPase activity that is not revealed when assayed with a catalytic domain fusion protein. The in vitro assays, however, are still non-physiological in many ways, including the use of PIPs with short-chain fatty acids (for solubility) and the absence of a lipid bilayer environment. Rather than attempt to recreate in vitro assay conditions that faithfully recreate conditions in vivo, we elected to pursue further evaluations in vivo where we could correlate effects on membrane PIPs with physiologic effects on insulin secretion. In control INS-1 cells we detected PI(4,5)P2 in the plasma membrane, consistent with published data that PI(4,5)P2 is most abundant in the plasma membrane and not detectable in secretory vesicles (66). We detected endogenous phogrin and phogrin-myc in the intracellular compartment, co-localizing with insulin, probably in DCV membranes as reported by other investigators (4, 6, 8, 10, 11). When we reduced levels of endogenous phogrin using siRNA, we observed increased PI(4,5)P2 levels (Fig. 5). This suggests that endogenous phogrin normally plays a significant role in maintaining cell PI(4,5)P2 levels. Expression of increasing levels of phogrin-myc reduced PI(4,5)P2 levels in the plasma membrane (Fig. 4 and supplemental Fig. S1C) with the highest phogrin-myc expression levels reducing PI(4,5)P2 by about 70%. Expression of phogrin(C934S)-myc increased PI(4,5)P2 staining in all experiments, although the increase was not statistically significant. Taylor et al. (53) reported that overexpression of the analogous catalytically inactive mutant of the PI(3)P 3-phosphatase myotubularin increased cell PI(3)P content in HEK293 cells and speculated that catalytically inactive forms of PIPases may bind and protect PIP substrates from hydrolysis by active PIPases. The ability of phogrin-myc, present largely in DCVs, to affect levels of PI(4,5)P2 in the plasma membrane could result from the transient exposure of phogrin-myc to plasma membrane PI(4,5)P2 during the membrane fusion events of exocytosis (67) or could reflect the presence of a small fraction of the phogrin-myc that is constitutively “mis-localized” to the plasma membrane. When we expressed phogrin-myc in HEK293 cells, which do not have DCVs to retain phogrin, the reduction of plasma membrane PI(4,5)P2 was greater than when phogrin-myc was expressed in INS-1 cells. This supports the hypothesis that phogrin is a PIPase whose activity against plasma membrane PIPs in INS-1 cells is limited, in part, by limited access to that pool.
Role of PIPs and Phogrin in Insulin Secretion
A large body of evidence suggests that PI(4,5)P2 plays a role in regulation of both exocytosis and endocytosis (27, 28, 33, 36, 37), including secretion of insulin from pancreatic beta cells (31, 38, 39). PI(4)P and PI(4,5)P2 are involved in “priming,” during which DCVs become competent for glucose-triggered exocytosis of peptide hormones (29, 30, 32, 37, 38). Olsen et al. (38) proposed that PI 4-kinase acts as a “metabolic sensor” for insulin release, such that elevated glucose decreases intracellular ADP levels, which relieves inhibition of PI 4-kinase, and increases synthesis of PI(4)P that is subsequently phosphorylated to PI(4,5)P2. Several PI(4,5)P2-binding proteins potentially involved in stages of vesicle exocytosis have been identified, including rabphilin (26), Ca2+-dependent activator protein for secretion (CAPS (35)), secretory carrier membrane protein 2 (SCAMP2 (34)), and synaptotagmins (25). Bai et al. (25) demonstrated that an increase in intracellular calcium alters binding of synaptotagmin to PI(4,5)P2 such that it penetrates into PI(4,5)P2-containing membranes. The gradient of PI(4,5)P2 (low in DCV and high in target plasma membrane), thus, creates a direction for vesicle fusion mediated by synaptotagmin in the DCV membrane.
