Abstract
The accumulation of the intermediate filament protein, glial fibrillary acidic protein (GFAP), in astrocytes of Alexander disease (AxD) impairs proteasome function in astrocytes. We have explored the molecular mechanism that underlies the proteasome inhibition. We find that both assembled and unassembled wild type (wt) and R239C mutant GFAP protein interacts with the 20 S proteasome complex and that the R239C AxD mutation does not interfere with this interaction. However, the R239C GFAP accumulates to higher levels and forms more protein aggregates than wt protein. These aggregates bind components of the ubiquitin-proteasome system and, thus, may deplete the cytosolic stores of these proteins. We also find that the R239C GFAP has a greater inhibitory effect on proteasome system than wt GFAP. Using a ubiquitin-independent degradation assay in vitro, we observed that the proteasome cannot efficiently degrade unassembled R239C GFAP, and the interaction of R239C GFAP with proteasomes actually inhibits proteasomal protease activity. The small heat shock protein, αB-crystallin, which accumulates massively in AxD astrocytes, reverses the inhibitory effects of R239C GFAP on proteasome activity and promotes degradation of the mutant GFAP, apparently by shifting the size of the mutant protein from larger oligomers to smaller oligomers and monomers. These observations suggest that oligomeric forms of GFAP are particularly effective at inhibiting proteasome activity.
Keywords: Chaperones, Cytoskeleton/Intermediate Filaments, Neurobiology/Neuroscience, Protein/Degradation, Proteasome, Alexander Disease, Astrocyte
Introduction
Alexander disease (AxD)2 is a rare but fatal disease of the central nervous system characterized by the presence in astrocytes of Rosenthal fibers, cytoplasmic protein aggregates that contain the intermediate filament protein, glial fibrillary acidic protein (GFAP), ubiquitinated proteins, and small heat shock proteins (shsps) (1). Because of the loss of myelin and oligodendrocytes and a variable loss of neurons, AxD has been historically thought of as a leukodystrophy or neurodegenerative disorder. AxD is, however, a disease of astrocyte dysfunction, as mutations in GFAP are found in the large majority of AxD patients (2). All mutations detected so far are heterozygous coding mutations, and most cases of AxD disease arise through de novo, dominant, GFAP mutations (2). Most of the AxD mutations reside in the 1A, 2A, and 2B segments of the conserved central rod domain of GFAP, but several are located in the tail region (2). Although GFAP mutations must underlie the pathogenesis of AxD, the mechanisms by which GFAP mutations are toxic have not been fully elucidated.
The Arg-239 residue is a “hot spot” for GFAP mutagenesis, as substitutions at this arginine are the most frequent mutations found in AxD and appear among different ethnic groups (2). Our previous work has concentrated on the most common substitution, R239C, and has demonstrated that overexpressing GFAP, either wild type (wt) or the R239C mutant, leads to the formation of intracellular protein aggregates (3–5). GFAP accumulation also inhibited proteasome protease activity and increased levels of ubiquitinated GFAP species (5). Although overexpressing wt GFAP can lead to intracellular protein aggregates, the R239C mutation exacerbates GFAP accumulation and aggregation and aggravates the effects of GFAP accumulation in inducing intracellular stress responses and in inhibiting proteasome activities (5).
In the present study we have explored the mechanisms that underlie the impaired proteasome function by GFAP accumulation. We asked if GFAP interacted with proteasomes in cells and in vitro and further asked if this interaction resulted in proteasome inhibition. Finally, we examined if αB-crystallin, a shsp and molecular chaperone, could reverse the GFAP-mediated proteasome inhibition. Both assembled (10-nm wide polymers) and unassembled (soluble at 80,000 × g) GFAP protein, both wild type and mutant, interact with the 20 S proteasome complex. The unassembled GFAP includes a mix of monomeric and oligomeric forms. Compared with the wt GFAP, the unassembled R239C GFAP showed a marked shift toward larger oligomers. In addition, the unassembled R239C had a far greater inhibitory effect on proteasome proteolytic activity in vitro than did the wt GFAP. The addition of αB-crystallin, which accumulates massively in AxD astrocytes, reversed the inhibitory effects of the R239C GFAP on proteasome activity. αB-Crystallin also changed the oligomer-monomer distribution of the R239C GFAP to a point at which it appeared identical to that of the wt GFAP. Our observations direct a focus on oligomeric forms of GFAP as effective proteasome inhibitors and suggest mechanisms that underlie the proteasome inhibition in astrocytes of the AxD brain. In addition, because the R239C GFAP aggregates could bind components of the ubiquitin-proteasome system, they may deplete the cytosolic stores of these proteins.
EXPERIMENTAL PROCEDURES
Reagents
Cell culture medium (Dulbecco's modified Eagle's medium) and molecular biological reagents were obtained from Invitrogen and Qiagen. Primary antibodies included anti-GFAP polyclonal antibody (pAb, DAKO), anti-GFAP monoclonal antibody (mAb), anti-αB-crystallin pAb, anti-ubiquitin mAb (Chemicon), anti-glyceraldehyde-3-phosphate dehydrogenase mAb (Encor), and anti-20 S pAb (Biomol). The fluorescence-conjugated and horseradish peroxidase-conjugated secondary antibodies were from Chemicon and Amersham Biosciences. Imidazole-HCl, EDTA, MG132, lactacystin, camptothecin, anti-FLAG M2-agarose affinity gel and protein G-Sepharose were from Sigma.
Brain Tissue
Tissues stored at −80 °C from cerebral hemispheric white matter of control subjects or AxD cases with different mutations were obtained postmortem. For immunohistochemistry, the tissue was fixed in formalin and then embedded in paraffin. All AxD cases were diagnosed based on histopathological examination and confirmed by the molecular genetic analysis for GFAP mutation. Controls included frozen central nervous system tissue from two children, one with no neurological disease (Control I) and one with Werdnig-Hoffman disease (non-AxD, non-leukodystrophy neurological disease without Rosenthal fibers) (Control II).
Cell Cultures and Transfection
cDNA clones encoding wt GFAP or R239C mt GFAP were inserted into EGFP-C1 expression vector for expressing a GFP-GFAP fusion protein. To generate permanent cell lines, U251 astrocytoma cells were transfected with the GFAP-GFP plasmid using calcium phosphate. The transfected cells were then cultured in Dulbecco's modified Eagle's medium supplement with 10% fetal calf serum, antibiotics, and 500 μg/ml G418. Clonal lines of stably transfected cells were isolated and confirmed by fluorescence-activated cell sorter sorting or Western blot analysis. MG132 (10 μm) and lactacystin (10 μm) were added to cultures at 24 h post-transfection and incubated for a further 24 h, with DMSO as the vehicle control.
Immunoprecipitation and Western Blotting
For protein extraction and fractionation, brain tissue or cells were lysed in total cell lysis buffer (2% w/v SDS, 6.25% mm Tris, pH 7.5, 5 mm EDTA supplemented with a Roche Applied Science protease inhibitor mixture tablet) for total protein or in fractional lysis buffer to separate soluble and insoluble fraction. Briefly, cells or brain tissues were recovered in S1 buffer (0.5% v/v Triton X, 2 mm Tris, 2 mm EDTA, and sucrose implemented with Complete Mini Roche Applied Science protease inhibitor mixture). After centrifugation for 20 min at 15,000 rpm at 4 °C, the supernatant was collected as the “soluble” fraction, and the pellet (“insoluble” fraction) was recovered in S2 buffer (2% w/v SDS, 2 mm Tris, 2 mm EDTA). Protein concentration was determined using a Bio-Rad protein assay kit according to the manufacturer's instructions.
