Abstract
The differentiated phenotype of chondrocyte is lost in pathological situations and after interleukin (IL)-1β challenge. Wnt proteins and the inorganic pyrophosphate (PPi) transporter Ank regulate the differentiation process in many cell types. We investigated the possible contribution of Ank and/or PPi to the maintenance of the differentiated chondrocyte phenotype with special care to Wnt signaling. Primary articular chondrocytes lost their phenotype upon IL-1β challenge, with cessation of type II collagen and Sox-9 expression. Ank expression and PPi transport were strongly reduced by IL-1β, whereas Wnt-5a was the only Wnt protein increased. Transient overexpression of Ank counteracted most of IL-1β effects on Type II collagen, Sox-9, and Wnt-5a expression. When resting chondrocytes were transfected with a siRNA against Ank, this reproduced the phenotype induced by IL-1β. In both cases, no markers for hypertrophic chondrocytes were detected. The conditioned supernatant from chondrocytes knocked-down for Ank contained Wnt-5a, which activated Tcf/Lef reporter plasmids and promoted translocation of β-catenin into the nucleus without activating the c-Jun N-terminal kinase (JNK) pathway. Supplementation with PPi compensated for most effects of Ank deficiency on Type II collagen, Sox-9, and Wnt-5 expression, both in IL-1β and Ank knock-down conditions. Phenotype changes induced by IL-1β were also supported by activation of the JNK pathway, but this latter was not sensitive to PPi supplementation. Altogether our data demonstrate that the transport of PPi by ANK contributed to the maintenance of the differentiated phenotype of chondrocyte by controlling the canonical Wnt pathway in a Wnt-5a-dependent manner.
Keywords: Cell/Differentiation, Channels/Organic Anion, Diseases/Aging, Signal Transduction, Wnt pathway, ANK, Chondrocyte, Inorganic Pyrophosphate
Introduction
Articular chondrocyte phenotype is mainly characterized by the expression of genes coding for cartilage-specific extracellular matrix (ECM)3 molecules or their regulators, such as Type II collagen, Sox-9, and Aggrecan, responsible for the maintenance of cartilage anabolism (1, 2). There is a dominant influence of the ECM composition on the chondrocytic phenotype because changes in its composition, like depletion in proteoglycan content and destabilization of the supramolecular collagen network, result in chondrocyte proliferation and partial loss of the articular phenotype (3). Moreover, the balance between ECM synthesis and degradation is disturbed during rheumatic diseases, which are often associated with a progressive loss of the chondrocyte phenotype (dedifferentiation) and/or an enhanced cell apoptosis (4). Several factors, including the family of Wnt proteins, are thought to play a key role both in the occurrence and the maintenance of these articular hallmarks (5, 6).
Since the discovery of Wnt genes (7), 20 members have been described in the Wnt family, that act on the Frizzled receptors, to initiate signaling cascades resulting in the regulation of cell adhesion or the expression of genes of the homeobox family (8, 9). Wnt proteins activate cell transduction by two major ways: (i) activation of the β-catenin-T-cell factor (Tcf) pathway, referred as the canonical pathway, (ii) signaling via the cytoskeleton or the JNK pathway/protein kinase C (PKC) pathway, referred together as the non-canonical pathways (10–12). Activation of the canonical Wnt pathway induces nuclear translocation of β-catenin and its dimerization with Tcf/Lymphoid enhancer factor (Tcf/Lef), resulting in the displacement of co-repressors and the subsequent activation of transcription of target genes (13).
Wnt proteins and Frizzled receptors are thought to be involved in cartilage destruction, as they are increasingly expressed in cartilage from patients with osteoarthritis (OA) (14, 15). On the other hand, several exogenous recombinant Wnt proteins (including Wnt-3a and -7a) suppress a reporter gene expression driven by the Type II collagen promoter in a chondrogenic cell line (16). However, the cellular effect may depend on which Wnt protein is involved. Thus, expression of Wnt-5a blocked the differentiation of articular chondrocytes during development (17), whereas exposure to Wnt-7a caused chondrocyte dedifferentiation by stimulating β-catenin transcriptional activity (18). Moreover, chondrocytes express low basal levels of β-catenin, and overexpression of β-catenin caused dedifferentiation of chondrocytes (19). These observations suggest a major role of Wnt proteins in the control of chondrocyte phenotype, as well as a link between the loss of chondrocyte phenotype and the activation of the β-catenin pathway.
Interleukin (IL)-1β, which can be produced in diseased joints by resident or inflammatory cells, is a major pro-inflammatory cytokine involved in cartilage degradation and inhibition of ECM synthesis (20–22). As a consequence, IL-1β modifies cell-matrix interactions and promotes phenotypic changes in chondrocytes. Two recent studies demonstrated that Wnt-5a (18, 23) and Wnt-7a (18) were up-regulated by IL-1β, and contributed to loss of chondrocyte phenotype by activating the canonical (18) and non-canonical Wnt signaling pathways (18, 23). IL-1β was also described as a potent inhibitor of inorganic pyrophosphate (PPi) generation by chondrocytes (24), and we recently demonstrated that the PPi transporter ANK was the main contributor of ePPi production in this cell type (25). Interestingly, Ank mRNA is developmentally regulated in the cartilage of mouse embryo (26) whereas ANK dysfunction facilitates the occurrence of calcium pyrophosphate dihydrate (CPPD) deposition, which is often associated with OA (27). These observations support the key role of IL-1β in the loss of chondrocyte-differentiated phenotype and question the possible contribution of PPi and its transporter ANK in this process.
Our work aimed to investigate the role of Ank and ePPi in the maintenance of the differentiated chondrocyte phenotype and to elucidate the molecular mechanisms underlying this phenomenon. A reduced expression of Ank provoked either by IL-1β challenge or by RNA silencing was associated with a loss of collagen type II and Sox-9 expression. The effect of IL-1β on cartilage-specific markers was compensated by overexpression of Ank. The phenotypic loss of differentiated chondrocytes was driven mainly by the up-regulation of Wnt-5a, at the mRNA and protein levels, which triggered the nuclear translocation of β-catenin and activated the canonical, but not the non-canonical JNK-related Wnt pathway. Addition of exogenous PPi compensated for most of the phenotype loss induced by Ank knock-down, by decreasing activation of the Wnt canonical pathway. Exogenous PPi supplementation did not affect activation of the non-canonical Wnt pathway by IL-1β, but reduced its effect on the loss of cartilage-specific markers. Markers for hypertrophic chondrocytes including collagen type X and tissue nonspecific alkaline phosphatase (TNAP) failed to be detected throughout the study. These data demonstrate that Ank prevents the loss of the differentiated chondrocyte phenotype by controlling Wnt-5a release and secondary activation of the Wnt canonical pathway, i.e. β-catenin accumulation, through PPi exportation.
EXPERIMENTAL PROCEDURES
Chondrocyte Isolation and Culture
Articular cartilage was obtained from 6-week-old healthy male Wistar rats (130–150 g) that were killed under dissociative anesthesia (ketamine (Rhône-Mérieux) and acepromazine (Sanofi Santé Animale) in accordance with our local ethics committee and the national animal care guidelines. Articular cartilage pieces were aseptically dissected from femoral head caps and chondrocytes were obtained by sequential digestion with Pronase and collagenase B (Roche Molecular Biochemicals) as described previously (28). Cells were washed twice in PBS and cultured to confluence in 75-cm2 flasks at 37 °C in a humidified atmosphere containing 5% CO2. Cells were maintained in DME/F-12 supplemented with l-glutamine (2 mm), gentamicin (50 μg/ml), amphotericin B (0.5 μg/ml), and 10% heat-inactivated fetal calf serum (Invitrogen). In all the experiments, we used first passaged chondrocytes plated at 4 × 105 cells/well in 6-well plates. For nuclear extracts preparation, chondrocytes were seeded at 3 × 106 cells/75-cm2 flask.
Chemicals
All chemical reagents were obtained from Sigma, unless specified.
Study Design
First, we studied the changes in chondrocytes phenotype induced by IL-1β challenge (18). For that goal, chondrocytes were stimulated with 10 ng/ml of IL-1β (R&D Systems) in DME/F-12 containing 1% fetal calf serum. Expression of markers for fully differentiated or hypertrophic chondrocytes was studied by RT-qPCR at 36 h and ePPi level was assessed in culture supernatants at 72 h.