Overall, the studies reported here demonstrate that phogrin has biologically significant activity as a PIPase and that this activity plays some role(s) in regulating PI(4,5)P2 levels and glucose-stimulated insulin secretion. We focused most of our attention on PI(4,5)P2, but recombinant phogrin is also able to hydrolyze other PIPs, and it is possible that these are also physiologically significant substrates in vivo. We suggest that the PIPase activity of endogenous phogrin could contribute to optimal function of the insulin secretion machinery by maintaining low levels of PI(4,5)P2 in DCVs and recycling endosomes, but further studies will be needed to clarify the exact site(s) of action and most important physiological substrate(s) of the PIPase activity of phogrin.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant HL69066 (to D. F. B.-P.) and Training Grant HL007312 (to L. A. C.). This work was also supported by a grant from the Juvenile Diabetes Research Foundation (to D. F. B.-P.) and by a Samuel and Althea Stroum Endowed Diabetes Fellowship and Ruth L. Kirschstein National Research Service Award DK081307 (to L. A. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- DCV
- dense-core vesicle
- PTPase
- protein-tyrosine phosphatase
- PIP
- phosphatidylinositol phosphate
- PI
- phosphatidylinositol
- PI(3)P
- PI 3-phosphate
- PI(4,5)P2
- PI 4,5-diphosphate
- PI(3,4,5)P3
- PI 3,4,5-trisphosphate
- GST
- glutathione S-transferase
- PIPase
- phosphatidylinositol phosphatase
- pNPP
- p-nitrophenyl phosphate
- Ni-NTA
- nickel-nitrilotriacetic acid
- FITC
- fluorescein isothiocyanate
- TRITC
- tetramethylrhodamine isothiocyanate
- siRNA
- small interfering RNA.
REFERENCES
- 1.Bonifacio E., Lampasona V., Bingley P. J. (1998) J. Immunol. 161, 2648–2654 [PubMed] [Google Scholar]
- 2.Lan M. S., Wasserfall C., Maclaren N. K., Notkins A. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6367–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J., Li Q., Xie H., Chen Z. J., Borovitskaya A. E., Maclaren N. K., Notkins A. L., Lan M. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2307–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dirkx R., Jr., Hermel J. M., Rabin D. U., Solimena M. (1998) Adv. Pharmacol. 42, 243–246 [DOI] [PubMed] [Google Scholar]
- 5.Lan M. S., Lu J., Goto Y., Notkins A. L. (1994) DNA Cell Biol. 13, 505–514 [DOI] [PubMed] [Google Scholar]
- 6.Solimena M., Dirkx R., Jr., Hermel J. M., Pleasic-Williams S., Shapiro J. A., Caron L., Rabin D. U. (1996) EMBO J. 15, 2102–2114 [PMC free article] [PubMed] [Google Scholar]
- 7.Xie H., Notkins A. L., Lan M. S. (1996) Cancer Res. 56, 2742–2744 [PubMed] [Google Scholar]
- 8.Wasmeier C., Hutton J. C. (1996) J. Biol. Chem. 271, 18161–18170 [DOI] [PubMed] [Google Scholar]
- 9.Torii S., Saito N., Kawano A., Zhao S., Izumi T., Takeuchi T. (2005) Traffic 6, 1213–1224 [DOI] [PubMed] [Google Scholar]
- 10.Wasmeier C., Bright N. A., Hutton J. C. (2002) Traffic 3, 654–665 [DOI] [PubMed] [Google Scholar]
- 11.Wasmeier C., Burgos P. V., Trudeau T., Davidson H. W., Hutton J. C. (2005) Traffic 6, 474–487 [DOI] [PubMed] [Google Scholar]
- 12.Henquin J. C., Nenquin M., Szollosi A., Kubosaki A., Notkins A. L. (2008) J. Endocrinol. 196, 573–581 [DOI] [PubMed] [Google Scholar]
- 13.Kubosaki A., Nakamura S., Notkins A. L. (2005) Diabetes 54, S46–S51 [DOI] [PubMed] [Google Scholar]
- 14.Kubosaki A., Gross S., Miura J., Saeki K., Zhu M., Nakamura S., Hendriks W., Notkins A. L. (2004) Diabetes 53, 1684–1691 [DOI] [PubMed] [Google Scholar]
- 15.Doi A., Shono T., Nishi M., Furuta H., Sasaki H., Nanjo K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torii S., Saito N., Kawano A., Hou N., Ueki K., Kulkarni R. N., Takeuchi T. (2009) Diabetes 58, 682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y., Nishimura T., Zhang A., Notkins A. L. (2009) Diabetes 58, e8. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald L. R., Walton K. M., Dixon J. E., Largent B. L. (1997) J. Neurochem. 68, 1820–1829 [DOI] [PubMed] [Google Scholar]
