Abstract
Eukaryotes utilize fatty acids by β-oxidation, which occurs in the mitochondria and peroxisomes in higher organisms and in the peroxisomes in yeast. The AMP-activated protein kinase regulates this process in mammalian cells, and its homolog Snf1, together with the transcription factors Adr1, Oaf1, and Pip2, regulates peroxisome proliferation and β-oxidation in yeast. A constitutive allele of Adr1 (Adr1c) lacking the glucose- and Snf1-regulated phosphorylation substrate Ser-230 was found to be Snf1-independent for regulation of peroxisomal genes. In addition, it could compensate for and even suppress the requirement for Oaf1 or Pip2 for gene induction. Peroxisomal genes were found to be regulated by oleate in the presence of glucose, as long as Adr1c was expressed, suggesting that the Oaf1/Pip2 heterodimer is Snf1-independent. Consistent with this observation, Oaf1 binding to promoters in the presence of oleate was not reduced in a snf1Δ strain. Exploring the mechanism by which Adr1c permits Snf1-independent peroxisomal gene induction, we found that strength of promoter binding did not correlate with transcription, suggesting that stable binding is not a prerequisite for enhanced transcription. Instead, enhanced transcriptional activation and suppression of Oaf1, Pip2, and Snf1 by Adr1c may be related to the ability of Adr1c to suppress the requirement for and enhance the recruitment of transcriptional coactivators in a promoter- and growth medium-dependent manner.
Keywords: AMP-activated Kinase (AMPK), Peroxisomes, Transcription, Transcription Coactivators, Transcription Factors, Adr1, Oaf1, Pip2, Snf1, Gene Regulation
Introduction
β-Oxidation of fatty acids is an important metabolic pathway in eukaryotes, from yeast to humans. In mammalian cells, this process occurs primarily in the mitochondria, but in yeast it is carried out in specialized organelles, the peroxisomes (1). Yeast preferentially undergo fermentative metabolism in the presence of glucose, and most genes not essential for fermentation are turned off in its presence, a process known as glucose repression. Upon glucose exhaustion, yeast change their metabolism to aerobic respiration leading to the derepression of numerous genes involved in respiration and stress responses. This process involves the Snf1 protein kinase and its dependent transcription factors, Adr1 and Cat8 (2). The newly induced genes include those responsible for peroxisome proliferation and β-oxidation.
In the presence of fatty acids, these genes are further induced leading to active metabolism of exogenous fatty acids and peroxisome biogenesis (3–5). The mammalian AMP-activated protein kinase and its yeast homolog, Snf1 protein kinase, are important for both peroxisome proliferation in the presence of fatty acids, as well as β-oxidation (6). In yeast, the pathway further requires the coordinate action of three transcription factors, Adr1, Oaf1, and Pip2 (6–8). The Oaf1/Pip2-dependent response is analogous to the response of the superfamily of nuclear hormone receptors in higher eukaryotes that regulate transcription in response to a high fat diet or exposure to other peroxisome proliferators (9–11). Oaf1 binding to oleate potentiates the binding to the Gal11 component of Mediator, a tail subunit that is essential for growth on oleate (12). A fourth transcription factor, Oaf3, has been identified as a transcriptional repressor in the presence of oleate (13). How these transcription factors interact with each other and with the upstream signaling pathways for peroxisome proliferation is unknown.
The AMP-activated protein kinase/Snf1 kinase is considered the “energy sensor of the cell” because it responds to low cellular energy stores, often manifest as reduced AMP levels, by activating transcription factors and metabolic enzymes required for metabolism of alternative sugars and nonfermentable substrates (14). Snf1 is required for promoter binding of Adr1 (15), a process regulated by phosphorylation of Ser-98 in the DNA binding domain (16) and by acetylation of promoter nucleosomes (17, 18).
Rare semi-dominant alleles of ADR1, known as ADR1-constitutive (ADR1c), that constitutively express ADH2 on glucose and lead to higher levels of derepression (19, 20) in the absence of glucose, have been identified. Cloning and molecular analysis of these alleles showed they were in the open reading frame between amino acids 226 and 239 with the serine at position 230 being especially important (21, 22).
Adr1 is phosphorylated at Ser-230 by an unknown protein kinase under repressing conditions, when it is inactive (23). Activation of Adr1 in derepressing conditions is accompanied by Snf1-dependent dephosphorylation of Ser-230, consistent with Ser-230 phosphorylation contributing to inactivation of Adr1 (23). Thus, Snf1 is clearly not the Ser-230 kinase because phosphorylation of Ser-230 increases when Snf1 is absent. Moreover, the phenotype of a snf1 mutant is opposite to mutation of Ser-230 to a nonphosphorylatable allele. snf1 mutants are non-derepressible, i.e. inactive, whereas a S230A mutant is constitutively active and hyper-derepressible.
How Snf1 contributes to Ser-230 dephosphorylation is unknown (23). It could activate a protein phosphatase that dephosphorylates Ser-230 in derepressing conditions. Alternatively, it could inactivate a protein kinase or cause some other alteration that indirectly influenced the level of Ser-230 phosphorylation.
At least 14 genes involved in peroxisome proliferation and β-oxidation, including PEX11, CTA1, FOX2, SPS19, and POX1, have two consensus binding sequences in their promoters: the upstream activating sequence 1 (UAS1) for Adr1 binding (24, 25) and the oleate-response element (ORE)2 (26–28) for Oaf1/Pip2 binding (29, 30). UAS1 and ORE are typically found in close proximity or even overlapping in promoters. Pip2 itself has both UAS1 and ORE in its promoter, and thus Adr1 and Oaf1 regulate expression in the presence of fatty acids (31). Oaf1 and Pip2 form heterodimers that bind OREs, and this binding was shown recently to depend on Adr1 (32, 33). Oaf1 appears to be more important than Pip2 because overexpression of Oaf1 leads to gene expression in a pip2Δ strain but not vice versa (34). It has been shown, similar to the ligand binding domains of nuclear hormone receptors, that fatty acids, such as oleate, bind to the ligand binding domain of Oaf1 but not of Pip2, leading to its activation (12, 35). Interestingly, Oaf1 has been reported to also repress some genes independently of Pip2 upon activation by oleate (13). Because genes with ORE in their promoters are not expressed in snf1Δ or snf4Δ strains, and the transcriptional activity of Oaf1/Pip2 is repressed by glucose, it has been suggested that Oaf1 and/or Pip2 might be phosphorylated by Snf1 (36). However, phosphorylation of Oaf1 at multiple sites was demonstrated upon oleate addition, even in the absence of SNF1, eliminating Snf1 as the Oaf1 kinase (33). Therefore, regulation of Oaf1 and Pip2 by Snf1 is not understood.
Understanding how Snf1 regulates peroxisome proliferation and β-oxidation would reveal an important function of this AMP-activated protein kinase homolog. Using the constitutive alleles of Adr1, we have determined the differences in Snf1 dependence between Adr1 and Oaf1/Pip2 and elucidated some of the reasons for Adr1c potency. We show that Adr1c can induce peroxisomal genes in a snf1Δ mutant and compensate for the lack of OAF1 or PIP2 for gene expression, implying that Snf1 does not activate Oaf1/Pip2 directly. This property of Adr1c is analogous to its ability to activate genes in derepressing conditions in the absence of Snf1 and Cat8 (23). We found that, in the presence of Adr1c, peroxisomal genes are expressed upon oleate addition, even in the presence of glucose, when Snf1 is not fully phosphorylated. Finally, we show that Oaf1 binds promoters even in the absence of SNF1 or an active Adr1. We found that Adr1c is capable of binding promoters and overcoming the requirement for some transcriptional coactivators as well as recruiting some coactivators in the presence of glucose. In contrast, Adr1 phosphorylated on Ser-230 showed lower promoter occupancy and lower recruitment of pol II compared with WT Adr1.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Culture Conditions
Strains used in the study are listed in Table 1. Yeast strains were grown in complete or synthetic media as described by Sherman (37). Repressing medium contained 5% glucose; derepressing medium contained 0.05% glucose with or without 3% glycerol; and oleate-inducing medium was made by adding 0.5% Tween 40 and 0.1% oleate to the derepressing medium. Epitope tagging, marker swapping and gene deletion were according to Knop et al. (38), Guldener et al. (39), and Cross (40), respectively.
TABLE 1.
