Abstract
Bacteria mostly live as multicellular communities, although they are unicellular organisms, yet the mechanisms that tie individual bacteria together are often poorly understood. The adhesin involved in diffuse adherence (AIDA-I) is an adhesin of diarrheagenic Escherichia coli strains. AIDA-I also mediates bacterial auto-aggregation and biofilm formation and thus could be important for the organization of communities of pathogens. Using purified protein and whole bacteria, we provide direct evidence that AIDA-I promotes auto-aggregation by interacting with itself. Using various biophysical and biochemical techniques, we observed a conformational change in the protein during AIDA-AIDA interactions, strengthening the notion that this is a highly specific interaction. The self-association of AIDA-I is of high affinity but can be modulated by sodium chloride. We observe that a bile salt, sodium deoxycholate, also prevents AIDA-I oligomerization and bacterial auto-aggregation. Thus, we propose that AIDA-I, and most likely other similar autotransporters such as antigen 43 (Ag43) and TibA, organize bacterial communities of pathogens through a self-recognition mechanism that is sensitive to the environment. This could permit bacteria to switch between multicellular and unicellular lifestyles to complete their infection.
Keywords: Membrane/Proteins, Organisms/Bacteria, Protein/Cell Surface, Protein/Protein-Protein Interactions, Protein/Conformation, Protein/Self-assembly, Autotransporter, Escherichia coli
Introduction
Many Gram-positive and Gram-negative bacterial species, including Escherichia coli (1), Bordetella pertussis (2), Staphylococcus aureus (3), or Streptococcus pyogenes (4), possess the ability to auto-aggregate. Auto-aggregation can be visualized microscopically by the clumping of bacterial cells and, in some cases, macroscopically by the settling of static cultures. It is thought that bacterial aggregates can enhance colonization, participate in biofilm formation, and confer resistance to host defenses (4–7). In addition, cell-cell aggregation is a characteristic of many multicellular behaviors such as swarming (8). Auto-aggregation is usually conferred by surface-exposed adhesins like curli (9), pili/fimbriae (1, 10, 11), or autotransporters (12–14). How bacteria achieve and control their multicellular lifestyle has recently become a major research area. With a few notable exceptions (15), however, little is known on the mechanisms of action of the molecules that promote tight association between identical bacterial cells. A recently described family of E. coli outer membrane proteins called self-associating autotransporters (SAAT)4 provides a desirable model system to study these interactions (6).
SAAT are involved in adhesion to epithelial cells, biofilm formation, and bacterial auto-aggregation (6). To date, this family comprises three proteins: the adhesin involved in diffuse adherence (AIDA-I) (16, 17), the auto-aggregation factor Ag43 (18, 19), and the TibA adhesin/invasin (20, 21). AIDA-I was originally discovered in an E. coli strain isolated from a case of infantile diarrhea (16). Since then it has been associated with a high percentage of pathogenic strains of E. coli involved in neo-natal and post-weaning diarrhea in piglets (22–26). Experimental infections have confirmed the importance of AIDA-I in the pathogenesis of AIDA-expressing strains (27). Like all autotransporters (28), AIDA-I comprises an N-terminal signal sequence, a large extracellular domain, and a C-terminal membrane-embedded domain (see Fig. 1A). The extracellular domain of AIDA-I is mainly composed of imperfect repeats of a 19-amino acid sequence that is characteristic of SAAT (6). AIDA-I is cleaved after insertion into the outer membrane (29). The extracellular fragment mature AIDA-I and the membrane-embedded fragment AIDAc remain strongly associated despite cleavage (30). Mature AIDA-I can be released from the bacterial surface by a brief heat treatment (16), which denatures mature AIDA-I (31).
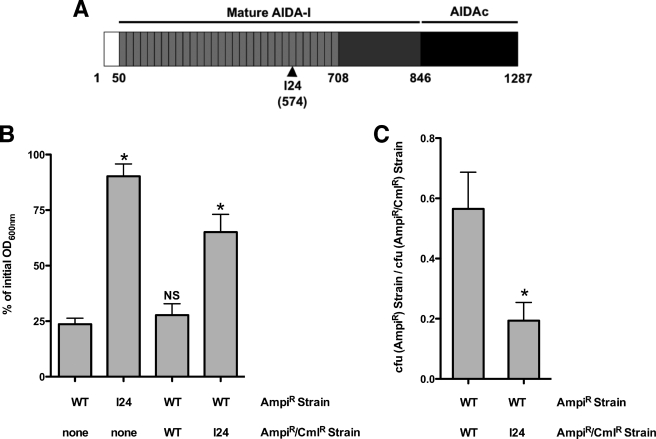
FIGURE 1.
Auto-aggregation of bacterial cells expressing AIDA-I. A, schematic representation of AIDA-I and the location of an auto-aggregation defective mutant, I24. The white box represents the signal sequence, the light gray boxes represent the imperfect 19-amino acids repeats, and the dark box represents the membrane-embedded fragment, AIDAc. B and C, auto-aggregation in mixes of bacteria expressing AIDA-I or I24. Overnight cultures of bacteria expressing wild-type AIDA-I (WT) or the I24 mutant were left standing in culture tubes for 180 min. The cultures have resistance against ampicillin (AmpiR) or against ampicillin and chloramphenicol (AmpiR/CmlR), respectively. In B, the OD600 nm values at the top of the cultures were compared with the values at the beginning of the assay. The columns were compared with the WT control by performing a one-way ANOVA and Dunnett's post tests. *, statistical significance (p < 0.05); NS, not significant. In C, the colony forming units (cfu) of bacteria bearing the different combinations of markers were evaluated, and a ratio was calculated. The ratios were compared by a Student's t test and were found to be statistically different. *, p < 0.05. The assays were performed three times in duplicate.
Based on observation of the aggregation of fluorescently labeled bacteria and other evidence, it was hypothesized that SAAT-mediated auto-aggregation is caused by direct SAAT-SAAT interaction (17, 18, 21). But direct biochemical evidence of this interaction is lacking. This is problematic because interaction of a surface adhesin from one cell with a different surface molecule of an adjacent cell can still permit specific auto-aggregation. For instance, yeast cells expressing the FLO1 adhesin selectively auto-aggregate, yet FLO1 does not bind to itself but rather to the cell wall of neighboring cells (32). It is therefore important to clearly ascertain whether SAAT are truly “self-associating” and to characterize the interaction these proteins can engage. For instance, whether environmental cues affect these interactions is unknown.