Second, to investigate the contribution of Ank to the maintenance of chondrocyte phenotype, we transfected chondrocytes either with a plasmid overexpressing ANK or with siRNA directed against Ank. Expression of chondrocytes markers was analyzed again by RT-qPCR, and immunocytochemistry was performed for ANK, Type II collagen, and Wnt-5a.
Third, the supernatant (referred to as conditioned supernatant) harvested from cells transfected for 72 h either with scramble or Ank siRNA A was used on chondrocytes monolayers to check for its ability to activate the canonical (electroporation of Tcf/Lef reporter plasmids and Western blotting of nuclear β-catenin) and non-canonical (Western blotting of JNK phosphorylation) Wnt pathways. These experiments were repeated, respectively, with mock and neutralizing Wnt-5a antibodies to define the contribution of Wnt-5a in these processes.
In another set of experiment, chondrocytes monolayers were stimulated with conditioned supernatant from cells transfected with Ank siRNA in the presence and the absence of 0.1 mm of exogenous PPi, to compensate for the loss of Ank expression. Expression of cartilage-specific genes was analyzed by RT-qPCR and the Tcf/Lef canonical pathway was checked with a plasmid reporter assay. PPi was freshly prepared as a mixture of Na2H2P2O7 and Na4P2O7 (pH 7.4).
Last, the impact of exogenous PPi on the IL-1β-induced activation of JNK was studied by Western blotting. Moreover, the regulation of cartilage-specific genes expression induced by the IL-1β challenge was analyzed in the presence and the absence of the JNK inhibitor SP600125 (5 μm), because activation of JNK was shown to inhibit Type II collagen expression in chondrocyte (23).
Extracellular PPi (ePPi) Assay
ePPi levels were measured using the differential adsorption of UDP-[6-3H]glucose (GE Healthcare), and its reaction product 6-phospho-[6-3H]gluconate on activated charcoal, as previously described (29). The standards, ranging from 10 to 400 pmol of PPi, were included in each assay. After adsorption of the reaction mixture on charcoal, and centrifugation at 16,000 × g for 10 min, 100 μl of the supernatant was counted for radioactivity in 5 ml of Bio-Safe II (Research Products International Corp.). Results were expressed as pmol of ePPi per microgram of total cell proteins (quantified by bicinchonic acid assay).
RNA Extraction and RT-PCR Analysis
Total RNA was isolated using RNeasy plus mini kit® (Qiagen), which allows the total removal of genomic DNA with an on-column DNase. 500 ng of total RNA were reverse-transcribed for 90 min at 37 °C in a 20-μl reaction mixture containing 2.5 mm dNTP, 5 μm random hexamer primers, 1.5 mm MgCl2, and 200 units of Moloney Murine Leukemia Virus reverse transcriptase (Invitrogen). Amplification of generated cDNA was performed in the Mastercycler gradient thermocycler (Eppendorf).
RT-qPCR
To quantify mRNA expression of target genes, a RT-qPCR was performed using the Lightcycler® (Roche) technology. PCR was done with SYBRgreen master mix system (Qiagen). The gene-specific primer pairs are provided in Table 1. Melting curve was performed to determine the melting temperature of the specific PCR products and after amplification the product size was checked on a 1% agarose gel stained with ethidium bromide (0.5 μg/ml). Each run included positive and negative reaction controls. The mRNA level of the gene of interest and of S29, chosen as housekeeping gene, was determined in parallel for each sample. The S29 gene, which codes for a ribosomal protein, was shown previously to be invariable in chondrocytes challenged with IL-1β (30) or transforming growth factor-β1 (TGF-β1) (25). Quantification was determined using a standard curve made for each assayed gene, from a purified PCR product, with concentrations ranging from 10−3 to 10−9 ng/ml. Results were expressed as the ratio of the mRNA level of each gene of interest over S29 gene.
TABLE 1.
Gene-specific primer pairs used in real-time quantitative PCR
| Gene | Sense | Antisense | Amplicon length (pb) | GenBankTM accession number |
|---|---|---|---|---|
| Aggrecan | 5′-ACA CCC CTA CCC TTG CTT CT-3′ | 5′-AAA GTG TCC AAG GCA TCC AC-3′ | 124 | NM_022190 |
| Ank | 5′-CAA GAG AGA CAG GGC CAA AG-3′ | 5′-AAG GCA GCG AGA TAC AGG AA-3′ | 173 | NM_053714 |
| PC-1 | 5′-TAT GCC CAA GAA AGG AAT GG-3′ | 5′-GCA GCT GGT AAG CAC AAT GA-3′ | 165 | NM_053535 |
| Runx-2 | 5′-TAT TCC CGT AGA TCC GAG CA-3′ | 5′-GCT CAC GTC GCT CAT CTT G-3′ | 82 | NM_053470 |
| S29 | 5′-AAG ATG GGT CAC CAG CAG CTC TAC TG-3′ | 5′-AGA CGC GGC AAG AGC GAG AA-3′ | 67 | NM_012876 |
| Sox-9 | 5′-CTG AAG AAG GAG AGC GAG GA-3′ | 5′-GGT CCA GTC ATA GCC CTT CA-3′ | 84 | AB 073720 |
| TNAP | 5′-GAA CGT CAA TTA ACG GCT GA-3′ | 5′-CAG ATG GGT GGG AAG AGG T-3′ | 50 | NM_013059 |
| Type IA2 collagen | 5′-TTG ACC CTA ACC AAG GAT GC-3′ | 5′-CAC CCC TTC TGC GTT GTA TT-3′ | 197 | NM_053356 |
| Type II collagen | 5′-TCC CTC TGG TTC TGA TGG TC-3′ | 5′-CTC TGT CTC CAG ATG CAC CA-3′ | 161 | NM_012929 |
| Type X collagen | 5′-ATA TCC TGG GGA TCC AGG TC-3′ | 5′-TGG GTC ACC CTT AGA TCC AG-3′ | 241 | AJ131848 |
| Wnt-3a | 5′-ACT GCA CCA CTG TCA GCA AC-3′ | 5′-CAT GGA CAA AGG CTG ACT CC-3′ | 81 | NM_001107005 |
| Wnt-5a | 5′-AAA TAG GCA GCC GAG AGA CA-3′ | 5′-GGC TCA TGG CAT TTA CCA CT-3′ | 69 | AB 244721 |
| Wnt-5b | 5′-AAC GTG GAG TAT GGC TAC CG-3′ | 5′-TGA TCC CTT GGC AAA GTT CT-3′ | 78 | NM_001100489 |
| Wnt-7a | 5′-CAC AAT TCC GAG AGC TAG GC-3′ | 5′-ACG GCC TCG TTG TAT TTG TC-3′ | 52 | NM_009527 |
| Wnt-11 | 5′-CTG ACC TCA AGA CCC GCT AC-3′ | 5′-TAG GCC GGT GTA CCA CTT TC-3′ | 51 | NM_080401 |
RNA Silencing Experiments
siRNA sequences (Table 2) were designed by Eurogentec. Transfections were carried out with each siRNA at a final concentration of 10 nm, using INTERFERinTM (Polyplus-transfection SA). siRNA were diluted in serum-free medium, INTERFERinTM was then added to the mix for a short incubation at room temperature. Cells were then washed with PBS and placed in serum-free medium. The siRNA-INTERFERinTM mix was then added to the culture for 24 h. After this time, cells were stimulated or not with 0.1 mm of PPi for 48 h.
TABLE 2.
siRNA sequences used in transitory knock-down experiments
| Name | Sense | Antisense |
|---|---|---|
| Scramble | 5′-CGA UGG GUU CGU GUC GUU U-3′ | 5′-AAA CGA CAC GAA CCC AUC G-3′ |
| siRNA Ank | 5′-CUG GCC AAC ACG AAC AAC A-3′ | 5′-UGU UGU UCG UGU UGG CCA G-3′ |
| siRNA Ank bis | 5′-GGG UAC UAC AUC AUC AAC A-3′ | 5′-UGU UGA UGA UGU AGU ACC C-3′ |
Preparation of Conditioned Supernatants
Supernatants from chondrocytes transfected either with scramble or Ank siRNA were harvested and clarified by centrifugation at 12,000 × g for 5 min, then concentrated 50-fold using Microcon (Millipore) centrifugation devices with a 30-kDa molecular mass cut-off. In one set of experiments, these conditioned supernatants were incubated for 1 h at 37 °C either with anti-β actin (mock) or anti-Wnt-5a (H-58, sc-30224, Santa Cruz Biotechnology) rabbit polyclonal antibodies, each used at 0.4 μg/ml, prior being clarified and concentrated as described above.