Saccharomyces cerevisiae strains used in the study
| Strain | Genotype | Source |
|---|---|---|
| CKY19 | W303-1A MATaade2 cam1-100 his3-11,15 leu2-13, 112 trp1-1 ura3-1 | Yeast stock center |
| EAY14 | W303-1α adr1Δ::kanMX ada1Δ::hygMX | This study |
| EAY12 | W303-1A adr1Δ::kanMX gcn5Δ:: hygMX | This study |
| EAY15 | W303-1A adr1Δ::kanMX snf2Δ:: hygMX | This study |
| KKTY04 | W303-1A snf5Δ::natMX | This study |
| TYY540 | W303-1A adr1Δ1::LEU2 ADH2::YIpADH2/lacZ(trp1::HIS3) gal11::natMX | This study |
| TYY541 | W303-1A adr1Δ1::LEU2 ADH2::YIpADH2/lacZ(trp1::HIS3) med2Δ:: kanMX | This study |
| TYY542 | W303-1A adr1Δ1::LEU2 ADH2::YIpADH2/lacZ(trp1::HIS3) pgd1Δ:: kanMX | This study |
| TYY543 | W303-1A adr1Δ1::LEU2 ADH2::YIpADH2/lacZ(trp1::HIS3) sin4Δ:: kanMX | This study |
| CKY13 | W303-1A adr1Δ::kanMX | This study |
| CKY26 | W303-1A adr1Δ::natMX snf1Δ::kanMX | This study |
| CKY7 | W303-1A snf1Δ::kanMX | This study |
| SRY1 | W303-1A oaf1Δ::kanMX | This study |
| SRY3 | W303-1A pip2Δ::kanMX | This study |
| SRY2 | W303-1A adr1Δ::kanMX oaf1Δ::natMX | This study |
| SRY5 | W303-1A adr1Δ::kanMX pip2Δ::natMX | This study |
| SRY67 | W303-1A adr1Δ::hygMX ADA1-myc-TRP1 | This study |
| RBY34 | W303-1A adr1Δ::LEU2 CAT8-TAP-HA::kanMX GCN5-myc-TRP1 | 43 |
| RBY36 | W303-1A adr1Δ::LEU2 CAT8-TAP-HA::kanMX SNF2-myc-TRP1 | 43 |
| Z1603 | W303-1A OAF1-myc-TRP1 | 57 |
The ADR1 and ADR1c expression plasmids used (15) were modified in some cases by introducing an epitope tag at the C terminus of Adr1 as described below. For gene expression studies, CEN-TRP1 plasmids with the native ADR1 promoter controlling expression of either wild type ADR1 (pKD16) or ADR1 alleles S230A (pKD14), R228K (pKD27), or Δ3, a deletion that removes Adr1 amino acids 226–233 (pKD26), were used. The plasmids pKD14H and pKD16H, where TRP1 was replaced by HIS3, were used in strains with myc-TRP1-tagged coactivators. For chromatin immunoprecipitation, plasmids expressing wild type ADR1 (pKD16-HA::kanMX (TRP1)) and the ADR1-S230A allele (pKD14-HA::kanMX (TRP1)) tagged with an HA epitope were employed.
Real Time Quantitative PCR (qPCR)
For expression analysis, RNA was isolated by hot phenol extraction (41) and converted to cDNA with a SuperScript III reverse transcriptase kit (Invitrogen) according to the manufacturer's directions. cDNA were quantified by real time qPCR (18) with an MJ Research Chromo4 system, using Quantace SYBR-Green Master Mix, according to the manufacturers' instructions. Primer sequences are available on request. The mRNA level of ACT1 was used as reference for normalization. Experiments were done in biological triplicates with two technical replicates per sample. The average values with standard deviations of the biological replicates are shown.
Chromatin Immunoprecipitation (ChIP)
ChIP was carried out using ethylene glycol-bis(succinimidylsuccinate) (Pierce) and formaldehyde as cross-linking agents as described previously (42). For immunoprecipitation, monoclonal antibodies against c-Myc (9E10) and HA (F7) epitopes and anti-pol II antibody against RNA polymerase II (8WG16) and Ser-5 C-terminal domain-specific antisera were purchased from Santa Cruz Biotechnology. Quantitation was done by real time qPCR as described above using primers designed to cover the promoter regions of specific genes. Primer sequences are available on request. Binding or recruitment was calculated as the ratio of the ChIP qPCR value, calculated from a standard curve, to the input value at a specific locus, relative to the same ratio for a negative control telomeric region (43). Sequential ChIP was performed as described by Tachibana et al. (18). Experiments were done in biological duplicates or triplicates with two technical replicates per sample, and results depict the average values with standard deviations.
RESULTS
Adr1c and Adr1 Overexpression Overcome the Lack of OAF1 or PIP2 and SNF1 to Induce Gene Expression on Oleate
Adr1c and overexpression of WT Adr1 led to the expression of Adr1-, Cat8-co-dependent genes on glucose (constitutive expression), as well as increased induction upon derepression, compared with WT Adr1 (15, 21–23, 43). To determine whether this was the case for peroxisomal genes, we used an adr1Δ strain (CKY13) transformed with CEN plasmids expressing Adr1c or Adr1 from a native promoter or WT Adr1 on a high copy 2-μm plasmid under the ADH1 promoter. We found that both Adr1c and WT Adr1 overexpression led to constitutive expression of peroxisomal genes (Table 2). Adr1c more efficiently activated peroxisomal genes than overexpressed Adr1, in contrast to expression of nonperoxisomal genes, such as ADH2, where the reverse was true. After 1 h of oleate induction, gene expression was significantly higher with Adr1c or overexpressed Adr1 as activator compared with single copy Adr1, but overexpressed Adr1 was able to activate most genes about 2-fold higher than Adr1c in the presence of oleate. The highly efficient activation by Adr1c is not because of increased stability, because Adr1c protein levels are much lower than those of WT Adr1 for unknown reasons (15, 44).
TABLE 2.
Constitutive gene expression and higher oleate induction with Adr1c and Adr1 overexpression
Values are average and standard deviation of two biological replicates expressed as fold overexpression with wtAdr1.
| Adr1a/conditionb | ADH2 | CTA1 | FAA2 | FOX2 | POT1 | POX1 | SPS19 |
|---|---|---|---|---|---|---|---|
| Adr1c R | 110 ± 1.8 | 19 ± 7.3 | 17 ± 0.66 | 21 ± 5.1 | 28 ± 1.2 | 53 ± 4.7 | 26 ± 7.3 |
| Adr1c DRI | 4.8 ± 0.45 | 6.6 ± 4.9 | 3.0 ± 1.6 | 3.5 ± 1.9 | 2.0 ± 0.69 | 6.7 ± 1.8 | 3.2 ± 1.9 |
| 2-μm Adr1 R | 390 ± 37 | 8.6 ± 3.4 | 6.0 ± 0.23 | 4.6 ± 1.5 | 6.0 ± 0.59 | 32 ± 8.2 | 8.1 ± 2.4 |
| 2-μm Adr1 DRI | 33 ± 3.9 | 16 ± 9.4 | 8.4 ± 1.6 | 9.0 ± 2.9 | 4.5 ± 1.4 | 16 ± 8.8 | 13 ± 3.6 |
a CEN plasmids expressing Adr1 (pKD16), Adr1c (pKD14), or 2-μm plasmid expressing Adr1 (pKD17) were transformed in CKY13.
b Repression (R) was in 5% glucose for 18 h; oleate induction (derepressed-induced, DRI) was for 1 h in 0.1% oleate + 3% glycerol.
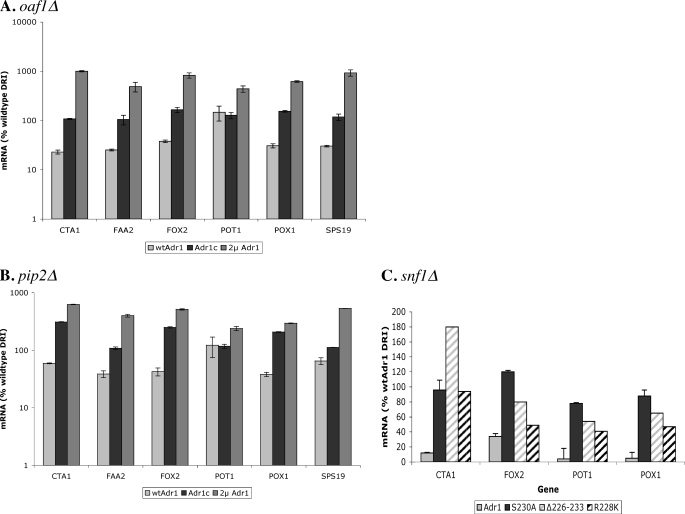
We have reported previously that expression of Adr1c or overexpression of WT Adr1 leads to the normal induction of genes that are co-dependent on Adr1 and Cat8 in a cat8Δ strain (23, 43). Because many peroxisomal genes are under the regulation of Adr1 and Oaf1/Pip2, we tested gene expression in adr1Δoaf1Δ and adr1Δpip2Δ mutants carrying the plasmids expressing Adr1 or Adr1c from a CEN plasmid or Adr1 from a 2-μm plasmid. Both Adr1c and Adr1 overexpression were able to fully overcome the absence of Oaf1 or Pip2 for oleate induction of peroxisomal genes (Fig. 1, A and B). When Adr1c was the activator, the genes were expressed fully in oaf1Δ and were highly induced in pip2Δ. This is likely to be because Pip2 is known to have a smaller role than Oaf1 in gene regulation, and Oaf1 can form homodimers in the absence of Pip2 to regulate genes (33, 45). In addition, Oaf1 is involved in the regulation of Pip2, so an oaf1Δ strain is doubly impaired in peroxisomal gene induction (31). In case of Adr1 overexpression, most genes tested were induced a few-fold higher than with Adr1c. This is similar to the behavior seen in the OAF1 PIP2 strain in Table 2.