Here we provide a detailed characterization of the auto-aggregation mediated by AIDA-I. We first confirmed that the extracellular domain of AIDA-I mediates auto-aggregation. Using purified protein or whole bacteria, we then directly showed that AIDA-I interacts with itself, giving the first unequivocal proof that AIDA-I is self-associating. Finally, and unexpectedly, we demonstrated that AIDA-AIDA interactions and bacterial auto-aggregation can be modulated through induction of reversible conformational change in AIDA-I. This suggests that SAAT allow self-recognition of a pathogenic strain, but in a specific and highly controllable fashion, which could be a way to quickly adapt to a new environment once in the host.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The E. coli K-12 strain C600 (F- thr-1 leuB6 thi-1 lacY1 supE44 rfbD1 fhuA21) obtained from New England Biolabs was used in this study and does not contain the locus coding for AIDA-I. The plasmid pAgH has been described previously (31). The plasmid expresses AIDA-I with a six-histidine and glycine tag (HisG) in the N-terminal part of mature AIDA-I and Aah, the specific glycosyltransferase of AIDA-I. Bacterial cultures were grown at 30 °C on LB agar plates or in LB medium containing 100 μg/ml of ampicillin. The expression of AIDA-I was induced overnight when bacteria reached an OD600 nm of 0.8 with 10 μm of isopropyl-β-d-thiogalactopyranoside, unless indicated otherwise. This low level of induction results in the production of an amount of AIDA-I that is slightly less than the maximal amount produced in the wild-type strain 2787, as checked by quantitative PCR and Western blotting (data not shown).
Purification of Proteins and Preparation of Cellular Lysates and Membrane Fractions
The purification of His-tagged AIDA-I was performed essentially as previously described (33). Briefly, 1 liter of culture of C600 expressing glycosylated AIDA-I, either wild-type or mutated, was grown to an OD600 nm of 0.8 and induced overnight with 10 μm isopropyl-β-thiogalactopyranoside. The cells were harvested and resuspended in 40 ml of Tris-buffered saline (TBS; 50 mm Tris-HCl, pH 8, 150 mm NaCl) containing lysozyme (50 μg/ml final concentration) and EDTA (10 mm final concentration). The cells were lysed using a French press and an ultrasonic processor. After a low speed centrifugation to clear cellular debris, the total lysates were recovered. The lysates were centrifuged for 50 min in an ultracentrifuge at 250,000 × g. The pellets containing bacterial membranes were resuspended in TBS. For AIDA-I purification purposes, Triton X-100 (1% final concentration) was added to the membrane fractions, which were incubated for 1 h on ice and centrifuged again for 50 min in an ultracentrifuge. The supernatant, containing the HisG-tagged AIDA-I, was purified by immobilized metal affinity chromatography using a His Trap HP column (Amersham Biosciences). The purity of the purified protein was evaluated by SDS-PAGE followed by Coomassie Blue staining and was consistently over 95%. The concentration of the protein was measured with the BCA protein assay kit (Pierce). The purified protein was dialyzed in TBS overnight prior to use in all assays to eliminate all detergent. To purify heat-extracted protein, 1 liter from overnight culture of C600 expressing pAgH was grown as above. The cells were harvested and resuspended in 15 ml of 10 mm sodium phosphate buffer, pH 7, and heated at 60 °C for 20 min. The cells were centrifuged for 15 min at 16,000 × g, and the supernatant, containing the HisG-tagged heat-extracted AIDA-I, was purified as described before (31).
Auto-aggregation Assays
The assays were performed as previously described (31). Overnight cultures of C600 harboring an empty vector or the plasmid pAgH were normalized in 1.5 ml of LB to an OD600 nm of 1 to 1.5 and left standing at 4 °C in deep well 96-well plates. Samples were taken at the surface every 30 min, and the OD600 nm was measured. Alternatively, 20 μl of each suspension was spotted on a microscope slide and incubated at 4 °C, and aggregates of bacteria were examined after 3 h of incubation with a phase contrast microscope. For the inhibition experiments, purified protein in TBS, cell lysates, or membrane preparations were added to the wells at the start of the assay. To study the effect of salt on autoaggregation, the cultures were harvested and resuspended in TBS at different NaCl or sodium deoxycholate concentrations. In dose-response curves, the concentration required to achieve 50% inhibition was obtained by nonlinear regression fitting to a sigmoidal dose-response curve, using Prism 4.0 (GraphPad Software). All other statistical comparisons (one-way and two-way ANOVA, Student's t test) were performed using Prism 4.0.
Bead Aggregation Assays
Purified AIDA-I (0.15 mg/ml) was incubated for 4 h at room temperature with 0.8 μg of sulfate-modified polystyrene green fluorescent beads (average size, 1 μm; Sigma) in 500 μl of TBS. Control beads were prepared by incubation in TBS. After incubation, the beads were centrifuged, washed with TBS containing 1% (w/v) bovine serum albumin, and resuspended in 500 μl of TBS, 1% bovine serum albumin. For sedimentation assays, the beads were diluted four times in TBS in a final volume of 500 μl and left standing at room temperature overnight. In some experiments 1 m NaCl or 0.1% sodium deoxycholate (final concentrations) was added. The samples were taken at the surface at the beginning and end of the assay, and fluorescence at 515 nm (excitation at 485 nm) was measured with a fluorometer (Jasco Spectroscopic Co. Ltd.; model FMO-427). The samples were taken at the bottom of the same overnight solutions and visualized by fluorescence microscopy with a Leica DMI4000B microscope (Meyer Instruments) equipped with epifluorescence and UV excitation modules (Chroma Technology Corporation). Alternatively, the beads were diluted five times in TBS in a final volume of 500 μl and left standing for 6 h at room temperature before flow cytometry acquisition and analysis using Cellquest Pro software on a FACScalibur instrument (BD Biosciences).
Surface Plasmon Resonance
Using a Biacore 3000 instrument (GE Healthcare), SPR experiments were performed on research grade CM5 sensor chips at 25 °C using filtered (0.2 μm) and degassed HBS-EP (10 mm Hepes, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% (v/v) Tween 20) for the immobilization procedure. Immobilized AIDA-I surfaces (1000–6000 RU) were prepared using the Biacore amine coupling kit as recommended; corresponding bovine serum albumin reference surfaces were prepared in the similar manner. To assess binding specificity, the kinetic analyses were performed in which AIDA-I (0–1000 nm), diluted in TBS containing 2% (w/v) bovine serum albumin, was injected over immobilized surface at 30 μl/min using KINJECT mode (5 min of association and 8 min of dissociation). In additional trials, AIDA-I (200 nm) was heated at 60 °C for 20 min prior to injection, or AIDA-I (150 nm) was injected in the presence of increasing NaCl concentrations (150, 300, and 500 mm). To assess binding affinity, equilibrium analyses were performed in which AIDA-I was injected in a similar manner at 10 μl/min (15 min of association and 8 min of dissociation). Steady-state binding responses were plotted as a function of AIDA-I concentration and then subjected to nonlinear regression analysis (GraphPad Prism software). For all SPR assays, the surfaces were regenerated between sample injections using 60-s pulses of 1 m NaCl at 50 μl/min. All of the binding data are representative of duplicate injections acquired from at least three independent trials.