Immunocytochemistry
Chondrocytes were seeded at 104 cells per chamber in Lab-Tek II chamber slide (Nunc GmbH & Co.KG, Germany). Immunocytochemistry was processed for 15 min at 37 °C using LSAB+ System-HRP kit (DakoCytomation) on cells fixed with a 4% PBS-PFA solution (pH 7.4). Cell peroxidases were blocked by covering monolayers with a 3% H2O2 solution for 5 min. After two washing steps with PBS, chondrocytes were incubated for 30 min with primary antibodies against ANK, Type II collagen (MAB1330, mouse monoclonal, Millipore) or Wnt-5a (H-58, rabbit polyclonal, sc-30224, Santa Cruz Biotechnology) used at 1:100 in PBS, 1% bovine serum albumin. Purified rabbit polyclonal antibody against ANK was designed by Eurogentec, using a keyhole limpet hemocyanin-coupled peptide with the following sequence NH2-AEEVTDIVEMREENE-COOH. After washing with PBS, cells were incubated for 30 min with biotinylated anti-rabbit or anti-mouse IgG. After PBS washing, chondrocytes were incubated for 15 min with streptavidin conjugated to horseradish peroxidase. Cells were then stained using a solution containing DAB as a chromogen and H2O2. Counterstaining was done using Harris Hematoxylin. Images were acquired in air medium at room temperature using DMD108 microimaging device, connected to a 3 million pixel charge-coupled device camera (Leica Microsystems GmbH) and software Leica Acquisition Suite. Magnification was 10× and numerical aperture was 0.25.
Nuclear Extracts Preparation
To study the translocation of β-catenin into the nucleus, nuclear fractions were isolated from cells stimulated for 48 h either with conditioned supernatants or 10 mm of LiCl used as a positive control. Briefly, chondrocytes were harvested and placed in a cold buffer (pH 7.9) containing 10 mm Hepes, 10 mm KCl, 0.1 mm EGTA, 0.1 mm EDTA, 1 mm dithiothreitol, and one tablet of complete mini protease inhibitor mixture (Roche). After 15 min of incubation at 4 °C, 0.6% Igepal CA-630 was added to the cell suspension, which was then homogenized. Nuclei were pelleted by centrifugation for 5 min at 4 °C and 200 × g. Supernatant, containing the cytosolic fraction was harvested. Nuclei were lysed in a cold buffer containing 20 mm Hepes, 0.4 m NaCl, 1 mm EGTA, 1 mm EDTA, 1 mm dithiothreitol, 0.25% Igepal CA-630, and one tablet of Complete mini protease inhibitor mixture (Roche). Lysate was then centrifuged at 12,000 × g for 10 min at 4 °C. Cytosolic and nuclear proteins were quantified by bicinchonic assay and used for Western blotting of β-actin and β-catenin, respectively.
Western Blot Analyses
Concentrated supernatants were placed in final concentration of 1× Laemmli buffer. For JNK analyses, chondrocytes were harvested and lysed in 1× Laemmli buffer. Samples were run on SDS-PAGE (10%) and transferred onto a polyvinylidene fluoride membrane. After 2 h in blocking buffer (TBS-Tween-5% nonfat dry milk), membranes (Immobilon, Waters) were washed three times with TBS-Tween and incubated overnight at 4 °C with antibodies against either phospho and total JNK (Cell Signaling Technology) used at 1:500, or against Wnt-5a (Santa Cruz Biotechnology) and β-catenin (Affinity BioReagents) used at 1:1000 or against β-actin (Sigma) used at a dilution of 1:4000. After three washing steps with TBS-Tween, each blot was incubated for 1 h at room temperature with anti-rabbit IgG conjugated with horseradish peroxidase (Cell Signaling Technology) at a 1:2000 dilution in blocking buffer. After four washing steps with TBS-Tween, protein bands were detected by chemiluminescence with the Phototope DetectionTM system according to manufacturer's recommendations (Cell Signaling Technology). The band intensities were quantified by densitometry with a computerized image processing system (Gnome, Syngene).
Plasmid Electroporation
Chondrocytes were electroporated as previously described (25), in two set of experiments. In the mRNA expression study, cells were transfected with 6 μg of either empty pcDNA3.1 (Invitrogen) used as control, or a plasmid encoding for wild-type Ank (pANK). For that goal, Ank coding DNA sequence was cloned from chondrocytes (GenBankTM accession number NP_446166) in pcDNA3.1. Cells were then stimulated or not with 10 ng/ml of IL-1β for 48 h before RNA extraction. Plasmid pmaxGFPTM (Amaxa Biosystems), encoding a GFP was used to determine the transfection efficiency. In the Tcf/Lef reporter plasmid study, 3 μg of either TOPFlash (wild-type Lef-binding site) or FOPFlash (control, mutated Lef-binding site) (Millipore), and 600 ng of pCMV-Renilla (Promega) were electroporated in chondrocytes.
Reporter Gene Assay
Luciferase activities were measured using Dual-Luciferase Reporter Assay kit (Promega). Briefly, electroporated cells were stimulated for 48 h with conditioned supernatant. After two washing steps with PBS, cells were lysed in 1× passive lysis buffer for 15 min at room temperature. The cell lysate was placed in LAR II (substrate for firefly luciferase) and luminescence was read with a Sirius luminometer (Berthold Technologies). Afterward, Stop & Glo reagent was added to the sample (substrate for Renilla luciferase), and luminescence was acquired by the same way. Cells stimulated with LiCl (10 mm) for 48 h were used as a positive control for Tcf/Lef signaling. Results were expressed as mean ratio of luciferase activity, firefly/Renilla.
Statistical Analysis
Results are expressed as the mean ± S.D. of at least three independent assays. Comparisons were made by ANOVA, followed by Fisher's t post-hoc test, using the StatviewTM 5.0 software (SAS Institute Inc). A value of p < 0.05 was considered significant.
RESULTS
IL-1β Induces the Loss of the Differentiated Phenotype of Articular Chondrocytes
As shown in Table 3, IL-1β reduced Type II collagen and Sox-9 expression by 9.5-fold and 3-fold, respectively. A concomitant 2.9-fold decrease of Aggrecan mRNA level was observed. In these experimental conditions, Ank and PC-1 expression were reduced by 3.4-fold and 2.8-fold, respectively, whereas Wnt-5a mRNA level increased by 2.5-fold. No expression of either type I or X collagens, TNAP or runx-2 was detected in basal or IL-1β conditions. Wnt-3a and Wnt5b also failed to be detected whereas Wnt-7a and Wnt-11 mRNA level was detectable in basal conditions but remained unaffected in response to IL-1β challenge.
TABLE 3.
Effects of IL-1β on the relative mRNA expression of genes involved in the PPi metabolism, extracellular matrix synthesis, and Wnt pathway
| Gene | Control | +IL-1β | Fold range |
|---|---|---|---|
| Aggrecan | 6.40 ± 0.70 | 2.20 ± 0.20*a | −2.9 |
| Ank | 4.10 ± 0.20 | 1.20 ± 0.05* | −3.4 |
| PC-1 | 2.50 ± 0.60 | 0.90 ± 0.10* | −2.8 |
| Runx-2 | N.D.b | N.D. | N.D. |
| Sox-9 | 0.60 ± 0.05 | 0.20 ± 0.01* | −3 |
| TNAP | N.D. | N.D. | N.D. |
| Type IA2 collagen | N.D. | N.D. | N.D. |
| Type II collagen | 9.50 ± 1.80 | 1.00 ± 0.40* | −9.5 |
| Type X collagen | N.D. | N.D. | N.D. |
| Wnt-3a | N.D. | N.D. | N.D. |
| Wnt-5a | 1.00 ± 0.25 | 2.50 ± 0.15* | +2.5 |
| Wnt-5b | N.D. | N.D. | N.D. |
| Wnt-7a | 0.10 ± 0.02 | 0.09 ± 0.02 | N.S. |
| Wnt-11 | 0.30 ± 0.05 | 0.32 ± 0.07 | N.S. |
a Statistically significant differences from the control, p < 0.05.
b N.D., not detected.