For immunoprecipitation, Cos7 cells were transiently transfected with FLAG-tagged wt and R239C GFAP or vector only. Immunoprecipitations were performed 48 h post-transfection. Equal amounts of protein from whole cell extract soluble fractions were diluted in immunoprecipitation lysis buffer (6) and were incubated with anti-FLAGM2-agarose gel or anti-20 S antibody and protein G-Sepharose overnight at 4 °C. Immunoprecipitates were then separated by SDS-PAGE and subjected to Western blotting.
Proteasome Activity Assay
Brain tissues and cells were homogenized in ice-cold proteolysis buffer (10 mm Tris-HCl, pH 7.2, 0.035% SDS, 5 mm MgCl2, 5 mm ATP, and 0.5 mm dithiothreitol). The homogenates were centrifuged at 13,000 × g for 10 min, and the resulting supernatant was incubated with proteasome substrate II (S-II: benzyloxycarbonyl-Leu-Leu-Glu-amidomethylcoumarin) or III (S-III: Suc-Leu-Leu-Val-Tyr-amidomethylcoumarin) to detect the peptidylglutamyl peptide hydrolase (PGPH) and chymotrypsin-like peptidase activities, respectively. Fluorescence was monitored at 360-nm excitation and 450-nm emission in a fluorescence plate reader (Bio-Tek FL600).
Immunocytochemistry and Immunohistochemistry
Immunostaining of cultured cells was performed at 48 h after transfection as described before (4). A cytoskeleton buffer (PHEM buffer: 100 mm Pipes, 2 mm EGTA, 1 mm MgCl2, 0.5% Triton X-100, pH 6.8) was utilized to characterize the cytoskeleton. Cells were visualized by a Zeiss LSM510 confocal microscope.
Immunohistochemistry was performed according to general protocols. 10-μm-thick consecutive sections were prepared. Sections were blocked in Tris-buffered saline-Tween, 5% bovine serum albumin for 1 h and subsequently incubated with antibodies against the 20 S proteasome complex (1:100), αB-crystallin (1:100) ubiquitin (1:100), and GFAP (1:200) overnight at 4 °C. Negative control sections were held in phosphate-buffered saline during the primary incubation. Sections were then incubated with horseradish peroxidase-labeled secondary antibody 1 h at room temperature and developed with 3,3′-diaminobenzidine.
In Vitro GFAP Assembly and Co-sedimentation Assay
Recombinant human wt and R239C GFAP were purified from Escherichia coli BL21 (DE3) pLysS and assembled in vitro as before (7). The assembled wt or R239C GFAP filaments were incubated with purified 20 S proteasomes (8) at room temperature for 1.5 h. Samples were then separated into pellet and supernatant fractions by ultracentrifugation at 80,000 × g for 30 min at 4 °C and assayed by Western blotting.
Statistical Analysis
Results are expressed as the mean ± S.D. Statistical analysis was performed using ANOVA. p < 0.05 was considered statistically significant.
RESULTS
GFAP Aggregation Decreases Proteasome Activity and Depletes Cytosolic Proteasomes
We first assessed the proteasome protease activities in the U251 astrocytoma cell line stably transfected with control, wt GFAP, and R239C GFAP expression vectors. In comparing cells transfected with vector only (used as control and set to 100% activity), the PGPH activity was reduced in cells expressing wt GFAP (p < 0.05) and further in those expressing R239C GFAP (p < 0.01), whereas the chymotrypsin-like peptidase activity showed a non-significant reduction in wt and a modest reduction in R239C cells (p < 0.01, Fig. 1A). The decreased proteasome activity due to GFAP accumulation was not caused by the loss or reduction of proteasome complexes as the total amount of the 20 S complex remained the same in the vector-transfected cells and the GFAP-expressing cells (Fig. 1B). However, we observed a shift of 20 S complex from a Triton X-100-soluble fraction to the insoluble fraction (Fig. 1C).
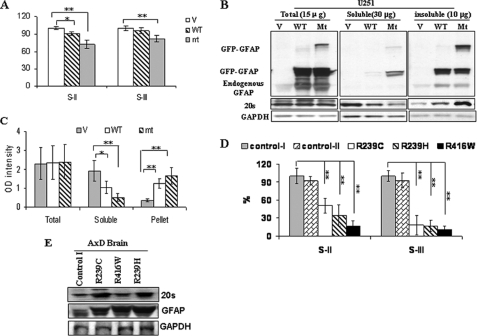
FIGURE 1.
GFAP accumulation decreases proteasome activity. A, proteasome proteolytic activity in U251 cells stably expressing EGFP vector (V), wt GFAP (wt), and R239C GFAP (mt) is shown. 48 h after plating, cells were lysed, and the proteasome PGPH and chymotrypsin-like protease activities were analyzed by using the fluorescent substrates S-II and S-III, respectively. Protease activities in cells expressing EGFP-C1 vector were set to 100%. The results are the average from five independent experiments. *, p < 0.05, **, p < 0.001, by two-tailed ANOVA. B, shift of 20 S proteasome from soluble pool to insoluble pool in U251 cells stably expressing wt or mt GFAP is shown. The U251 cells were harvested 48 h after plating at 80% confluency. Triton X-100 soluble (30 μg), insoluble (10 μg), or total protein (15 μg) was analyzed by Western blotting with an anti-20 S antibody and, after stripping, with an anti-GFAP and finally with an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody to normalize for loading. C, shown is quantitation of levels of 20 S in total, soluble, and insoluble fractions of cell lysates from U251 cells expressing EGFP-C1 vector (V), wt (WT), and mt GFAPs (mt). In comparing cells transfected with vector only as control, protein levels of 20 S in wt or mt GFAP-expressing cells were reduced markedly in the soluble fraction but enhanced significantly in the insoluble fraction. *, p < 0.05, **, p < 0.001, by two-tailed ANOVA. D, shown is decreased proteasome proteolytic activity in AxD brain with R239C, R239H, and R416W mutations. Protease activities in control subject I was set to 100%. Each result represents a mean ± S.D. of five independent experiments. *, p < 0.05, **, p < 0.001, by two-tailed ANOVA. E, shown is Western blotting analysis of 20 S proteasome in brains of AxD patients (control subject II not included).
We detected significantly decreased PGPH and chymotrypsin-like protease activities in white matter from AxD patients with the R239C, R416W, and R239H mutations (Fig. 1D), although 20 S complex protein levels were at the same levels or increased compared with that in white matter without Rosenthal fibers from a non-AxD subject (Fig. 1E).
The 20 S proteasome complex has been reported to be associated with aggresomes, intracellular protein aggregates formed by misfolded proteins (9). In the U251 cells, the GFAP inclusions were labeled with antibodies to the proteasomal 20 S complex and to ubiquitin (Fig. 2A). Many more of the cells expressing the R239C GFAP showed inclusions compared with cells expressing wt GFAP (Fig. 2D). Treatment of cells with proteasome inhibitors increased GFAP levels and increased the proportion of cells that contained inclusions (Fig. 2, C and D). When we examined sections from the brains of AxD patients, we found a similar distribution of GFAP, the 20 S proteasome and Rosenthal fibers (Fig. 2B). Because this similar distribution can only suggest, but cannot prove physical interactions among these components, we looked at cell and in vitro systems (see below).