Far-UV CD and Intrinsic Fluorescence Spectroscopy
Far-UV CD spectra of the protein were recorded on a spectropolarimeter (Jasco Spectroscopic Co. Ltd.; model J-810) using a 0.1-cm-path length cuvette. The far-UV CD spectra of AIDA-I (125–200 μg/ml in TBS) were recorded between 200 and 260 nm. For each spectrum, five to ten accumulations were averaged, and the contribution of buffer to the measured ellipticity was subtracted. To study the effect of salts, the protein samples was diluted in TBS with different concentrations of NaCl or sodium deoxycholate prior to measurement. Ellipticities were converted to mean residual ellipticities. Intrinsic fluorescence was recorded in the same spectropolarimeter equipped with a fluorescence emission monochromator (Jasco Spectroscopic Co. Ltd.; model FMO-427). The spectra were recorded between 300 and 400 nm after excitation at 290 nm. Five accumulations were averaged, and the contribution of the buffer was subtracted.
Chemical Cross-linking
Overnight cultures of C600 harboring the different plasmids were diluted 1:100 in fresh medium and grown to an OD600 nm of 0.4, at which point the expression of AIDA-I was induced for 3 h with 10 μm isopropyl-β-thiogalactopyranoside. The cultures were normalized to the same OD600 nm of 1 and washed in phosphate-buffered saline. The samples (25 μl) were incubated for 30 min at room temperature with different concentrations of bis[sulfosuccinimidyl] suberate (BS3; Pierce), which is a membrane-impermeable NHS-ester reagent reacting with amino groups to form stable amide bonds. The best reactions were found to occur at 300 μm BS3. The reaction was stopped by incubating for an additional 15 min with 50 mm Tris-HCl, pH 7.5. A similar reaction was conducted with purified proteins at a concentration of 0.2 mg/ml in 1 mm Tris-HCl, pH 8, 150 mm NaCl. The samples were diluted in 2× SDS-PAGE loading buffer containing β-mercaptoethanol and denatured by heating at 100 °C for 10 min. The samples were then separated by SDS-PAGE at 4 °C on 7% acrylamide gels and analyzed by immunoblotting. Immunodetection was performed with a serum raised against heat-extracted mature AIDA-I diluted 1:50,000 in blocking buffer (5% skim milk, 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% Triton X-100). A goat anti-rabbit horseradish peroxidase-conjugated antibody (Sigma) was used as a secondary antibody according to the instructions of the manufacturer. Immune complexes were revealed using a 3,3′,5,5′-tetramethylbenzidine solution for membranes (Sigma).
Analytical Ultracentrifugation
Purified AIDA-I in TBS was diluted in TBS alone, in TBS containing NaCl so as to reach a final concentration of 1 m, or in TBS containing sodium deoxycholate so as to reach a final concentration of 0.5%. All of the samples contained ∼0.69 mg/ml of protein, which gave an absorbance of ∼0.7 at 280 nm. Sedimentation velocity experiments were performed on the samples using a Beckman XL-I analytical ultracentrifuge. The reference and sample sectors of a 1.2-cm-path length double-sector ultracentrifuge cell were filled with 400 μl of the appropriate buffer and 390 μl of protein, respectively. Sedimentation velocity runs were performed at either 20,000 or 30,000 rpm, with absorbance scans monitored at 280 nm in 10-min intervals over a total spin time of 4–6 h at 20 °C. Molecular mass, buffer viscosity, density, and hydration of the protein were calculated using the software SEDNTERP. Molecular mass and hydration of the protein were calculated assuming 20 heptose residues/molecule. Sedimentation velocity data were analyzed by the computer program SEDFIT (41). Sedimentation profiles were obtained by fitting the absorbance data to the continuous c(S) model.
RESULTS
Evidence That AIDA-I Mediates Auto-aggregation via Direct Self-interaction
As shown in previous reports (6, 17, 31), we observed that bacteria from standing cultures of E. coli expressing AIDA-I form large aggregates of cells that can be observed by phase contrast microscopy (supplemental Fig. S1). This was not the case with bacteria bearing a control empty plasmid. These aggregates settle at the bottom of culture tubes, which can be easily followed with a spectrophotometer by measuring optical density at the top of the bacterial cultures (supplemental Fig. S1 and Fig. 1B).
We isolated in a previous study (33) a mutant of AIDA-I, I24, bearing an insertion of a 5-amino acid sequence at amino acid 574 in the C-terminal region of the imperfect repeats of AIDA-I (Fig. 1A). As observed before, this mutant is unable to mediate auto-aggregation (Fig. 1B). When we mixed equal amounts of bacteria expressing wild-type AIDA-I and bacteria expressing I24, we observed an intermediate amount of settling. Furthermore, at the top of the cultures, there were fewer bacteria expressing WT compared with bacteria expressing I24 (Fig. 1C). Thus, AIDA-I is able to mediate auto-aggregation preferentially between bacteria expressing the wild-type protein and exclude the bacteria expressing I24. This argues that AIDA-I functions by interacting with itself, directly or indirectly.
We decided to exclude the possibility that AIDA-I engage another molecule to mediate auto-aggregation using another approach. As indicated above, strain-specific auto-aggregation of yeast mediated by the FLO1 adhesin involves interaction of FLO1 with neighboring cell wall molecules (32). We therefore added different subcellular extracts of bacteria harboring an empty vector or expressing AIDA-I. We observed that auto-aggregation is totally inhibited by the addition of a lysate or a membrane extract of bacteria expressing AIDA-I but, by contrast, the addition of an identical amount of cell lysate or a membrane extract of bacteria harboring an empty vector had no significant effect (Fig. 2A). This experiment suggested that AIDA-I does not mediate auto-aggregation by interacting with another cell surface molecule.
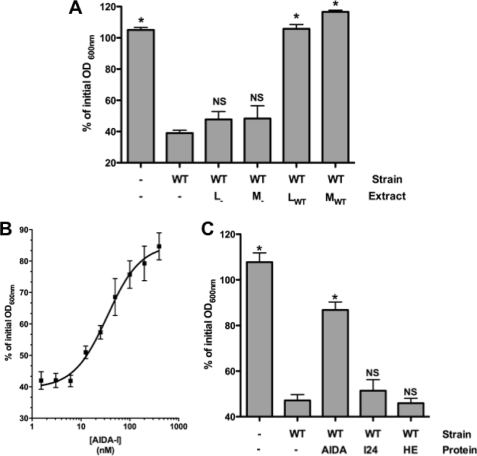
FIGURE 2.