FIGURE 1.
Adr1c and Adr1 overexpression can regulate peroxisomal genes upon oleate induction in oaf1Δ, pip2Δ, or snf1Δ. A and B, gene expression was measured by qPCR in adr1Δoaf1Δ or adr1Δpip2Δ carrying plasmids for Adr1 (pKD16), Adr1c (S230A, pKD14) or Adr1 overexpression (pKD17) after subjecting to glucose-derepressing, oleate-inducing conditions (0.1% oleate + 3% glycerol (derepressing-inducing, DRI)) for 1 h. C, adr1Δsnf1Δ strain carrying plasmids for either WT Adr1 (pKD16) or one of three Adr1c alleles (S230A, pKD14; Δ226–233, pKD26; and R228K, pKD27) was assayed for gene expression by qPCR after subjecting to glucose-derepressing, oleate-inducing conditions (0.1% oleate + 3% glycerol (DRI)) for 1 h. Values are expressed as percent wild type expression; error bars indicate standard deviation of three biological samples assayed in duplicate.
We showed previously that Adr1c, specifically the S230A allele, is not Snf1-dependent for gene regulation because it lacks a phosphorylatable residue at position 230 (23). Hence, we asked whether Adr1c was Snf1-independent for the expression of peroxisomal genes on oleate. The adr1Δsnf1Δ mutant was transformed with CEN plasmids expressing three different Adr1c alleles, S230A, Δ3(226–233), and R228K, or with WT Adr1, and transcription of peroxisomal genes was assayed after 1 h of incubation in glycerol-oleate. Although WT Adr1 was unable to induce the expression of peroxisomal genes in the absence of Snf1, all three Adr1c alleles were able to overcome the Snf1 deficiency (Fig. 1C). Similar to previous results of Snf1 independence upon derepression (23), the S230A allele and the partial deletion allele restored WT levels of peroxisomal gene expression. The R228K allele was not as efficient, possibly because it still carries a phosphorylatable Ser-230. Thus, Adr1c can overcome the requirement for its activating kinase Snf1 and the co-regulators of peroxisomal gene expression, Oaf1 and Pip2.
We also tested whether Adr1c could suppress the requirement for Oaf1 for growth on oleate. A strain with deletions of OAF1 and ADR1 was transformed with plasmids expressing either WT Adr1 or Adr1c, and transformants were tested for their ability to grow with various carbon sources in the medium. Both WT and Adr1c transformants grew equally well with glucose in the medium, but neither WT Adr1 nor Adr1c transformants could grow with oleate as the sole carbon source. Thus, Adr1c cannot suppress the requirement for Oaf1 for growth in oleate-inducing conditions.
Oaf1-dependent Transcription and Its Promoter Binding Are Independent of Snf1
The phosphorylation of Oaf1 upon oleate addition is Snf1-independent (33), and we showed that Adr1c can overcome Snf1 dependence (23). Thus, we tested if the transcriptional regulation of peroxisomal genes by Oaf1 depends on Snf1 in strains carrying Adr1c. For this purpose, we measured gene expression in glucose-repressed cultures, in which Snf1 is inactive, grown for 7–8 h in the presence of oleate. We found that 8 h after oleate addition, in cultures that still had repressing levels of glucose, Adr1c was able to induce expression of peroxisomal genes as effectively as after 1 h of growth in glucose-derepressing, oleate-inducing conditions. The control strain with WT Adr1 did not show significant peroxisomal gene expression (Table 3A).
TABLE 3.
Oleate induction of peroxisomal genes occurs in glucose in the presence of Adr1c
| A. Oleate induction occurs in repressing conditions in the presence of Adr1c | ||||
|---|---|---|---|---|
| Adr1a | CTA1 | FOX2 | POT1 | POX1 |
| Wild type | 8.0 ± 0.09 | 7.0 ± 0.2 | 2.0 ± 0.02 | 8.0 ± 0.03 |
| Adr1c | 99 ± 15 | 100 ± 43 | 82 ± 6 | 61 ± 27 |
| B. Adr1c-dependent oleate induction in repressing conditions occurs in the absence of Snf1 but is dependent on Oaf1/Pip2 | ||||
|---|---|---|---|---|
| Genotypeb | CTA1 | FOX2 | POT1 | POX1 |
| snf1Δ | 38 ± 0.04 | 71 ± 14 | 79 ± 0.08 | 66 ± 7 |
| oaf1Δ | 26 ± 5.0 | 52 ± 7.0 | 5 ± 0.8 | 5 ± 0.2 |
| pip2Δ | 37 ± 0.03 | 68 ± 9.0 | 9.0 ± 0.6 | 10 ± 2 |
a CEN-TRP1 plasmids expressing wild type Adr1 (pKD16) or Adr1c (pKD14; S230A) were transformed in CKY13 (adr1Δ). Gene expression was measured by qPCR 8 h after addition of 0.1% oleate to glucose-repressed cultures. The data are presented as the percent of expression with Adr1c in glucose-derepressing, oleate-inducing conditions; errors indicate standard deviation (as percent) of three biological samples.
b The adr1Δ strains employed are listed in Table 1. Gene expression in strains carrying Adr1c on pKD14 with deletion of SNF1, OAF1, or PIP2 was assayed by qPCR in glucose-repressed cultures 8 h after 0.1% oleate induction (repressing). Values are expressed as percent of expression in the same conditions, but with WT SNF1, OAF1, and PIP2; errors indicate standard deviation (as percent) of three biological samples.
The Snf1 independence of peroxisomal gene expression in the presence of Adr1c was confirmed by analyzing peroxisomal gene expression in the snf1Δ mutant in repressing-inducing (glucose-oleate) conditions. In the absence of Snf1, Adr1c allowed levels of expression close to the WT SNF1 strain for three of the four tested genes (FOX2, POT1, and POX1) (Table 3B). Thus, the induction of peroxisomal genes, which depends on active Snf1, Adr1, and Oaf1/Pip2, appeared to be Snf1-dependent only because of Snf1 regulation of Adr1 and not Snf1 regulation of Oaf1/Pip2.
Because Adr1c allowed oleate induction of peroxisomal genes in the absence of Oaf1 or Pip2, we asked if it could similarly overcome the deficiency of either of these transcription factors on glucose-oleate. However, we found that Adr1c still required Oaf1 and Pip2 for the induction of most peroxisomal genes on glucose-oleate; FOX2 was an exception. It was expressed as well in the oaf1 and pip2 mutants as in strains with WT OAF1 and PIP2 (Table 3B). In summary, Adr1c suppressed the Snf1, Oaf1, and Pip2 requirement of peroxisomal gene expression for derepression and induction, but not in the unusual condition of oleate induction in the presence of glucose.
Having shown that Snf1-dependent peroxisomal gene expression is not mediated by Oaf1, we predicted that promoter binding of Oaf1 would be independent of Snf1. We employed WT and snf1Δ strains in which Oaf1 was tagged with a Myc epitope to measure Oaf1 promoter binding by ChIP. Almost no Oaf1 binding was observed in glucose-repressed cells (data not shown). After 1 h of induction in glycerol-oleate, we found that Oaf1 bound to the promoters of all peroxisomal genes tested, irrespective of the presence or absence of Snf1 (Table 4). Because we have observed a decrease in ADH2 expression in oaf1Δ strains (data not shown), we tested and demonstrated Oaf1 binding to the ADH2 promoter (Table 4).
TABLE 4.