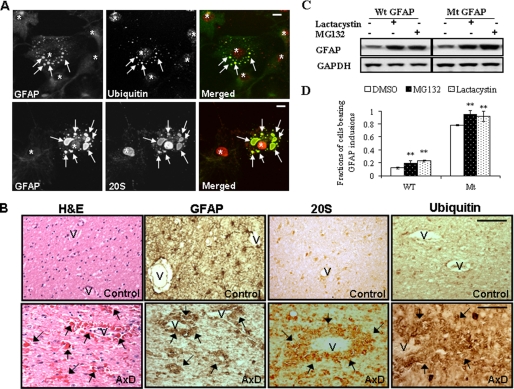
FIGURE 2.
R239C GFAP aggregates recruit ubiquitin proteasome system components, and proteasome inhibitors regulate GFAP accumulation. A, GFAP inclusions in R239C mutant GFAP cells were labeled with antibodies to ubiquitin (upper) and the proteasomal 20 S complex (lower). U251 cells stably expressing mutant GFAP were treated with Triton X-containing cytoskeletal buffer and fixed with 4% paraformaldehyde, then were immunostained for GFAP, 20 S, or ubiquitin. Merged images on the right show 20 S or ubiquitin in red, GFAP-GFP in green, and the overlap in yellow. Aggregates are indicated by arrows, and nuclei are indicated by asterisks. Note that in each panel cells without aggregates were included for control. After removal of soluble cytosolic protein, most of the proteasome components remain in cell nuclei. Scale bar, 10 μm. B, immunohistochemistry of brain tissue of an AxD patient and a control using antibodies against GFAP, 20 S, and ubiquitin are shown. Hematoxylin and eosin staining of the AxD brain showed extensive, brightly eosinophilic Rosenthal fibers (arrows), some of which surround vessels (hematoxylin and eosin (H&E), 40×). No Rosenthal fibers are present in control white matter. GFAP immunostaining of control white matter shows cells with typical, process-bearing, astrocyte morphology and many thin astrocyte processes, whereas in AxD, GFAP immunostains highlight the Rosenthal fibers. Correlated with the GFAP immunostaining, the signal for both 20 S and ubiquitin was localized to Rosenthal fibers. V, blood vessel; arrows, Rosenthal fibers. Scale bar, 100 μm. C, effects are shown of proteasome inhibition on GFAP accumulation. U251 cells stably expressing vector control (V), wt GFAP (WT), and R239C GFAP (mt) were incubated with medium with or without MG132 or lactacystin for 12 h. Total cell lysates were subjected to SDS-PAGE and Western blotting analysis with antibodies against GFAP and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). D, shown is the percentage of cells with aggregates after exposure to proteasome inhibitors. Results are the average ± S.D. from three independent experiments, with at least 400 cells counted in each. A larger percentage of the R239C GFAP-expressing cells, compared with wt expressing cells, contained aggregates under all treatment conditions. Different treatments were then compared within the wt or R239C GFAP expressing group using DMSO as control. **, compared with DMSO control, by two-tailed ANOVA, p < 0.001.
Associations between 20 S Proteasome and Non-assembled GFAPs
Based on the fact that proteasomes associate with different types of intermediate filaments, including keratin, desmin, and vimentin (10, 11), we inferred that, as a type III intermediate filament, GFAPs may interact with 20 S proteasome complexes, and the interaction of proteasome complexes with the accumulated GFAP might interrupt the normal proteasome function. To address this possibility, we performed a series of co-immunoprecipitations in Cos7 cells transiently transfected with FLAG-tagged wt or R239C GFAP. Because it has been reported that some proteasomes are present in the Triton X-100-soluble membrane and cytosolic fractions (12), we looked for GFAP-20 S complexes in the Triton X-100-soluble fractions of Cos7 cells (Fig. 3A). Reciprocal co-immunoprecipitations using anti-20 S antibodies and anti-FLAG antibodies revealed that both wt and R239C GFAPs were present in complexes with 20 S proteasomes. Protein A-Sepharose beads without anti-20 S antibodies were used as a background control, as were non-transfected Cos7 cells and Cos7 cells transfected with the empty vector. Note that the polymerized GFAP intermediate filaments were excluded from this assay, as they are part of the Triton X-100-insoluble, cytoskeletal network, which is also insoluble in immunoprecipitation buffer. However, when we treated the cells first with a cytoskeleton buffer (PHEM, containing 0.5% Triton X), which extracts soluble proteins, double immunofluorescence examination showed that proteasomes continued to associate with GFAP inclusions even when soluble, cytoplasmic proteins were removed (Fig. 3B), suggesting that some of the 20 S subunits associated with cytoskeletal GFAP.
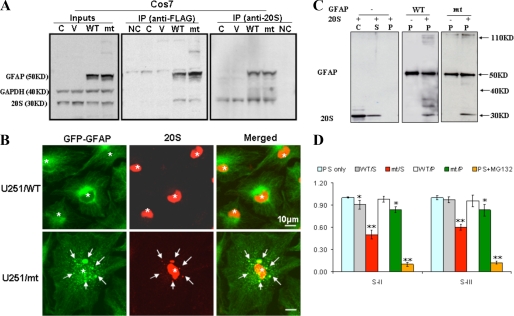
FIGURE 3.
20 S proteasome associates with soluble and aggregated GFAPs. A, co-immunoprecipitation (IP) of 20 S proteasome and GFAP revealed that both wt and R239C mutant GFAPs were present in complexes with 20 S proteasomes. Cos7 cells were transiently transfected with pcDNA3-FLAG vector, vectors expressing wt, and R239C GFAP. Forty-eight hours after transfection, cells were harvested and lysed with 1× radioimmune precipitation assay buffer. GFAP was immunoprecipitated with anti-FLAG M2-agarose affinity gel, and the 20 S proteasome complex was precipitated by protein A-Sepharose beads conjugated with an anti-20 S antibody. C, untransfected cells; V, pcDNA3-FLAG vector-transfected cells; wt and mt, cells expressing FLAG-tagged wt or mt GFAP; NC, protein A-Sepharose beads without GFAP and 20 S antibody. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, 20 S proteasome associates with Triton-insoluble GFAP aggregates in U251 cells. U251 cells stably expressing wt or mt GFAPs were treated with Triton X-containing cytoskeletal buffer and fixed with 4% paraformaldehyde and then examined for GFAP-GFP (green) and 20 S (red) fluorescence. GFAP aggregates are indicated by arrows, and nuclei are indicated by asterisks. Scale bar, 10 μm. C, 20 S proteasome associates with insoluble, filamentous GFAPs in vitro. Both wt and R239C GFAP were assembled in vitro as described (7) with or without purified 20 S proteasomes. Without GFAP (left panel), the 20 S proteasomes remain in the soluble (S) fraction, and none appeared in the pellet fraction (P). C is a control lane loaded with 20 S proteasome subunits. With reassembled wt and R239C GFAP (middle and right panels), the 20 S subunits were recovered in the pellet fraction (P). Western blots were performed with both anti-GFAP and anti-20 S antibodies. D, both soluble and aggregated forms of GFAP inhibit proteasome function. Purified proteasomes were incubated without (PS only) or with soluble non-assembled (S) or assembled (P) wt and R239C (RC) GFAPs. One hundred μg of soluble (S) or assembled (P) GFAPs were incubated with proteasome (100 μg) for 30 min in a total volume of 200 μl. Proteasome activity was then analyzed using fluorescent substrates (100 μm) with a fluorescence microplate reader at an excitation wavelength of 360 nm and an emission wavelength of 440 nm. Compared with those exposed to substrates only (PS only), both soluble mt GFAP (RC/S) and pellet R239C GFAP (RC/P) significantly inhibited proteasome PGPH and chymotrypsin-like proteolytic activities. **, compared with PS only, by two-tailed ANOVA, p < 0.001.