Inhibition of auto-aggregation by purified AIDA-I and not by cellular extracts. A, cultures bearing an empty vector (-) or expressing AIDA-I (WT) were incubated in deep well 96-wells plates with or without total lysates (L) or membrane extracts (M) prepared from cultures harboring the empty vector or expressing AIDA-I and left standing for 90 min. The columns were compared with the wild-type control by performing a one-way ANOVA and Dunnett's post tests. *, statistical significance (p < 0.05); NS, not significant. B, auto-aggregation assays of cultures of bacteria expressing AIDA-I were performed with various concentrations of purified AIDA-I present. The points were fitted by nonlinear regression to a sigmoidal dose-response. C, normalized cultures of bacteria harboring an empty vector or expressing AIDA-I were incubated with or without 200 nm wild-type protein, mutated protein I24, or heat-extracted purified protein (HE). The columns were compared with the wild-type vector control by performing a one-way ANOVA and Dunnett's post tests. *, statistical significance (p < 0.05); NS, not significant.
We then added purified whole AIDA-I to standing cultures of bacterial cells expressing AIDA-I. As shown in Fig. 2B, we observed that the protein can inhibit auto-aggregation in a dose-dependent and saturable manner with 50% inhibition (IC50) reached at a concentration of ∼34 nm. Such a low concentration strongly suggests that this effect is specific. By contrast, neither the purified I24 mutant nor the heat-extracted portion of the extracellular domain of AIDA-I were able to prevent auto-aggregation (Fig. 2C). This is consistent with the inability of I24 to mediate auto-aggregation even with wild-type AIDA-I and with the fact that the heat-extracted extracellular domain of AIDA-I is denatured (31). In addition, the fact that heat-denatured AIDA-I did not prevent auto-aggregation suggests that the inhibition is not due to a putative heat-resistant contaminant in our protein preparation.
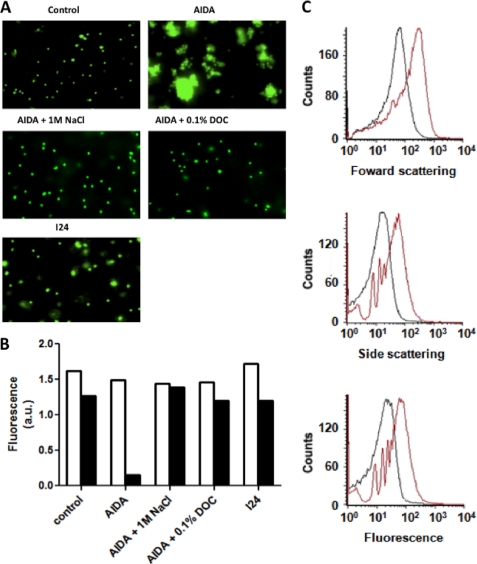
To finally prove that AIDA-I mediates auto-aggregation by direct self-interaction, we coated fluorescent polystyrene beads with purified protein. As shown in Fig. 3A, we observed that beads coated with wild-type AIDA-I could auto-aggregate, whereas control empty beads or beads coated with the purified I24 mutant did not. This phenomenon could be quantified using a sedimentation assay similar to the one used with whole bacteria (Fig. 3B) or by flow cytometry analysis of particle fluorescence and scattering (Fig. 3C). These assays clearly show the magnitude of the aggregation mediated solely by AIDA-I, and the I24 control ascertains that this is a specific effect. In conclusion, taken together our results represent, to our knowledge, the first direct proof that SAAT-mediated auto-aggregation is due to direct self-interaction.
FIGURE 3.
Auto-aggregation of fluorescent beads mediated by purified AIDA-I. A, solutions containing empty fluorescent polystyrene beads (Control), AIDA-coupled beads (AIDA), and I24-coupled beads (I24) were left standing overnight, and the samples were taken at the bottom of the tube and examined by fluorescent microscopy. The experiments were performed with AIDA-coupled beads in the presence of 1 m NaCl or 0.1% sodium deoxycholate (DOC). B, samples were taken on top of the solutions containing the beads at the beginning of the assay (white bars) and after overnight incubation (black bars), and fluorescence at 515 nm was measured. C, the solutions containing empty beads (black) or AIDA-coupled beads (red) solutions were left standing at room temperature for 6 h and then analyzed by flow cytometry. All of the experiments were repeated three times; typical results are shown.
Direct Interaction of the Extracellular Domain of AIDA-I in Vitro
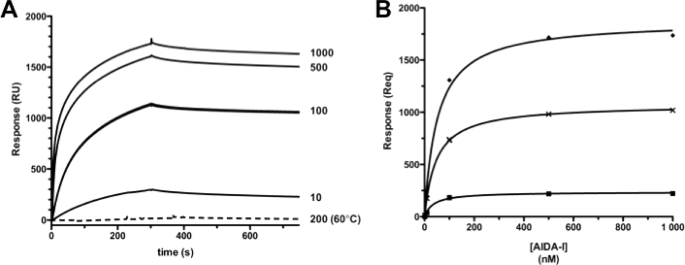
To directly demonstrate that AIDA-I can associate with itself, we used SPR. Reference and purified AIDA-I surfaces were immobilized on an SPR chip. A series of soluble AIDA-I concentrations were then titrated over the prepared surfaces in tandem. Typical kinetic analyses exhibited specific, dose-dependent binding of soluble AIDA-I to immobilized AIDA-I (Fig. 4A). No detectable binding responses were observed, however, when AIDA-I was heated at 60 °C for 20 min prior to injection. This result of heat treatment, known to denature the mature portion of AIDA-I but not AIDAc (31), strongly suggests that the binding observed by SPR involves interactions between the mature AIDA-I extracellular fragments. To assess binding affinity, the extremely slow off rates observed precluded fitting of the kinetic titrations to a simple 1:1 binding model (i.e. simultaneous on and off rate determinations). Alternatively, equilibrium analyses were performed in which the steady-state binding responses were plotted as a function of AIDA-I concentration to determine the overall equilibrium dissociation constant, KD (Fig. 4B). The apparent KD predicts an interaction of high affinity (48 ± 10 nm) and was not dependent on the level of protein immobilized on the chip. The latter confirms that the SPR signal is specific. Additional SPR experiments with different chips, buffers, and conditions yielded highly similar results (supplemental Fig. S2), strengthening the notion that AIDA-AIDA interaction is specific. Interestingly, the KD of the interaction is in excellent agreement with the IC50 determined in our inhibition experiment with purified AIDA-I (34 nm; Fig. 2B). Our results therefore show that AIDA-I can interact with itself in vitro with a high affinity and, to our knowledge, constitutes the first direct observation of a SAAT-SAAT interaction.
FIGURE 4.
