Abstract
The lymphatic endothelial hyaluronan (HA) receptor Lyve-1 is a member of the Link protein superfamily most similar to the leukocyte HA receptor CD44. However, the structure of Lyve-1 and the nature of its interaction with ligand are obscure. Here we present new evidence that Lyve-1 is functionally distinct from CD44. Using truncation mutagenesis we confirm that Lyve-1 in common with CD44 contains an extended HA-binding unit, comprising elements flanking the N and C termini of the consensus lectin-like Link module, bridged by a third conserved disulfide linkage that is critical for HA binding. In addition, we identify six essential residues Tyr-87, Ile-97, Arg-99, Asn-103, Lys-105, and Lys-108 that define a compact HA-binding surface on Lyve-1, encompassing the epitope for an adhesion-blocking monoclonal antibody 3A, in an analogous position to the HA-binding surface in CD44. The overtly electrostatic character of HA binding in Lyve-1 and its sensitivity to ionic strength (IC50 of 150 mm NaCl) contrast markedly with CD44 (IC50 > 2 m NaCl) in which HA binding is mediated by hydrogen bonding and hydrophobic interactions. In addition, unlike the extended Link module in CD44, which binds HA efficiently when expressed as a soluble monomer (Kd = 65.7 μm), that of Lyve-1 requires artificial dimerization, although the full ectodomain is active as a monomer (Kd = 35.6 μm). Finally, full-length Lyve-1 did not form stable dimers in binding-competent 293T transfectants when assessed using bioluminescent resonance energy transfer. These results reveal that elements additional to the extended Link module are required to stabilize HA binding in Lyve-1 and indicate important structural and functional differences with CD44.
Keywords: Extracellular Matrix/Hyaluronan, Methods/Site-directed Mutagenesis, Protein/Ligand Binding, Protein/Structure, Receptors/Extracellular Matrix, Receptors/Structure-function, Lyve-1, Lymphatic
Introduction
The lymphatic system handles the uptake of fluid and dissolved macromolecules that constantly leak from the blood to the tissues, coordinating their subsequent return to the circulation via the thoracic duct and subclavian veins (1, 2). In fulfilling this function, the lymphatics also provide a discrete compartment for the turnover and catabolism of interstitial matrix components such as the large abundant glycosaminoglycan hyaluronan (GlcNAcβ1–4GlcUAβ1–3)n, which undergoes terminal degradation in lymph nodes, liver, and spleen by means of the endocytic HARE (hyaluronan receptor on endothelium) receptor (3, 4), an arrangement that may prevent buildup of potentially inflammatory oligosaccharides in the tissues (5, 6). The lymphatics are also a major limb of the immune system, through which antigen-presenting cells and other leukocytes traffic from the tissues to the draining lymph nodes during the course of immune surveillance and the generation of primary T-cell responses (7–9). Some steps in these latter processes such as the migration of cells within the lymph nodes involve specific interactions with hyaluronan (10), which forms a substratum for leukocyte adhesion and migration via the primary leukocyte HA receptor CD44, a member of the Link protein superfamily (11, 12). In addition, the vessels of the lymphatic system selectively express another hyaluronan receptor termed lymphatic vessel endothelial receptor-1 (Lyve-1)3 in place of CD44 (13–15), suggesting that HA engages in yet further interactions within the lymphatic compartment that relate specifically to lymphatic endothelium.
Like CD44, Lyve-1 is an integral membrane glycoprotein and a member of the HA-binding Link protein superfamily (13). Each representative of this group contains a single copy of the consensus Link module (also known as the proteoglycan tandem repeat (16, 17)), a conserved domain resembling the C-type lectin fold composed of two antiparallel β sheets made from a total of six β strands (β1–6) and two α helices stabilized by a pair of conserved disulfide bridges (18, 19). In CD44 the Link fold is known to be extended by four additional β strands (β0, β7–9) contributed by flanking N- and C-terminal sequences and a third disulfide linkage that stabilizes a unique extended Link domain (20). As deduced from x-ray crystallography and NMR spectroscopy, this extension appears to mediate a conformational change in CD44 necessary for transition between the non-binding “off” state in normal resting leukocytes and the HA-binding “on” state in antigen or inflammatory cytokine activated leukocytes (21, 22). Curiously, some of these features are also anticipated within the Lyve-1 hyaluronan-binding domain (HABD), which shares 51% similarity with that of CD44, including the presence of a third conserved disulfide linkage (15, 23). Thus far, however, the failure to generate a high resolution Lyve-1 crystal structure has hindered further confirmation of predicted similarities, and no detailed structure function analyses have yet been reported for this protein.
The Lyve-1 molecule was originally shown to bind and internalize high molecular weight HA in studies of the recombinant receptor expressed in HEK293T cells (14). Like CD44, the receptor showed specific saturable binding to HA that could be blocked selectively by appropriate Lyve-1 mAbs and that was not diminished by competition with other glycosaminoglycans such as chondroitin or heparan sulfates (13, 14). More recent studies of native endogenous Lyve-1 in primary lymphatic endothelial cells and tissue lymphatics indicate that the receptor is constitutively inactivated in vivo through terminal sialylation (24), ostensibly the same mechanism that can maintain CD44 in its low affinity state in non-activated leukocytes. However, unlike CD44, which can be activated by inflammatory agents such as tumor necrosis factor-α and lipopolysaccharide in cells expressing the appropriate sialidase activity (25–27), the factors required for unmasking Lyve-1 remain elusive. These various observations suggest that Lyve-1, although evidently similar to CD44, may have quite distinct properties, possibly explaining why two such related receptors might have evolved such markedly segregated patterns of expression.
Here we have carried out the first detailed structure-function analysis of the Lyve-1 HABD. We provide confirmation that the domain shares significant structural similarity with that of CD44 insofar as it requires contributions from critical N- and C-terminal flanking regions and a third conserved disulfide linkage for function. Furthermore, the predicted HA-binding patch is more compact than that of CD44 despite sharing a similar location and arrangement of critical binding residues. However, unlike CD44, in which HA binding is dominated by mixed hydrophobic and hydrogen bonding character, the Lyve-1 binding patch appears to be dominated by charged residues that support mainly electrostatic interactions. Moreover, in contrast to CD44, where soluble monomeric forms of the extended Link module bind HA with appreciable affinities (Kd = 10–65 μm) (28), the equivalent unit in Lyve-1 is shown to be inactive unless either dimerized as an Fc fusion protein or expressed as a full-length Lyve-1 ectodomain monomer (Kd = 35.6 μm). Finally, we present evidence from physical studies indicating that the full-length functional receptor in transfected 293T cells is almost entirely monomeric. These characteristics indicate unexpected differences in HA binding properties between Lyve-1 and CD44 that are likely to have important consequences for biological function.