20 S Proteasome Bind to Assembled GFAP Filaments in Vitro
To determine whether GFAP intermediate filaments will associate directly with proteasomes, we turned to in vitro analyses, first using a co-sedimentation assay (7). Purified 20 S proteasome complexes were incubated with in vitro assembled wt or R239C GFAP filaments at a weight ratio of 1:1 or without GFAP (as a control) for 1.5 h. The samples were centrifuged at 80,000 × g in 40% sucrose, washed with assembly buffer and the PHEM buffer, and finally resolved by SDS-PAGE. Nitrocellulose blots were probed with an anti-20 S antibody and then re-probed with an anti-GFAP antibody. The 20 S proteasomes incubated without GFAP could not be recovered in the pellet fraction (Fig. 3C). In contrast, when incubated with assembled wt or R239C GFAP filaments, a fraction of the 20 S proteasomes co-sedimented with the GFAP (Fig. 3C). This result revealed a direct association between proteasomes and GFAP filaments. Note that the R239C mutation apparently does not interfere with this binding.
Both Soluble and Aggregated Forms of GFAP Inhibit Proteasome Function
Our data above indicated a direct interaction between GFAP and proteasome complexes, but we could not discern if the interaction interrupts proteasome function. To address this possibility, we performed an in vitro ubiquitin-independent proteasome degradation assay by examining the activity of purified 20 S proteasomes in the presence of recombinant wt GFAP, R239C GFAP, or MG132, again using fluorescent substrates to monitor proteasomal protease activity. We first examined how soluble wt and R239C GFAPs (separated from pelleted, assembled wt and R239C GFAPs by centrifugation in 40% sucrose at 80,000 × g as described above) affected proteasome function. wt and R239C GFAPs were isolated from the top layers and were incubated at identical concentrations with purified 20 S proteasome for 30 min. We then added fluorescent substrates and 30 min later analyzed the samples. Compared with those incubated with substrates only, 20 S proteasome complexes exposed to R239C GFAP showed a significantly decreased activity. In contrast, wt GFAP showed less inhibitory effects on 20 S proteasome activity (Fig. 3D). In the positive control, MG132-treated proteasomes were almost completely inhibited.
We also assessed the effect of assembled wt and R239C GFAPs on 20 S proteasome activity. In vitro assembled wt or R239C GFAP filaments were separated from soluble GFAP by centrifugation and preincubated with purified 20 S proteasome complexes. The fluorescent substrates were added to examine proteasomal proteolytic activity. Compared with MG132-treated samples and the negative control, the R239C GFAP filaments exhibited a significant inhibitory effect on proteasomes, whereas the wt GFAP filaments only slightly inhibited PGPH protease activity (Fig. 3D). However, these inhibitory effects were far less than those produced by non-assembled GFAP on a weight/weight basis. This result suggested that the inhibition of proteasomes by R239C GFAP might be predominantly attributed to soluble GFAPs.
In all of the proteasome inhibition experiments we kept the GFAP concentration between 0.25 mg/ml (50 μg/200 μl) and 1 mg/ml (200 μg/200 μl), which we believe is within the physiological range given the following estimates. GFAP represents about 1.0% of total brain protein in the adult rat (13). If brain is ∼11% protein and 9% protein by wet weight in adult rat and human, respectively (14), or about 90 mg/ml, the GFAP concentration would be 0.90 mg/ml. If astrocytes take up about 10% of brain volume (15) and as GFAP is only found in astrocytes, the GFAP concentration in astrocytes is in the order of 9.0 mg/ml (may be higher in white matter astrocytes, as their GFAP content is greater than that of gray matter astrocytes). The central nervous system in patients with AxD contains markedly increased amounts of GFAP, up by at least 10-fold, thus, giving a GFAP concentration of 90 mg/ml. The soluble GFAP is operationally defined as material that does not pellet at a specified g force and in a normal brain represents a small fraction of the total GFAP. Thus, in the adult rat brain after Triton X-100 solubilization and centrifugation at 180,000 × g, about 0.8% of the total GFAP appears in the soluble fraction (13). Thus, the soluble GFAP concentration in astrocytes would be around 0.72 mg/ml.
R239C GFAP Inhibits Proteasome Activity in a Non-competitive Manner
To understand how non-assembled GFAPs inhibit 20 S proteasome activity, we varied the concentration of fluorogenic substrates at a constant concentration of wt or R239C GFAP and measured proteolytic activities. Purified proteasomes were incubated with soluble or polymerized wt and R239C GFAPs, and 20 min later, proteasome substrates II or III were added. For proteasomes unexposed to GFAPs, as a control, the degradation of proteasome substrates increased with increasing substrate concentration and reached a maximum activity at a concentration of 100 μm. With a further increase in substrate concentration, the proteasome activity decreased slightly (Fig. 4A), suggesting that increasing substrate concentration beyond a threshold may inhibit proteasome proteolytic activities. In the range of 100 μm substrate concentration, the activity of 20 S proteasome exposed to filamentous or unassembled wt GFAP increased with increasing substrate concentrations (Fig. 4A), similarly to the activity without GFAP, although there appeared to be some small inhibition at higher SII substrate levels. These data indicate that the proteasomal inhibition that we observed before was not a false positive result from substrate binding and substrate sequestration. Thus, GFAP seems to inhibit the proteasome activity via an interaction with the proteasome rather than by binding substrate peptide. The R239C GFAP produced a significant inhibition of proteasome activity, with the unassembled (soluble) GFAP exhibiting the greatest effect (Fig. 4A). Interestingly, proteasome activity did not continue to increase with increasing concentrations of substrate, suggesting that the interaction between soluble R239C GFAP and proteasomes produced a non-competitive inhibition of proteasome activity.
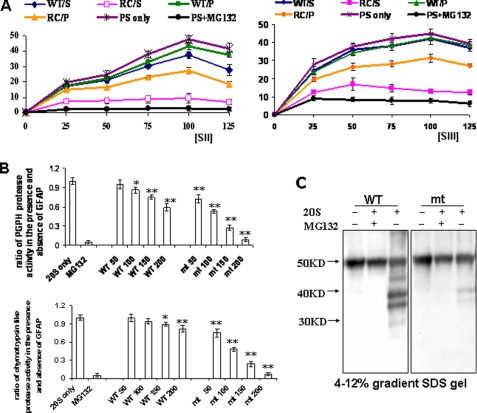
FIGURE 4.
R239C GFAP inhibited proteasome activity in a non-competitive manner. A, R239C mt GFAPs inhibited proteasome activity in a non-competitive manner. Purified proteasomes (100 μg) were incubated without (PS only) or with soluble (S) or polymerized (P) wt and R239C (RC) GFAPs (100 μg). 20 min later proteasome substrate II or III was added at different concentrations (25, 50, 75, 100, 125 μm), and proteasome proteolytic activity was analyzed. B, shown are PGPH and chymotrypsin-like protease activities at different GFAP concentrations (50, 100, 150, 200 μg/200 μl), with the fluorescent peptide substrate concentration fixed at 100 μm. Compared with 20 S only, *, p < 0.05; **, p < 0.001, by two-tailed ANOVA. C, shown is proteasome degradation of wt and R239C GFAP. Soluble wt and R239C GFAPs were incubated with or without 20 S proteasomal subunits and with or without MG132, electrophoresed on SDS gels, and immunoblotted with anti-GFAP antibody. This experiment was repeated three times with similar results.