In vitro evidence of AIDA-AIDA interactions. A, representative SPR for kinetic titrations of soluble AIDA-I (0–1000 nm) binding to 6,700 RU amine-coupled AIDA-I at 30 μl/min. The dashed line indicates 200 nm injection of heat-treated AIDA-I (60 °C for 20 min). B, representative SPR for equilibrium titrations of soluble AIDA-I (0–1000 nm) binding to immobilized AIDA-I at 10 μl/min. The symbols represent RU at equilibrium (Req), and the lines represent nonlinear regression analysis. The average of three independent trials yielded apparent KD constants of 36 nm (top curve, 6700 RU AIDA-I), 59 nm (middle curve, 2500 RU AIDA-I), and 52 nm (bottom curve, 1000 RU AIDA-I).
Observation of AIDA-I Oligomers in Vitro and in Whole Bacteria
To characterize the oligomers made by AIDA-I, we used analytical ultra centrifugation (AUC). The sedimentation profile of the purified protein showed two discrete peaks (Table 1 and supplemental Figs. S3 and S4). When the same experiment was performed with the purified I24 mutant, we observed that the protein was monomeric, with a sedimentation coefficient similar to the one of the wild-type monomer. This is consistent with the fact that I24 does not allow auto-aggregation and thus most likely remains monomeric. The computed molecular masses of the first peak observed with wild-type AIDA-I and of the peak of I24 are consistent with the predicted molecular mass of AIDA-I. However, the computed molecular mass of the second peak observed with wild-type AIDA-I is too small for a dimer. There is little doubt that this second peak comes from a dimeric species, however. The aberrant computed mass could be explained if the two species have different shapes and therefore different frictional coefficients, whereas the fitting program uses the same coefficient for both to make its calculations. When constraining the molecular masses of the two species as a monomer and a dimer of AIDA-I, a solution could be found with two different frictional coefficients (Table 1). This shows that the peaks could be interpreted as a monomer and a dimer if these two species have different conformations. More evidence confirming this hypothesis is presented below. All of the calculated frictional coefficients indicate that the overall shape of the AIDA-I protein is elongated. This is in agreement with structural prediction (34) and the known structures of other extracellular domains of autotransporters, which mainly consists of elongated β-helices (35). Overall, this experiment confirms that AIDA-I is able to oligomerize in solution. It is intriguing to see more of the monomer form of AIDA-I than the dimer form, despite the protein concentration being far above the KD measured by SPR. This could be explained if dimers themselves were associating into a range of higher order oligomers, which would be too big and heterogeneous to be observed in this AUC experiment. Evidence for this is provided below.
TABLE 1.
Analytical ultracentrifugation of purified AIDA-I
Sedimentation velocity experiments were performed with purified wild-type or mutant AIDA-I diluted in 50 mm Tris-HCl, pH 8, containing 150 mm NaCl, 1 m NaCl, or 150 mm NaCl and 0.5% sodium deoxycholate. Sedimentation (c) and frictional (f/f0) coefficients and molecular mass were then determined by fitting the data from the sedimentation profiles, and the root mean square deviation (RMSD) from the best solution is indicated.
| Protein | Condition | c(S) | Molecular mass | f/f0 | RMSD |
|---|---|---|---|---|---|
| kDa | |||||
| Wild-type | Peak 1 with 150 mm NaCl | 5.65 ± 0.234 | 133 ± 10 | 1.95a | 0.00434 |
| Peak 2 with 150 mm NaCl | 7.27 ± 0.36 | 189 ± 12 | 1.95a | 0.00434 | |
| 1 m NaCl | 4.375 ± 0.17 | 124 ± 8 | 1.77 | 0.00561 | |
| 0.5% sodium deoxycholate | 5.85 ± 0.2 | 135 ± 8 | 1.66 | 0.00542 | |
| I24 | 150 mm NaCl | 5.13 ± 0.25 | 133 ± 10 | 1.89 | 0.00537 |
a This is an average frictional coefficient for the two species assumed to have similar shape; if the two species are constrained to be a monomer and a dimer of AIDA-I during the fitting, then two separate frictional coefficient can be calculated and are 1.69 for peak 1 and 2.08 for peak 2.
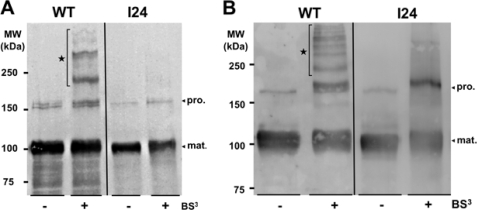
To test for direct interactions between AIDA-I molecules in whole bacteria, we performed cell surface cross-linking experiments with BS3, a membrane-impermeable NHS-ester reagent bridging primary amino groups of proteins. We added a low amount of BS3 to cultures of bacteria expressing AIDA-I. By immunoblotting with an antibody raised against mature AIDA-I, we observed the 100- and 150-kDa bands of mature extracellular AIDA-I and pro-protein, respectively (Fig. 5A). In addition, we observed bands of ∼200 and 300 kDa as well as fainter bands of even higher molecular masses. These bands could correspond to dimers and higher order oligomers of mature extracellular domains of AIDA-I. None of these oligomers can be observed when bacteria expressed the I24 mutant, showing that the cross-links observed with the wild-type protein are indeed specific and that AIDA-I interacts with itself in whole bacteria through its mature AIDA-I portion. Similar patterns of cross-linking were observed with purified proteins in solution (Fig. 5B) and confirm that AIDA-I is making similar higher order oligomers in solution. As stated above, this would explain the unexpected monomer:dimer ratio observed during sedimentation in analytical ultracentrifugation. In summary, these results characterize AIDA-I interactions with itself, both in vitro and in vivo, and show that they involve heterogeneous oligomers.
FIGURE 5.
Chemical cross-linking of AIDA-I. A, cultures of bacteria expressing wild-type AIDA-I (WT) or the I24 mutant were incubated with or without 300 μm BS3 cross-linker for 30 min. The samples were diluted in 2× SDS-PAGE loading buffer and immunoblotted with an antibody directed against the extracellular part of AIDA-I. B, purified proteins (0.2 mg/ml in 1 mm Tris, pH 8, 150 mm NaCl) were cross-linked in the same conditions, and the samples were treated by SDS-PAGE and immunoblotting similarly. pro., pro-protein; mat., mature extracellular AIDA-I.