- 19.Lu J., Notkins A. L., Lan M. S. (1994) Biochem. Biophys. Res. Commun. 204, 930–936 [DOI] [PubMed] [Google Scholar]
- 20.Magistrelli G., Toma S., Isacchi A. (1996) Biochem. Biophys. Res. Commun. 227, 581–588 [DOI] [PubMed] [Google Scholar]
- 21.Jiang S., Tulloch A. G., Kim T. A., Fu Y., Rogers R., Gaskell A., White R. A., Avraham H., Avraham S. (1998) Gene 215, 345–359 [DOI] [PubMed] [Google Scholar]
- 22.Drake P. G., Peters G. H., Andersen H. S., Hendriks W., Møller N. P. (2003) Biochem. J. 373, 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y. F., Zhang H. L., Cai T., Harashima S., Notkins A. L. (2005) Diabetologia 48, 2576–2581 [DOI] [PubMed] [Google Scholar]
- 24.Mziaut H., Kersting S., Knoch K. P., Fan W. H., Trajkovski M., Erdmann K., Bergert H., Ehehalt F., Saeger H. D., Solimena M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai J., Tucker W. C., Chapman E. R. (2004) Nat. Struct. Mol. Biol. 11, 36–44 [DOI] [PubMed] [Google Scholar]
- 26.Chung S. H., Song W. J., Kim K., Bednarski J. J., Chen J., Prestwich G. D., Holz R. W. (1998) J. Biol. Chem. 273, 10240–10248 [DOI] [PubMed] [Google Scholar]
- 27.Cremona O., De Camilli P. (2001) J. Cell Sci. 114, 1041–1052 [DOI] [PubMed] [Google Scholar]
- 28.Di Paolo G., Moskowitz H. S., Gipson K., Wenk M. R., Voronov S., Obayashi M., Flavell R., Fitzsimonds R. M., Ryan T. A., De Camilli P. (2004) Nature 431, 415–422 [DOI] [PubMed] [Google Scholar]
- 29.Gong L. W., Di Paolo G., Diaz E., Cestra G., Diaz M. E., Lindau M., De Camilli P., Toomre D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5204–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grishanin R. N., Kowalchyk J. A., Klenchin V. A., Ann K., Earles C. A., Chapman E. R., Gerona R. R., Martin T. F. (2004) Neuron 43, 551–562 [DOI] [PubMed] [Google Scholar]
- 31.Gromada J., Bark C., Smidt K., Efanov A. M., Janson J., Mandic S. A., Webb D. L., Zhang W., Meister B., Jeromin A., Berggren P. O. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10303–10308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hay J. C., Fisette P. L., Jenkins G. H., Fukami K., Takenawa T., Anderson R. A., Martin T. F. (1995) Nature 374, 173–177 [DOI] [PubMed] [Google Scholar]
- 33.James D. J., Khodthong C., Kowalchyk J. A., Martin T. F. (2008) J. Cell Biol. 182, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao H., Ellena J., Liu L., Szabo G., Cafiso D., Castle D. (2007) Biochemistry 46, 10909–10920 [DOI] [PubMed] [Google Scholar]
- 35.Loyet K. M., Kowalchyk J. A., Chaudhary A., Chen J., Prestwich G. D., Martin T. F. (1998) J. Biol. Chem. 273, 8337–8343 [DOI] [PubMed] [Google Scholar]
- 36.Martin M. M., Victor X., Zhao X., McDougall J. K., Elton T. S. (2001) Mol. Cell. Endocrinol. 183, 81–91 [DOI] [PubMed] [Google Scholar]
- 37.Milosevic I., Sørensen J. B., Lang T., Krauss M., Nagy G., Haucke V., Jahn R., Neher E. (2005) J. Neurosci. 25, 2557–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen H. L., Hoy M., Zhang W., Bertorello A. M., Bokvist K., Capito K., Efanov A. M., Meister B., Thams P., Yang S. N., Rorsman P., Berggren P. O., Gromada J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waselle L., Gerona R. R., Vitale N., Martin T. F., Bader M. F., Regazzi R. (2005) Mol. Endocrinol. 19, 3097–3106 [DOI] [PubMed] [Google Scholar]
- 40.Malecz N., McCabe P. C., Spaargaren C., Qiu R., Chuang Y., Symons M. (2000) Curr. Biol. 10, 1383–1386 [DOI] [PubMed] [Google Scholar]
- 41.Kinoshita S., Su L., Amano M., Timmerman L. A., Kaneshima H., Nolan G. P. (1997) Immunity 6, 235–244 [DOI] [PubMed] [Google Scholar]
- 42.Taylor G. S., Dixon J. E. (2001) Anal. Biochem. 295, 122–126 [DOI] [PubMed] [Google Scholar]
