Abstract
Macroautophagy (autophagy) is a process wherein bulk cytosolic proteins and damaged organelles are sequestered and degraded via the lysosome. Alterations in autophagy-associated proteins have been shown to cause neural tube closure defects, neurodegeneration, and tumor formation. Normal lysosome function is critical for autophagy completion and when altered may lead to an accumulation of autophagic vacuoles (AVs) and caspase activation. The tumor suppressor p53 is highly expressed in neural precursor cells (NPCs) and has an important role in the regulation of both autophagy and apoptosis. We hypothesized that altered lysosome function would lead to NPC death via an interaction between autophagy- and apoptosis-associated proteins. To test our hypothesis, we utilized FGF2-expanded NPCs and the neural stem cell line, C17.2, in combination with the lysosomotropic agent chloroquine (CQ) and the vacuolar ATPase inhibitor bafilomycin A1 (Baf A1). Both CQ and Baf A1 caused concentration- and time-dependent AV accumulation, p53 phosphorylation, increased damage regulator autophagy modulator levels, caspase-3 activation, and cell death. Short hairpin RNA knockdown of Atg7, but not Beclin1, expression significantly inhibited CQ- and Baf A1-induced cell death, indicating that Atg7 is an upstream mediator of lysosome dysfunction-induced cell death. Cell death and/or caspase-3 activation was also attenuated by protein synthesis inhibition, p53 deficiency, or Bax deficiency, indicating involvement of the intrinsic apoptotic death pathway. In contrast to lysosome dysfunction, starvation-induced AV accumulation was inhibited by either Atg7 or Beclin1 knockdown, and Atg7 knockdown had no effect on starvation-induced death. These findings indicate that Atg7- and Beclin1-induced autophagy plays a cytoprotective role during starvation but that Atg7 has a unique pro-apoptotic function in response to lysosome dysfunction.
Keywords: Autophagy, Cell Death, Lysosomes, Neurodegeneration, Neuron, p53
Introduction
Neural stem cell death is critical for normal development of the nervous system (7, 8). Programmed cell death in the nervous system is classically defined as either apoptotic (type I) or autophagic (type II) depending on the ultrastructural features of the degenerating cell (9). Apoptosis is regulated by Bcl-2 family members, which have either pro- or anti-apoptotic properties depending in part on the number and types of Bcl-2 homology domains they possess. At base line, the pro-survival Bcl-2 family members such as Bcl-2 and Bcl-xL, negatively regulate the multi-Bcl-2 homology domain pro-apoptotic proteins Bax and Bak by direct interaction (10, 11). A subclass of Bcl-2 proteins called the Bcl-2 homology 3 domain-only proteins may be up-regulated in response to specific death stimuli and shift the balance between multidomain Bcl-2 family proteins leading to apoptosis (12–15).
Autophagy is a homeostatic process that is responsible for sequestering long-lived proteins and damaged organelles for subsequent degradation upon fusion with the lysosome (16). Alterations in basal autophagy may cause “autophagic stress,” where the number of AVs3 is increased over basal conditions (17). Autophagic stress can result from either increased synthesis or decreased lysosomal breakdown of AVs. Autophagic stress may lead to autophagic cell death, which is defined by the presence of large numbers of AVs in a cell concomitant with features of apoptosis and/or necrosis (18). Autophagy is regulated by a series of Atg proteins including Atg5 and Atg7. Neuron specific gene disruptions of atg7 and atg5 in mice result in defective autophagosome formation, increased neuronal ubiquitin aggregates, and neurodegeneration (19, 20). Beclin1, the mammalian homolog of atg6 and a Bcl-2 homology 3-only protein, is an important component in the initiation of the pre-autophagosome membrane. Beclin1 interacts with vacuolar protein sorting 34 (VPS34), a class III phosphoinositide 3-kinase (PI3K), and knockdown of its expression has been shown to inhibit AV accumulation and in some cases, inhibit cell death (21, 22). Beclin1-deficient mice die early in embryogenesis, whereas beclin1+/− embryonic stem cells display defects in autophagy (1). In addition, Ambra1-(activating molecule in Beclin1-regulated autophagy)-deficient mice have increased ubiquitinated proteins, excessive cell death, and defects in neural tube closure and autophagy (2), suggesting autophagy has a prominent role during nervous system development.
The tumor suppressor p53 is a potent inducer of NPC apoptosis (23, 24) and has recently been suggested to be an important regulator of autophagy. p53 has been implicated in both the suppression and activation of autophagy depending on the cell type and stimulus. p53-mediated gene transcription has been shown to induce AV synthesis, whereas cytoplasmic p53 can suppress AV formation (6). A newly discovered p53 target gene, DRAM, was found to be important for p53 effects on AV formation through both direct and indirect mechanisms (25).
Starvation is a potent stimulus for autophagy induction and efficient AV formation, and clearance may promote cell survival when nutrients are limited. In contrast, defective lysosome degradation of AV content can lead to massive AV accumulation and promote cell death. To explore the potential interaction between autophagic and apoptotic pathways in neural stem cells under different autophagic stress inducing conditions, we compared the response of neural stem cells to starvation versus lysosome “dysfunction” caused by either CQ or Baf A1. CQ, a weak base, localizes to acidic vesicles, leading to an increase in their intracellular pH and lysosomal dysfunction (10). Baf A1, a macrolide antibiotic, causes lysosomal dysfunction by inactivating lysosomal vacuolar ATPases with a resultant increase in lysosomal pH (26). We found that both starvation and lysosome dysfunction were able to increase Beclin1 and LC3-II protein levels and that AV accumulation was Atg7-dependent. In addition, starvation-induced, but not CQ-induced AV accumulation was Beclin1-dependent. Lysosomal dysfunction-induced NPC death occurred through an Atg7-, Bax-, p53-, and caspase-dependent pathway, whereas starvation-induced death was Atg7- and caspase-independent. In total, these studies indicate that lysosomal dysfunction may promote cell death in part via Atg7-dependent macroautophagy induction.
MATERIALS AND METHODS
Animals
C57BL/6J mice were used in all experiments. Mice were housed and cared for according to the National Institutes of Health Guide for the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. The generation of Bax-deficient mice have been described previously (27). p53+/− mice were purchased from Taconic (Germantown, NY). The genotypes for gene-disrupted mice were determined from tail DNA extracts by PCR.