EXPERIMENTAL PROCEDURES
Lyve-1 Antibodies
The mouse mAbs 3A and 8C were generated against the human Lyve-1 (hLyve-1 soluble ectodomain Fc fusion protein Δ232) as described previously (24, 29). Polyclonal antisera recognizing the human Lyve-1 ectodomain were prepared (13) and affinity-purified by chromatography on hLyve-1Δ232Fc-Sepharose.
HA Preparations
High molecular weight hyaluronan derived from rooster comb (Sigma) was used either without modification or after conjugation with fluorescein or biotin using methods described previously (21, 22). Size-fractionated hyaluronan (300 kDa) modified by addition of a single biotin at the reducing terminus (end-labeled hyaluronan) was kindly provided by Prof. Paul DeAngelis (Hyalose, LLC). Purified hyaluronan 4- to 22-mer oligosaccharides (21, 22) were obtained from Prof. Tony Day (University of Manchester, UK).
Generation of Full-length Lyve-1 Site-directed Mutants
Individual mutations were introduced into the hyaluronan-binding domain of Lyve-1 by means of the QuikChangeTM (Stratagene) system using full-length human Lyve-1 cDNA as template in the eukaryotic expression vector pRcCMV (13). A total of 44 mutants S31A, I32V, Q33A, R37A, L43A, K46A, K46Q, K46R, K47A, K47Q, N49A, N49D, Q50A, Q50E, Q51A, Q51E, N53A, R62A, K71A, K71Q, F82L, F82Y, S86R, Y87F, Y87S, I97T, R99A, N103D, K105Q, K108A, N109A, L114R, L114T, W116A, W116Y, K117A, R122A, R122Q, Q123A, Q123R, F124L, F124Y, N130A, and N130D were prepared. Individual constructs were transiently transfected into human embryonic kidney 293T fibroblasts using calcium phosphate. All mutants were assessed for both surface expression and authentic folding by immunocytochemical staining with polyclonal Lyve-1 Ig and with the conformation sensitive Lyve-1 mAbs 3A and 8C followed by quantitative flow cytometry as described below.
Generation of Soluble Dimeric Lyve-1 and CD44 Ectodomain Truncation Mutants
The hLyve-1Δ232Fc fusion protein containing all but six residues of the extracellular domain and hCD44Δ267 containing all but one residue of the extracellular domain have been described previously (13, 21). Three additional truncation mutants hLyve-1Δ130, hLyve-1Δ141, and hLyve-1Δ172 (numbered as in Ref. 13) were also prepared, which lacked residues 131–238, 142–238, and 173–238 of the extracellular domain, respectively, from the region predicted to form a C-terminal extension to the consensus Link module. Mutants were generated by appropriate PCR amplification (see below) using full-length Lyve-1 cDNA as template in pRcCMV and cloned into unique HindIII/BamHI site in the expression vector pCDM7Ig for fusion with the hinge, CH2, and CH3 regions of human IgG1 (termed Lyve-1Fcs). PCR reactions used the common forward primer hLyve-1 1F Hind (5′-3′) CGCGAAGCTTGGGTAGGCACGATG GCCAG and the reverse primers (5′-3′) hLyve-1Δ130Fc CGCGGATCCTTGTAACAATAGGCTGCAAACTG, hLyve-1Δ141Fc CGCGGATCCGGAATGCACAGTTAGTCCAA, and hLyve-1Δ172Fc CGCGGATCCGCCACCGAGTAGGTACTGTCA. Restriction sites are underlined, and a schematic of the mutants is shown in Fig. 1B. Fusion proteins were generated by transfection of HEK 293T fibroblasts with appropriate constructs using calcium phosphate and purified by affinity chromatography on protein A-Sepharose as described previously (13).
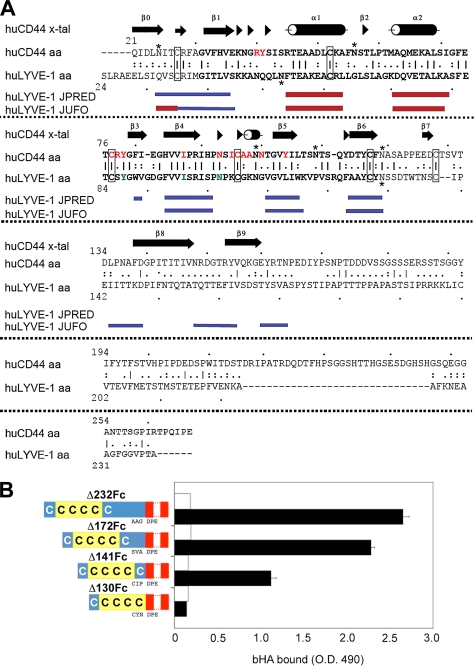
FIGURE 1.
Delineation of an extended HA-binding Link domain in Lyve-1 by truncation mutagenesis. A, primary sequence and secondary structural elements of hCD44 (21) aligned above those of hLyve-1 as predicted using the programs JPRED (47) and JUFO (48); regions corresponding to β strands are colored blue, and α helices are colored red. Conserved cysteines involved in disulfide bonds are boxed. Lines between residues in the amino acid sequences indicate identity. Paired or single dots indicate conservative or semi-conservative substitutions, respectively. Red and green lettering indicates residues critical for HA binding in CD44 and conserved equivalents in Lyve-1. B, effect of graded truncations within the Lyve-1 membrane proximal region on HA binding. Soluble ectodomain Fc fusion proteins representing the full-length extracellular domain (hLyve-1Δ232Fc), and successively greater truncations of the membrane proximal region (hLyve-1Δ172Fc, Δ141Fc, and Δ130Fc) were assayed for capacity to bind biotinylated HA (black bars) either alone or in the presence of the Lyve-1 mAb 3A included as a control for specificity (white bars). In the diagrams to the left of the figure the Lyve-1 extracellular domain is shaded blue and the consensus Link module containing the four conserved cysteine residues is yellow. The hIg hinge region and Fc domain are depicted in red. The amino acid sequences at the Lyve-1/hinge region junctions for each construct are indicated. Data shown are from an experiment that was repeated three times.