Oaf1 binding but not pol II recruitment is Snf1-independent
| Genotypea | ADH2 | CTA1 | FOX2 | POT1 | POX1 |
|---|---|---|---|---|---|
| Oaf1 binding | |||||
| Wild type | 3.1 ± 0.047 | 2.9 ± 0.16 | 3.7 ± 0.9 | 4.0 ± 0.082 | 2.8 ± 0.061 |
| snf1Δ | 3.4 ± 0.3 | 2.9 ± 0.1 | 4.1 ± 0.17 | 3.0 ± 0.2 | 3.6 ± 0.079 |
| pol II recruitmentb | |||||
| Wild type | 4.3 ± 0.08 | 4.0 ± 0.8 | 5.4 ± 1.0 | 5.0 ± 1.0 | 3.3 ± 0.6 |
| snf1Δ | 1.4 ± 0.1 | 1.5 ± 0.3 | 3.5 ± 0.7 | 1.0 ± 0.2 | 2.2 ± 0.4 |
a Oaf1 binding to promoters was detected by ChIP-qPCR as described under “Materials and Methods” in WT and snf1Δ strains with Myc-tagged Oaf1 after 1 h of oleate induction in glycerol medium (derepressed-induced). Values are the ratio of binding upon oleate induction over binding on glucose and represent the average and standard deviations of two biological samples.
b RNA polymerase recruitment upon oleate induction was determined at specific promoters by ChIP with anti-pol II antibody in WT and snf1Δ. Values are the ratio of binding after 1 h of oleate induction over binding on glucose; errors depict the standard deviations of two biological samples.
Peroxisomal gene expression is very low in the snf1Δ strain with WT Adr1 despite the presence of Oaf1 at the promoter (Fig. 1C). To test whether Oaf1 had recruited a preinitiation complex in these conditions, we assayed for the recruitment of RNA polymerase in the same extracts. As expected, whereas pol II was found at promoters in the strain with WT SNF1, it was significantly reduced in snf1Δ cells (Table 4), explaining the reduced transcription in a snf1Δ mutant with WT Adr1. Thus, Oaf1 binding is independent of Snf1 activity, but its binding is insufficient for pol II recruitment. pol II recruitment and induced transcription of peroxisomal genes requires the activity of Snf1 because Snf1 is required for Adr1 binding and coactivator and pol II recruitment (15, 17, 18).
Adr1c Binds Promoters of Some Genes More Efficiently than WT Adr1
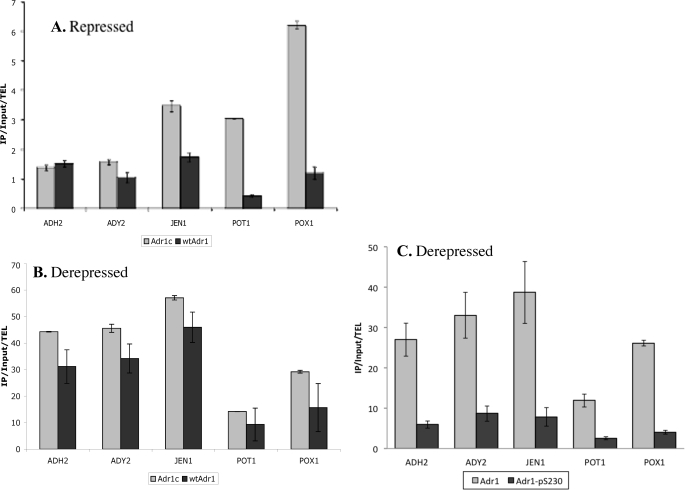
Having found that Adr1c could induce Snf1-independent expression at a variety of genes, we investigated why Adr1c is a better activator than WT Adr1 by measuring the binding efficiencies of the two activators by ChIP, using HA-tagged proteins. Under repressing conditions, where WT Adr1 is not detected at promoters (18, 43), Adr1c was detected at the promoters of JEN1, POT1, and POX1 (Fig. 2A). Surprisingly, Adr1c did not bind the ADH2 promoter better than WT Adr1, even though it causes constitutive expression of ADH2. Possibly, binding at the ADH2 promoter was too weak to be detected by ChIP, or Adr1c levels were transiently low at 4 h when the samples were collected, because the constitutive regulation of ADH2 is unlikely to be driven without promoter binding. Similarly, although better binding of Adr1c at POT1 and POX1 promoters might explain their constitutive expression (Table 2), JEN1 is not constitutively expressed (data not shown) despite Adr1c binding. Therefore, it appears that promoter binding alone is not sufficient to explain constitutive gene expression. It should be noted that Adr1c promoter binding on glucose is still far lower than that under derepressing conditions (Fig. 2B). On glycerol, Adr1c was found to bind ADH2, ADY2, JEN1, and POX1 promoters consistently better than WT Adr1. However, its binding was only 1.2–1.5-fold higher than WT binding, although its transcription activation was 2–6.7-fold higher (Table 2). Thus, it seems unlikely that the higher gene expression driven by Adr1c is due solely to its better promoter binding.
FIGURE 2.
Adr1c binds some gene promoters better than WT Adr1 under repressing conditions, and Ser-230-phosphorylated Adr1 binds less well in derepressing conditions. Adr1 binding at promoters was detected by ChIP under repressing (A, 5% glucose) and derepressing (B, 3% glycerol) conditions using adr1Δ transformed with plasmids pKD14HA (Adr1c-HA) or pKD16HA (wtAdr1-HA). C, binding of total Adr1 and Adr1 phosphorylated at Ser-230 was determined under derepressing (3% glycerol) conditions using anti-HA and anti-Ser(P)-230Adr1 antibodies in an adr1Δ strain carrying pKD16HA (wtAdr1-HA). Values were normalized to input and binding at telomere (TEL); error bars represent standard deviations of two biological samples. IP, immunoprecipitation.
Although most of Adr1 is phosphorylated at Ser-230 on glucose, a significant proportion is dephosphorylated upon derepression (23). This and the constitutive activity of Adr1c, which cannot be phosphorylated at Ser-230 (46), led to the hypothesis that phosphorylation at Ser-230 is inhibitory to Adr1 activity (23, 46). Therefore, we asked whether phosphorylated Adr1 could bind promoters using an antibody specific for phosphorylated Ser-230 of Adr1.
Ser-230-phosphorylated Adr1 was detected at all Adr1-dependent promoters tested in derepressing conditions, although its binding appeared to be lower than nonphosphorylated Adr1 (Fig. 2C). Adr1c, which cannot be phosphorylated at Ser-230 and is not recognized by the anti-phospho-Ser-230 antisera (47), was used as control, and the ChIP with anti-phospho-Ser-230 antibody did not detect binding above background (data not shown). In related experiments, we showed that promoter occupancy by Ser-230-phosphorylated Adr1 increased at a slower rate than for total Adr1 after removal of glucose, an observation that is consistent with a lower binding efficiency.3 In summary, nonphosphorylatable Adr1 (Adr1c) appears to have enhanced activity primarily due to a post-promoter binding function of the activator. In contrast, Ser-230-phosphorylated Adr1 is competent to bind DNA but appears to do so less well than nonphosphorylated Adr1.
Adr1c Overcomes the Requirement for Some Transcriptional Coactivators for Gene Expression
To investigate the possibility that phosphorylation of Ser-230 inhibited coactivator recruitment, we asked whether Adr1c might enable transcription without the recruitment of all coactivators required by WT Adr1 (43, 48). To test this, we measured gene expression in mutants lacking one of the coactivators required by, and recruited to, Adr1-dependent genes (43, 48). The coactivator mutants tested included mutants in the tail genes for the mediator complex (MED15, MED2, MED3, and MED16), in two genes encoding components of the SAGA complex (GCN5 and ADA1), and in two subunits of the Swi/Snf chromatin remodeling complex (SNF2 and SNF5). CoactivatorΔ adr1Δ strains were transformed with CEN plasmids expressing either WT Adr1 or Adr1c, and mRNA levels of several Adr1-dependent genes were measured after growth in glucose, glycerol, or oleate.
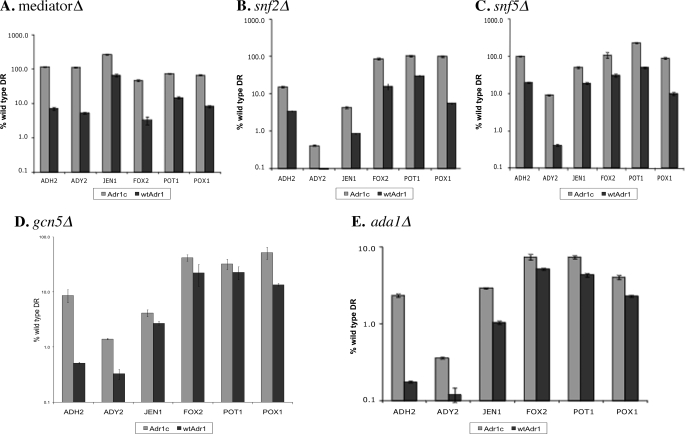
Under glucose-repressing conditions, deletion of components of Mediator or Swi/Snf did not affect constitutive expression with Adr1c (Table 5). The data are shown for two genes, ADH2 and POX1, that display high constitutive expression in the presence of Adr1c. In the snf5Δ strain, ADH2 was expressed constitutively even by WT Adr1. However, the deletion of either the histone acetyltransferase (Gcn5) or the structural scaffold (Ada1) of the SAGA complex greatly reduced the constitutive activity with Adr1c. Thus, on glucose, gene activation by Adr1c appears to be independent of the Mediator complex and Swi/Snf but is dependent on SAGA for constitutive gene expression.