Cell Culture
Fibroblast growth factor-2 (FGF2) has been previously demonstrated to stimulate NPC proliferation while maintaining their undifferentiated state. NPC cultures were obtained from the cerebellum of postnatal day 6–7 mice as previously published (28). Cells were incubated at 37 °C in humidified 5% CO2, 95% air atmosphere which allowed glia and post-mitotic neurons to adhere, whereas non-adherent cell populations floated in the supernatant. 24h later, non-adherent cells were transferred to a poly-l-lysine (Sigma) and laminin (BD Biosciences)-coated flask. Poly-l-lysine/laminin allows cerebellar NPCs to form an adherent monolayer. Media were replaced with fresh FGF2 every 3–4 days. Cells were plated once 75% confluent, and a small aliquot of cells was stained with trypan blue and counted. 30,000 cells were plated per well in a 48-well plate and were allowed to grow for 2–4 days in FGF2-containing media. CQ and Baf A1 were added to FGF2-containing media for experimentation.
C17.2 Cells
The C17.2 cell line was a generous gift from Dr. Evan Y. Snyder. Generation of these cells was described previously (29). C17.2 cells were cultured in high modified Dulbecco's modified Eagle's medium (Invitrogen) containing 1% penicillin/streptomycin, 1% l-glutamine (Sigma), 5% horse serum, and 10% fetal calf serum (Invitrogen). C17.2 cells were passaged every other day by trypsinization and the addition of a tenth of the cells to a new flask. Cells were plated on 48-well plates at a density of 15,000 cells per well. Cultures were incubated for 1 day before being used because C17.2 cells have a doubling rate of 24 h. CQ and Baf A1 were dissolved in 3% (horse and fetal calf serum) media to keep C17.2 cells in a proliferative state. For serum starvation, NPCs were washed twice with 1× phosphate-buffered saline (PBS) and kept in Dulbecco's modified Eagle's medium minus serum.
Cell Transfection and LAMP1-cherry Generation
C17.2 cells were transfected with Lipofectamine 2000 (Invitrogen) and DNA constructs containing LC3 fused to a green fluorescent protein (GFP) and a cherry-labeled lysosome-associated membrane protein 1 (LAMP1). The GFP-LC3 was a generous gift from Dr. Mizushima. The LAMP1fusion protein was generated by PCR amplification of lamp1 from a mouse brain cDNA library. The lamp1 product was inserted into the CherryN1 (Clontech) vector at EcoR1 and BamH1 restriction sites.
Cell Viability and Caspase Assays
Cell viability assays were performed as follows. Cells were washed in Locke's buffer (154 mm NaCl, 5.6 mm KCl, 3.6 mm NaHCO3, 2.3 mm CaCl2, 1.2 mm MgCl2, 5.6 mm glucose, 5 mm HEPES, pH 7.4), and 5 μm calcein AM (Molecular Probes, Eugene, OR) was diluted in Locke's buffer and incubated with the cells at 37 °C for 30 min. Calcein AM conversion was measured using a fluorescence plate reader (excitation 488 nm, emission 530 nm). Casapase-3-like enzymatic activity can be detected in vitro by cleavage assays on AMC-labeled DEVD. After treatment, cells were lysed in 100 μl of buffer A (10 mm HEPES, pH 7.4, 42 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, 0.5% CHAPS, 10% sucrose, 1 mm phenylmethylsulfonyl fluoride, and 1 μg/ml leupeptin) and 150 μl of buffer B (25 mm HEPES, pH 7.4, 1 mm EDTA, 0.1% CHAPS, 10% sucrose, and 3 mm dithiothreitol) containing 10 μm DEVD-AMC (Biomol, Plymouth Meeting, PA) followed by incubation at 37 °C for 30 min. Production of the fluorescent AMC caspase-3 product was measured with a fluorescent plate reader (excitation 360 nm, emission 460 nm). Both assays were expressed in comparison to untreated controls.
Immunocytochemistry
NPC cultures were mixed in 4% paraformaldehyde for 20 min at 4 °C followed by a PBS wash. Cells were permeabilized with PBS-blocking buffer (PBS-BB: PBS with 0.1% bovine serum albumin, 0.3% Triton X-100, and 0.2% nonfat powdered milk) for 30 min at room temperature. Primary antibodies were incubated overnight at 4 °C in PBS-BB without detergent. The primary antibodies used were rabbit anti-Atg5 (FL-275) (Santa Cruz, Santa Cruz, CA) and rabbit anti-microtubule-associated protein light chain-3 (2775), anti-rabbit cleaved caspase-3 (9661) (both from Cell Signaling Technologies, Danvers, MA), and anti-mouse LC3 (Nanotools). Plates were washed with 1× PBS three times, and secondary antibody was applied, either horseradish peroxidase-conjugated horse anti-rabbit SuperPicture (Zymed Laboratories Inc., South San Francisco, CA) or goat anti-mouse Vector Impress (Vector Laboratories, Burlingame, CA) in PBS-BB without Triton for 1 h at room temperature. Plates were washed with PBS, and immunostaining was detected using the Tyramide Signal Amplification system (PerkinElmer Life Sciences, Boston, MA). After PBS washes, cells were counterstained with bisbenzimide (2 μg/ml; Hoechst 33 258; Sigma). Plates were examined with a Zeiss Axioskop microscope equipped with epifluorescence. Images were captured using Axiovision® software.
Confocal Laser Scanning Microscopy
For dual labeling of LC3 and Atg5, immunocytochemical (IC) analysis was performed as described above. After Tyramide Signal Amplification detection of mouse anti-LC3 (0260/LC3-2G6, nano Tools), 0.3% H2O2 was added for 10 min to destroy any residual horseradish peroxidase activity followed by a PBS wash. Cells were then incubated for 30 min in PBS-BB containing Triton followed by diluted rabbit anti-Atg5 (FL-275, Santa Cruz) in PBS-BB without Triton overnight. The next day, goat anti-rabbit Vector Impress (Vector Laboratories) was applied for 1 h followed by Tyramide Signal Amplification detection (PerkinElmer Life Sciences). Chamber slides were stained with bisbenzimide, coverslipped, and imaged using a Leica Confocal TCS SP1 UV unit with a Coherent Laser Group Enterprise UV laser for blue fluorochromes, argon laser for green fluorochromes, and helium/neon laser for far red fluorochromes. Filter sets for fluorescein and Cy3 were used (Chroma Technology Corp., Brattleboro VT). Fluorochrome excitation and emission was controlled by using an accusto optical tunable filter (AOTF) and prism spectrophotometer. The 100× objective and Leica confocal software were used to acquire images from UT- and CQ-treated C17.2 cells.
Lysate Preparation
Preparation of whole cell lysates was performed as follows. Briefly, cells were suspended in lysis buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 10% glycerol) with 1% phenylmethylsulfonyl fluoride, 1% protease inhibitor mixture (Sigma), and 1% phosphatase inhibitor mixture protease and incubated on ice for 30 min with vortexing every 10 min. Samples were centrifuged at 13,000 × g for 15 min. The supernatant was transferred to a fresh tube, and the protein concentrations were determined via Pierce BCA assay kit (Pierce).