Ank Overexpression Reduces IL-1β-induced Changes in Chondrocyte Phenotype Markers
The data obtained with scramble-transfected chondrocytes confirmed the inhibitory effect of IL-1β on the mRNA level of cartilage-specific markers, as well as the inverse variation of Wnt-5a (Fig. 1A). Consistently with its inhibitory effect on Ank expression, IL-1β reduced ePPi generation by 3-fold in these cells, although a concomitant 2.5-fold decrease of PC-1 mRNAs was noted (Fig. 1B).
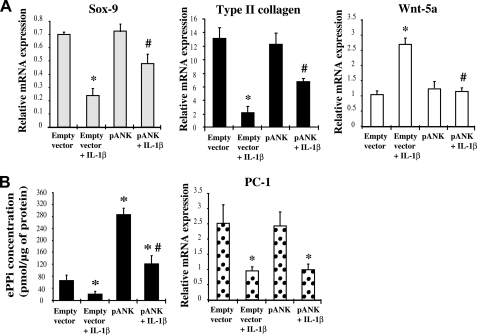
FIGURE 1.
Influence of Ank overexpression on IL-1β-induced changes in chondrocytes markers. A, effect of Ank overexpression on Sox-9, Type II collagen, and Wnt-5a expression. Total RNA was extracted from chondrocytes electroporated either with empty vector or with Ank-overexpressing plasmid (pANK) and stimulated with 10 ng/ml of IL-1β, then subjected to RT-qPCR analysis. The levels of mRNA of interest were normalized to that of S29 (reference gene). Results are presented in histograms as means (± S.D.) over S29 value (n = 3). B, influence of Ank overexpression on ePPi generation and PC-1 expression. ePPi was assayed in supernatants of chondrocytes supernatants of chondrocytes electroporated either with empty vector or with Ank-overexpressing plasmid (pANK), and stimulated with 10 ng/ml of IL-1β for 72 h, and normalized to cell proteins content (n = 6). Data are expressed as mean (± S.D.) in picomoles per microgram of protein. Total RNA was extracted from chondrocytes electroporated either with empty vector or with Ank overexpressing plasmid (pANK) and stimulated with 10 ng/ml of IL-1β, then subjected to RT-qPCR analysis. The level of PC-1 mRNA was normalized to that of S29 (reference gene). Results are presented in histograms as means (± S.D.) over S29 value (n = 3). Statistically significant differences from control/empty vector are indicated as *, p < 0.05, and from empty vector + IL-1β as #, p < 0.05.
A control experiment confirmed the efficiency of our method of transfection and the functionality of the plasmid used because Ank mRNA level was ∼50-fold higher in resting chondrocytes transfected with pANK than with an empty vector (data not shown). In chondrocytes electroporated with this Ank-overexpressing plasmid, ePPi production was around 4-fold higher than in cells transfected with the empty vector, further demonstrating the activity of our construct (Fig. 1B). Moreover, the inhibitory effect of IL-1β on ePPi generation was totally counteracted in cells transfected with pANK whereas the IL-β1-induced decrease of PC-1 expression remained unaffected (Fig. 1B). This result demonstrated that the restoration of PPi exportation was supported only by the correction of Ank deficiency. In these conditions, the effect of IL-1β on Wnt-5a expression was almost completely suppressed, whereas the mRNA level of Sox-9 and Type II collagen were diminished by 33 and 47% instead of 65 and 85%, respectively (Fig. 1A). These data demonstrated that ANK overexpression counteracted most of the ability of IL-1β to provoke loss of the differentiated phenotype in chondrocytes.
Ank Silencing Mimics IL-1β Effect on Chondrocyte Phenotype Markers
We previously demonstrated that our siRNA sequence reduced the basal expression of Ank by at least 90% in chondrocytes without affecting that of the housekeeping gene S29 (25). As shown in Fig. 2A, the basal mRNA level of Ank was strongly reduced in the presence of Ank siRNA compared with scramble-transfected chondrocytes. The silencing of Ank reduced Type II collagen expression by 50% and Sox-9 expression by 35% while increasing Wnt-5a mRNA level by 2.3-fold. Similar results were obtained with the alternative siRNA Ank bis sequence (data not shown). Type I or X collagens, TNAP or runx-2 mRNA remained undetectable in siRNA-transfected chondrocytes (data not shown). Immunocytochemical analysis confirmed that siRNA reduced efficiently the number of chondrocytes staining positively for Ank and Type II collagen, while increasing the number of Wnt-5a-positive cells (Fig. 2B). The efficiency of Ank silencing was also demonstrated at the functional level because it reduced the basal production of ePPi by 3.6-fold (Fig. 2C). Altogether, these observations indicated that the transient knock-down of Ank stimulated Wnt-5a expression and promoted the loss of cartilage-specific markers without switching chondrocyte toward a hypertrophic phenotype.
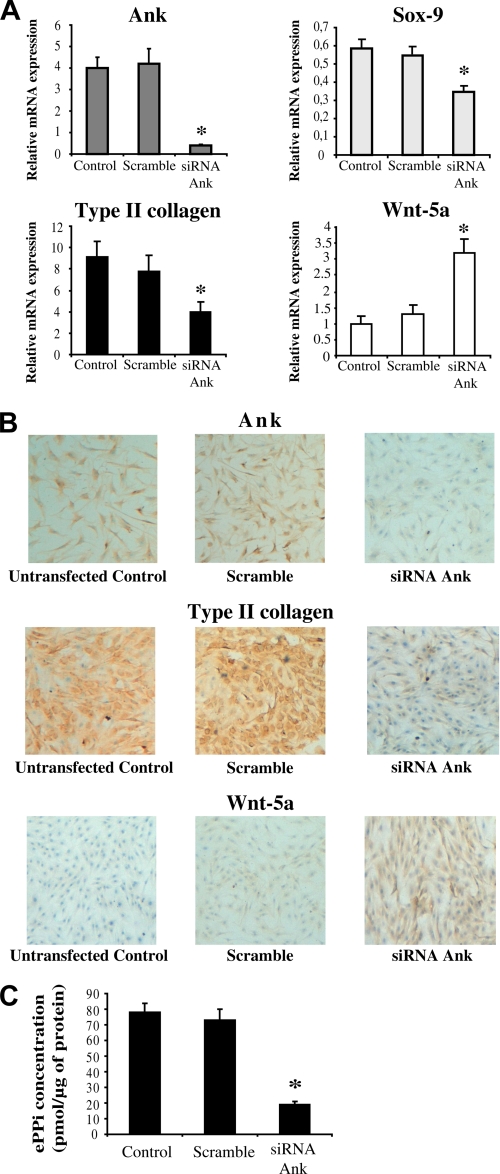
FIGURE 2.
Influence of Ank knock-down on chondrocyte phenotype markers. A, effect of Ank siRNA on Ank, Sox-9, Type II collagen, and Wnt-5a mRNA expression. Total RNA was extracted from chondrocytes transfected (or not, depicted as “control”) with Ank siRNA, and subjected to RT-qPCR analysis. The levels of mRNA of interest were normalized to that of S29 (used as reference gene). Results are presented in histograms as means (± S.D.) over S29 value (n = 3). Statistically significant differences from the Scramble are indicated as *, p < 0.05. B, effect of Ank siRNA on Ank, Type II collagen, and Wnt-5a protein expression. Chondrocytes were fixed with a 4% PBS-PFA, and then incubated with anti-Ank, anti-Type II collagen, or anti-Wnt-5a antibodies. Biotinylated anti-rabbit and anti-mouse IgG were then used, followed by incubation with streptavidin conjugated to horseradish peroxidase. Cells were stained using a solution containing DAB chromogen and H2O2. Counterstaining was done using Harris hematoxylin. Magnification is ×10. Images presented are representative of three independent experiments. C, influence of transitory Ank knock-down on ePPi generation. ePPi was assayed in supernatants of chondrocytes supernatants of chondrocytes transfected (or not, depicted as “control”) with Ank siRNA, and normalized to cell proteins content (n = 6). Data are expressed as mean (± S.D.) in picomoles per microgram of protein. Statistically significant differences from the Scramble are indicated as *, p < 0.05.