TABLE 5.
Adr1c compensates for the loss of some transcriptional coactivators in repressed gene expression
mRNA levels are average and standard deviation of two biological replicates expressed relative to fold overexpression in a WT coactivator strain (CKY13) carrying pKD16 (wtAdr1) after overnight growth in 5% glucose.
| Coactivator mutationa | Adr1b | ADH2 | POX1 |
|---|---|---|---|
| None | Adr1c | 96 ± 13 | 60 ± 13 |
| Adr1 | 1.0 ± 1.0 | 1.0 ± 0.20 | |
| MediatorΔ | Adr1c | 53 ± 7.6 | 85 ± 15 |
| Adr1 | 3.6 ± 1.1 | 1.5 ± 0.30 | |
| snf2Δ | Adr1c | 50 ± 2.4 | 11.5 ± 0.85 |
| Adr1 | 8.6 ± 0.40 | 1.0 ± 0.10 | |
| snf5Δ | Adr1c | 165 ± 3.1 | 13 ± 0.10 |
| Adr1 | 35 ± 0.055 | 1.5 ± 0.15 | |
| gcn5Δ | Adr1c | 7.6 ± 0.38 | 0.50 ± 0.05 |
| Adr1 | 0.2 ± 0.014 | 0.50 ± 0.15 | |
| ada1Δ | Adr1c | 5.2 ± 0.48 | 0.50 ± 0.10 |
| Adr1 | 1.2 ± 0.06 | 0.50 ± 0.010 |
a None of the strains possess chromosomal ADR1. The strains used are listed in Table 1. MediatorΔ is the average expression from individual mutants of the Mediator complex (med2Δ, med3Δ, med15Δ, and med16Δ).
b pKD14 and pKD16 expressing Adr1c and Adr1, respectively, were transformed into the relevant strains.
In derepressing conditions, Adr1c was a stronger activator than WT Adr1 in all of the coactivator mutants (Fig. 3). A striking effect was observed in a mutant lacking Gcn5, the histone acetyltransferase component of the SAGA complex. WT Adr1 showed the expected strong dependence on Gcn5 (Fig. 3D), whereas Adr1c activated expression of most genes to nearly WT levels. In contrast, deleting ADA1, a scaffold component of SAGA, severely reduced gene activation by Adr1c, although it was still significantly higher than activation by WT Adr1. Thus, an intact SAGA complex is required for efficient activation by the Adr1c activator, but its histone acetyltransferase activity is dispensable. The suppression of the coactivator defects by Adr1c for the ADH2 gene was confirmed by in-gel staining of alcohol dehydrogenase activity and reporter gene assays (supplemental Fig. 1 and data not shown).
FIGURE 3.
Adr1c partially suppresses coactivator defects in derepressing conditions. mRNA levels were measured after 4 h of incubation in 3% glycerol by qPCR in strains deleted for specific coactivator components and ADR1, carrying either plasmid pKD14 for Adr1c or pKD16 for Adr1. A, data for individual mutants of the Mediator complex (med2Δ, med3Δ, med15Δ, and med16Δ) were combined and averaged; B, snf2Δ; and C, snf5Δ; D and E, SAGA mutants gcn5Δ and ada1Δ. Values are expressed as percentage of WT expression; error bars indicate standard deviations of three biological samples. DR, derepressing.
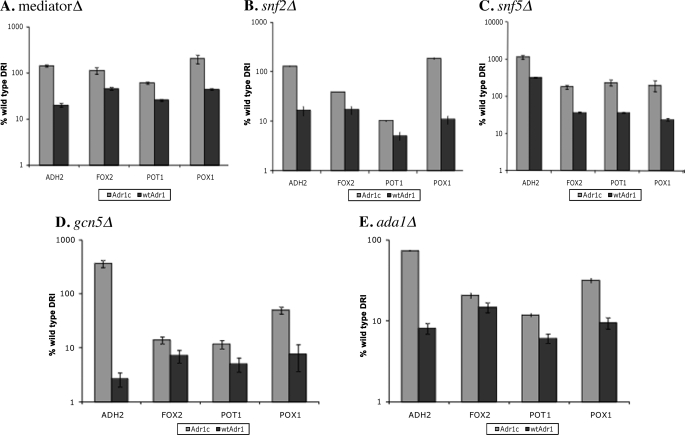
Upon oleate induction, coactivator dependence was decreased for Adr1c relative to WT Adr1 as in repressing and derepressing conditions with some differences (Fig. 4). Mediator components were more dispensable than in derepressing conditions, and gene expression was restored fully by Adr1c (Fig. 4). The Swi/Snf complex was important for gene expression induced by WT Adr1, but Adr1c could overcome its loss and induce full expression for all genes except FOX2 and POT1 in the snf2Δ mutant. In mutants of the SAGA complex, expression of peroxisomal genes was very low when either WT Adr1 or Adr1c was the activator. However, Adr1c restored expression of ADH2 to WT levels in the SAGA mutants. This may be due to the fact that ADH2 is regulated mainly by glucose repression rather than by oleate induction. The inability of Adr1c to activate the peroxisomal genes in oleate-inducing conditions in the absence of an active SAGA complex suggests that SAGA recruitment by Adr1c may have an important role in its suppression of an oaf1Δ defect in peroxisomal gene expression. In summary, Adr1c suppresses loss of coactivators in a complex manner that is affected by both the specific promoter examined and the growth conditions.
FIGURE 4.
Adr1c partially suppresses coactivator defects during oleate induction. mRNA levels were measured after 1 h of incubation in 0.1% oleate + 3% glycerol by qPCR in strains deleted for ADR1 and specific coactivator components as well as carrying either plasmid pKD14 for Adr1c or pKD16 for Adr1. A, average expression in individual mutants of the Mediator complex (med2Δ, med3Δ, med15Δ, and med16Δ); in Swi/Snf mutants: snf2Δ (B), and snf5Δ (C); and in SAGA mutants: gcn5Δ (D) and ada1Δ (E). Values are expressed as percentage of WT expression; error bars indicate standard deviations of three biological samples. DRI, derepressing-inducing.
Differential Recruitment of Transcriptional Coactivators and pol II by Adr1c and Ser-230-phosphorylated Adr1
Because Adr1c could overcome the loss of some coactivators for gene expression, we tested whether Adr1c could recruit coactivators differently than WT Adr1. Strains deleted for ADR1 and containing Myc-tagged coactivators were transformed with the Adr1 or Adr1c CEN plasmids. We focused on Swi/Snf and SAGA because Adr1c suppressed these defects most effectively under derepressing or oleate-inducing conditions. We had observed that SAGA was important for gene expression activated by WT Adr1 under all three growth conditions, and Adr1c compensated for the loss of Gcn5, but not Ada1, under glucose-derepressing conditions (Table 5; Figs. 3 and 4; and supplemental Fig. 1).
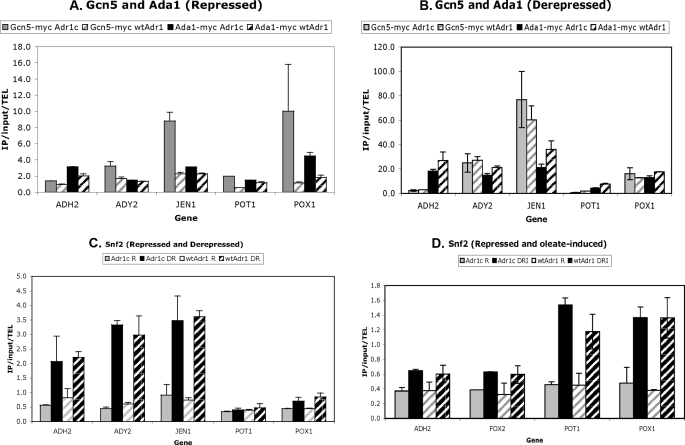
The recruitment of these coactivators was studied by quantitative ChIP analysis (Fig. 5). Under glucose-repressing conditions, Adr1c recruited Gcn5-myc better than Adr1 at all promoters tested, except ADH2 (Fig. 5A). The relatively better recruitment of SAGA components by Adr1c at POX1, but not POT1, under repressing conditions suggests that recruiting SAGA may overcome its Swi/Snf dependence (Fig. 3) by recruiting an alternative chromatin remodeling complex, SAGA. Ada1 was recruited more effectively by Adr1c at some promoters (Fig. 5A). However, under glucose-derepressing conditions, there was no significant difference in the recruitment of either SAGA component by Adr1c or Adr1 (Fig. 5B).