Preparation of Soluble Monomeric Lyve-1 and CD44 Ectodomains
Human Lyve-1Δ232 and the murine Lyve-1 constructs mLyveΔ165 and mCD44Δ172 were constructed as ectodomain Fc fusion proteins in the vector pCDM7IgTEV containing an optimized cleavage site (ENLYFQG) at the N terminus of the Fc region for cleavage by tobacco etch virus (TEV) protease (30) (Invitrogen). After transfection of 293T cells, Fc fusion protein dimers were buffer-exchanged into 50 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, 1 mm dithiothreitol and digested with one unit of enzyme per 3 μg of protein for 30 min at 20 °C. Cleaved Fc fragments and undigested Fc fusion protein were removed by adsorption to protein A-Sepharose (Sigma), and histidine-tagged TEV protease was removed by adsorption to nickel-nitrilotriacetic acid-agarose (Invitrogen). The integrity of cleaved monomers was assessed by SDS-PAGE and enzyme-linked immunosorbent assay, using Lyve-1- or CD44-specific mAbs as appropriate.
Three-dimensional Structure Modeling
A model of human Lyve-1 was constructed using Modeler 9v6. The coordinates of human CD44 (PDB code 1UUH) were used as a template. A CLUSTAL alignment of the sequence of human Lyve-1 to human CD44 was optimized manually by introduction of gaps between hLyve-1 residues 146/147 and 166/167. The resulting alignment and the template structure were provided to the automodel algorithms of Modeler. The resulting model had a GA341 score of 0.9911. This score assumes a value from 0 to 1, with a value of 1.0 indicating a native-like fold.
Immunocytochemical Staining and Flow Cytometry
Transfectants were detached in ice-cold phosphate-buffered saline, pH 7.5, containing 5 mm EDTA, followed by gentle pipetting and stained with the human Lyve-1-specific mAbs 8C or 3A (5 μg/ml) or with affinity-purified Lyve-1 polyclonal Ig prior to washing in FACS buffer (5% v/v fetal calf serum, 0.1% w/v sodium azide in phosphate-buffered saline, pH 7.5) and incubation with phycoerythrin-conjugated anti-mouse IgG (Dako, UK) or phycoerythrin-conjugated goat anti-rabbit IgG as appropriate (from Sigma). Cells were then washed (×3) in FACS buffer and resuspended in FACS fix (2% v/v formalin, 0.02% w/v sodium azide in phosphate-buffered saline, pH 7.5) and analyzed on a BD Biosciences FACSCaliburTM flow cytometer with CellQuestTM software.
Hyaluronan Binding Assays
Binding of hyaluronan to Lyve-1 site-directed mutants in HEK 293T cells and to soluble Lyve-1 ectodomain truncation mutants was assessed using either a flow cytometric or a plate binding assay, respectively.
Flow Cytometric Binding Assay
Cells were double stained with fluorescein-labeled HA (Fl-HA) and the Lyve-1-specific (non HA-blocking) mAb 8C followed by phycoerythrin-conjugated anti-mouse IgG-specific antibody (24). Fluorescence was quantitated using a FACSCaliburTM flow cytometer, and data were analyzed with CellQuestTM software. To determine HA binding as a function of Lyve-1 surface density, the phycoerythrin channel was divided into six equal gates for estimation of Fl-HA mean fluorescence intensity during flow cytometric analysis essentially as described in a previous study (24). The HA binding capacity of mutants was scored by comparison with that of wild-type Lyve-1 and categorized as being either 75–100% (+++), 50–75% (++), 25–50% (+), or 0–25% (−) at the highest level of expression. Lyve-1 mutants that demonstrated levels of HA binding that were 50% or less of the wild-type value were considered significantly perturbed.
Plate Binding Assay
Truncation mutants were coated on Nunc MaxiSorp plates at 2.5 pmol/well. After blocking, biotinylated HA (5 μg/ml) was added either alone or in combination with affinity-purified human Lyve-1 polyclonal Ig (final concentration, 50 μg/ml) and incubated for 1 h at 20 °C. Detection of bound HA was carried out as described previously (21, 24). Coating efficiency was determined by a parallel enzyme-linked immunosorbent assay in which MaxiSorp plates were coated with either hLyve-1Δ232Fc or appropriate truncation mutants, and the amount bound was determined using the Lyve-1-specific mAb 8C (5 μg/ml) followed by a horseradish peroxidase-conjugated anti-mouse IgG antibody (1/2000 v/v, Pierce). Plates were developed by adding orthophenylenediamine substrate before quenching with one volume of 2.5 m H2SO4. Absorbance values were read at 490 nm. Amounts coated on the plate were found to be equivalent for the three truncations and the hLyve-1Δ232Fc fusion protein (data not shown).
Competition with HA Oligosaccharides
hLyve-1Δ232Fc was coated on Nunc MaxiSorp plates (2.5 pmol/well). After blocking, biotinylated HA (5 μg/ml) was added either alone or in combination with HA oligosaccharides (4- to 22-mer) or high molecular weight HA (calculated as 22-mer equivalents) over a range of concentrations from 0 to 1000 μg/ml (0–0.66 mm) and incubated for 1 h at 20 °C. Detection of bound HA was achieved by addition of horseradish peroxidase-conjugated streptavidin (Dako, 1/500 v/v). Plates were developed by adding orthophenylenediamine substrate before quenching with one volume of 2.5 m H2SO4. Absorbance values at 490 nm were measured in a spectrophotometer.