Wnt5a Contributes to the Loss of Phenotype Induced by Conditioned Supernatant of Chondrocytes Knocked-down for Ank
As depicted in Fig. 3A, the conditioned supernatant of chondrocytes transfected with Ank siRNA contained a significant amount of Wnt-5a, which was released only marginally in the conditioned supernatant of cells transfected with scramble When these supernatants were added to chondrocytes monolayers, they promoted a loss of phenotype markers, which was unaffected by a prior neutralization with mock antibody but was totally prevented by an incubation with anti-Wnt-5a antibody (Fig. 3B). This suggested that most effects of supernatants were supported by Wnt5a.
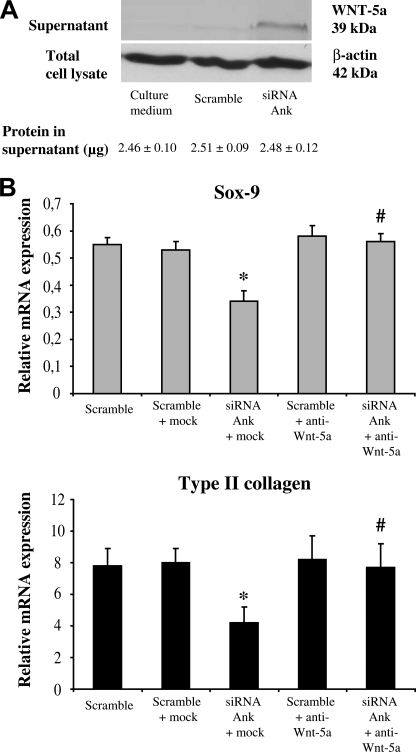
FIGURE 3.
Influence of transitory knock-down of Ank on Wnt-5a production. A, Western blot analysis of conditioned supernatant. Supernatants were subjected to Western blotting. Quantification of protein content of the loaded volumes of supernatant (in micrograms) was done using the bicinchonic acid method. Images presented are representative of three independent experiments. B, effect of conditioned supernatant on Sox-9 and type II collagen expression. Conditioned supernatants were neutralized or not for 1 h at 37 °C, using either mock (anti-β-actin) or anti-Wnt-5a antibodies. Total RNA was extracted from chondrocytes transfected with either scramble or Ank siRNA, and subjected to RT-qPCR analysis. The levels of mRNA of interest were normalized to that of S29 (used as reference gene). Results are presented in histograms as means (± S.D.) over S29 value (n = 3). Statistically significant differences from the Scramble are indicated as *, p < 0.05, and from mock as #, p < 0.05.
Wnt5a Accounts for the Potency of Conditioned Supernatant to Activate the Canonical Wnt Pathway in Chondrocyte Monolayers
As depicted in Fig. 4A, the conditioned supernatant of chondrocytes transfected with Ank siRNA activated the Tcf/Lef (TOPFlash) reporter plasmid by 2.3-fold compared with those collected from scramble-transfected cells. A control experiment with 10 mm LiCl, which is known to cause β-catenin accumulation by inhibiting glycogen synthase kinase (GSK)-3β activity, confirmed that the TOPFlash plasmid was responsive. In addition, the defective FOPFlash construct failed to be activated either by LiCl or by conditioned supernatants (Fig. 4A). The activation of the Tcf/Lef reporter plasmid was unaffected by a prior neutralization with mock antibody but was totally prevented by the Wnt-5a antibody (Fig. 4B). As expected, neither of both antibodies affected the responsiveness of the FOPFlash reporter plasmid. Finally, Western blotting demonstrated that the conditioned supernatant of chondrocytes transfected with Ank siRNA increased the translocation of β-catenin into the nucleus, as was the case for the LiCl positive control (Fig. 4C). However, the supernatant was unable to induce the phosphorylation of JNK, which was obvious in IL-1β-stimulated cells chosen as a positive control (Fig. 4D). Taken together, these data indicated that the supernatant of chondrocytes transfected with Ank siRNA activated the canonical, but not non-canonical, Wnt pathway in a Wnt-5a-dependent manner.
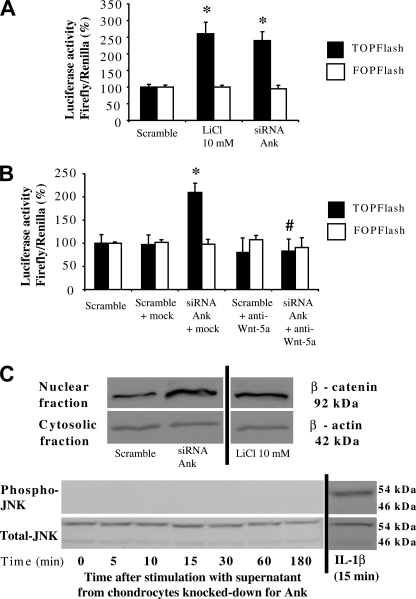
FIGURE 4.
Influence of transitory knock-down of Ank on the canonical and non-canonical Wnt pathways activation. A, effect of conditioned supernatant on Tcf/Lef pathway activation. Chondrocytes were electroporated either with active (TOPFlash) or with inactive (FOPFlash) Tcf/Lef reporter genes, and with CMV-Renilla reporter gene before stimulation with supernatant from cells transfected with Ank siRNA for 48 h. Cells were stimulated with LiCl (10 mm) as a positive control. Results are presented in histograms as mean luciferase activity ratio of firefly/Renilla (± S.D.) (n = 4). B, effect of neutralized supernatant on Tcf/Lef pathway activation. Conditioned supernatants were neutralized or not for 1 h at 37 °C, using either mock (anti-β-actin) or anti-Wnt-5a antibodies. Chondrocytes transfected with reporter genes were stimulated with these supernatants for 48 h. Results are presented in histograms as mean luciferase activity ratio of firefly/Renilla (± S.D.) (n = 4). Statistically significant differences from Scramble are indicated as *, p < 0.05, and from mock as #, p < 0.05. C, effect of conditioned supernatants on the nuclear translocation of β-catenin and on the activation of the non-canonical Wnt pathways. Chondrocytes were stimulated with supernatants from cells transfected with either scramble or Ank siRNA for 48 h or the indicated times. Cells stimulated with 10 mm LiCl were used as a positive control for nuclear translocation of β-catenin, and cells stimulated with 10 ng/ml of IL-1β for 15 min were used as a positive control for JNK activation. Proteins were extracted and subjected to Western blotting using either anti-β-catenin, or anti-phospho- and anti-total-JNK antibodies. The relative abundance of β-catenin was normalized to that of β-actin protein. Images presented are representative of three independent experiments.
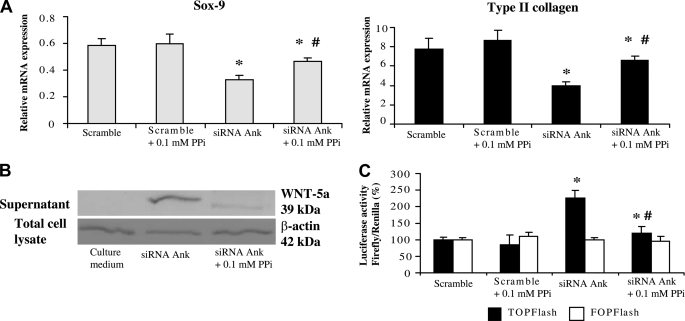
Exogenous PPi Compensates for the Loss of Phenotype Induced by Conditioned Supernatant of Chondrocytes Knocked-down for Ank
As shown in Fig. 5A, the inhibitory effect of conditioned supernatant of Ank knocked-down chondrocytes on Sox-9 mRNA level was reduced from 1.6-fold to 1.3-fold when 0.1 mm of exogenous PPi was added to culture medium. The expression of Type II collagen displayed the same profile, with an inhibitory effect switching from 2–1.3-fold after addition of exogenous PPi (Fig. 5A). In these experimental conditions, 0.01 mm PPi remained ineffective, whereas 1 mm PPi worsened the loss of cartilage-specific markers (data not shown). Western blot analysis further demonstrated that addition of 0.1 mm exogenous PPi diminished the potency of chondrocytes transfected with Ank siRNA to release Wnt-5a into culture supernatant (Fig. 5B). Moreover, exogenous PPi reduced by 1.9-fold the ability of this conditioned supernatant to activate the Tcf/Lef reporter plasmid (Fig. 5C). These data indicated that exogenous PPi compensated for most effects of Ank silencing on the expression of cartilage-specific markers, Wnt-5a release, and subsequent activation of the canonical Wnt signaling pathway.