Modulation of AIDA-AIDA Interaction and Bacterial Auto-aggregation
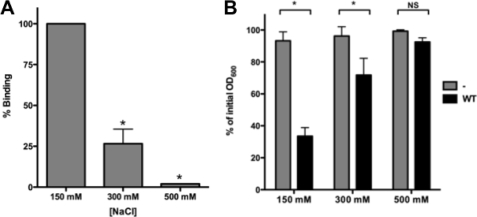
Because ionic interactions are often important in protein-protein interactions, we assessed AIDA-AIDA interactions at different NaCl concentrations. Typical SPR binding profiles showed that AIDA-AIDA interaction is salt-sensitive, and the association decreased dramatically with increasing NaCl concentrations (Fig. 6A). The AUC sedimentation profile of purified AIDA-I in the presence of 1 m NaCl also showed that AIDA-I is a single species (Table 1). In addition, the presence of 1 m NaCl prevented the aggregation of beads coated with AIDA-I (Fig. 3, A and B). Together, these experiments show that high salt concentrations disrupt AIDA-AIDA interactions. We investigated the effect of ionic strength on the settling of cultures of bacterial cells expressing AIDA-I by resuspending bacteria in buffers at different NaCl concentrations (Fig. 6B). We observed a significant decrease in the settling of bacterial cells at 300 mm NaCl and no aggregation at 500 mm NaCl. These results correlate well with the in vitro measurements of AIDA-AIDA interactions, thereby reinforcing the notion that direct AIDA-AIDA interactions mediate auto-aggregation. As previously noted (31), the extracellular domain of AIDA-I is extremely tightly associated to AIDAc and did not dissociate even in the presence of up to 2 m NaCl (data not shown). In summary, these results suggest that electrostatic interactions are involved in the self-association of AIDA-I in vitro and in whole bacteria.
FIGURE 6.
Modulation of AIDA-AIDA interaction and bacterial auto-aggregation. A, kinetic SPR analysis in which purified AIDA-I (150 nm) was injected in the presence of increasing NaCl concentration. The experiments were performed three times. The average responses observed were normalized to the response observed at 150 mm NaCl. The columns were compared by performing a one-way ANOVA and Dunnett's post tests. *, statistical significance (p < 0.05). B, overnight cultures of bacteria expressing AIDA-I (WT) or harboring an empty vector were resuspended in 50 mm Tris-HCl pH 8 buffers at different ionic strengths and were left standing at 4 °C. The OD600 nm at the top of the culture was measured at the end of the assay after 180 min and compared with the value at the beginning of the assay. The assays were performed three times in duplicate, and the columns were compared by a two-way ANOVA and Bonferroni post-tests. *, statistical significance (p < 0.05); NS, not significant.
Modulation of AIDA-AIDA Interaction Involves a Reversible Conformation Change
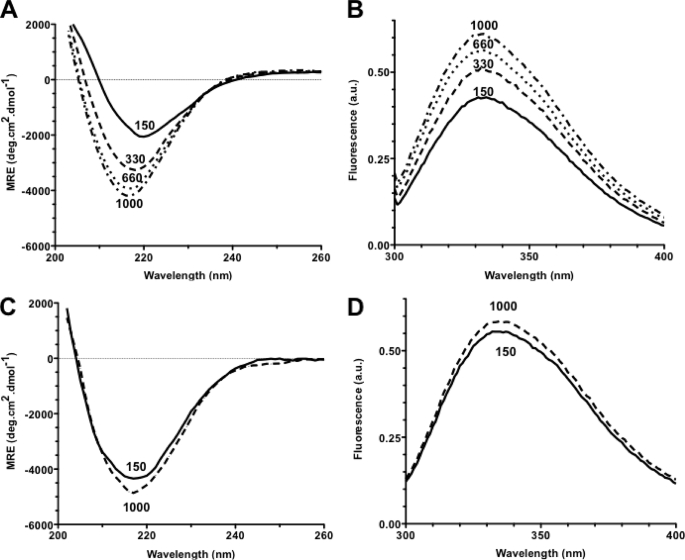
As indicated above, AUC suggested that monomeric and oligomeric AIDA-I have different conformations. We performed far-UV CD spectroscopy on purified AIDA-I at different ionic strengths to directly assess the influence of oligomerization on the conformation of AIDA-I. The CD spectrum of AIDA-I shows a clear single minimum of ellipticity at 218 nm (Fig. 7A), a characteristic of β-strands, in agreement with the predicted structure of AIDA-I and our previous observations (31, 34). Upon increase of NaCl concentration, the CD spectrum of AIDA-I changed dramatically, with an important decrease in ellipticity and the minimum of ellipticity shifting toward a lower wavelength. This result signals important conformational changes. Measuring the intrinsic fluorescence of purified AIDA-I in the presence of various concentrations of NaCl showed that high salt concentrations also caused a dramatic increase in intrinsic fluorescence (Fig. 7B), another indicator of conformational changes. Interestingly, I24 had the same CD and intrinsic fluorescence spectra at low and high NaCl concentrations, and these spectra were similar to the ones of AIDA-I in the presence of high salt concentration (Fig. 7, C and D). This is consistent with the fact that I24 is unable to mediate auto-aggregation, does not form complexes by chemical cross-linking, and is monomeric as seen in AUC. Taken together, these experiments suggest that the conformational change observed with increasing NaCl concentration corresponds to a transition toward the conformation of a monomeric protein and that the monomer and oligomers have different conformations. The difference in frictional coefficients between the two species observed in AUC is also in agreement with these results. Strikingly, regardless of the method, we observed that these conformational changes are completely reversible and the protein can repeatedly switch back and forth between conformations, strengthening the notion that these changes are very specific.
FIGURE 7.
Conformation change in AIDA-I. A, far-UV CD spectra of purified wild-type AIDA-I when sodium chloride was added to the buffer to yield a final concentration of 150, 330, 660, or 1000 mm. The signal was converted to mean residual ellipticity (MRE). B, intrinsic fluorescence of purified AIDA-I after excitation at 290 nm was recorded in the same buffers and reported in arbitrary units (a.u.). C and D, far-UV CD spectra (C) and intrinsic fluorescence (D) of the purified I24 mutant were obtained as in A and B, in the presence of 150 mm or 1 m NaCl.
Bile Salt Can Modulate AIDA-AIDA Interaction and Auto-aggregation
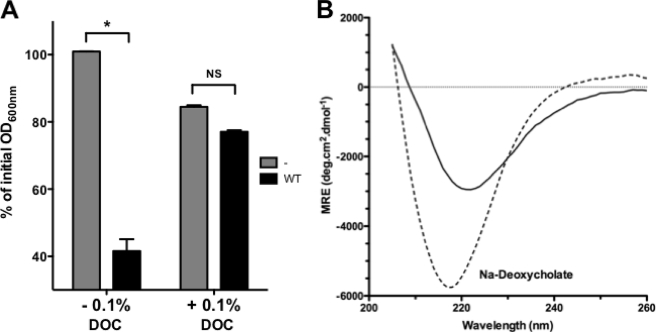
We wondered whether another physiologically relevant condition could alter AIDA-AIDA interaction and bacterial auto-aggregation. Because AIDA-I is expressed in diarrheagenic strains of E. coli, it is bound to encounter the bile secreted in the intestine (36, 37). We therefore tested the effects of sodium deoxycholate, a typical bile salt, on AIDA-I conformation and oligomerization. Strikingly, we observed that bacteria expressing AIDA-I could not auto-aggregate when incubated in the presence of sodium deoxycholate (Fig. 8A). The sedimentation of AIDA-I observed by AUC in the presence of deoxycholate showed that it is monomeric (Table 1). The CD spectrum of AIDA-I in these conditions (Fig. 8B) was also similar to the one observed in monomeric AIDA-I. Lastly, aggregation of beads coated with AIDA-I was also disrupted by sodium deoxycholate (Fig. 3, A and B). Taken together, these results show that a physiological cue is able to prevent AIDA-AIDA interaction as well as auto-aggregation.