Next, we assayed the PGPH and chymotrypsin-like protease activities at different GFAP concentrations (50, 100,150, 200 μg/200-μl total reaction volume), with the fluorescent peptide substrate concentration fixed at 100 μm. The R239C GFAP inhibited PGPH and chymotrypsin-like activity more than the wt GFAP at all GFAP concentrations (Fig. 4B). For example, at the 100 μg/200 μl, the R239C GFAP inhibited almost 50% of proteasome activity, whereas the wt inhibited only 10% of total activity; at 200 μg/200 μl, the R239C GFAP produced a 90% percent inhibition, whereas the wt GFAP was far less inhibitory. MG132, as a positive control, blocked the protease activity markedly (Fig. 4B).
Soluble wt GFAP, but Not R239C GFAP, Is Partially Degraded by the 20 S Proteasome
We then asked whether the wt and the R239C GFAPs could be efficiently degraded by 20 S proteasomes via a ubiquitin-independent manner in vitro. Soluble wt and R239C GFAPs were incubated with 20 S proteasomes, and products were analyzed by Western blotting. After exposure to 20 S proteasomes, a fraction of the wt GFAP was broken down into smaller fragments (Fig. 4C). In contrast, the R239C GFAP was degraded only minimally (Fig. 4C). In the presence of MG132, as a positive control, neither wt nor R239C GFAP was degraded. Thus, although both wt and R239C GFAP can interact with proteasome 20 S complexes, the R239C GFAP cannot be efficiently degraded.
The Molecular Chaperone αB-Crystallin Restores Proteasome Proteolytic Activity That Was Reduced by R239C GFAP
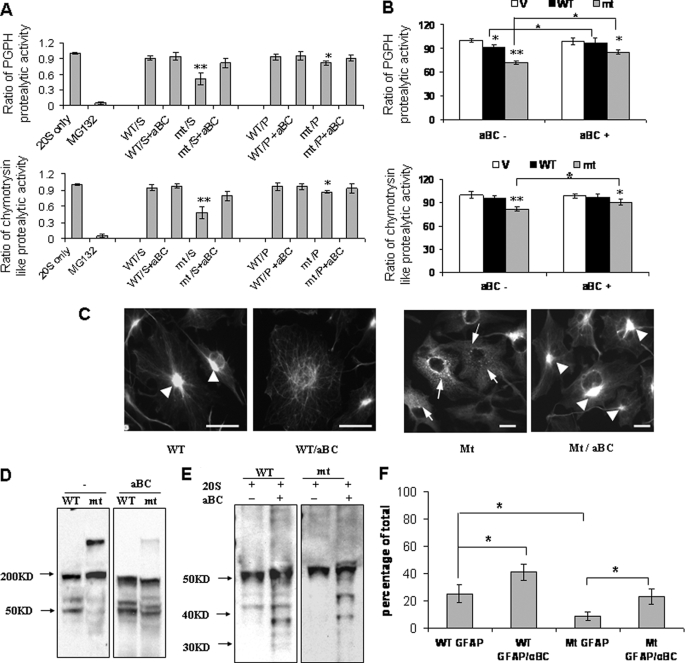
We then asked if αB-crystallin could rescue proteasome function from the R239C GFAP toxicity. We incubated purified 20 S proteasomes with soluble or aggregated wt or R239C mt GFAPs in the presence or absence of αB-crystallin (at a weight/weight ratio of GFAP to αB-crystallin of 1:2) and then analyzed the PGPH and chymotrypsin-like proteasome proteolytic activities. At the GFAP concentration used here (100 μg/200 μl), the soluble R239C GFAP inhibited both PGPH and chymotrypsin activities, and the pelleted R239C GFAP produced a milder inhibition (Fig. 5A). The introduction of recombinant αB-crystallin attenuated the toxicity of R239C GFAP on proteasome activities (Fig. 5A).
FIGURE 5.
αB-Crystallin restores proteasome proteolytic activity reduced by R239C GFAPs. A, in vitro preincubation of R239C GFAP with αB-crystallin (aBC) reversed the inhibitory activity of the R239C GFAP. 20 S only, unexposed proteasomes; MG132, MG 132 control treatment; S, soluble; P, polymerized. *, p < 0.05; **, p < 0.001. B, adenoviral introduction of αB-crystallin increased proteasome activity in U251 cells expressing R239C GFAP. Protease activities in cells expressing EGFP-C1 vector (V) are set to 100%. The results are the average from five independent experiments. Adenoviral infection did not affect the proteasome activity. αB-Crystallin expression increased PGPH proteolytic activity in both wt and R239C GFAP-expressing U251 cells and chymotrypsin-like proteolytic activity in R239C GFAP-expressing cells. *, p < 0.05; **, p < 0.001. C, morphological changes of GFAP-expressing U251 cells. U251 cells stably expressing wt or R239C GFAP were infected with αB-crystallin-expressing adenovirus, then fixed and processed for GFP fluorescence. αB-Crystallin dispersed the GFAP aggregates in wt-expressing cells and shifted them into filamentous structures. GFAP aggregates (arrowheads), whereas αB-crystallin expression reversed the formation of small GFAP inclusions, shifting the GFAP to large perinuclear aggregates and filaments. Scale bar, 20 μm. D, shown is oligomerization a profile analysis of soluble wt and R239C GFAP preincubated with (right panel) or without αB-crystallin (left panel). The same amounts of soluble wt and R239C GFAP in the absence or the presence of αB-crystallin were subjected to non-SDS, non-reducing, 8% PAGE gel electrophoresis and then immunoblotted for GFAP. E, αB-crystallin promotes degradation of the R239C GFAP by the 20 S proteasome. The same amounts of soluble wt and R239C GFAP (100 μg) in the absence or the presence of αB-crystallin (200 μg) were incubated with 20 S proteasome (100 μg). The incubation products were then subjected to 4–12% SDS gradient gel electrophoresis and Western blotting. After preincubation with αB-crystallin, the degree of proteolytic breakdown of the R239C GFAP was similar to that of the wt GFAP. F, the optical density of degraded GFAP protein bands in E was quantitated and plotted to analyze the effects of αB-crystallin on GFAP protein degradation. Compared with wt GFAP, R239C GFAP was less susceptible to proteasome degradation (p < 0.05). Preincubation with αB-crystallin promoted proteasome degradation of both wt (p < 0.05) and R239C GFAP (p < 0.05).
To validate the protective effects of αB-crystallin on proteasome function in vivo, we increased intracellular αB-crystallin levels using a recombinant adenovirus in U251 cells stably expressing R239C GFAP. By determining the proteasome activity using fluorescent substrates, we found that the overexpression of exogenous αB-crystallin attenuated the proteasome toxicity induced by R239C GFAP (Fig. 5B). As visualized by immunofluorescence, the overexpressed αB-crystallin dispersed the GFAP aggregates and shifted them into filamentous structures (Fig. 5C).