FIGURE 5.
Influence of ePPi on the loss of phenotype induced by supernatants from chondrocytes transfected with Ank siRNA. A, effect of Ank siRNA, in the presence and the absence of 0.1 mm ePPi, on Type II collagen, and Sox-9 expression. Total RNA was extracted from chondrocytes transfected with Ank siRNA and subjected to RT-qPCR analysis. The levels of mRNA of interest were normalized to that of S29 (used as reference gene). Results are presented in histograms as means (± S.D.) over S29 value and are representative of three independent experiments. B, Western blot analysis of conditioned supernatant. Supernatants were concentrated and subjected to Western blotting. Images presented are representative of three independent experiments. C, effect of conditioned supernatant, in the presence and the absence of 0.1 mm PPi, on Tcf/Lef pathway activation. Chondrocytes were electroporated either with active (TOPFlash) or inactive (FOPFlash) Tcf/Lef reporter genes (1 μg/well of 6-well plate), and with CMV-Renilla reporter gene (200 ng/well of 6-well plate). Cells were then stimulated with conditioned supernatant from siRNA-transfected cells challenged or not with 0.1 mm ePPi for 48 h. Data are expressed as the mean luciferase activity ratio of firefly/Renilla (± S.D.) (n = 3). Statistically significant differences from supernatant produced by cells transfected with scramble are indicated as *, p < 0.05, and from cells transfected with Ank siRNA as #, p < 0.05.
Exogenous PPi Failed to Affect the Non-canonical Wnt Pathway in the Il-1β-induced Loss of Chondrocyte Phenotype
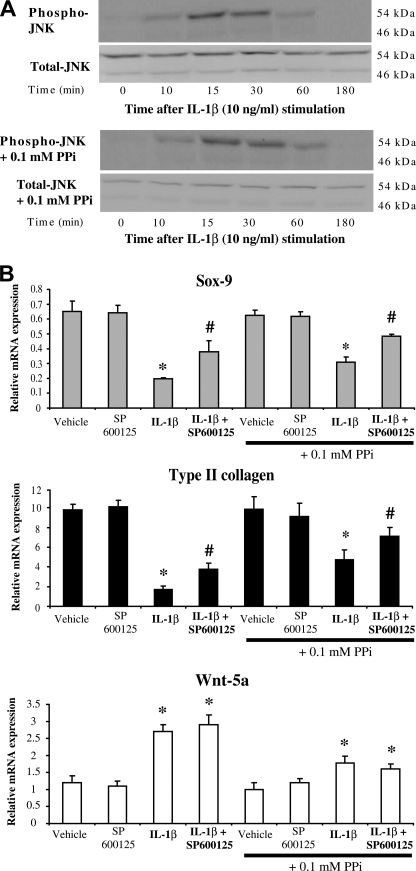
As shown in Fig. 6A, IL-1β induced the phosphorylation of JNK with a maximal activation at 15 min. Addition of 0.1 mm of exogenous PPi did not modify the pattern of JNK activation by IL-1β.
FIGURE 6.
Influence of ePPi on the loss of phenotype induced by IL-1β. A, effect of ePPi on the activation of JNK pathway by IL-1β. Chondrocytes were stimulated with 10 ng/ml of IL-1β, in the presence and the absence of 0.1 mm of ePPi for the indicated times. Proteins were extracted and subjected to Western blotting using anti-phospho- and anti-total-JNK antibodies. Images presented are representative of three independent experiments. B, effect of SP600125 and ePPi on the dedifferentiating effects of IL-1β. Total RNA was extracted from chondrocytes challenged for 1 h with SP600125 (5 μm), then stimulated with 10 ng/ml of IL-1β in the presence and absence of 0.1 mm of ePPi. Total RNA was subjected to RT-qPCR analysis. The levels of mRNA of interest were normalized to that of S29 (used as reference gene). Results are presented in histograms as means (± S.D.) over S29 value (n = 3). Statistically significant differences from vehicle or vehicle + PPi are indicated as *, p < 0.05, and from cells stimulated with IL-1β and from cells stimulated with IL-1β + PPi as #, p < 0.05.
The inhibitory potency of IL-1β on Sox-9 mRNA level was reduced from 3.3-fold to 2-fold in the presence of the selective JNK inhibitor SP600125 (Fig. 6B). Addition of exogenous PPi attenuated the IL-1β inhibitory effect from 2.1-fold in the absence to 1.3-fold in the presence of SP600125. Similarly, the inhibitory potency of IL-1β on Type II collagen mRNA level was reduced from 5.3- to 2.5-fold in the presence of SP600125 (Fig. 6B). Once again, the addition of exogenous PPi diminished the inhibitory effect of IL-1β from 2.2-fold in the absence to 1.3-fold in the presence of SP600125. In contrast, the stimulating effect of IL-1β on Wnt-5a mRNA level was not affected by the presence of SP600125 (Fig. 6B). However, addition of exogenous PPi diminished this stimulating potency from 2.2-fold to ∼1.7-fold. Altogether, these data demonstrated that IL-1β promoted the loss of the differentiated phenotype of chondrocyte by two distinct mechanisms: a JNK-dependent PPi-independent pathway and a Wnt-5a PPi-dependent pathway. This suggests that the Ank deficiency provoked by IL-1β contributes by its own to the changes of chondrocyte phenotype seen under cytokine challenge.
DISCUSSION
IL-1β or TNF-α has been favored as the main trigger of experimental arthritis although with a variable contribution depending on the model used. Nonetheless, in vitro and in vivo studies have suggested that IL-1β is a major driver of cartilage damage and possibly exceeds the effects of TNF-α (31). More recently, IL-1β was even demonstrated to support cartilage damage in human TNF-α-transgenic mice possibly due to its regulatory role on matrix-degrading enzymes (32). At the cellular level, IL-1β was shown to be a potent modulator of the chondrocyte phenotype, as it down-regulated the expression of some cartilage-specific genes including those encoding Types IX (33), XI, and II (34) collagens and aggrecan (34). Most of these effects are thought to be supported by the reduced expression of the transcription factor Sox-9 (35), which is able to activate chondrocyte-specific enhancer elements in Coll2a1 and Coll1a2 genes (36). Therefore, IL-1β has a well-known pathological relevance to cartilage degradation occurring in rheumatic diseases, and to the accompanying loss of differentiated phenotype that affects chondrocytes under stress conditions.
In the present study, we demonstrate that IL-1β induced chondrocytes to express less cartilage specific markers without favoring the expression of markers for hypertrophic chondrocytes, such as collagen type X and runx-2. TNAP was also neither detected at the mRNA level nor as an enzymatic activity in resting chondrocytes, consistently with the fully differentiated phenotype of our cell population (25). TNAP expression remained undetectable in response to IL-1β as it was previously the case when chondrocytes were challenged with TGF-β1 (25). These data are in good agreement with previous findings (18–22) and support that IL-1β causes a loss of the differentiated phenotype of articular chondrocytes and not the progression of differentiated chondrocytes to a hypertrophic state.
The IL-1β-induced loss of differentiated phenotype paralleled the reduced expression of the transporter Ank whereas an opposite pattern of expression was observed for Wnt-5a. The up-regulation of Wnt-5a confirmed previous data in rabbit articular (23) or temporomandibular joint condylar chondrocytes (37), although two different patterns of expression have been reported in response to IL-1β (23, 37). Similarly, Wnt-3a remained insensitive to cytokine challenge (18), albeit it was reported to contribute to chondrocyte dedifferentiation (38). In contrast, neither induction of Wnt-7a nor down-regulation of Wnt-11 by IL-1β was detected in our experimental system despite their reported variations in rabbit chondrocytes (23). Such discrepancy may reflect differences in chondrocytes sources and/or experimental conditions. However, our data confirmed that, among the Wnt family, Wnt-5a was representative of the chondrocyte response to IL-1β. This finding is particularly interesting when considering that several domains are evolutionary conserved between the rodents and human Wnt-5a promoters (39), which makes sense to use rat cells for experiments. The inverse variation between Wnt-5a and Type II collagen or Sox-9 expression was highly consistent with the previous demonstration of an inhibitory role of Wnt-5a on cartilage-specific components in articular chondrocytes (23).