- 43.Andersen J. N., Mortensen O. H., Peters G. H., Drake P. G., Iversen L. F., Olsen O. H., Jansen P. G., Andersen H. S., Tonks N. K., Møller N. P. (2001) Mol. Cell. Biol. 21, 7117–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chi Y., Zhou B., Wang W. Q., Chung S. K., Kwon Y. U., Ahn Y. H., Chang Y. T., Tsujishita Y., Hurley J. H., Zhang Z. Y. (2004) J. Biol. Chem. 279, 44987–44995 [DOI] [PubMed] [Google Scholar]
- 45.Guo S., Stolz L. E., Lemrow S. M., York J. D. (1999) J. Biol. Chem. 274, 12990–12995 [DOI] [PubMed] [Google Scholar]
- 46.Schmid A. C., Wise H. M., Mitchell C. A., Nussbaum R., Woscholski R. (2004) FEBS Lett. 576, 9–13 [DOI] [PubMed] [Google Scholar]
- 47.Maehama T., Dixon J. E. (1998) J. Biol. Chem. 273, 13375–13378 [DOI] [PubMed] [Google Scholar]
- 48.Oganesian A., Poot M., Daum G., Coats S. A., Wright M. B., Seifert R. A., Bowen-Pope D. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7563–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maehama T., Taylor G. S., Dixon J. E. (2001) Annu. Rev. Biochem. 70, 247–279 [DOI] [PubMed] [Google Scholar]
- 50.Whisstock J. C., Wiradjaja F., Waters J. E., Gurung R. (2002) IUBMB Life 53, 15–23 [DOI] [PubMed] [Google Scholar]
- 51.Watt S. A., Kular G., Fleming I. N., Downes C. P., Lucocq J. M. (2002) Biochem. J. 363, 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwasaki H., Murata Y., Kim Y., Hossain M. I., Worby C. A., Dixon J. E., McCormack T., Sasaki T., Okamura Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7970–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor G. S., Maehama T., Dixon J. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Begley M. J., Dixon J. E. (2005) Curr. Opin. Struct. Biol. 15, 614–620 [DOI] [PubMed] [Google Scholar]
- 55.Das S., Dixon J. E., Cho W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7491–7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Georgescu M. M., Kirsch K. H., Kaloudis P., Yang H., Pavletich N. P., Hanafusa H. (2000) Cancer Res. 60, 7033–7038 [PubMed] [Google Scholar]
- 57.Lee J. O., Yang H., Georgescu M. M., Di Cristofano A., Maehama T., Shi Y., Dixon J. E., Pandolfi P., Pavletich N. P. (1999) Cell 99, 323–334 [DOI] [PubMed] [Google Scholar]
- 58.Campbell R. B., Liu F., Ross A. H. (2003) J. Biol. Chem. 278, 33617–33620 [DOI] [PubMed] [Google Scholar]
- 59.Georgescu M. M., Kirsch K. H., Akagi T., Shishido T., Hanafusa H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10182–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murata Y., Iwasaki H., Sasaki M., Inaba K., Okamura Y. (2005) Nature 435, 1239–1243 [DOI] [PubMed] [Google Scholar]
- 61.Murata Y., Okamura Y. (2007) J. Physiol. 583, 875–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker S. M., Downes C. P., Leslie N. R. (2001) Biochem. J. 360, 277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guipponi M., Tapparel C., Jousson O., Scamuffa N., Mas C., Rossier C., Hutter P., Meda P., Lyle R., Reymond A., Antonarakis S. E. (2001) Hum. Genet 109, 569–575 [DOI] [PubMed] [Google Scholar]
- 64.Kumánovics A., Levin G., Blount P. (2002) FASEB J. 16, 1623–1629 [DOI] [PubMed] [Google Scholar]
- 65.Wu Y., Dowbenko D., Pisabarro M. T., Dillard-Telm L., Koeppen H., Lasky L. A. (2001) J. Biol. Chem. 276, 21745–21753 [DOI] [PubMed] [Google Scholar]
- 66.Holz R. W., Hlubek M. D., Sorensen S. D., Fisher S. K., Balla T., Ozaki S., Prestwich G. D., Stuenkel E. L., Bittner M. A. (2000) J. Biol. Chem. 275, 17878–17885 [DOI] [PubMed] [Google Scholar]
- 67.Taraska J. W., Almers W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wasmeier C., Hutton J. C. (1999) Biochem. J. 341, 563–569 [PMC free article] [PubMed] [Google Scholar]
- 69.Wasmeier C., Hutton J. C. (2001) J. Biol. Chem. 276, 31919–31928 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.