Western Blot
Equal amounts of whole cell lysates (25 μg) were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Blots were blocked for 1 h at room temperature, 5% milk in wash buffer (200 mm Tris base, 1.37 m NaCl, 1% Tween 20, pH 7.6) followed by overnight incubation with primary antibodies. Blots were probed for either p53 (2524), phosho-p53 (9284), and cleaved caspase-3 (9661) (Cell Signaling), DRAM (4033, ProSci, Poway, CA), and LC3 (AP-1802, Abgent, San Diego, CA) or Beclin (H-300, Santa Cruz), p62/SQSTM1 (Abnova, Jhongli, Taiwan), and Apg7/Atg7 (Abcam, Cambridge, MA) with β-tubulin (sc9104, Santa Cruz) serving as a loading control. After primary antibody incubation, all blots were washed with 1× Tris-buffered saline containing 0.1% Tween 20, then incubated with secondary antibody, either goat anti-rabbit IgG (Bio-Rad) or anti-mouse (Cell Signaling), for 1 h at room temperature and washed. Signal was detected using Supersignal (Pierce) or ECL (Amersham Biosciences) chemiluminescence. Western blots were scanned into Adobe Photoshop and digitized for quantification with the UN-SCAN-IT software, Version 6.1 (Orem, UT).
RNAi
Lentiviral shRNA (Atg7 and Beclin1) constructs were purchased from Open Biosystems (RMM4534_019584 and RMM4534_028835). shRNAs were cotransfected into 293FT cells together with packaging plasmids by following the manufacturer's protocol (Invitrogen ViraPowerTM Lentiviral Expression Systems kit, Carlsbad, CA). C17.2 cells were passaged and plated in a 6-well plate and allowed to adhere for 24 h before infection. C17.2 cells were subjected to lentiviral infection in the presence of Polybrene overnight, and the following day media were replaced by fresh media. After 24 h cells were selected by treating with media containing 1.5 μg/ml puromycin. Protein levels and experiments were assessed after 72 h.
Statistics
For experiments involving quantification, the S.E. was determined from at least three independent experiments, with an n of 1 representing one gene disrupted mouse accompanied by 1 wild-type littermate control. Effects of genotype were analyzed for significance using two-way ANOVA followed by Bonferroni's test for all pairwise comparisons. In all cases, a p value of ≤0.05 was considered significant.
RESULTS
Lysosome Dysfunction and Cell Starvation Induce an Increase in Autophagy-associated Proteins
To determine whether lysosomal dysfunction results in increased AV accumulation in NPCs, we monitored the appearance of LC3, the mammalian homolog to the yeast Atg8 gene, via Western blots, GFP-LC3 transfection, and IC analysis. During autophagic stress, the 16-kDa cytosolic form (LC3-I) is cleaved and post-translationally modified (LC3-II, 14 kDa) and translocated to the autophagosome. Whole cell lysates from C17.2 neural stem cells treated with CQ or Baf A1 were subjected to Western blot analysis to confirm AV accumulation. C17.2 cells treated with either CQ or Baf A1 demonstrated a time-dependent increase in LC3-II compared with untreated cells (Fig. 1A and data not shown). CQ treatment resulted in increased LC3-II levels as early as 6 h after addition and then continued to increase over a 24-h period. Inhibition of Beclin1 expression by siRNA knockdown or 3-methyladenine inhibition of VPS34 has been previously shown to inhibit AV production (1, 30). Western blot analysis revealed a significant increase in Beclin1 levels at 12 and 24 h after CQ treatment, suggesting AV induction (Fig. 1B).
FIGURE 1.
CQ, Baf A1, and starvation result in elevated autophagy-associated proteins. A, whole cell lysates from C17.2 cells were changed to fresh media with or without CQ for 6, 12, and 24 h followed by Western blot analysis. CQ exposure resulted in increased LC3-II levels as early as 6 h and continued to increase over time compared with UT cells. B, Beclin1 levels were assessed via Western blot analysis on C17.2 whole cell lysates that were treated with CQ as described above. CQ-treated cells at 24 h contained elevated Beclin1 protein levels compared with UT, 6-h, and 12-h lysates. Western blots were digitized by UN-SCAN-IT software, averages for LC3-II/β-tubulin and Beclin1/β-tubulin pixel totals were calculated, and the data points were expressed in -fold change. Data points in A and B represent the mean ± S.E., with n = 4. *, p < 0.01 by one-way ANOVA/Bonferroni's post hoc test versus untreated controls. C, C17.2 NPCs were co-transfected with LC3-GFP and LAMP1-cherry and treated for 6 h with lysosomal dysfunction-inducing agents CQ or Baf A1 or subjected to serum starvation for 3 h. Autophagic stress leads to the accumulation of autophagosomes as evidenced by the presence of intense, punctate GFP-LC3 in NPCs treated with CQ, Baf A1, or starvation (Stv) compared with the weak, diffuse GFP-LC3 signal observed in UT cells. CQ and starvation resulted in increased dual LC3-GFP and LAMP1-cherry co-labeled autophagolysosomes (white arrowheads) in contrast to Baf A1 and untreated NPCs. Scale bar, 20 μm.
LC3-GFP and LC3 immunoreactivity (IR) appears diffuse under normal conditions but during autophagic stress becomes clumped or punctate, suggesting an increase in the autophagosome-associated LC3-II form (31). For autophagy completion to occur, AVs must first fuse with the lysosome to form an autophagolysosome before degradation of AV content. To detect AV and lysosome co-localization after experimental treatment, suggesting the accumulation of autophagolysosomes, C17.2 cells were co-transfected with GFP-LC3 and LAMP1-cherry labeled fusion proteins. GFP-LC3 was diffuse in appearance in untreated cells, whereas LAMP1-cherry, a lysosome marker, appeared punctate. CQ concentrates in lysosomes and inhibits the degradation of AVs that have reached the lysosome, whereas Baf A1 has been reported to prevent the fusion of AVs with the lysosome. CQ, Baf A1, and starvation exposure resulted in increased punctate GFP-LC3 in C17.2 neural stem cells compared with untreated cells (Fig. 1C). CQ treatment and starvation resulted in increased co-localization of LC3 and LAMP1 fluorescence, suggesting increased numbers of autophagolysosomes. In contrast, the addition of Baf A1 resulted in minimal co-localization of LC3 and LAMP1 fluorescence, suggesting a failure to form autophagolysosomes (Fig. 1C).
Lysosome Dysfunction and Cell Starvation Lead to Increased Caspase-3-like Activity and NPC Death
To delineate the pathways associated with lysosomal dysfunction and starvation-induced cell death in NPCs, NPCs were exposed to CQ, Baf A1, or serum starvation, and cell viability was assessed. CQ, Baf A1, and starvation all produced a time-dependent decrease in cell viability (Fig. 2, A and B).