A major finding was that overexpression of Ank suppressed the stimulating effect of IL-1β on Wnt-5a while counterbalancing most of its inhibitory potency on Sox-9 and Type II collagen mRNA levels. An additional demonstration of the contribution of Ank to the maintenance of the differentiated phenotype of chondrocyte was that Ank RNA silencing induced an “IL-1β-like” phenotype in resting cells. Based on the marginal changes of the cell phenotype after transfection with a scramble sequence and on the efficiency of the silencing technology used (25), our data validate that the loss of the differentiated phenotype was closely related to Ank deficiency. Although the Ank gene is expressed in several non skeletal tissues, it has been shown to be developmentally regulated in the cartilage of mouse embryo (26) and to be expressed in an oxygen-dependent manner (40) in the hypertrophic zone of the growth plate, where it plays a regulatory role in the mineralization process (41). However, Ank was also found in the superficial zone of articular cartilage (41), consistently with the basal expression we detected in primary chondrocytes, and its expression was shown to extent to the deepest zones of cartilage in osteoarthritic samples (42). We showed that the expression of Wnt-5a varied inversely to those of Ank in chondrocytes whereas no markers of hypertrophy occurred when Ank was expressed at a very low level. We demonstrated further that neutralization of Wnt-5a in culture supernatant prevented the loss of phenotype induced by Ank deficiency. Therefore, Ank contributed to the maintenance of the differentiated chondrocyte phenotype by controlling Wnt-5a expression.
As Ank is a membrane transporter, we next investigated whether phenotype changes depended or not on PPi export. We demonstrated that the loss of differentiated phenotype, i.e. the reduced expression of Sox-9 and Type II collagen, induced either by IL-1β or by RNA silencing was reversed consistently by addition of PPi. This suggests that most effects of Ank were mediated by its transporter function. Very few data are available on how ePPi could regulate gene expression in chondrocytes. In contrast, several evidences support an important role for inorganic phosphate (Pi) as an early differentiation factor in the ATDC5 cell line (43) and other mineralizing cells (44, 45). However, mineralizing cells have a high TNAP activity which was not the case of our mature chondrocytes. Therefore, a Pi-mediated mechanism resulting from hydrolysis of PPi by TNAP seems highly unlikely. Among possible ways to regulate Wnt-5a, PPi could activate ERK1/2 and p38 MAPK signaling pathways, as was demonstrated in osteoblasts (46). Alternatively, expression of Wnt-5a could be increased in a STAT-3-dependent manner (47) although the biological responses could vary with the cell type. That addition of PPi failed to compensate entirely for the loss of phenotype induced by IL-1β was not surprising, because this cytokine is known to activate many signaling pathways in chondrocytes (48, 49). We showed that IL-1β activated JNK independently of the Wnt cascade because the kinetics of JNK phosphorylation was too short to account for the production of Wnt-5a. In addition, inhibition of JNK pathway with the selective inhibitor SP600125 left Wnt-5a mRNA level unchanged. Therefore, IL-1β promoted the loss of the differentiated phenotype of chondrocyte in a JNK-dependent PPi-insensitive pathway in addition to a Wnt-5a PPi-dependent mechanism.
The Wnt family of proteins has been classically separated into the Wnt-1 class, which includes Wnt-1, -3a, -7a, and -8, and the Wnt-5a class, which includes Wnt-4, -5a, and -11 (50). The Wnt-1 class is thought to activate the canonical Wnt/β-catenin pathway whereas the Wnt-5a class is expected to signal independently of β-catenin by activating protein kinase C, calmodulin-dependent kinase II or JNK. In accordance with the general classification, Wnt-5a was shown previously to activate the non-canonical Wnt pathway in IL-1β-stimulated chondrocytes (23) and to promote the expression of several MMPs by these alternative ways (37). However, recent data have raised the complexity of subdivision of the Wnt family which may be dependent on the receptor profile (51). Thus Wnt-1 does not always signal through the canonical Wnt pathway (52) whereas β-catenin has been implicated in the Wnt-4 signaling pathway (53). Several lines of evidence support that Wnt-5a activated the canonical Wnt pathway in our experimental system since conditioned supernatant of cells knocked-down for Ank: (i) activated Tcf/Lef reporter plasmids; (ii) increased the nuclear translocation of β-catenin; (iii) failed to activate reporter plasmids after prior neutralization with an Wnt-5a antibody. In addition, such conditioned supernatant was unable to induce JNK phosphorylation in resting chondrocytes, further demonstrating that this signaling pathway was not involved. As our data are inconsistent with previous findings in rabbit chondrocytes (23, 37), further studies are required to investigate whether this may reflect differences in the expression pattern of Frizzled receptors rather than in experimental conditions. However, among the Wnt receptors expressed in the rat (54, 55), Wnt-5a was described to activate the canonical Wnt pathway through a particular Rfz-2 signaling (56). Finally, activation of the canonical Wnt pathway by Wnt-5a, secondary to the reduction of Ank expression, was highly consistent with the major role of β-catenin in the phenotypic loss of differentiated chondrocytes reported after IL-1β challenge (19).
In summary, we have demonstrated here that the PPi transporter Ank preserved the differentiated phenotype of chondrocyte by regulating the canonical Wnt pathway in a Wnt-5a-dependent manner. Our results provide evidence that change in Ank expression may contribute to the loss of phenotype that can affect articular chondrocytes under pathological conditions.
Acknowledgment
We thank Dr. Pascal Reboul for helpful comments and remarks concerning the writing of the manuscript.
This work was supported by grants from the Fondation pour la Recherche Médicale (FRM) and from the University Hospital of Nancy (CPRC/PHRC), the Région Lorraine, the Conseil Général 54, and the Communauté Urbaine du Grand Nancy.
- ECM
- extracellular matrix
- IL-1β
- interleukin-1β
- JNK
- c-Jun N-terminal kinase
- PC-1
- plasma cell membrane glycoprotein-1
- PKC
- protein kinase C
- ePPi
- extracellular PPi
- Tcf/Lef
- T-cell factor/lymphoid enhancer factor
- TGF-β1
- transforming growth factor-β1
- TNAP
- tissue nonspecific alkaline phosphatase
- TNF-α
- tumor necrosis factor-α
- PBS
- phosphate-buffered saline
- RT-qPCR
- real-time quantitative RT-PCR.