FIGURE 5.
Adr1c recruits some coactivator components better than WT Adr1. Recruitment of coactivator components was measured by ChIP-qPCR in strains deleted for ADR1 but carrying either plasmid pKD14 for Adr1c or pKD16 for Adr1 and with Myc-tagged coactivator components. Samples were collected under repressing (R; 5% glucose), derepressing (DR; 3% glycerol) and inducing (DRI; 0.1% oleate + 3% glycerol) conditions. The recruitment of SAGA components (Gcn5 and Ada1) was determined upon repression (A) and derepression (B). C and D show Snf2 recruitment under derepression and oleate induction, respectively, compared with repressed conditions. Error bars indicate standard deviations of two biological samples. IP, immunoprecipitation.
Because Gcn5 was dispensable for transcription by Adr1c under derepressing conditions, we asked whether Adr1c could recruit other histone acetyltransferases, such as Taf1 of the TFIID complex, or Esa1 of the NuA4 complex. However, ChIP for Taf1 or the NuA4 components, Esa1 and Epl1, showed very low recruitment and no difference between Adr1 and Adr1c (data not shown). This confirms previous results indicating that these coactivators are not recruited to ADH2 by WT Adr1 (43) and suggests that Adr1c does not compensate for loss of Gcn5 by recruiting either of these histone acetyltransferase activities.
The recruitment of Snf2 was tested because, under glucose-repressing conditions, Adr1c partially overcame the loss of either Swi/Snf component at several promoters (Table 5). However, Snf2 was not found at the promoter of any gene tested under repressing conditions (Fig. 5, C and D). This could be due to the relatively low level of gene expression in repressing conditions or because the dependence on Snf2 is indirect.
In derepressing conditions, Snf2 was recruited similarly by both Adr1 and Adr1c to the nonperoxisomal genes but was not recruited by either to peroxisomal genes. Snf2 was previously found to be more important for the expression of nonperoxisomal than peroxisomal genes (Fig. 3), and its absence was suppressed by Adr1c. The ChIP analysis thus provides an explanation for these observations because Swi/Snf does not appear to be recruited to the peroxisomal genes.
Under oleate-inducing conditions, gene expression in the snf2Δ strain partly corresponded to Snf2 recruitment, in that Adr1c restored ADH2 and FOX2 expression (Fig. 4B), but neither Adr1 nor Adr1c recruited Snf2 to these promoters (Fig. 5D). Both activators recruited Snf2 equally to the POT1 promoter, in agreement with its expression in the snf2Δ strain. However, although Adr1c restored POX1 expression in a snf2Δ mutant, both Adr1 and Adr1c recruited Snf2 similarly. Thus, enhanced recruitment by Adr1c of the coactivators tested does not appear to explain the ability of Adr1c to suppress defective transcription in individual coactivator mutants.
We studied the recruitment of coactivators and RNA pol II by Ser-230-phosphorylated Adr1 and total Adr1 (both phosphorylated and nonphosphorylated) in derepressing conditions using sequential ChIP. Sequential ChIP was performed with either anti-Adr1-Ser(P)-230 or anti-HA antibody (for total Adr1-HA) for the primary ChIP. This first step would pull down promoters bound by either phosphorylated or total Adr1. Anti-Myc antibody was used in the second ChIP to determine the subpopulation of promoters that was also bound by Myc-tagged coactivators. Anti-pol II antibody and anti-pol II-Ser(P)-5 antibody were used to identify promoters occupied by pol II and Ser-5(P)-pol II, respectively, and the activators, and anti-Tbp antibody was used to identify Tbp.
The coactivators recruited by Ser-230-phospho-Adr1 and total Adr1 by sequential ChIP included TATA-binding protein, SAGA (Gcn5 and Ada1), Swi/Snf (Snf2), and Mediator (Gal11), total pol II, and Ser-5-phosphorylated pol II. All of these, with the possible exception of Ser-5-phosphorylated pol II, were found at levels equivalent to promoter binding by the respective forms of Adr1 (supplemental Fig. 2). This result suggests that Ser-230-phosphorylated Adr1 recruits these complexes as well as nonphosphorylated Adr1. However, the relative enrichment of Ser-230-phosphorylated Adr1 at promoters was low (Fig. 2), making the signal-to-noise ratio weak in the first ChIP. In addition, most promoters have multiple Adr1-binding sites, and the promoters could be occupied by both Ser(P)-230 and nonphosphorylated Adr1. As a consequence, the coactivators present at promoters containing Ser(P)-230-Adr1 could also be occupied by nonphosphorylated Adr1.
In conclusion, the results suggest that Ser-230-phosphorylated Adr1 is able to recruit coactivators and pol II. If this conclusion is confirmed by future studies, it would indicate that phosphorylation blocks a post-recruitment step in activation by Adr1. This would be consistent with the observation that the strong Adr1c activator is not significantly better at coactivator recruitment than WT Adr1 in derepressing conditions but activates transcription to significantly higher levels.
DISCUSSION
We have shown that when Adr1 is constitutively active because of mutation of Ser-230 (Adr1c), Snf1 is not necessary for peroxisomal gene expression. This conclusion is derived from the observation that oleate induction of peroxisomal genes occurs in the absence of Snf1 if Adr1c is the activator but not if WT Adr1 is the activator. These results were obtained both in the presence of glucose, when Snf1 is inactive, and under derepressing conditions in a snf1Δ strain. Because nonphosphorylated Snf1 may retain some activity on glucose, the results obtained in the snf1Δ strain provide the most compelling evidence that Adr1c suppresses Snf1 dependence. The observations that Oaf1 promoter binding is Snf1-independent and that peroxisomal gene induction in oleate-inducing conditions can occur in a snf1Δ strain are convincing evidence that Snf1 regulates peroxisomal gene expression primarily through activation of Adr1. This conclusion pertains only to the peroxisomal genes we analyzed, which are primarily in the β-oxidation pathway. Other peroxisomal genes, particularly the PEX genes that are involved in peroxisome biogenesis, may be regulated differently. We suspect this may be the case because Adr1c could not suppress the Oaf1 requirement for growth on oleate.
These studies confirm that Ser-230 in the regulatory domain is an important target through which Snf1 regulates the activity of Adr1 (23). Snf1 is not the Ser-230 kinase because the phenotype of snf1Δ is opposite to that of an S230A mutation in Adr1. Loss of Snf1 inactivates Adr1, whereas the S230A mutation activates Adr1. More importantly, loss of Snf1 causes an increased level of Ser-230 (and Ser-98) phosphorylation rather than a decreased level, as would be expected if Snf1 were the S230 (or Ser-98) kinase (16, 23). Thus, it appears that Snf1 inhibits the activity of a Ser-230 kinase or activates a phospho-Ser-230 protein phosphatase. We emphasize this point because recent reports assume that Snf1 activates Adr1 by phosphorylating it, whereas the opposite is true. Snf1 is important for dephosphorylating both Ser-98 in the DNA binding domain and Ser-230 in the regulatory domain (33, 36).
Although the Snf1 dependence of peroxisomal gene expression was found to be primarily through Adr1, snf1Δ is known to lack peroxisomal structures, whereas adr1Δ develops small single peroxisomes (6). If Snf1 acts solely through Adr1, the same phenotype should be observed in both mutants. This apparent discrepancy could be because another target of Snf1 is necessary for full peroxisome biogenesis, such as the numerous PEX genes, or post-transcriptional modifications that are important for structural changes of peroxisomes (49). This interpretation is consistent with our observation that Adr1c could suppress the Oaf1 requirement for expression of genes encoding enzymes of the β-oxidation pathway (Fig. 1) and a few other peroxisomal genes such as CTA1 and PEX11,4 but it could not suppress the requirement for Oaf1 for growth on oleate.
How Adr1 and Oaf1/Pip2 interact to promote peroxisomal gene expression is unknown. A promoter containing both UAS1 and ORE is better regulated than a promoter with either element alone on oleate addition (50), indicating synergistic transcriptional activation.
Mutual stabilization at promoters may be another way that Adr1 and Oaf1/Pip2 interact to induce peroxisomal genes. Adr1, Oaf1, and Pip2 have been reported to be at least partially dependent on each other for promoter binding (33, 34). This could explain the Snf1 dependence of peroxisomal genes, where promoter binding by Adr1 is Snf1-dependent (51), but Oaf1 phosphorylation (33) and promoter binding are not (Table 4). This also suggests that a Snf1-activated Adr1 is not required for Oaf1 binding. Baumgartner et al. (34) used constitutively expressed Oaf1 and Pip2 to study the oleate response on glucose and found that Oaf1 responds to oleate only in the absence of glucose. We interpret this result as due to Snf1 inactivity on glucose that keeps Adr1 inactive (23). When Adr1c was the activator, we observed Oaf1-dependent gene expression in glucose-oleate (Table 3). Similarly, none of the nonessential kinase or phosphatase deletions led to oleate induction of genes in the presence of glucose (52). This could be because the Glc7 phosphatase that regulates Snf1 kinase is an essential gene, whereas the deletion of its regulatory subunit, Reg1, is known to activate Adr1 (15).