αB-Crystallin Changes the Oligomerization State of R239C GFAP and Promotes GFAP Proteolysis
The R239C mutant GFAP can assemble into 10-nm filaments, like the wt GFAP (4). However, a fraction of the GFAP remains in suspension after centrifuging the reassembled GFAP at 80,000 × g, as noted above. This soluble GFAP fraction was electrophoresed through a non-SDS, non-reducing 8% PAGE gel and then immunoblotted to reveal the different GFAP species. The soluble wt GFAP resolved as a mix of monomers (50 kDa) and higher molecular mass bands, the two main ones representing dimers and tetramers (Fig. 5D, left panel). In contrast, the R239C GFAP showed a pronounced shift to higher molecular mass forms (∼200–300 kDa, perhaps reflecting 4–6 GFAP molecules). When we added αB-crystallin to the soluble R239C GFAP and then electrophoresed the protein, we found a pattern of GFAP monomers and oligomers much more similar to that of the wt GFAP (Fig. 5D, right panel). We then asked if αB-crystallin would promote more proteolytic degradation of the R239C GFAP by the 20 S proteasome. When the soluble R239C GFAP was preincubated with αB-crystallin, we found that the degree of proteolytic breakdown of the GFAP was now similar to that of the wt GFAP (Fig. 5, E and F). Thus, αB-crystallin not only shifts the monomer-oligomer equilibrium of the R239C GFAP, but it also allows proteolysis.
DISCUSSION
GFAP Accumulation Inhibits Proteasome Activity
In this study we investigated how GFAP accumulation in the astrocytes of AxD might affect the ubiquitin-proteasome system. Using fluorogenic peptide assays, we demonstrated decreased proteasome activities in U251 astrocytoma cells stably expressing GFAP and in AxD white matter. These results corroborated our previous findings with a GFP-U proteasome reporter system, where overexpressing GFAP impaired proteasome function (5). Both wt and R239C GFAP accumulation inhibited proteasome activity, but the R239C GFAP exhibited a greater inhibitory effect. R239C GFAP tended to aggregate into inclusions (5, 16), which were associated with components of the proteasome system, such as proteasome 20 S core complexes, ubiquitinated proteins, and small heat shock proteins, as evidenced by immunostaining of cells and of Rosenthal fibers in AxD brains. Furthermore, we found that ubiquitinated GFAP protein accumulated in GFAP-overexpressing U251 cells and in AxD brain tissues and that GFAP further accumulated upon treatment with MG132 and lactacystin, two proteasome inhibitors. These observations point to a role of the ubiquitin-dependent proteasome pathway in the removal of excess GFAP. The inhibition of proteasome appear, in turn, to aggravate GFAP accumulation and aggregation, as we observed an increase in GFAP protein levels and in the percentage of cells bearing GFAP aggregates (5).
A recent study (17) showed in astrocytoma cells that proteasome inhibitors down-regulate GFAP gene transcription and thereby decrease GFAP protein levels. Furthermore, proteasome inhibition prevents GFAP accumulation in reactive astrocytes in a model in which a microdialysis probe through which an inhibitor was infused was placed in the rat central nervous system. The trauma of probe placement itself produced increased GFAP immunostaining, which was prevented by inhibitor. However, this effect only occurred during the early increase in GFAP but not after 7 days, suggesting that GFAP gene expression is initially susceptible to proteasome inhibition but is no longer susceptible at the higher levels of GFAP protein that had accumulated by the later stages of astrogliosis. We do not know if this model applies to the proteasome inhibition in AxD. For example, in mouse models of AxD, the levels of GFAP mRNA and GFAP protein are both elevated (18).
Proteasomes Bind to Filamentous GFAP
Our data demonstrated a direct association between filamentous GFAP and the 20 S proteasome complex. The association of proteasomes with intermediate filament network of the keratin type has been reported in PtK1 and HeLa cells (10). In addition, intracellular localization of proteasomes overlapped with the vimentin network in proliferating human fibroblasts and with desmin intermediate filament in myoblasts of the mouse myogenic cell line C2.7 (11). A later study also revealed a direct interaction between proteasomes and the actin network (12). In the present study we identified an association between proteasomes and the GFAP intermediate filament network in U251 astrocytoma cells. This co-localization was particularly visible after treatment with Triton X-100, which facilitates observation of GFAP, as it eliminates most of the soluble proteins. In vitro co-sedimentation experiments further showed a direct interaction between 20 S proteasome and assembled GFAP filaments. This was consistent with previous reports on the interaction of proteasomes and reconstituted intermediate filaments of the vimentin type (12).
Whether this interaction of proteasomes with filamentous GFAP interferes with proteasome activity is not fully resolved. For example, we show that a larger proportion of the 20 S subunit associates with the “pellet” fraction of U251 cells than in control cells (Fig. 1B). This association would deplete proteasomes from the soluble fraction and, thus, might account for the loss of proteasome activity measured in a soluble fraction in the cells (Fig. 1A) simply because there are fewer proteasomes in this soluble fraction.
The in vitro activity assays showed that the filamentous forms of GFAP did not inhibit proteasome proteolytic activity as much as the soluble forms on a weight/weight basis, although the filamentous R239C GFAP was more inhibitory than the wt GFAP.
The Interaction between Proteasomes and Soluble GFAP Inhibits Proteasome Activity
We demonstrated a direct association between Triton X-100-soluble GFAP and 20 S proteasome complexes by reciprocal immunoprecipitation from cell lysates. With this result in mind, we tested the effects of the soluble fraction of the reassembled GFAP on proteasome activity in vitro. The soluble fraction consisted of a mix of monomeric and oligomeric forms. We use the term “oligomer” to denote a molecule larger than the monomeric GFAP but smaller than the polymerized intermediate filament. However, we know neither the exact structure of these larger soluble forms nor whether they are normal intermediates in the polymerization of R239C GFAP, which will assemble into normal-appearing 10-nm filaments (4), or abnormal intermediates separate from the pathway of filament assembly. There was a marked shift to larger oligomeric forms with the R239C mutation, suggesting that these larger intermediates played a critical role in the inhibition. Indeed, the greater ability of soluble, unassembled R239C GFAP compared with assembled R239C GFAP to inhibit proteasome activity implies that AxD mutant GFAP inhibits proteasome function primarily through the accumulation of monomeric and/or oligomeric forms. We would consider the oligomer forms to be inhibitory given that decreasing oligomer levels with αB-crystallin restored proteasome proteolytic activity. These observations add to the recent view in protein misfolding disorders that oligomeric protein species may be biologically more toxic than larger, aggregated fibrils (19, 20).
However, we cannot conclude that wt GFAP is innocuous and can never cause proteasome inhibition. wt GFAP will inhibit proteasome activity but required higher concentrations than R239C GFAP. One of the effects of increasing wt GFAP concentrations would be to increase the concentration of wt oligomers, but we would suggest that, because of the differences in equilibrium, one needs a far higher concentration of wt GFAP than R239C to reach the same oligomer concentration. Alternatively, larger amounts of wt GFAP monomers may simply provide a competitive inhibition, as proteasome inhibition might occur at higher rates of substrate degradation, and a competition between the excess GFAP and other cellular proteins for degradation might occur.