FIGURE 8.
Effect of bile salt on AIDA-AIDA interaction and auto-aggregation. A, overnight cultures of bacteria expressing AIDA-I (WT) or harboring an empty vector were resuspended in TBS with or without 0.1% sodium deoxycholate and were left standing at 4 °C. The OD600 nm at the top of the culture was measured at the end of the assay after 180 min and compared with the value at the beginning of the assay. The assays were performed three times in duplicate, and columns were compared by a two-way ANOVA and Bonferroni post-tests. *, statistical significance (p < 0.05); NS, not significant. B, far-UV CD spectrum of purified AIDA-I was recorded in TBS without (solid line) or with (dashed line) 0.1% sodium deoxycholate (DOC).
DISCUSSION
The autotransporter adhesins of E. coli AIDA-I, TibA and Ag43, have been grouped into the family of self-associating autotransporters (6). As the name implies, preliminary evidence suggested that these proteins mediate bacterial auto-aggregation through self-interaction (17, 21, 38). The assumption that a SAAT interacts with itself was never actually directly tested, however. In this study we bring proof that AIDA-I does indeed interact with itself, using in vitro and in vivo approaches. Our experiments also show that this self-interaction is directly responsible for auto-aggregation. In particular, we observed that purified protein could trigger auto-aggregation of beads and compete with auto-aggregation of bacteria. In addition we observed that high salt concentrations were able to disrupt AIDA-AIDA interaction as well as auto-aggregation. The latter result suggests that charged or polar residues contribute to AIDA-AIDA interactions. A similar effect of high salt concentrations was observed on bacterial aggregation mediated by Ag43 (18), consistent with the idea that the self-interaction mechanism might be common in all SAAT. Interestingly, we observed that AIDA-I interactions result in the formation of a range of oligomers beyond dimers. This suggests that AIDA-AIDA interactions involve proteins on the same bacterium as well as proteins on adjacent bacteria. Additional experiments suggest that this is indeed the case.5
We observed that high salt concentrations also caused a substantial change in AIDA-I conformation, suggesting that the AIDA-I protein has significantly different conformations depending on its oligomeric state. Large conformational changes are also observed in the mammalian cell adhesion surface proteins cadherins (39), allowing them to play their role as fine-tuned and modulable organizers of all tissues. Their conformations are calcium-dependent, and the conformational changes act as a switch modulating the engagement of cell-cell interactions. We observed that sodium deoxycholate, a bile salt, was able to disrupt auto-aggregation and AIDA-AIDA interaction, as well as change AIDA-I conformation. Because the osmolality of the intestine is usually high (∼300 mm) and in the range of the NaCl concentration we tested and because the concentration of bile salt we tested is also in the physiological range, it is tempting to speculate that AIDA-I is ideally suited to allow bacteria to sense the intestinal environment and modulate cell-cell interactions. Similarly, zinc released from immune cells was recently hypothesized to induce conformational changes and auto-association of Staphylococcal adhesins (15). It should also be noted that the osmolality and bile salt content of the intestine are not constant spatially and temporally. Thus, environmental regulation of AIDA-I self-interaction could help bacteria to adapt to a specific niche and/or a specific time. In this respect it is interesting to note that diarrheagenic strains of E. coli bearing AIDA-I are most often isolated in cases of diarrhea occurring post-weaning, a situation where the animals are stressed and undergo significant changes in intestinal homeostasis.
It has been previously observed that some enteropathogenic and enteroaggregative strains of E. coli are able to make bacterial aggregates through the expression of pili or fimbriae and that these aggregates are important for pathogenesis (11, 40). The roles these bacterial aggregates could play are not precisely known, but they could enhance colonization, participate in biofilm formation, and/or confer resistance to host defenses. Additionally, auto-aggregation could be construed as a way to discriminate “self” from “nonself.” This distinction could be advantageous by ensuring that pathogens only associate with their own kin, maximizing their infectivity by increasing their local density as well as by optimizing their pathogenic potential in bringing together their virulence factors. In this view, AIDA-I would represent a peculiarly simple and versatile example of such a tag defining self. In the previous examples of pathogenic E. coli forming aggregates described above, a dedicated protein can disperse the aggregates, and this dispersion was also shown to be required for efficient pathogenesis (11, 40). Although the reason why dispersion is needed is unclear, it was hypothesized that disaggregation is required to establish new foci of infection (11) or penetrate the mucous layer of the intestinal epithelium (40). Similarly, but in a considerably simpler and faster manner, bile salts could disperse AIDA-mediated aggregates and allow efficient infection. In addition, SAAT, like other autotransporters promoting auto-aggregation (13, 14), are multifunctional proteins and can mediate adhesion to host cells (6). Regulation of self-interaction could also act as a switch mechanism between these multiple activities.
In conclusion, we have shown here that AIDA-I can directly mediate auto-aggregation through self-interaction and that the environment could potentially modulate this interaction through specific conformation change. We thus propose that AIDA-I and similar other autotransporters such as Ag43 and TibA represent a versatile and simple family of tunable bacterial adhesion molecules enabling specific social behavior at an appropriate time. Further work on this type of adhesins is warranted to thoroughly test and pursue the implications of this hypothesis.
Supplementary Material
Acknowledgments
We thank M. Ramjeet, J.-M. Ghigo, and C. Beloin for critical reading of the manuscript and insightful comments. We are indebted to M.-A. Albert for help with the flow cytometry experiments.
This work was supported by Canadian Institutes for Health Research Grant 84578, funds from the Groupe de Recherche et d'Etudes sur les Maladies Infectieuses du Porc, Canada Research Chair and Canada Foundation for Innovation Grant 201414, and a research resource grant from Canadian Institutes for Health Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
J.-P. Côté, F. Berthiaume, and M. Mourez, unpublished results.