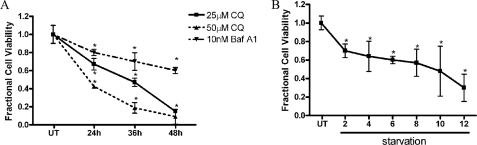
FIGURE 2.
Lysosomal dysfunction and serum starvation cause decreased cell viability. A, C17.2 cells were treated with 25 and 50 μm CQ or 10 nm Baf A1 in 3% fetal bovine serum for 12, 24, 36, and 48 h. Viability was assessed by measuring the fluorescence produced from the cleavage of calcein AM by intracellular esterases. CQ and Baf A1 caused a time-dependent decrease in cell viability. B, C17.2 cells exposed to Dulbecco's modified Eagle's medium minus fetal bovine serum showed decreased cell viability as early as 2 h after serum withdraw, and viability progressively declined over time. Data points for A and B represent the mean ± S.E., with n = 6. *, p < 0.01 by one-way ANOVA/Bonferroni's post hoc test versus untreated controls.
Lysosomal dysfunction has recently been shown to cause apoptosis in neuronal and non-neuronal cell types (10, 26); therefore, IC analysis was performed to determine whether CQ, Baf A1, and starvation induced caspase activation in NPCs. C17.2 cells were treated with CQ or Baf A1 (12 h) or starved (3 h), and cleaved caspase-3 IR was assessed. C17.2 cells exhibited increased cleaved caspase-3 IR after treatment with lysosomal dysfunction-inducing agents and starvation compared with control cells (Fig. 3A). A DEVD-AMC cleavage assay for caspase-3-like enzymatic activity revealed an increase in caspase-3-like activity after CQ, Baf A1, and starvation, which was confirmed by Western blot analysis of cleaved caspase-3 levels (Fig. 3B and data not shown). To determine whether cell death was caspase-dependent, we treated C17.2 neural stem cells with CQ, Baf A1, or starvation alone and in combination with a broad caspase inhibitor, t-butoxycarbonyl-aspartyl-(OMe)-fluoromethyl ketone. Our results revealed that broad caspase inhibition significantly attenuated CQ but not starvation-induced cell death (Fig. 3C).
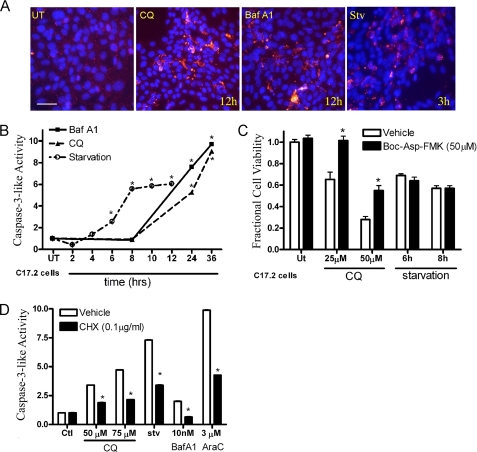
FIGURE 3.
CQ- and Baf A1- but not starvation-induced death is partially caspase-dependent. A, lysosomal dysfunction and starvation led to increased cleaved caspase-3 IR (red) in NPCs treated with CQ, Baf A1, and starvation in comparison to UT controls. B, C17.2 cells were treated as above and then tested for caspase-3-like enzymatic activity by measuring fluorescence produced by DEVD-AMC cleavage. CQ (25 μm), Baf A1 (10 nm), and starvation caused a time-dependent increase in caspase-3-like activity. *, p < 0.01 by one-way ANOVA/Bonferroni's post hoc test versus untreated controls. C, C17.2 cells were treated with CQ (25 or 50 μm for 24 h) or starvation (6 and 8 h) alone or in the presence of a broad caspase inhibitor (t-butoxycarbonyl-Asp-fluoromethyl ketone (Boc-Asp-FMK)). Broad caspase inhibition significantly attenuated CQ-induced NPC death but not starvation-induced NPC death. *, p < 0.01 by two-way ANOVA/Bonferroni post-test for CQ-treated versus CQ in the presence of t-butoxycarbonyl-Asp-fluoromethyl ketone. D, protein synthesis inhibition significantly attenuated caspase-3-like activation after 12 h of CQ, Baf A1, starvation, or AraC exposure. *, p < 0.01 by two-way ANOVA/Bonferroni post-test treated versus treated in the presence of cycloheximide (CHX).
Previous studies from our laboratory have shown that caspase-3 activation in NPCs can occur through either transcription-dependent or -independent pathways depending on the death stimulus (3, 4). To test whether lysosome dysfunction- or starvation-induced caspase-3 activation requires new protein synthesis, we treated C17.2 neural stem cells with CQ, Baf A1, or starvation alone or in the presence of the protein synthesis inhibitor cycloheximide for 12 h. AraC was used as a positive control because it induces caspase-3 in a protein synthesis-dependent fashion (12). We measured the in vitro enzymatic cleavage of DEVD-AMC, which indicated lysosomal dysfunction-, starvation-, and AraC-induced caspase-3-like activity were all attenuated by inhibiting new protein synthesis (Fig. 3D).
Lysosomal Dysfunction-induced NPC Death Requires Bax
Autophagic cell death may display markers of both apoptotic and necrotic cell death (32). CQ- and starvation-induced NPC death of immature neurons have previously been shown to be Bax-dependent (4, 10). To determine whether lysosomal dysfunction or starvation activate the intrinsic apoptotic death pathway, we treated wild-type and Bax-deficient NPCs with CQ (25 μm) or Baf A1 (5 nm) for 24 h or starvation (12 h) and assessed cell death. Bax deficiency significantly attenuated lysosomal dysfunction- and starvation-induced NPC death compared with wild-type treated NPCs (Fig. 4A). Furthermore, Bax deficiency significantly attenuated CQ and Baf A1 caspase-3-like activity, which was confirmed by Western blot analysis (Fig. 4B).
FIGURE 4.