Effect of Ionic Strength on HA Binding
hLyve-1Δ232Fc and hCD44Fc were coated on Nunc MaxiSorp plates at a concentration of 2.5 pmol/well. After blocking, 250 ng of Fl-HA was added to each well in 50 mm phosphate buffer, pH 7.5, containing NaCl at concentrations ranging from 50 to 1600 mm, and plates were incubated at room temperature for 1 h prior to washing with 50 mm phosphate buffer, pH 7.5, containing the appropriate NaCl concentration (×3). Bound Fl-HA was detected using an automated dual probe fluorescence plate reader (Biotek Synergy HT) with an excitation wavelength of 488 nm. The means of triplicate determinations for binding at each ionic strength value were expressed as a percentage of the maximum value.
For experiments with intact cells, HEK 293T stable transfectants were suspended in 20 mm phosphate buffer, pH 7.3, 5 mm glucose, 5 mm KCl supplemented with NaCl over the range 0–250 mm with constant osmolarity (500 mosm) maintained by appropriate addition of sucrose. Cell suspensions were then mixed with Fl-HA (5 μg/ml final) in the presence or absence of Lyve-1-blocking mAb 3A (50 μg/ml) and incubated at 4 °C for 30 min followed by washing and fixation prior to analysis by flow cytometry.
Determination of HA Binding Affinity by Surface Plasmon Resonance
Experiments were performed on a Biacore 3000 in HEPES-EP running buffer at a constant flow rate of 10 μl/min. Prior to analysis, dimeric and monomeric Lyve-1Fc and CD44Fc fusion proteins were subjected to gel filtration using a HiLoad 26/60 Superdex 200 chromatography column (Amersham Biosciences) to remove aggregates. End-biotinylated 300-kDa HA (Hyalose) was coupled to streptavidin-coated Biacore (SA) chips. The binding analyses for dimeric hLyve-1Δ232Fc and dimeric hCD44Δ267Fc were performed using maximum protein concentrations of 50 μm and 349 μm, respectively. The maximum protein concentration was 195 μm for hLyve-1Δ232 monomer and 1 mm for mLyve-1Δ165 monomer. Controls for nonspecific binding were performed in each case by subtracting responses from a parallel flow cell coated with biotinylated chondroitin 4-sulfate. Values for Kd were obtained by non-linear curve fitting of the data to a Langmuir isotherm.
BRET Analysis
For quantitative analysis of Lyve-1 self-association at the cell surface, full-length hLYVE-1 and hCD44 (standard form) cDNAs were ligated into the bioluminescence resonance energy transfer (BRET) vectors prLuc-N3 and pGFP-N3, fusing either luciferase (Luc) or GFP with the end of the cytoplasmic tail. Additional Luc and GFP fusion constructs for CD86 (constitutive monomer) and CTLA-4 (constitutive dimer) were donated by Prof. Simon Davis (University of Oxford). Constructs were introduced into HEK 293T cells by transfection with FuGENE (Roche Applied Science). In each case, Luc:GFP ratios for the transfectants ranged from 2:1 to 1:39, whereas the total amount of transfected plasmid was constant (31). Cells were lifted 24 h post transfection and collected by centrifugation at 600 × g for 3 min before re-suspension (∼1.5 × 106/ml) in minimal essential medium. For measurement of BRET, the luciferase substrate DeepBlueC (PerkinElmer) was added (10 μm final concentration) to a 100-μl sample of cells and light emission in the 370–450 nm (BRET-A) and 500–530 nm (BRET-B) spectral regions was collected over 1 s on a Fusion microplate analyzer (PerkinElmer). To determine actual GFP and Luc expression levels for each transfection ratio, separate 100-μl samples of cells were excited at 425 nm, and the emission was collected at 500–530 nm (fluorescence units) before addition of coelenterazine-h (10 μm final concentration) and acquisition of the total light emission. BRETeff was calculated as BRET-B/BRET-A after background subtraction (untransfected cells) and correction for luciferase expression alone (typically 7% of LU-A). The values from the fluorescence and luminescence units measurements were converted to concentrations of GFP, and Luc and resulting ([GFP]/[Luc]) ratios were plotted against BRETeff as described previously (31). Data points were tested for optimal fit to either a hyperbolic or a linear relationship (for [GFP]/[Luc] > 2.0) by the method of least squares analysis (see also supplemental Fig. S1) to determine behavior of receptors as dimers or monomers according to previously described criteria (31).
RESULTS
Evidence That Lyve-1 Contains an Extended HA-binding Domain
Previously we demonstrated that an Ig Fc fusion protein comprising residues 1–232 of the hLyve-1 extracellular domain displayed similar HA binding properties to the intact membrane-anchored molecule when assayed in vitro (13, 14). To date, however, no further delineation of the HA-binding unit within this domain has been reported. As illustrated by the alignment in Fig. 1A, the primary sequence of Lyve-1 (residues 54–130) encodes a consensus Link module similar to that of CD44 comprising four key conserved cysteine residues (Cys-61, Cys-85, Cys-106, and Cys-128) together with the six β strands (β1–β6) and two α helices that adopt the characteristic lectin fold. As previously reported (13), this unit is bracketed by an additional conserved cysteine pair (Cys-36/Cys-139) equivalent to the Cys-28/Cys-129 pair that makes a critical third disulfide bridge in CD44 linking the N- and C-terminal extensions (beta strands β0, β7, β8, and β9) that are essential for correct folding and HA binding in that receptor. To assess whether similar structural elements might be encoded within the Lyve-1 HABD we analyzed the sequence flanking the Link module using the secondary structure prediction programs JPRED and JUFO (Fig. 1A). Despite low sequence homology between Lyve-1 and CD44 in these regions (particularly the C-terminal flanking region) the JUFO program identified potential elements of beta structure in areas corresponding to the β0/β1 junction (residues 36–45) the β7/β8 region (residues 143–148) and the β8/β9 region (residues 157–164/168–172). To determine whether these might contribute to bona fide structural extensions of the Lyve-1 HA-binding domain we prepared a series of soluble ectodomain truncation mutants hLyve-1Δ172Fc, hLyve-1Δ141Fc, and hLyve-1Δ130Fc, designed to delete appropriate segments of the C-terminal region, including the critical cysteine, Cys-139. Comparison of the HA binding properties of these mutants using a plate binding assay with bHA revealed a graded reduction in HA binding with increasing truncation for hLyve-1Δ172Fc and hLyve-1Δ141Fc (Fig. 1B). However, the most dramatic loss of HA binding was seen upon deletion of Cys-139 in hLyve-1Δ130Fc. These results are consistent with the presence of additional elements of the Lyve-1 HABD, C-terminal to the consensus Link module, including a third essential disulfide, and constitute important evidence for an extended binding domain analogous to that of CD44.