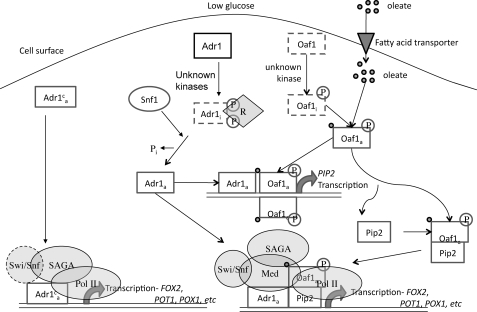
Thus, our findings agree with a model in which Adr1 is constitutively expressed but requires Snf1-dependent activation (in part by indirectly dephosphorylating Ser-230 and Ser-98 (23)), and Oaf1 is constitutively expressed but requires activation by binding to fatty acids (Fig. 6). Activation of both Adr1 and Oaf1 leads to Pip2 expression and its dimerization with Oaf1. When oleate is taken up by cells it binds to and activates Oaf1 (35). Active Adr1 and Oaf1 bind to the PIP2 promoter leading to its transcription (31). Oaf1 and Pip2 form heterodimers (32) and, together with Adr1, activate genes involved in β-oxidation and peroxisomal proliferation (5, 8, 13, 25, 28, 29, 53). The Oaf1/Pip2 heterodimer binds promoters upon glucose derepression, and binding only moderately increases upon oleate induction, suggesting that ligand binding by fatty acids probably activates transcription but not binding (33). Adr1c short-circuits this process in an unknown manner because it activates the peroxisomal genes in the absence of either Snf1 or Oaf1. The independence of Adr1c activation from some coactivators suggests that altered PIC formation or activity may be involved in allowing Adr1c to activate transcription in the absence of Snf1 and Oaf1.
FIGURE 6.
Model for the interaction of Snf1, Adr1, Oaf1, and Pip2 at co-dependent promoters. In this model, Snf1 controls the activity of Adr1 by indirectly stimulating its dephosphorylation (23). The inactive transcription factors are shown in rectangles with dashed borders and are denoted with an i; the active transcription factors are enclosed in solid rectangles and are denoted with an a. Adr1c does not require Snf1 for activation because it lacks that phosphorylatable Ser-230. Swi/Snf1 was recruited to Adr1c-activated peroxisomal genes, but it is shown with a dashed border because their transcription was less sensitive to its loss when Adr1c was the activator (Table 5; Figs. 3 and 4). Mediator was not assessed at peroxisomal genes because their transcription was relatively insensitive to its loss when Adr1c was the activator. Adr1c-activated transcription of peroxisomal genes was dependent on SAGA but was less sensitive to the loss of the histone acetyltransferase component Gcn5 than to the loss of the structural component Ada1 (Table 5; Figs. 3 and 4).
Although ADH2 does not encode a component of the β-oxidation pathway, it is the gene that was most highly expressed in a global analysis of oleate induction (5). We confirmed that ADH2 expression is oleate-induced and that it decreased during oleate induction of oaf1Δ and pip2Δ mutants (data not shown). The requirements for OAF1 and PIP2 may have been missed in the global transcriptome experiments because of the homology between ADH1 and ADH2. The ChIP data indicating that Oaf1 binds to the ADH2 promoter during oleate induction (Table 4) suggests that Oaf1 binding may be a direct effect of Oaf1/Pip2 activation. Oaf1 and Pip2 are known to transcribe some nonperoxisomal genes, such as CIT1 (28). Although the ADH2 promoter is not known to contain a consensus ORE, it appears to possess at least two putative ORE half-sites with CGGN3TN(A/R)N8–12CCG consensus sequence (54) at position −298 and at −386. OREs in other promoters such as CTA1 and POX1 overlap with or are adjacent to Adr1-binding sites (30). Adr1 and Cat8 interact at UAS1-UAS2/carbon source response element to synergistically activate ADH2 expression in derepressing conditions (42, 55, 56). The binding of Oaf1 in the ADH2 promoter might allow an analogous interaction between Adr1 and Oaf1 during oleate induction. However, further work will be required to identify the Oaf1-binding site in the ADH2 promoter, as some Oaf1/Pip2-regulated genes such as PXA1 do not possess a consensus ORE (4, 28, 33).
What is the mechanism that enhances transcription by Adr1c and allows it to suppress loss of upstream signaling by Snf1? Although Adr1c does not have enhanced DNA binding activity in vitro (44), a significant increase in binding was detectable by ChIP at some, but not all, promoters (Fig. 2). However, the increase in binding was not always associated with increased transcription. The lack of correlation between binding and activation by Adr1c suggests that mutation of the regulatory domain primarily affects a post-DNA binding step. A post-DNA binding role for the regulatory domain is also suggested by the observation that Adr1c partially suppressed, in a promoter- and condition-dependent manner, loss of several coactivators. It strongly suppressed the effects of loss of Mediator tail components (Table 5; Fig. 3A, and Fig. 4A, and supplemental Fig. 1), which are particularly important for oleate induction (52). In addition, Adr1c recruited Gcn5 and Ada1 more efficiently than WT Adr1 at some promoters but only in repressing growth conditions. Suppression of the requirement for Gcn5 did not appear to be due to recruitment of the histone acetyltransferase activity of Taf1 or NuA4.
Thus, enhanced recruitment of coactivators does not seem sufficient to explain the enhanced and Snf1- and Oaf1-independent activation by Adr1c. Additional possibilities include direct recruitment of RNA pol II or an effect on a post-binding step in initiation.
In contrast to promoters occupied by Adr1c, which cannot be phosphorylated on Ser-230, promoters that were occupied by Ser-230-phosphorylated Adr1 appeared to recruit lower levels of coactivators. The recruitment of pol II phosphorylated on Ser-5 in the C-terminal domain was particularly reduced by Ser-230 phosphorylation. We conclude that Ser-230-phosphorylated Adr1 is able to occupy promoters and recruit coactivators and pol II, but it may be defective in a later step of transcription initiation, consistent with our conclusion that the nonphosphorylatable Adr1c allele enhances a post-DNA binding step in transcription.
Partial independence from coactivators or more efficient coactivator recruitment may play a general role in Snf1-independent activation of Adr1 target genes. At Adr1- and Cat8-dependent promoters, the two transcription factors act both independently and cooperatively to recruit coactivators and form a preinitiation complex (42, 43). Adr1 and Oaf1 may act in a similar manner, and Adr1c might suppress the loss of Oaf1 by enhancing recruitment of Oaf1-dependent coactivators.
Supplementary Material
Acknowledgments
We thank Chris Tachibana and Virginia Price for valuable comments on the manuscript and other members of the laboratory for interest and encouragement. Erin Arms, Rhiannon Biddick, Kenneth Dombek, and Catherine Keil constructed some of the mutant strains, and Steve Hahn generously provided anti-Tbp antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant GM26079 (to E. T. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
N. Kacherovsky, unpublished data.
S. Ratnakumar and E. T. Young, unpublished data.
- ORE
- oleate-response element
- pol II
- RNA polymerase II
- ChIP
- chromatin immunoprecipitation
- qPCR
- quantitative PCR
- WT
- wild type
- HA
- hemagglutinin
- SAGA
- Spt-Ada-Gcn5-acetyltransferase complex.