Lysosomal dysfunction-induced NPC death and caspase activation is Bax-dependent. A, FGF2-expanded NPCs from wild-type and Bax-deficient mice were treated with CQ or Baf A1 for 24 h or starvation for 12 h. CQ-, Baf A1-, and starvation-induced death was significantly attenuated in Bax-deficient compared with wild-type NPCs (*, p < 0.01 by one-way ANOVA/Bonferroni's post hoc test versus untreated controls or **, p < 0.01, by two-way ANOVA/Bonferroni post hoc test of treated Bax-deficient versus treated wild-type groups). B, wild-type and Bax-deficient NPCs were treated with standard media (UT) or 25 μm CQ for 24 h. Bax-deficient NPCs exposed to CQ revealed a significant decrease in cleaved caspase-3 compared with wild-type NPCs. The data represent mean ± S.E., with n = 5. *, p < 0.01 by two-way ANOVA/Bonferroni's post hoc test Bax-deficient versus the wild-type treated group. Stv, starvation.
p53 Regulates Lysosomal Dysfunction-induced Caspase Activation and NPC Death
p53 is an important cell death regulator of genotoxic- and staurosporine-induced NPC death (12, 28). To determine whether CQ caused p53 stabilization, Western blot analysis was performed to assess phosphorylation of p53 at serine 15, an indicator of p53 activation, and total p53 levels. p53 phosphorylation was significantly increased at 12 and 24 h compared with starvation and untreated cells (Fig. 5A and data not shown). Total p53 levels remained unchanged relative to untreated cells (data not shown). To test whether p53 regulates autophagic stress-induced NPC death, we measured the viability of wild-type and p53-deficient NPCs after CQ, Baf A1, or AraC exposure. p53-deficient NPCs showed significant protection from CQ-, Baf A1-, and AraC-induced death compared with wild-type NPCs (Fig. 5B). The CQ- and Baf A1-induced increase in caspase-3-like activity was attenuated in p53-deficient NPCs relative to wild-type treated NPCs (Fig. 5C). p53-deficient NPCs were not protected from serum starvation-induced caspase-3 activation or death (Fig. 5).
FIGURE 5.
p53 plays a role in lysosomal dysfunction-induced NPC apoptosis. A, whole cell lysates from C17.2 cells in normal media (UT) or subjected to CQ for 6, 12, and 24 h were prepared. Immunoblot detection revealed a significant increase in p53 phosphorylation at serine 15 at 12 and 24 h compared with UT cells. There was no change detected in total p53 levels after CQ exposure (not shown), and β-tubulin was used as a loading control. B, wild-type and p53-deficient NPCs were treated with CQ, Baf A1, and AraC for 24 h, and cell viability was assessed. p53 deficiency significantly attenuated CQ-, Baf A1-, and AraC-induced NPC death (*, p < 0.01 by two-way ANOVA/Bonferroni's post hoc test, treated wild-type versus p53−/−). C, whole cell lysates were prepared from wild-type and p53-deficient NPCs treated with 25 μm CQ or control media for 24 h. CQ treatment results in increased levels of cleaved caspase-3, which is attenuated in p53-deficient versus wild-type NPCs. Western blots for A and C were digitized by UN-SCAN-IT software, and data represent the mean ± S.E., with n = 3. *, p < 0.01 by two-way ANOVA/Bonferroni's post hoc test, wild-type-treated versus the p53-deficient treated group. D, Western blot analysis showed similar LC3-II levels on whole cell lysates from both wild-type (WT) and p53-deficient NPCs treated with CQ for 24 h. *, p < 0.01 by one-way ANOVA/Bonferroni's post hoc test versus untreated wild type. ko, knock-out.
Recent studies have identified p53 as a potential inducer of AV accumulation; therefore, we tested whether p53 deficiency would attenuate LC3-II levels after CQ exposure via Western blot analysis. Our results revealed p53 deficiency had no effect on LC3-I or LC3-II levels after CQ exposure (Fig. 5D).
CQ Induces p53-dependent Up-regulation of DRAM
To better understand the role of p53 in the response to lysosomal dysfunction, we measured the levels of the p53 target gene DRAM, a potential mediator of p53-induced cell death. Western blot analysis was performed on lysates from control NPCs and NPCs treated with CQ (25 μm). CQ exposure resulted in a 4-fold increase in DRAM protein levels compared with untreated NPCs (Fig. 6A). p53-deficient NPCs exhibited significantly decreased DRAM levels after CQ treatment compared with wild-type NPCs (Fig. 6A), indicating that DRAM induction was p53-dependent. We detected the monomeric (16 kDa) and dimeric (32 kDa) forms of DRAM in NPCs in contrast to the C17.2 cells, which only exhibited the monomeric form. CQ also induced significant and temporal increases in DRAM protein levels in C17.2 cells (Fig. 6B).
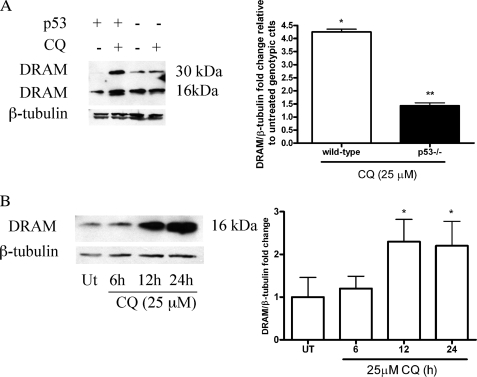
FIGURE 6.
p53-dependent up-regulation of DRAM in response to CQ. A, whole cell lysates were processed from wild-type and p53-deficient NPCs maintained in media with or without 25 μm CQ for 24 h. CQ caused an increase in both the DRAM dimer (32 kDa) and monomer (16 kDa) compared with untreated controls. Western blots were digitized by UN-SCAN-IT software, and data represent the mean ± S.E., with n = 3; *, p < 0.01 by one-way ANOVA/Bonferroni's post hoc test control versus the treated group. DRAM levels were significantly attenuated in p53-deficient NPCs compared with wild-type CQ-treated group. **, p < 0.01 by two-way ANOVA/Bonferroni post-test compared with the wild-type-treated group and the knock-out-treated group. ctl, control. B, C17.2 cells were treated with CQ for 6, 12, and 24 h, and DRAM protein levels were assessed via Western blot. DRAM protein levels were significantly increased at both 12 and 24 h after CQ exposure. Western blots were digitized as described above, and data represent the mean ± S.E., with n = 3. *, p < 0.01 by one-way ANOVA/Bonferroni's post hoc test control versus the treated group.
Atg7 Plays an Important Role in Lysosome Dysfunction-induced AV Accumulation and NPC Death
To test the hypothesis that Atg7 or Beclin1 is critical for CQ and starvation-induced AV accumulation and caspase-3 cleavage, lentiviruses containing Atg7 or Beclin1 shRNAs were generated. C17.2 cells were infected with lentiviruses containing either the control vector PLKO, Atg7 shRNA, or shRNA for Beclin1 followed by puromycin selection for 48 h. Atg7 lentivirus was generated, and Western blot analysis was performed to confirm Atg7 knockdown compared with PLKO controls (Fig. 7A). Atg7 knockdown significantly reduced CQ- and Baf A1-induced AV accumulation (Fig. 7, A and B, and data not shown). In addition, Atg7 knockdown and CQ resulted in increased p62 levels, a protein that has been shown to be mediated by autophagy (Fig. 7A) (33). Furthermore, Atg7 significantly attenuated phosphorylation of p53 and cleavage of caspase-3, suggesting p53-induced apoptosis occurs downstream of Atg7 and autophagic stress (Fig. 7, A, C, and D).