Identification of Putative HA Contact Residues by Site-directed Mutagenesis
The recent crystallographic analysis of a murine CD44 HA8 complex identified thirteen contact residues located within the loop regions between the β1–α1, α2–β3, and β4–β5 strands of the consensus Link module that together constitute a shallow HA-binding groove on one face of the CD44 HABD (22). Inspection of the Lyve-1 amino acid sequence within the equivalent regions indicates that only three of these residues (excluding conserved cysteines), specifically Tyr-87, Ile-97, and Asn-103 (equivalent to hCD44 residues Tyr-79, Ile-88, and Asn-94) are fully conserved in Lyve-1. Interestingly, some of the key contact residues of CD44 such as Arg-41, Tyr-42, and Arg-78 and the aliphatic side-chain amino acids Ile-96 and Ala-98 in the conserved β4/β5 hook-like region have no obvious counterparts in Lyve-1. To assess whether corresponding non-conserved residues might form an HA-binding site in Lyve-1 we mutated 29 individual amino acids distributed mostly within the β1/α1 α2/β3, β4/β5, and β5/β6 loop regions deduced from the sequence alignment in Fig. 1 and a Lyve-1 structural model based on crystal coordinates of human CD44 (corresponding to the so-called type A structure (22); see below). In all, some 43 individual site-directed mutants carrying non-conservative or semi-conservative substitutions were generated, and each was assayed for its capacity to bind Fl-HA after expression on the surface of transfected 293T cells, with assessment for correct folding by the conformation-sensitive 8C mAb. The results as summarized in Table 1 (see supplemental Fig. S1 for quantitative data) show that non-conservative mutation of eight residues in particular (R37A, Y87S/Y87F, I97T, R99A, N103D, K105D/K105Q, K108A, and K117A) led to significant perturbation of HA binding, although one mutant (K117A), which failed to bind the 8C mAb, was discounted on the basis of incorrect folding. Among these residues, Tyr-87, Ile-97, and Asn-103 show exact conservation with the HA-binding counterparts Tyr-79, Ile-88, and Asn-94 in hCD44, whereas Lys-105 and Lys-108 occupy the same positions as HA-binding residues Ile-96 and Ala-99 in hCD44. These features are discussed further in the following section.
TABLE 1.
Functional characterization of the panel of Lyve-1 site-directed mutants
| Mutation | Cell surface expressiona | Fl-HA bindingb | Reactivity with mAb 8Cc | Reactivity with mAb 3Ac |
|---|---|---|---|---|
| S31A | +++ | +++ | ++ | +++ |
| I32V | +++ | +++ | ++ | +++ |
| Q33A | +++ | +++ | ++ | +++ |
| R37A | +++ | + | +++ | +++ |
| L43A | +++ | ++ | +++ | ++ |
| K46A | +++ | +++ | +++ | +++ |
| K46Q | +++ | ++ | ++ | +++ |
| K46R | +++ | +++ | ++ | +++ |
| K47A | +++ | +++ | − | +++ |
| K47Q | +++ | +++ | − | +++ |
| N49A | ++ | +++ | ++ | +++ |
| N49D | ++ | ++ | ++ | +++ |
| Q50A | ++ | ++ | ++ | +++ |
| Q50E | ++ | ++ | ++ | ++ |
| Q51A | ++ | ++ | ++ | ++ |
| Q51E | +++ | ++ | ++ | +++ |
| N53A | +++ | ++ | ++ | − |
| R62A | ++ | ++ | ++ | +++ |
| K71A | +++ | ++ | ++ | +++ |
| K71Q | +++ | ++ | ++ | +++ |
| F82L | ++ | ++ | ++ | +++ |
| F82Y | +++ | ++ | ++ | +++ |
| S86R | +++ | ++ | + | ++ |
| Y87F | +++ | +++ | + | ++ |
| Y87S | ++ | − | +++ | − |
| I97T | +++ | + | +++ | +++ |
| R99A | ++ | − | ++ | − |
| N103D | +++ | + | +++ | +++ |
| K105Q | +++ | − | +++ | +++ |
| K108A | ++ | + | +++ | +++ |
| N109A | +++ | ++ | ++ | +++ |
| L114R | ++ | +++ | + | + |
| L114T | ++ | ++ | ++ | +++ |
| W116Y | +++ | ++ | +++ | − |
| K117A | + | − | − | − |
| R122A | +++ | +++ | ++ | − |
| R122Q | +++ | +++ | +++ | − |
| Q123A | +++ | +++ | ++ | +++ |
| Q123R | +++ | +++ | ++ | ++ |
| F124L | +++ | ++ | +++ | ++ |
| F124Y | +++ | +++ | ++ | +++ |
| N130A | +++ | + | +++ | +++ |
| N130D | +++ | + | +++ | +++ |
a Expression of mutated Lyve-1 on the surface of transfected 293T cells. Mean fluorescence was measured after staining samples with an affinity-purified polyclonal antibody specific for human Lyve-1 and compared with the value for wild-type transfectants, where +++ = 75–100% (or above), ++ = 50–75%, + = 25–50%, and − = 0–25% of the mean fluorescence value of the wild type.
b Fl-HA binding for mutated full-length human Lyve-1-transfected 293T cells compared to binding to wild type. Scores used are as described in footnote a.
c Reactivity of mutated Lyve-1-transfected 293T cells with the human Lyve-1-specific monoclonal antibodies 8C and 3A compared to binding to the wild type. Symbols used are as described in footnote a. All mutagenesis experiments were repeated three times. Representative quantitative data are provided in supplemental Fig. S1.