REFERENCES
- 1.Hiltunen J. K., Mursula A. M., Rottensteiner H., Wierenga R. K., Kastaniotis A. J., Gurvitz A. (2003) FEMS Microbiol. Rev. 27, 35–64 [DOI] [PubMed] [Google Scholar]
- 2.Schüller H. J. (2003) Curr. Genet. 43, 139–160 [DOI] [PubMed] [Google Scholar]
- 3.Veenhuis M., Mateblowski M., Kunau W. H., Harder W. (1987) Yeast 3, 77–84 [DOI] [PubMed] [Google Scholar]
- 4.Smith J. J., Marelli M., Christmas R. H., Vizeacoumar F. J., Dilworth D. J., Ideker T., Galitski T., Dimitrov K., Rachubinski R. A., Aitchison J. D. (2002) J. Cell Biol. 158, 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kal A. J., van Zonneveld A. J., Benes V., van den Berg M., Koerkamp M. G., Albermann K., Strack N., Ruijter J. M., Richter A., Dujon B., Ansorge W., Tabak H. F. (1999) Mol. Biol. Cell 10, 1859–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon M., Binder M., Adam G., Hartig A., Ruis H. (1992) Yeast 8, 303–309 [DOI] [PubMed] [Google Scholar]
- 7.Luo Y., Karpichev I. V., Kohanski R. A., Small G. M. (1996) J. Biol. Chem. 271, 12068–12075 [DOI] [PubMed] [Google Scholar]
- 8.Rottensteiner H., Kal A. J., Filipits M., Binder M., Hamilton B., Tabak H. F., Ruis H. (1996) EMBO J. 15, 2924–2934 [PMC free article] [PubMed] [Google Scholar]
- 9.Ishii H., Fukumori N., Horie S., Suga T. (1980) Biochim. Biophys. Acta 617, 1–11 [DOI] [PubMed] [Google Scholar]
- 10.Reddy J. K., Krishnakantha T. P. (1975) Science 190, 787–789 [DOI] [PubMed] [Google Scholar]
- 11.Näär A. M., Thakur J. K. (2009) Genes Dev. 23, 419–432 [DOI] [PubMed] [Google Scholar]
- 12.Thakur J. K., Arthanari H., Yang F., Chau K. H., Wagner G., Näär A. M. (2009) J. Biol. Chem. 284, 4422–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith J. J., Ramsey S. A., Marelli M., Marzolf B., Hwang D., Saleem R. A., Rachubinski R. A., Aitchison J. D. (2007) Mol. Syst. Biol. 3, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardie D. G., Carling D., Carlson M. (1998) Annu. Rev. Biochem. 67, 821–855 [DOI] [PubMed] [Google Scholar]
- 15.Dombek K. M., Young E. T. (1997) Mol. Cell. Biol. 17, 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kacherovsky N., Tachibana C., Amos E., Fox D., 3rd, Young E. T. (2008) PLoS ONE 3, e3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verdone L., Wu J., van Riper K., Kacherovsky N., Vogelauer M., Young E. T., Grunstein M., Di Mauro E., Caserta M. (2002) EMBO J. 21, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tachibana C., Biddick R., Law G. L., Young E. T. (2007) J. Biol. Chem. 282, 37308–37315 [DOI] [PubMed] [Google Scholar]
- 19.Bemis L. T., Denis C. L. (1988) Mol. Cell. Biol. 8, 2125–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciriacy M. (1979) Mol. Gen. Genet. 176, 427–431 [DOI] [PubMed] [Google Scholar]
- 21.Denis C. L., Gallo C. (1986) Mol. Cell. Biol. 6, 4026–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denis C. L., Fontaine S. C., Chase D., Kemp B. E., Bemis L. T. (1992) Mol. Cell. Biol. 12, 1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratnakumar S., Kacherovsky N., Arms E., Young E. T. (2009) Genetics 182, 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon M., Adam G., Rapatz W., Spevak W., Ruis H. (1991) Mol. Cell. Biol. 11, 699–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young E. T., Dombek K. M., Tachibana C., Ideker T. (2003) J. Biol. Chem. 278, 26146–26158 [DOI] [PubMed] [Google Scholar]
- 26.Wang T., Luo Y., Small G. M. (1994) J. Biol. Chem. 269, 24480–24485 [PubMed] [Google Scholar]
- 27.Einerhand A. W., Voorn-Brouwer T. M., Erdmann R., Kunau W. H., Tabak H. F. (1991) Eur. J. Biochem. 200, 113–122 [DOI] [PubMed] [Google Scholar]
- 28.Karpichev I. V., Small G. M. (1998) Mol. Cell. Biol. 18, 6560–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurvitz A., Wabnegger L., Rottensteiner H., Dawes I. W., Hartig A., Ruis H., Hamilton B. (2000) Mol. Cell. Biol. Res. Commun. 4, 81–89 [DOI] [PubMed] [Google Scholar]
- 30.Gurvitz A., Hamilton B., Hartig A., Ruis H., Dawes I. W., Rottensteiner H. (1999) Mol. Gen. Genet 262, 481–492 [DOI] [PubMed] [Google Scholar]
- 31.Rottensteiner H., Wabnegger L., Erdmann R., Hamilton B., Ruis H., Hartig A., Gurvitz A. (2003) J. Biol. Chem. 278, 27605–27611 [DOI] [PubMed] [Google Scholar]
- 32.Rottensteiner H., Kal A. J., Hamilton B., Ruis H., Tabak H. F. (1997) Eur. J. Biochem. 247, 776–783 [DOI] [PubMed] [Google Scholar]
- 33.Karpichev I. V., Durand-Heredia J. M., Luo Y., Small G. M. (2008) J. Biol. Chem. 283, 10264–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baumgartner U., Hamilton B., Piskacek M., Ruis H., Rottensteiner H. (1999) J. Biol. Chem. 274, 22208–22216 [DOI] [PubMed] [Google Scholar]
- 35.Phelps C., Gburcik V., Suslova E., Dudek P., Forafonov F., Bot N., MacLean M., Fagan R. J., Picard D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7077–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurvitz A., Rottensteiner H. (2006) Biochim. Biophys. Acta 1763, 1392–1402 [DOI] [PubMed] [Google Scholar]
- 37.Sherman F. (1991) Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 38.Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. (1999) Yeast 15, 963–972 [DOI] [PubMed] [Google Scholar]
- 39.Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. (1996) Nucleic Acids Res. 24, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cross F. R. (1997) Yeast 13, 647–653 [DOI] [PubMed] [Google Scholar]
- 41.Collart M., Oliveiro S. (1993) in Current Protocols in Molecular Biology (Ausubel F. M., Brent R., Kingston R. T., Moode D. D., Seidman J., Smith J. A., Struhl K. eds) pp. 13.12.1–13.12.5, Greene/Wiley Interscience, New York [Google Scholar]
- 42.Tachibana C., Yoo J. Y., Tagne J. B., Kacherovsky N., Lee T. I., Young E. T. (2005) Mol. Cell. Biol. 25, 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biddick R. K., Law G. L., Young E. T. (2008) PLoS ONE 3, e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor W. E., Young E. T. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 4098–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trzcinska-Danielewicz J., Ishikawa T., Miciałkiewicz A., Fronk J. (2008) Biochem. Biophys. Res. Commun. 374, 763–766 [DOI] [PubMed] [Google Scholar]
- 46.Cherry J. R., Johnson T. R., Dollard C., Shuster J. R., Denis C. L. (1989) Cell 56, 409–419 [DOI] [PubMed] [Google Scholar]
- 47.Dombek K. M., Camier S., Young E. T. (1993) Mol. Cell. Biol. 13, 4391–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biddick R. K., Law G. L., Chin K. K., Young E. T. (2008) J. Biol. Chem. 283, 33101–33109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith J. J., Sydorskyy Y., Marelli M., Hwang D., Bolouri H., Rachubinski R. A., Aitchison J. D. (2006) Mol. Syst. Biol. 2, 2006.0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gurvitz A., Hiltunen J. K., Erdmann R., Hamilton B., Hartig A., Ruis H., Rottensteiner H. (2001) J. Biol. Chem. 276, 31825–31830 [DOI] [PubMed] [Google Scholar]
- 51.Young E. T., Kacherovsky N., Van Riper K. (2002) J. Biol. Chem. 277, 38095–38103 [DOI] [PubMed] [Google Scholar]
- 52.Saleem R. A., Knoblach B., Mast F. D., Smith J. J., Boyle J., Dobson C. M., Long-O'Donnell R., Rachubinski R. A., Aitchison J. D. (2008) J. Cell Biol. 181, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurvitz A., Rottensteiner H., Hiltunen J. K., Binder M., Dawes I. W., Ruis H., Hamilton B. (1997) Mol. Microbiol. 26, 675–685 [DOI] [PubMed] [Google Scholar]
- 54.Rottensteiner H., Hartig A., Hamilton B., Ruis H., Erdmann R., Gurvitz A. (2003) Eur. J. Biochem. 270, 2013–2022 [DOI] [PubMed] [Google Scholar]
- 55.Donoviel M. S., Kacherovsky N., Young E. T. (1995) Mol. Cell. Biol. 15, 3442–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walther K., Schüller H. J. (2001) Microbiology 147, 2037–2044 [DOI] [PubMed] [Google Scholar]
- 57.Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., Gerber G. K., Hannett N. M., Harbison C. T., Thompson C. M., Simon I., Zeitlinger J., Jennings E. G., Murray H. L., Gordon D. B., Ren B., Wyrick J. J., Tagne J. B., Volkert T. L., Fraenkel E., Gifford D. K., Young R. A. (2002) Science 298, 799–804 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.