FIGURE 7.
Atg7 knockdown attenuates AV accumulation and molecular markers for apoptosis. A, C17.2 cells were infected with lentiviruses containing PLKO (control (Ctl)) empty vector or Atg7 shRNA followed by puromycin selection for 72 h. Control and Atg7 shRNA-transformed cells were treated with CQ followed by Western blot analysis of phosphorylated p53 (serine 15), p62, Atg7, LC3-II, and cleaved caspase-3. B, C, and D, LC3-II, cleaved caspase-3, and phospho-p53 levels were significantly decreased in Atg7 knockdowns in contrast to PLKO controls. Western blots were digitized by UN-SCAN-IT software, LC3-II, cleaved caspase-3, and phospho-p53/β-tubulin pixel totals were determined, and values were expressed as -fold change. The data represent the mean ± S.E., with n = 3. *, p < 0.01 by two-way ANOVA/Bonferroni post-test versus the untreated control. **, p < 0.01 by two-way ANOVA/Bonferroni post test, Atg7 shRNA-treated versus controls.
The Beclin1 and Class III PI3K Interaction Has Been Shown Previously to Induce the Formation of the Developing Autophagosome (1, 22, 34). To test the hypothesis that Beclin1 is critical for CQ and starvation-induced AV accumulation, lentivirus containing Beclin1 shRNA was generated. C17.2 cells containing Beclin1 knockdown were treated with CQ (24 h) followed by Western blot analysis. In contrast to Atg7 knockdown, our results revealed Beclin1 knockdown did not attenuate CQ-induced LC3-II accumulation. To detect whether Beclin1 or Atg7 was critical for starvation-induced LC3-II accumulation, Atg7 or Beclin1 knockdown cells were subjected to starvation and assessed for LC3-II accumulation in comparison to control (serum-enriched) treatment. Both Beclin1 and Atg7 knockdown attenuated starvation-induced LC3-II accumulation compared with control treated cells (Fig. 8, A and B).
FIGURE 8.
Role of Atg7 and Beclin1 in starvation- and CQ-induced LC3-II accumulation and NPC death. A, C17.2 cells were infected with lentiviruses containing PLKO (control (Ctl)) empty vector or shRNAs for Beclin1 or Atg7 followed by puromycin selection for 72 h. Control, Beclin1, and Atg7 shRNA- transformed cells were subjected to starvation for 3 h. Starvation-induced LC3-II accumulation was attenuated by Beclin1 and Atg7 knockdown compared with controls. Control and Beclin1 knockdowns were also subjected to CQ treatment for 24 h. Ctl and Beclin1 knockdowns exhibited robust LC3-II levels after 24 h of CQ treatment. B, Western blots were digitized by UN-SCAN-IT software, LC3-II/β-tubulin pixel totals were determined, and values were expressed as -fold change. The data represent the mean ± S.E., with n = 4. *, p < 0.05 by two-way ANOVA/Bonferroni's post hoc test versus untreated controls. **, p < 0.01 by two-way ANOVA/Bonferroni post-test compared with the control-treated group. C, control cells or Atg7 knockdowns were treated with CQ (25 μm), Baf A1 (10 nm) for 24 h, or starvation (STV) for 12 h, and viability was assessed. Atg7 knockdown cells were protected against lysosomal dysfunction-induced NPC death compared with treated control cells. Beclin1 knockdowns and control cells subjected to 25 μm CQ and 10 nm Baf A1 for 24 h contained similar amounts of cell death. Control, Beclin1, and Atg7 knockdown cells all contained similar amounts of cell death after starvation. The data represent the mean ± S.E., with n = 8. *, p < 0.05 by two-way ANOVA/Bonferroni's post hoc test versus untreated controls. **, p < 0.01 by two-way ANOVA/Bonferroni post-test compared with the control-treated group.
Induction of autophagy has been proven to be survival-promoting under starvation conditions (35). To determine whether Atg7 or Beclin1 regulates lysosomal dysfunction- or starvation-induced death, C17.2 control cells or cells containing Atg7 or Beclin1 shRNAs were subjected to CQ (25 μm) or Baf A1 (10 nm) for 24 h, or 12 h starvation and viability was assessed. Knockdown of Atg7 but not Beclin1 attenuated lysosomal dysfunction-induced death compared with control-treated cells (Fig. 8C). However, neither Atg7 nor Beclin1 knockdown inhibited starvation-induced NPC death (Fig. 8C). These results suggest a unique pro-apoptotic role for Atg7 in lysosomal dysfunction-induced death.
DISCUSSION
In this report we investigated the role of apoptosis and autophagy-associated proteins in regulating lysosomal dysfunction-and starvation-induced NPC death. We showed that treatment of NPCs with lysosomal dysfunction-inducing agents or starvation resulted in the accumulation of AVs followed by caspase activation. In addition, we found that broad caspase or new protein synthesis inhibition attenuated caspase activation and/or death after lysosomal dysfunction. Lysosome dysfunction-induced autophagic stress led to an Atg7-, p53-, Bax-, and caspase-dependent apoptotic NPC death. Our data further revealed Atg7-dependent but Beclin1-independent accumulation of AVs after lysosome dysfunction with subsequent p53 phosphorylation, caspase activation, and NPC death. Atg7 knockdown attenuated starvation-induced AV accumulation but had no effect on starvation-induced cell death. Our results reveal a novel death-promoting effect of Atg7 in response to lysosome dysfunction.
Relative to other cell types, neural cells are more susceptible to nutrient stress, as their energy consumption needs rely heavily on oxidative metabolism. Therefore, neuronal starvation models have proved useful in defining the interaction between autophagic and apoptotic pathways. It is well established that PI3K signaling, Beclin1, Atg5, and Atg7 are important for AV formation in many cell types after serum or growth factor deprivation (34, 36). However, the role of these Atg proteins in the regulation of neural cell death is not fully understood. It has been shown that nutrient starvation increases protein levels of Bcl-2-associated death protein levels, which resulted in decreased interaction between Beclin1 and Bcl-2 and the promotion of both apoptosis and autophagy (37). Another study showed 3-methyladenine was capable of inhibiting serum deprivation-induced neuronal cell death, suggesting that PI3K may play a role in initiating caspase-3 activation and apoptotic cell death (38). However, PI3K inhibitors may affect both the class I and class III PI3K, which are involved in many biological processes (39). Therefore, we generated knockdowns for Atg7 or Beclin1, which attenuated starvation-induced AV accumulation but not starvation-induced death, suggesting PI3K inhibitors may inhibit starvation-induced cell death via an alternative mechanism.