The kinetic analysis showed that the proteolysis of fluorogenic substrates was saturable at a constant proteasome concentration (Fig. 4). The soluble R239C GFAP produced a marked inhibition, with a relatively flat activity curve, suggesting a non-competitive component to the inhibition. This could reflect, for example, R239C oligomer binding to the proteasome. The oligomer would not be degraded, but its binding would inhibit the proteolysis of other substrates. The mechanistic model of proteasome degradation proposes that substrates are directed into proteasomes and degraded in the inner channel of 20 S proteasomes, where the active sites of the catalytic subunits are located. However, only unfolded monomeric peptides are able to slide into the peptide channel, which functions by size exclusion (21). One implication of this model is that proteins that are in oligomeric assemblies will not be efficiently degraded but will only be degraded after depolymerization into a monomeric form. Because monomeric and oligomeric forms of GFAP must exist in equilibrium, any changes to the protein that shift the equilibrium toward oligomeric forms will slow proteasomal degradation. One of the dramatic effects of the R239C mutation is the shift in molecular mass size in the soluble GFAP pool toward a larger class of oligomers. We do not yet know why this equilibrium is so shifted. Presumably the larger oligomers of the R239C GFAP are more stable than those of the wt GFAP, but the structural basis for this stability is unclear. These observations suggest that further studies of monomer-oligomer equilibrium with the various GFAP mutations of Alexander disease are worth pursuing.
Ubiquitin-mediated and Ubiquitin-independent Degradation of GFAP
The 20 S proteasome particle catalyzes proteasomal hydrolysis, which can take two forms; ubiquitin-dependent protein degradation, representing the major fraction, relies on the 26 S proteasomes, and ubiquitin-independent proteolysis of certain unfolded or modified proteins can be carried out by the single 20 S proteasome (22, 23). Our data suggest that GFAP can be degraded via both ubiquitin-dependent and ubiquitin-independent processes. The ubiquitin-independent GFAP protein degradation has been demonstrated above. We also found that in cell cultures and AxD brains, GFAPs were ubiquitinated. Upon proteasome inhibition, there was a further accumulation of these ubiquitinated species (5). This result was consistent with our previous observations that defects in proteasome degradation occurred when GFAP were overexpressed in the GFP-U proteasome reporter system, suggesting that overloaded GFAP proteins also damaged the ubiquitin-dependent proteasomal degradation. We also do not exclude the possibilities that the R239C GFAP may interact with 26 S proteasome at other sites, such as the proteasome subunit α4 (24) and the 19 S proteasomal component S6 (25). These sites exert important roles in mediating the toxicity of α-synuclein and parkin (25, 26). However, whether these sites were occupied by R239C GFAPs still await further study.
The R239C GFAP probably exerted its inhibition by direct binding to the proteasome. Thus, we found that the protein degradation of proteasomes preincubated with the R239C GFAP was inhibited. The binding of R239C GFAP presumably prevented the fluorogenic peptide from entering the catalytic compartment of 20 S proteasome complexes either by binding at the entrance of the peptide channel or by interacting directly with the inner catalytic compartment of the proteasome in a manner suggested for the toxicity of amyloid β peptide in Alzheimer disease (27). Either external or internal binding could block the entrance of other substrates into the proteasome chamber or alter the conformation of the active proteolytic site.
αB-Crystallin Shifts Monomer-Oligomer Equilibrium of R239C GFAP and Reinstates Proteasome Degradative Function
αB-Crystallin normalized the equilibrium between oligomers and monomers to that which resembled wt GFAP. αB-Crystallin also mitigated the R239C GFAP-dependent proteasome inhibition, as measured both by fluorogenic substrate assays and by the degree of proteolysis of GFAP itself. Thus, a restoration of the normal GFAP size range correlated with a restoration of proteolysis. We presume these two phenomena are causally related.
The interactions between αB-crystallin and GFAP are complex. shsps, αB-crystallin, and hsp27 do bind intermediate filaments, both the soluble and the filamentous forms (7). αB-Crystallin can inhibit GFAP polymerization (28) and can prevent GFAP assembly in vitro as well as filament-filament interactions in vitro (7). In cultured astrocytes, αB-crystallin overexpression can reorganize abnormal intermediate filament aggregates into a normal filamentous network (3) and can spread filament bundles. In the present study, αB-crystallin overexpression mitigated the proteasome inhibition produced by the R239C GFAP. Furthermore, αB-crystallin associates with GFAP aggregates in AxD brains and with aggregates in cell models produced by overexpression of R239C GFAP (this study) and R416W (29).
How does αB-crystallin exert its effects on proteasome dysfunction? We think the most likely effect is to depolymerize/disaggregate the larger GFAP oligomers, presumably by binding to GFAP and interfering with GFAP assembly. It seems less likely that αB-crystallin relieves the proteasome inhibition by preventing the R239C GFAP from binding to proteasomes. The interaction between αB-crystallin and GFAP appears to be more critical than any possible direct interactions between αB-crystallin and the 20 s proteasome for two reasons. First, we preincubated αB-crystallin with GFAP and found a shift in the oligomer profile in the absence of proteasomes. Second, although αB-crystallin is reported to interact with the proteasome subunit, C8/α7, its interaction with the full 20 S proteasome appears much weaker (30). However, we cannot completely rule out the possibility that a binding of αB-crystallin to proteasomes could relieve the GFAP-induced inhibition, although it is difficult to formulate such a mechanism.
In cells with accumulated R239C GFAP, the overexpression of αB-crystallin restored proteasomal protein degradation to a normal level, suggesting that in the living cell αB-crystallin alters the distribution of soluble unassembled GFAP proteins between monomeric and oligomeric states and helps prevent an accumulation of toxic oligomers. One might ask, however, why there is such strong proteasomal inhibition in AxD, given the markedly increased levels of αB-crystallin in astrocytes. The degree of inhibition may be a matter of relative levels of GFAP and αB-crystallin. It is important to consider that although the large majority of cellular GFAP is cytoskeletal-associated (Triton X-100 insoluble), there is a soluble pool, and cells expressing the R239C GFAP contain a relatively larger soluble pool than cells expressing wt GFAP (31). A further complication is that αB-crystallin shifts from a largely cytosolic form to a cytoskeletal-associated form in the presence of GFAP accumulation, probably being bound up by the excess of filaments. Thus, there is less soluble αB-crystallin available in a soluble pool to bind to GFAP oligomers.
Can Proteasome Inhibition Explain Metabolic Changes Seen in AxD?
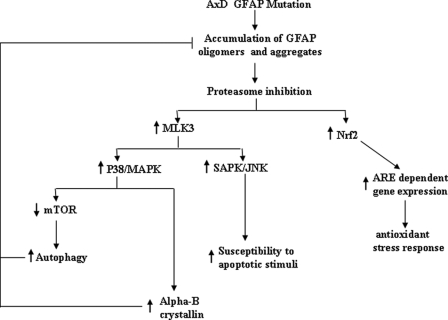
Proteasome inhibition would have a number of consequences for astrocyte function (Fig. 6). It could activate the SAPK/JNK stress pathway and compromise the astrocyte ability to withstand apoptotic stress stimuli (5). Meanwhile, cells will develop compensatory mechanisms to compensate for the dysfunction of proteasomes. These mechanisms include the induction of shsp gene transcription, the accumulation of shsps, and the induction of autophagy (16). Inhibiting proteasomes in several cell systems leads to an induction of oxidative response genes via the Nrf2-regulated transcriptional pathway (32). Indeed, as predicted, analysis of brain tissues in knock-in and transgenic mouse models of AxD showed a marked oxidative stress response and the induction of several oxidative stress response genes (18, 33).
FIGURE 6.
Proposed consequences of proteasome inhibition by the AxD GFAP R239C mutation. Accumulation of GFAP oligomers and aggregates inhibits proteasome proteolytic activity. This inhibition in turn activates mixed lineage kinase (MLK)/JNK/p38 stress kinase pathways, which up-regulate αB-crystallin transcription and activate autophagy. Both αB-crystallin and autophagy act to decrease GFAP protein levels, the former by inhibiting aggregate formation and reducing inhibitory GFAP oligomer levels and the latter by degrading GFAP aggregates. Proteasome inhibition also activates ARE-regulated gene expression via increased levels of Nrf2. MAPK, mitogen-activated protein kinase; ARE, anti-oxidant response element.