Modeling of an HA-binding Surface in Lyve-1
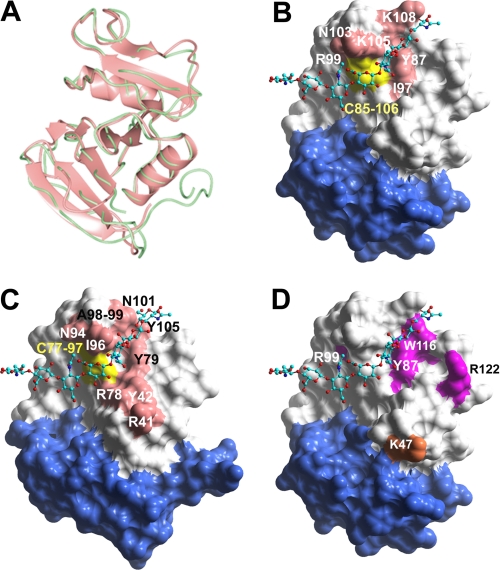
To visualize the HA-binding site in Lyve-1 we prepared a three-dimensional structural model based on the crystal coordinates of human CD44 (PDB code 1UUH). This represents the CD44 “type A” crystal structure (22), which differs subtly from the “type B” structure insofar as the critical HA contact residue Arg-45 within the β1/α1 loop is oriented away from the ligand and more likely represents the “off” or lower affinity state of the receptor. The Lyve-1 sequence can be threaded onto the CD44 structure without causing significant structural perturbations, with the exception of some minor variation in the loop region between β5 and β6 (Fig. 2A). Mapping of the seven candidate HA-contact residues onto a space-filling model of the Lyve-1 structure (Fig. 2B) reveals that all except Arg-37 come together to form a compact binding patch that encompasses the same characteristic hook-like region (centered on Lys-105) that is a prominent feature of the ligand-binding groove in CD44 (Fig. 2C) as identified recently by x-ray crystallography and NMR analysis of the mCD44/HA8 complex (22). The disruption of HA binding in the R37A mutant is not readily explained, but may signify a long range effect as reflected by the subtle change in mAb 8C reactivity (Table 1). The near identical location of the ligand interaction surface in the two receptors is highlighted by docking a model of an HA8-mer in its CD44-bound conformation into the putative binding groove of Lyve-1 (Fig. 2, B and C).
FIGURE 2.
Structure-based modeling of the extended Link domain and HA-binding surface in Lyve-1. A, overlay of Cα traces of the HABDs from human CD44 (green) and human Lyve-1 (pink) modeled using coordinates of the mCD44 high resolution crystal structure (structure B (22)). B, space-filled representation of the Lyve-1 HABD model with key residues comprising the predicted ligand-binding surface defined by site-directed mutagenesis (see Table 1) colored pink. The position of the Cys-85 to Cys-106 structural disulfide that may also contribute to HA binding is colored yellow. An HA octasaccharide docked onto the putative binding site is shown with individual atoms colored turquoise, red, or blue (backbone carbon, oxygen, and nitrogen, respectively). The region corresponding to the Link module extension in CD44 (supplementary lobe) is colored blue. C, space-filled representation of the HABD from human CD44 with contact residues colored pink and a bound HA oligosaccharide in green. D, space-filled representation of the Lyve-1 HABD model with residues identified as part of the epitopes for the Lyve-1-specific mAbs 3A and 8C (see Table 1) colored magenta and orange, respectively.
To further confirm the authenticity of the HA-binding patch, we probed each of the 43 site-directed mutants for binding to the Lyve-1 mAb 3A (supplemental Fig. S1), a reagent that was recently identified as a potent inhibitor of HA binding to both soluble and membrane-attached Lyve-1 (29). As summarized by the data in Table 1, non-conservative mutation of either Tyr-87, Arg-99, Trp-116, or Arg-122 drastically reduced mAb 3A binding but did not disrupt binding to mAb 8C. Plotting of these residues onto the model of Lyve-1 shows they lie within or in close proximity to the predicted HA-binding patch, thus providing further confirmation of its authenticity (Fig. 2D).
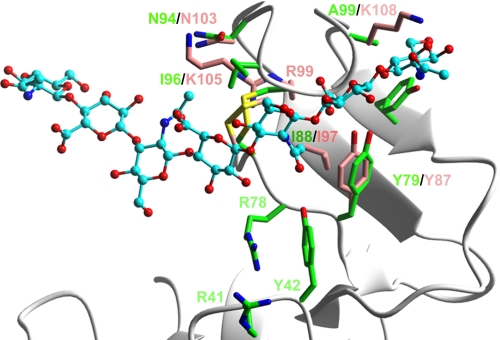
A more detailed view of the interaction site also emphasizes the highly conserved spatial arrangement of key HA-binding residues within this Lyve-1 groove, exemplified particularly by Tyr-87, Ile-97, and the Cys-85 to Cys-106 disulfide whose counterparts in CD44 constitute a hydrophobic “pocket” for the methyl group of GlcNAc (Fig. 3; see also Ref. 22). This suggests that the bound sugar lies in a similar orientation in both receptors. Interestingly, the total number of potential binding residues identified in Lyve-1 (seven) is considerably smaller than the 13 or more identified in CD44 by either site-directed mutagenesis or crystallography and the binding site is significantly more compact (see Fig. 2). Moreover, the character of the putative binding residues in Lyve-1 appears to differ from those of CD44. Most notably, of the six semi-contiguous HA-binding residues identified in Lyve-1, four have charged or polar side chains (Arg-99, Asn-103, Lys-105, and Lys-108), whereas those of CD44 are predominantly aliphatic (Ile-88, Ile-96, Cys-97, Ala-98, and Ala-99) or aromatic (Tyr-42, Tyr-79, and Tyr-105). Clearly these differences predict major dissimilarities in HA binding properties between Lyve-1 and CD44.
FIGURE 3.
Comparison of HA-binding surfaces in Lyve-1 and CD44. Ribbon diagram comparing identities and positions of key HA contact residues in the HABDs of Lyve-1 (carbon atoms colored pink) and CD44 (carbon atoms colored green), generated by overlay of the core region of the CD44 fold type A structure (22) and its equivalent in the Lyve-1 model from Fig. 2 (background ribbon diagram in gray). Sulfur atoms of cysteine residues within conserved C4–C5 (linking cysteines 85–106 in Lyve-1) disulfides are colored yellow. The docked HA8 is represented in ball and stick format with carbon atoms shaded turquoise. Oxygen and nitrogen atoms are colored red or blue, respectively.