Based on our findings and others, we conclude that autophagy and Atg proteins are typically survival-promoting during serum starvation (40). However, autophagy may prove to be detrimental to a cell that has altered lysosome function. For example, mice deficient in cathepsin D, an essential lysosomal protease, contain massive numbers of AVs and display increased neuronal death due to altered lysosome function (41). This neuronal phenotype is identical to that observed in human neuronal ceroid lipofuscinosis patients (Batten disease) (42). Our laboratory recently generated dual cathepsin D /Bax-deficient mice and found that Bax deficiency was capable of attenuating cleaved caspase-3 but not AV accumulation and neurodegeneration in cathepsin D-deficient brain (42). In addition, a npc1 gene-disrupted mouse model for Niemann-Pick Type C disease showed neurodegeneration, increased numbers of AVs, and increased levels of cathepsin D and cathepsin B in the brain, indicating that cholesterol accumulation results in altered lysosome function and neuronal cell death (43). These studies support our findings that lysosomal dysfunction results in reduced turnover of AVs leading to autophagic stress-induced neuronal death, an effect that can be attenuated by blocking AV synthesis.
Neonatal hypoxia ischemia (HI) has also been shown to cause an increase in neuronal AVs followed by increased markers for apoptosis (44). HI has been shown to cause both caspase-dependent and -independent neuronal cell death, and Koike et al. (45) showed central nervous system-specific Atg7-deficient mice subjected to neonatal HI were resistant to HI-induced caspase activation and pyramidal neuron death. Another recent study showed that ischemia leads to lysosome membrane permeabilization and cathepsin release followed by pyramidal neuron death (46). These results together with our findings suggest that HI may compromise normal lysosome function, thus, preventing autophagy completion resulting in caspase activation and cell death.
Investigating interactions between autophagic and apoptotic pathways has identified cross-talk between these pathways (47). Recent investigations have linked autophagy-associated proteins to apoptotic cell death. Atg5 cleavage was shown to promote cell death by translocating to the mitochondria where it interacts with Bcl-xL to trigger cytochrome c release (47). Similarly, the Beclin1/Bcl-2 or Bcl-xL interaction has been shown to be disrupted during starvation conditions (48, 49). However, this interaction may also interrupt the rheostat between Bcl-2 and Bax, thereby promoting apoptosis (50). In addition, endoplasmic reticulum stress induced by tunicamycin has been shown to result in an Atg5- and Beclin1-dependent cell death when combined with caspase-3 depletion (51). Ectopic Beclin1 expression may be survival-promoting, whereas Beclin1 knockdown sensitizes cells to apoptosis, indicating that Beclin1 plays an important role in regulating both autophagy and apoptosis.
Here we utilized potent inducers of autophagic stress, starvation, and lysosomal dysfunction-inducing agents to study cell death responses in NPCs. We found that Atg7, but not Beclin1, was important for lysosomal dysfunction-induced AV accumulation, p53 phosphorylation, caspase activation, and neural death. Atg7 is an important E1-activating enzyme that helps facilitate post-translational modifications in the ubiquitination-like process that is essential for autophagosome formation. Our results reveal Atg7 knockdown is important for lysosome dysfunction-induced phosphorylation of p53, suggesting Atg7 may have alternative functions and substrates outside of the autophagic signaling pathway. In addition, these studies show Atg7 may promote cell death when normal lysosome function is compromised.
We found that caspase-3 activation and NPC death after CQ or Baf A1 was attenuated by protein synthesis inhibition or p53 deficiency and that p53 up-regulates DRAM in response to autophagic stress. The neuroprotective effect of p53 deficiency in NPCs is interesting considering the recent investigations implicating p53 in autophagic cell death (52). Although in other systems p53 has been reported to regulate autophagy induction, it is noteworthy that p53-deficient NPCs showed robust LC3 IR and LC3-II accumulation on Western blots after CQ and starvation exposure, similar to what was observed in wild-type NPCs, suggesting that AV accumulation occurs upstream, not downstream, of p53. In addition, Atg7 knockdown attenuated phosphorylation of p53, suggesting autophagic stress triggers p53-dependent apoptosis.
The present study examined the role of autophagy-associated proteins in the NPC response to serum starvation- and lysosomal dysfunction-inducing agents. Our results indicate that both starvation and lysosomal dysfunction cause AV accumulation but that cell death is differentially regulated by these two stimuli. Unlike for starvation-induced cell death, Atg7 knockdown provided significant protection from lysosome dysfunction-induced caspase-3 activation and cell death, indicating a pro-apoptotic function for Atg7 under this condition. Thus, induction of autophagy in cells with diminished lysosomal capacity may convert this normally cytoprotective response into a potent death stimulus.
Acknowledgments
We thank Dr. Evan Snyder (Burnham Institute) for generously providing the C17.2 cell line and Cecelia B. Latham, the University of Alabama at Birmingham Neuroscience Core Facilities (National Institutes of Health Grants NS47466 and NS57098), Albert Tousson, Shawn Williams, and the University of Alabama at Birmingham Imaging Facility for technical assistance.
This work was supported by National Institutes of Health Grants NS35107, NS41962, and CA134773.
- AV
- autophagic vacuole
- NPC
- neural precursor cell
- CQ
- chloroquine
- PI3K
- phosphoinositide 3-kinase
- DRAM
- damage regulator autophagy modulator
- FGF2
- fibroblast growth factor-2
- GFP
- green fluorescent protein
- LAMP1
- lysosome-associated membrane protein 1
- PBS
- phosphate-buffered saline
- PBS-BB
- PBS-blocking buffer
- IC
- immunocytochemical
- UT
- untreated
- IR
- immunoreactivity
- HI
- hypoxia ischemia
- AMC
- amidomethylcoumarin
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- shRNA
- short hairpin RNA
- ANOVA
- analysis of variance.