Acknowledgments
We thank Prof. Albee Messing and Prof. Michael Brenner for helpful discussions and Prof. Tom K. Hei and Dr. Michael Partridge for help with our proteasome activity assay.
This work was supported, in whole or in part, by National Institutes of Health Grants PO1NS42803 and P30-HD03352.
- AxD
- Alexander disease
- GFAP
- glial fibrillary acidic protein
- shsp
- small heat shock protein
- wt
- wild type
- mt
- mutant
- PGPH
- peptidylglutamyl peptide hydrolase
- JNK
- c-Jun N-terminal kinase
- SAPK
- stress-activated protein kinase
- GFP
- green fluorescent protein
- Pipes
- 1,4-piperazinediethanesulfonic acid
- ANOVA
- analysis of variance.
REFERENCES
- 1.Brenner M., Johnson A. B., Boespflug-Tanguy O., Rodriguez D., Goldman J. E., Messing A. (2001) Nat. Genet. 27, 117–120 [DOI] [PubMed] [Google Scholar]
- 2.Li R., Johnson A. B., Salomons G., Goldman J. E., Naidu S., Quinlan R., Cree B., Ruyle S. Z., Banwell B., D'Hooghe M., Siebert J. R., Rolf C. M., Cox H., Reddy A., Gutiérrez-Solana L. G., Collins A., Weller R. O., Messing A., van der Knaap M. S., Brenner M. (2005) Ann. Neurol. 57, 310–326 [DOI] [PubMed] [Google Scholar]
- 3.Koyama Y., Goldman J. E. (1999) Am. J. Pathol. 154, 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsiao V. C., Tian R., Long H., Der Perng M., Brenner M., Quinlan R. A., Goldman J. E. (2005) J. Cell Sci. 118, 2057–2065 [DOI] [PubMed] [Google Scholar]
- 5.Tang G., Xu Z., Goldman J. E. (2006) J. Biol. Chem. 281, 38634–38643 [DOI] [PubMed] [Google Scholar]
- 6.Xu Z., Maroney A. C., Dobrzanski P., Kukekov N. V., Greene L. A. (2001) Mol. Cell. Biol. 21, 4713–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perng M. D., Muchowski P. J., van Den IJssel P., Wu G. J., Hutcheson A. M., Clark J. I., Quinlan R. A. (1999) J. Biol. Chem. 274, 33235–33243 [DOI] [PubMed] [Google Scholar]
- 8.Wilk S., Chen W. E. (2001) Curr. Protoc. Protein Sci. Chapter 21, Unit 21.6 [DOI] [PubMed] [Google Scholar]
- 9.Johnston J. A., Ward C. L., Kopito R. R. (1998) J. Cell Biol. 143, 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossi de Sa M. F., Martins de Sa C., Harper F., Olink-Coux M., Huesca M., Scherrer K. (1988) J. Cell Biol. 107, 1517–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olink-Coux M., Arcangeletti C., Pinardi F., Minisini R., Huesca M., Chezzi C., Scherrer K. (1994) J. Cell Sci. 107, 353–366 [PubMed] [Google Scholar]
- 12.Arcangeletti C., Sütterlin R., Aebi U., De Conto F., Missorini S., Chezzi C., Scherrer K. (1997) J. Struct. Biol. 119, 35–58 [DOI] [PubMed] [Google Scholar]
- 13.Malloch G. D., Clark J. B., Burnet F. R. (1987) J. Neurochem. 48, 299–306 [DOI] [PubMed] [Google Scholar]
- 14.Banay-Schwartz M., Kenessey A., DeGuzman T., Lajtha A., Palkovits M. (1992) Age 15, 51–54 [Google Scholar]
- 15.Reichenbach A., Wolburg H. (2005) in Neuroglia, (Kettenmann H., Ransom B. eds.) pp. 19–35, Oxford University Press, Oxford [Google Scholar]
- 16.Tang G., Yue Z., Talloczy Z., Hagemann T., Cho W., Messing A., Sulzer D. L., Goldman J. E. (2008) Hum. Mol. Genet. 17, 1540–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Middeldorp J., Kamphuis W., Sluijs J. A., Achoui D., Leenaars C. H., Feenstra M. G., van Tijn P., Fischer D. F., Berkers C., Ovaa H., Quinlan R. A., Hol E. M. (2009) FASEB J. 23, 2710–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagemann T. L., Gaeta S. A., Smith M. A., Johnson D. A., Johnson J. A., Messing A. (2005) Hum. Mol. Genet. 14, 2443–2458 [DOI] [PubMed] [Google Scholar]
- 19.Kristiansen M., Deriziotis P., Dimcheff D. E., Jackson G. S., Ovaa H., Naumann H., Clarke A. R., van Leeuwen F. W., Menéndez-Benito V., Dantuma N. P., Portis J. L., Collinge J., Tabrizi S. J. (2007) Mol. Cell. 26, 175–188 [DOI] [PubMed] [Google Scholar]
- 20.Caughey B., Lansbury P. T. (2003) Annu. Rev. Neurosci. 26, 267–298 [DOI] [PubMed] [Google Scholar]
- 21.Wenzel T., Baumeister W. (1995) Nat. Struct. Biol. 2, 199–204 [DOI] [PubMed] [Google Scholar]
- 22.Orlowski M., Wilk S. (2003) Arch Biochem. Biophys. 415, 1–5 [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Castelao B., Castaño J. G. (2005) FEBS Lett. 579, 4797–4802 [DOI] [PubMed] [Google Scholar]
- 24.Dächsel J. C., Lücking C. B., Deeg S., Schultz E., Lalowski M., Casademunt E., Corti O., Hampe C., Patenge N., Vaupel K., Yamamoto A., Dichgans M., Brice A., Wanker E. E., Kahle P. J., Gasser T. (2005) FEBS Lett. 579, 3913–3919 [DOI] [PubMed] [Google Scholar]
- 25.Snyder H., Mensah K., Theisler C., Lee J., Matouschek A., Wolozin B. (2003) J. Biol. Chem. 278, 11753–11759 [DOI] [PubMed] [Google Scholar]
- 26.Ghee M., Melki R., Michot N., Mallet J. (2005) FEBS J. 272, 4023–4033 [DOI] [PubMed] [Google Scholar]
- 27.Gregori L., Hainfeld J. F., Simon M. N., Goldgaber D. (1997) J. Biol. Chem. 272, 58–62 [DOI] [PubMed] [Google Scholar]
- 28.Nicholl I. D., Quinlan R. A. (1994) EMBO J. 13, 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Der Perng M., Su M., Wen S. F., Li R., Gibbon T., Prescott A. R., Brenner M., Quinlan R. A. (2006) Am. J. Hum. Genet. 79, 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boelens W. C., Croes Y., de Jong W. W. (2001) Biochim. Biophys. Acta 1544, 311–319 [DOI] [PubMed] [Google Scholar]
- 31.Tian R., Gregor M., Wiche G., Goldman J. E. (2006) Am. J. Pathol. 168, 888–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwak M. K., Itoh K., Yamamoto M., Kensler T. W. (2002) Mol. Cell. Biol. 22, 2883–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagemann T. L., Connor J. X., Messing A. (2006) J. Neurosci. 26, 11162–11173 [DOI] [PMC free article] [PubMed] [Google Scholar]