Distinctive Features of the Lyve-1 HA-binding Interaction
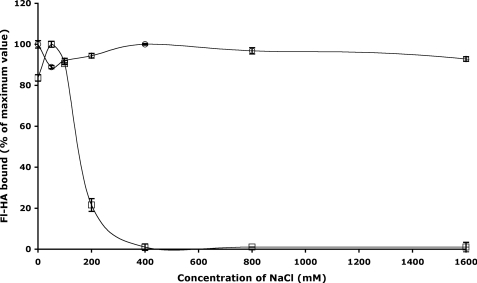
To assess whether the chemical nature of the HA-protein interaction in Lyve-1 was different to that of CD44, we determined the effect of ionic strength using the bHA plate binding assay. As shown in Fig. 4, binding of bHA to CD44Δ267Fc was largely independent of salt concentration up to and beyond 2 m NaCl. In contrast, however, binding to Lyve-1Fc was fully disrupted at NaCl concentrations as low as 250 mm. Importantly, this sensitivity was not due to an effect of ionic strength on the Fc partner, as similar findings were obtained in analyses with Lyve-1Δ232 monomers from which this Ig domain had been removed (data not shown). These results are consistent with a binding interaction that is mediated predominantly by charge interactions, in contrast to CD44, which binds HA primarily through H-bonds and hydrophobic interactions (22).
FIGURE 4.
Effect of ionic strength on HA binding to soluble Lyve-1 and CD44 ectodomain fusion proteins. Binding of Fl-HA to soluble hLyve-1Δ232Fc (open squares) or hCD44Δ267Fc (open circles) immobilized on microtiter plates was determined as a function of NaCl concentration (0–1600 mm) as described under “Experimental Procedures.” Values are the mean ± S.E. (n = 3) expressed as a percentage of maximum binding. Data shown are from an experiment that was repeated twice.
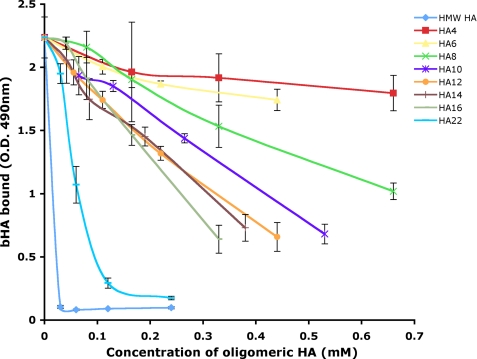
To further characterize the Lyve-1 HA interaction and the unit length of sugar polymer required for optimal binding, we measured the competition for high molecular weight bHA binding by unlabeled HA oligosaccharides of defined length using immobilized Lyve-1 ectodomain Fc fusion protein (Lyve-1Δ232). The results (Fig. 5) show the minimal effective saccharide unit was an 8-mer (IC50 0.55 mm). Furthermore, there was a sharp increase in potency as oligosaccharide length was increased from HA16 (IC50 0.25 mm) to HA22 (IC50 0.05 mm) that may indicate cooperativity. Again these properties of Lyve-1 are distinct from CD44 in which the minimal sugar unit is a 6-mer (21), an unexpected finding in light of the foregoing evidence that the area of the binding patch in Lyve-1 is smaller than that of CD44.
FIGURE 5.
Competition for Lyve-1 binding to high molecular weight HA by oligosaccharides of defined length. Binding of bHA to hLyve-1Δ232 Fc immobilized on a microtiter plate was determined in the presence of varying concentrations of HA 4- to 22-mer oligosaccharides or high molecular weight (HMW) HA, as described under “Experimental Procedures.” Values are the mean ± S.E. of triplicate determinations. Data shown are from an experiment that was repeated twice.
Analysis of Lyve-1 HA Binding Affinity by Surface Plasmon Resonance
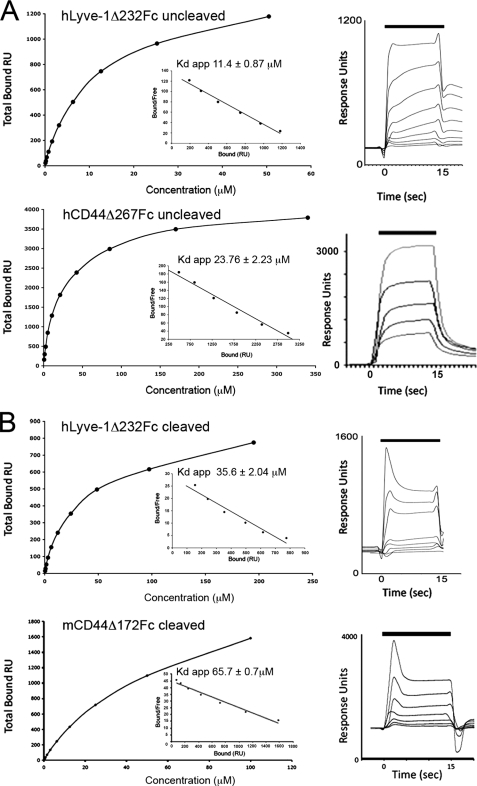
The relatively compact dimensions of the HA-binding surface in Lyve-1 and the marked sensitivity of the binding interaction to ionic strength are both suggestive of a low affinity receptor. To investigate this possibility more directly we estimated the apparent HA binding affinities of Lyve-1 and CD44 ectodomain Fc fusion proteins by means of surface plasmon resonance, using high molecular weight HA immobilized on the sensor chip and Lyve-1/CD44 as the analyte. Equilibrium binding analyses (Fig. 6A) revealed that both receptors have broadly similar apparent affinity for HA (Kdapp hLyve-1Δ232Fc, 11.4 μm; Kdapp hCD44Δ267Fc, 24 μm). Because Fc fusion proteins constitute covalent dimers linked by an interchain disulfide within the Ig hinge region, the measured binding affinities do not represent true monomeric binding constants. To generate appropriate monomers, we first transferred the hLyve-1Δ232Fc ectodomain fusion construct into the vector pCDM7IgTEV, which contains an engineered TEV protease cleavage motif within the Fc hinge region. As shown in Fig. 6 (A and B), the HA binding affinity was reduced to only a small extent (Kd hLyve-1Δ232 monomer, 35.6 μm; Kdapp hLyve-1Δ232 dimer, 11.4 μm). Likewise, a CD44 monomer comprising the extended Link module (mCD44Δ172) showed only a modest reduction in binding affinity compared with the dimeric receptor (Kd mCD44Δ172 monomer, 65.7 μm; Kdapp hCD44Δ267 dimer, 23.8 μm), consistent with the known capacity of this extended Link domain to form an independent folding unit. Curiously, however, an equivalent monomer comprising the extended Link module of Lyve-1 (mLyve-1Δ165Fc) exhibited little or no HA binding in the Biacore (data not shown). Possible explanations for this loss of binding include aggregation or partial unfolding of the monomerized ectodomains. Indeed, we have observed that similar hLyve-1Fc fusion constructs (e.g. Lyve-1Δ165) yielded poor recoveries on size-exclusion chromatography following monomerization. Nevertheless, the particular mLyve-1Δ165Fc monomers analyzed in this study eluted normally, retained normal reactivity with the conformation-sensitive mAbs C1/8 and B1/10 (29) in enzyme-linked immunosorbent assay, and yielded one-dimensional NMR spectra consistent with a folded structure (data not shown). Hence, we conclude that the HA-binding domain of Lyve-1 differs from that of CD44 in requiring additional elements from the C-terminal membrane proximal ectodomain for functional stability.