REFERENCES
- 1.Zhao Q., Eberspaecher H., Lefebvre V., De Crombrugghe B. (1997) Dev. Dyn. 209, 377–386 [DOI] [PubMed] [Google Scholar]
- 2.Goldring M. B. (2006) Best Pract. Res. Clin. Rheumatol. 20, 1003–1025 [DOI] [PubMed] [Google Scholar]
- 3.Aigner T., McKenna L. (2002) Cell Mol. Life Sci. 59, 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aigner T., Söder S., Gebhard P. M., McAlinden A., Haag J. (2007) Nat. Clin. Pract. Rheumatol. 3, 391–399 [DOI] [PubMed] [Google Scholar]
- 5.Goldring M. B., Tsuchimochi K., Ijiri K. (2006) J. Cell. Biochem. 97, 33–44 [DOI] [PubMed] [Google Scholar]
- 6.Pacifici M., Koyama E., Shibukawa Y., Wu C., Tamamura Y., Enomoto-Iwamoto M., Iwamoto M. (2006) Ann. N.Y. Acad. Sci. 1068, 74–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nusse R., Brown A., Papkoff J., Scambler P., Shackleford G., McMahon A., Moon R., Varmus H. (1991) Cell 64, 231. [DOI] [PubMed] [Google Scholar]
- 8.Wodarz A., Nusse R. (1998) Annu. Rev. Cell Dev. Biol. 14, 59–88 [DOI] [PubMed] [Google Scholar]
- 9.Yang Y. (2003) Birth Defects Res. C Embryo Today 69, 305–317 [DOI] [PubMed] [Google Scholar]
- 10.Huelsken J., Birchmeier W. (2001) Curr. Opin. Genet. Dev. 11, 547–553 [DOI] [PubMed] [Google Scholar]
- 11.Miller J. R., Hocking A. M., Brown J. D., Moon R. T. (1999) Oncogene 18, 7860–7872 [DOI] [PubMed] [Google Scholar]
- 12.Seidensticker M. J., Behrens J. (2000) Biochim. Biophys. Acta 1495, 168–182 [DOI] [PubMed] [Google Scholar]
- 13.Staal F. J., Noort Mv M., Strous G. J., Clevers H. C. (2002) EMBO Rep. 3, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura Y., Nawata M., Wakitani S. (2005) Am. J. Pathol. 167, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dell'accio F., De Bari C., Eltawil N. M., Vanhummelen P., Pitzalis C. (2008) Arthritis Rheum. 58, 1410–1421 [DOI] [PubMed] [Google Scholar]
- 16.Bergwitz C., Wendlandt T., Kispert A., Brabant G. (2001) Biochim. Biophys. Acta. 1538, 129–140 [DOI] [PubMed] [Google Scholar]
- 17.Topol L., Jiang X., Choi H., Garrett-Beal L., Carolan P. J., Yang Y. (2003) J. Cell Biol. 162, 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang S. G., Ryu J. H., Kim I. C., Jho E. H., Jung H. C., Kim K., Kim S. J., Chun J. S. (2004) J. Biol. Chem. 279, 26597–26604 [DOI] [PubMed] [Google Scholar]
- 19.Ryu J. H., Kim S. J., Kim S. H., Oh C. D., Hwang S. G., Chun C. H., Oh S. H., Seong J. K., Huh T. L., Chun J. S. (2002) Development 129, 5541–5550 [DOI] [PubMed] [Google Scholar]
- 20.Choy E. H., Panayi G. S. (2001) N. Engl. J. Med. 344, 907–916 [DOI] [PubMed] [Google Scholar]
- 21.Goldring M. B., Birkhead J. R., Suen L. F., Yamin R., Mizuno S., Glowacki J., Arbiser J. L., Apperley J. F. (1994) J. Clin. Invest. 94, 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martel-Pelletier J., Alaaeddine N., Pelletier J. P. (1999) Front Biosci. 4, D694–703 [DOI] [PubMed] [Google Scholar]
- 23.Ryu J. H., Chun J. S. (2006) J. Biol. Chem. 281, 22039–22047 [DOI] [PubMed] [Google Scholar]
- 24.Lotz M., Rosen F., McCabe G., Quach J., Blanco F., Dudler J., Solan J., Goding J., Seegmiller J. E., Terkeltaub R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10364–10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cailotto F., Bianchi A., Sebillaud S., Venkatesan N., Moulin D., Jouzeau J. Y., Netter P. (2007) Arthritis Res. Ther. 9, R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sohn P., Crowley M., Slattery E., Serra R. (2002) Osteoarthritis Cartilage 10, 482–490 [DOI] [PubMed] [Google Scholar]
- 27.Cheung H. S. (2005) Front Biosci. 10, 1336–1340 [DOI] [PubMed] [Google Scholar]
- 28.Kuettner K. E., Pauli B. U., Gall G., Memoli V. A., Schenk R. K. (1982) J. Cell Biol. 93, 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terkeltaub R., Rosenbach M., Fong F., Goding J. (1994) Arthritis Rheum. 37, 934–941 [DOI] [PubMed] [Google Scholar]
- 30.Bianchi A., Moulin D., Sebillaud S., Koufany M., Galteau M. M., Netter P., Terlain B., Jouzeau J. Y. (2005) Arthritis Res. Ther. 7, R1325–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Loo F. A., Joosten L. A., van Lent P. L., Arntz O. J., van den Berg W. B. (1995) Arthritis Rheum. 38, 164–172 [DOI] [PubMed] [Google Scholar]
- 32.Zwerina J., Redlich K., Polzer K., Joosten L., Krönke G., Distler J., Hess A., Pundt N., Pap T., Hoffmann O., Gasser J., Scheinecker C., Smolen J. S., van den Berg W., Schett G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11742–11747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldring M. B., Birkhead J., Sandell L. J., Kimura T., Krane S. M. (1988) J. Clin. Invest. 82, 2026–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robbins J. R., Thomas B., Tan L., Choy B., Arbiser J. L., Berenbaum F., Goldring M. B. (2000) Arthritis Rheum. 43, 2189–2201 [DOI] [PubMed] [Google Scholar]
- 35.Murakami S., Lefebvre V., de Crombrugghe B. (2000) J. Biol. Chem. 275, 3687–3692 [DOI] [PubMed] [Google Scholar]
- 36.de Crombrugghe B., Lefebvre V., Behringer R. R., Bi W., Murakami S., Huang W. (2000) Matrix Biol. 19, 389–394 [DOI] [PubMed] [Google Scholar]
- 37.Ge X., Ma X., Meng J., Zhang C., Ma K., Zhou C. (2009) Arthritis Rheum. 60, 2714–2722 [DOI] [PubMed] [Google Scholar]
- 38.Hwang S. G., Yu S. S., Lee S. W., Chun J. S. (2005) FEBS Lett. 579, 4837–4842 [DOI] [PubMed] [Google Scholar]
- 39.Katoh M., Katoh M. (2005) Int. J. Mol. Med. 15, 749–753 [PubMed] [Google Scholar]
- 40.Zaka R., Dion A. S., Kusnierz A., Bohensky J., Srinivas V., Freeman T., Williams C. J. (2009) J. Bone Miner Res. 24, 1869–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W., Xu J., Du B., Kirsch T. (2005) Mol. Cell. Biol. 25, 312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson K., Terkeltaub R. (2004) Osteoarthritis Cartilage 12, 321–335 [DOI] [PubMed] [Google Scholar]
- 43.Denison T. A., Koch C. F., Shapiro I. M., Schwartz Z., Boyan B. D. (2009) J. Cell. Biochem. 107, 155–162 [DOI] [PubMed] [Google Scholar]
- 44.Julien M., Magne D., Masson M., Rolli-Derkinderen M., Chassande O., Cario-Toumaniantz C., Cherel Y., Weiss P., Guicheux J. (2007) Endocrinology 148, 530–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foster B. L., Nociti F. H., Jr., Swanson E. C., Matsa-Dunn D., Berry J. E., Cupp C. J., Zhang P., Somerman M. J. (2006) Calcif. Tissue Int. 78, 103–112 [DOI] [PubMed] [Google Scholar]
- 46.Addison W. N., Azari F., Sørensen E. S., Kaartinen M. T., McKee M. D. (2007) J. Biol. Chem. 282, 15872–15883 [DOI] [PubMed] [Google Scholar]
- 47.Katoh M., Katoh M. (2007) Int. J. Mol. Med. 19, 273–278 [PubMed] [Google Scholar]
- 48.Badger A. M., Roshak A. K., Cook M. N., Newman-Tarr T. M., Swift B. A., Carlson K., Connor J. R., Lee J. C., Gowen M., Lark M. W., Kumar S. (2000) Osteoarthritis Cartilage 8, 434–443 [DOI] [PubMed] [Google Scholar]
- 49.Tew S. R., Hardingham T. E. (2006) J. Biol. Chem. 281, 39471–39479 [DOI] [PubMed] [Google Scholar]
- 50.Church V. L., Francis-West P. (2002) Int. J. Dev. Biol. 46, 927–936 [PubMed] [Google Scholar]
- 51.van Amerongen R., Mikels A., Nusse R. (2008) Sci. Signal 1, re9. [DOI] [PubMed] [Google Scholar]
- 52.Kengaku M., Capdevila J., Rodriguez-Esteban C., De La Peña J., Johnson R. L., Izpisúa Belmonte J. C., Tabin C. J. (1998) Science 280, 1274–1277 [DOI] [PubMed] [Google Scholar]
- 53.Hartmann C., Tabin C. J. (2000) Development 127, 3141–3159 [DOI] [PubMed] [Google Scholar]
- 54.Slusarski D. C., Corces V. G., Moon R. T. (1997) Nature 390, 410–413 [DOI] [PubMed] [Google Scholar]
- 55.Yang-Snyder J., Miller J. R., Brown J. D., Lai C. J., Moon R. T. (1996) Curr. Biol. 6, 1302–1306 [DOI] [PubMed] [Google Scholar]
- 56.Cong F., Schweizer L., Varmus H. (2004) Development 131, 5103–5115 [DOI] [PubMed] [Google Scholar]