REFERENCES
- 1.Yue Z., Jin S., Yang C., Levine A. J., Heintz N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fimia G. M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., Gruss P., Piacentini M., Chowdhury K., Cecconi F. (2007) Nature 447, 1121–1125 [DOI] [PubMed] [Google Scholar]
- 3.Walls K. C., Klocke B. J., Saftig P., Shibata M., Uchiyama Y., Roth K. A., Shacka J. J. (2007) Autophagy 3, 222–229 [DOI] [PubMed] [Google Scholar]
- 4.Shacka J. J., Klocke B. J., Shibata M., Uchiyama Y., Datta G., Schmidt R. E., Roth K. A. (2006) Mol. Pharmacol. 69, 1125–1136 [DOI] [PubMed] [Google Scholar]
- 5.Deleted in proof
- 6.Tasdemir E., Maiuri M. C., Galluzzi L., Vitale I., Djavaheri-Mergny M., D'Amelio M., Criollo A., Morselli E., Zhu C., Harper F., Nannmark U., Samara C., Pinton P., Vicencio J. M., Carnuccio R., Moll U. M., Madeo F., Paterlini-Brechot P., Rizzuto R., Szabadkai G., Pierron G., Blomgren K., Tavernarakis N., Codogno P., Cecconi F., Kroemer G. (2008) Nat. Cell Biol. 10, 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuida K., Zheng T. S., Na S., Kuan C., Yang D., Karasuyama H., Rakic P., Flavell R. A. (1996) Nature 384, 368–372 [DOI] [PubMed] [Google Scholar]
- 8.Kuida K., Haydar T. F., Kuan C. Y., Gu Y., Taya C., Karasuyama H., Su M. S., Rakic P., Flavell R. A. (1998) Cell 94, 325–337 [DOI] [PubMed] [Google Scholar]
- 9.Clarke P. G. (2002) Trends Pharmacol. Sci. 23, 308–309 [DOI] [PubMed] [Google Scholar]
- 10.Zaidi A. U., McDonough J. S., Klocke B. J., Latham C. B., Korsmeyer S. J., Flavell R. A., Schmidt R. E., Roth K. A. (2001) J. Neuropathol. Exp. Neurol. 60, 937–945 [DOI] [PubMed] [Google Scholar]
- 11.Motoyama N., Wang F., Roth K. A., Sawa H., Nakayama K., Nakayama K., Negishi I., Senju S., Zhang Q., Fujii S. (1995) Science 267, 1506–1510 [DOI] [PubMed] [Google Scholar]
- 12.Akhtar R. S., Geng Y., Klocke B. J., Roth K. A. (2006) Cell Death. Differ. 13, 1727–1739 [DOI] [PubMed] [Google Scholar]
- 13.Shacka J. J., Roth K. A. (2006) Cell Death. Differ. 13, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 14.Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 15.Noguchi K. K., Walls K. C., Wozniak D. F., Olney J. W., Roth K. A., Farber N. B. (2008) Cell Death Differ. 15, 1582–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Codogno P., Meijer A. J. (2006) Nat. Cell Biol. 8, 1045–1047 [DOI] [PubMed] [Google Scholar]
- 17.Chu C. T. (2006) J. Neuropathol. Exp. Neurol. 65, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shacka J. J., Klocke B. J., Roth K. A. (2006) Autophagy. 2, 228–230 [DOI] [PubMed] [Google Scholar]
- 19.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 20.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 21.Høyer-Hansen M., Bastholm L., Mathiasen I. S., Elling F., Jäättelä M. (2005) Cell Death. Differ. 12, 1297–1309 [DOI] [PubMed] [Google Scholar]
- 22.Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. (2001) EMBO Rep. 2, 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., Green D. R. (2004) Science 303, 1010–1014 [DOI] [PubMed] [Google Scholar]
- 24.Eizenberg O., Faber-Elman A., Gottlieb E., Oren M., Rotter V., Schwartz M. (1996) Mol. Cell. Biol. 16, 5178–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Z., Zhang H., Levine A. J., Jin S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boya P., González-Polo R. A., Casares N., Perfettini J. L., Dessen P., Larochette N., Métivier D., Meley D., Souquere S., Yoshimori T., Pierron G., Codogno P., Kroemer G. (2005) Mol. Cell. Biol. 25, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knudson C. M., Tung K. S., Tourtellotte W. G., Brown G. A., Korsmeyer S. J. (1995) Science 270, 96–99 [DOI] [PubMed] [Google Scholar]
- 28.Geng Y., Akhtar R. S., Shacka J. J., Klocke B. J., Zhang J., Chen X., Roth K. A. (2007) J. Neuropathol. Exp. Neurol. 66, 66–74 [DOI] [PubMed] [Google Scholar]
- 29.Snyder E. Y., Deitcher D. L., Walsh C., Arnold-Aldea S., Hartwieg E. A., Cepko C. L. (1992) Cell 68, 33–51 [DOI] [PubMed] [Google Scholar]
- 30.Canu N., Tufi R., Serafino A. L., Amadoro G., Ciotti M. T., Calissano P. (2005) J. Neurochem. 92, 1228–1242 [DOI] [PubMed] [Google Scholar]
- 31.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner M. (2005) Nat. Cell Biol. 7, 212. [DOI] [PubMed] [Google Scholar]
- 33.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 34.Tassa A., Roux M. P., Attaix D., Bechet D. M. (2003) Biochem. J. 376, 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 36.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 38.Guillon-Munos A., van Bemmelen M. X., Clarke P. G. (2005) Apoptosis 10, 1031–1041 [DOI] [PubMed] [Google Scholar]
- 39.Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kroemer G., Levine B. (2008) Nat. Rev. Mol. Cell Biol. 9, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saftig P., Hartmann D., Lüllmann-Rauch R., Wolff J., Evers M., Köster A., Hetman M., von Figura K., Peters C. (1997) J. Biol. Chem. 272, 18628–18635 [DOI] [PubMed] [Google Scholar]
- 42.Shacka J. J., Klocke B. J., Young C., Shibata M., Olney J. W., Uchiyama Y., Saftig P., Roth K. A. (2007) J. Neurosci. 27, 2081–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao G., Yao Y., Liu J., Yu Z., Cheung S., Xie A., Liang X., Bi X. (2007) Am. J. Pathol. 171, 962–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchiyama Y., Koike M., Shibata M. (2008) Autophagy. 4, 404–408 [DOI] [PubMed] [Google Scholar]
- 45.Koike M., Shibata M., Tadakoshi M., Gotoh K., Komatsu M., Waguri S., Kawahara N., Kuida K., Nagata S., Kominami E., Tanaka K., Uchiyama Y. (2008) Am. J. Pathol. 172, 454–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Windelborn J. A., Lipton P. (2008) J. Neurochem. 106, 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. U. (2006) Nat. Cell Biol. 8, 1124–1132 [DOI] [PubMed] [Google Scholar]
- 48.Levine B., Sinha S., Kroemer G. (2008) Autophagy 4, 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. (2008) Mol. Cell 30, 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho D. H., Jo Y. K., Hwang J. J., Lee Y. M., Roh S. A., Kim J. C. (2009) Cancer Lett. 274, 95–100 [DOI] [PubMed] [Google Scholar]
- 51.Gozuacik D., Bialik S., Raveh T., Mitou G., Shohat G., Sabanay H., Mizushima N., Yoshimori T., Kimchi A. (2008) Cell Death Differ. 15, 1875–1886 [DOI] [PubMed] [Google Scholar]
- 52.Crighton D., Wilkinson S., O'Prey J., Syed N., Smith P., Harrison P. R., Gasco M., Garrone O., Crook T., Ryan K. M. (2006) Cell 126, 121–134 [DOI] [PubMed] [Google Scholar]