- SAAT
- self-associating autotransporters
- AIDA-I
- adhesin involved in diffuse adherence
- AUC
- analytical ultracentrifugation
- BS3
- bis[sulfosuccinimidyl
- SPR
- surface plasmon resonance
- TBS
- Tris-buffered saline
- ANOVA
- analysis of variance
- RU
- response units.
REFERENCES
- 1.Nataro J. P., Deng Y., Maneval D. R., German A. L., Martin W. C., Levine M. M. (1992) Infect. Immun. 60, 2297–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menozzi F. D., Boucher P. E., Riveau G., Gantiez C., Locht C. (1994) Infect. Immun. 62, 4261–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDevitt D., Francois P., Vaudaux P., Foster T. J. (1994) Mol. Microbiol. 11, 237–248 [DOI] [PubMed] [Google Scholar]
- 4.Frick I. M., Mörgelin M., Björck L. (2000) Mol. Microbiol. 37, 1232–1247 [DOI] [PubMed] [Google Scholar]
- 5.Berge A., Kihlberg B. M., Sjöholm A. G., Björck L. (1997) J. Biol. Chem. 272, 20774–20781 [DOI] [PubMed] [Google Scholar]
- 6.Klemm P., Vejborg R. M., Sherlock O. (2006) Int. J. Med. Microbiol. 296, 187–195 [DOI] [PubMed] [Google Scholar]
- 7.Ochiai K., Kurita-Ochiai T., Kamino Y., Ikeda T. (1993) J. Med. Microbiol. 39, 183–190 [DOI] [PubMed] [Google Scholar]
- 8.Verstraeten N., Braeken K., Debkumari B., Fauvart M., Fransaer J., Vermant J., Michiels J. (2008) Trends Microbiol. 16, 496–506 [DOI] [PubMed] [Google Scholar]
- 9.Hammar M., Bian Z., Normark S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6562–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schembri M. A., Christiansen G., Klemm P. (2001) Mol. Microbiol. 41, 1419–1430 [DOI] [PubMed] [Google Scholar]
- 11.Bieber D., Ramer S. W., Wu C. Y., Murray W. J., Tobe T., Fernandez R., Schoolnik G. K. (1998) Science 280, 2114–2118 [DOI] [PubMed] [Google Scholar]
- 12.Girard V., Mourez M. (2006) Res. Microbiol. 157, 407–416 [DOI] [PubMed] [Google Scholar]
- 13.Alamuri P., Mobley H. L. (2008) Mol. Microbiol. 68, 997–1017 [DOI] [PubMed] [Google Scholar]
- 14.Felek S., Lawrenz M. B., Krukonis E. S. (2008) Microbiology 154, 1802–1812 [DOI] [PubMed] [Google Scholar]
- 15.Conrady D. G., Brescia C. C., Horii K., Weiss A. A., Hassett D. J., Herr A. B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19456–19461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benz I., Schmidt M. A. (1989) Infect. Immun. 57, 1506–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherlock O., Schembri M. A., Reisner A., Klemm P. (2004) J. Bacteriol. 186, 8058–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klemm P., Hjerrild L., Gjermansen M., Schembri M. A. (2004) Mol. Microbiol. 51, 283–296 [DOI] [PubMed] [Google Scholar]
- 19.Henderson I. R., Meehan M., Owen P. (1997) FEMS Microbiol. Lett. 149, 115–120 [DOI] [PubMed] [Google Scholar]
- 20.Elsinghorst E. A., Weitz J. A. (1994) Infect. Immun. 62, 3463–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherlock O., Vejborg R. M., Klemm P. (2005) Infect. Immun. 73, 1954–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha S. K., Choi C., Chae C. (2003) J. Vet. Diagn. Invest. 15, 378–381 [DOI] [PubMed] [Google Scholar]
- 23.Mainil J. G., Jacquemin E., Pohl P., Kaeckenbeeck A., Benz I. (2002) Vet. Microbiol. 86, 303–311 [DOI] [PubMed] [Google Scholar]
- 24.Ngeleka M., Pritchard J., Appleyard G., Middleton D. M., Fairbrother J. M. (2003) J. Vet. Diagn. Invest. 15, 242–252 [DOI] [PubMed] [Google Scholar]
- 25.Niewerth U., Frey A., Voss T., Le Bouguénec C., Baljer G., Franke S., Schmidt M. A. (2001) Clin. Diagn. Lab. Immunol. 8, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W., Zhao M., Ruesch L., Omot A., Francis D. (2007) Vet. Microbiol. 123, 145–152 [DOI] [PubMed] [Google Scholar]
- 27.Ravi M., Ngeleka M., Kim S. H., Gyles C., Berthiaume F., Mourez M., Middleton D., Simko E. (2007) Vet. Microbiol. 120, 308–319 [DOI] [PubMed] [Google Scholar]
- 28.Henderson I. R., Navarro-Garcia F., Desvaux M., Fernandez R. C., Ala'Aldeen D. (2004) Microbiol. Mol. Biol. Rev. 68, 692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suhr M., Benz I., Schmidt M. A. (1996) Mol. Microbiol. 22, 31–42 [DOI] [PubMed] [Google Scholar]
- 30.Charbonneau M. E., Janvore J., Mourez M. (2009) J. Biol. Chem. 284, 17340–17351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charbonneau M. E., Berthiaume F., Mourez M. (2006) J. Bacteriol. 188, 8504–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smukalla S., Caldara M., Pochet N., Beauvais A., Guadagnini S., Yan C., Vinces M. D., Jansen A., Prevost M. C., Latgé J. P., Fink G. R., Foster K. R., Verstrepen K. J. (2008) Cell 135, 726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charbonneau M. E., Mourez M. (2007) J. Bacteriol. 189, 9020–9029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kajava A. V., Steven A. C. (2006) J. Struct. Biol. 155, 306–315 [DOI] [PubMed] [Google Scholar]
- 35.Junker M., Schuster C. C., McDonnell A. V., Sorg K. A., Finn M. C., Berger B., Clark P. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Begley M., Gahan C. G., Hill C. (2005) FEMS Microbiol. Rev. 29, 625–651 [DOI] [PubMed] [Google Scholar]
- 37.Malik-Kale P., Parker C. T., Konkel M. E. (2008) J. Bacteriol. 190, 2286–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherlock O., Dobrindt U., Jensen J. B., Munk Vejborg R., Klemm P. (2006) J. Bacteriol. 188, 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leckband D., Prakasam A. (2006) Annu. Rev. Biomed. Eng. 8, 259–287 [DOI] [PubMed] [Google Scholar]
- 40.Sheikh J., Czeczulin J. R., Harrington S., Hicks S., Henderson I. R., Le Bouguénec C., Gounon P., Phillips A., Nataro J. P. (2002) J. Clin. Invest. 110, 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lebowitz J., Lewis M. S., Schuck P. (2002) Protein Sci. 11, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.