FIGURE 6.
Equilibrium binding analysis of Lyve-1 and CD44 to immobilized HA using surface plasmon resonance. A, plots of the maximum HA binding responses with varying concentrations of dimeric hLyve-1Δ232 ectodomain Fc fusion protein (upper) and dimeric hCD44Δ267 ectodomain Fc fusion protein (lower) with inset Scatchard plots. Overlays of the sensorgrams obtained over a range of fusion protein (analyte) concentrations for each dimeric receptor ectodomain fusion protein are shown to the left. Lyve-1Δ232Fc shows incomplete dissociation. B, binding data for monomeric hLyve-1Δ232 (upper) and monomeric mCD44Δ172 (lower) generated by controlled cleavage of dimers with TEV proteinase. Each experiment was repeated a minimum of two times.
Does Native Lyve-1 Require Self-association for HA Binding?
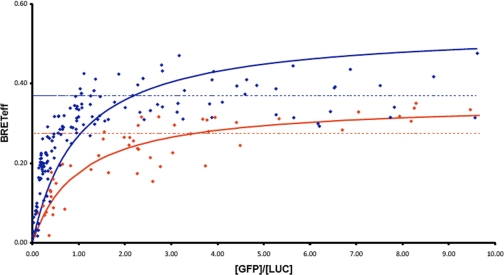
To explore the possibility of Lyve-1 self-association, we assayed full-length Lyve-1-transfected 293T cells for BRET, a sensitive technique that measures the proximity-dependent transfer of fluorescence between individual luciferase-tagged and GFP-tagged receptor pairs at the cell surface. For the purpose of comparison we carried out a parallel assay for self-association of CD44, a receptor that has been reported to undergo clustering at the cell surface (32, 33). As shown in Fig. 7, plots of BRETeff (GFP emission/Luciferase emission) for increasing ratios of Lyve-1 GFP/Lyve-1 luciferase and CD44 GFP/CD44 luciferase yielded broadly similar curves with the expected approach to a plateau at saturating levels of luciferase donor. Importantly, however, a root mean square analysis of residuals (supplemental Fig. S2) indicated a better fit of the Lyve-1 data to a linear rather than a hyperbolic relationship, implying no significant dimer formation. Moreover, BRETeff was not altered by addition of high molecular weight HA (not shown). In contrast, the CD44 data showed a closer approximation to a hyperbolic relationship, indicating the possibility of dimerization, even in the absence of ligand. Overall these data suggest that Lyve-1 undergoes neither constitutive nor ligand-induced dimerization in the plasma membrane.
FIGURE 7.
BRET analysis of Lyve-1 self-association at the surface of transfected 293T cells. Values for BRET efficiency (BRETeff) for HEK 293T cells transfected with appropriate full-length hLyve-1 (blue symbols) or hCD44 constructs (red symbols) were obtained over a range of fluorescent acceptor/donor ratios (GFP/Luc) while maintaining a constant level of total expression as described under “Experimental Procedures.” Lines of best fit to a hyperbolic or a linear relationship at [GFP]/[Luc] > 2.0 are depicted in solid or dotted style corresponding to formation of dimers or random interactions, respectively, according to the convention described in James et al. (31). A least-squares analysis of residual values is included in supplemental Fig. S2. Data shown are from an experiment that was repeated twice.
DISCUSSION
The analyses presented here uncover key functional differences that distinguish Lyve-1, the lymphatic endothelial HA receptor from CD44, the major HA receptor on leukocytes and mesenchymal cells. Until now these two receptors were assumed to be mutually redundant, both being members of the Link protein superfamily with single HA-binding Link modules located toward the N terminus of a highly N- and O-glycosylated extracellular domain. In terms of function, CD44 mediates leukocyte adhesion, as triggered by inflammatory mediators that exert dual control over synthesis of HA at the luminal surface of blood vessels, and the binding status of CD44, switching the receptor from an “off” state in resting leukocytes to an “on” state in activated leukocytes to promote recruitment from the circulation during inflammation (34–36). One of the mechanisms behind this switch in CD44 is the enzymatic cleavage of terminal sialic acid residues from N-glycans (25–27, 37) that block ligation of HA by steric hindrance or other effects on the binding site (21). In the case of Lyve-1, however, the physiological function is still open to conjecture, although evidence suggests the receptor could in principle mediate leukocyte-endothelial adhesion through binding HA presented by CD44 on the surface of trafficking leukocytes (13, 15, 38). In keeping with this notion, there is good evidence to suggest that Lyve-1 is also maintained in an off state in resting cells (i.e. lymphatic endothelium) through a consonant mechanism of sialylation, although unlike CD44, the identity of the factors responsible for activation are not yet known (24). Clearly a better knowledge of the structure and function of Lyve-1 is needed to distinguish its true physiological role from that of CD44.
These studies focused primarily on delineating the HA-binding domain in Lyve-1 and the character of the ligand-binding interaction. Specifically, truncation mutagenesis confirmed the critical importance for HA binding of a third disulfide linkage bracketing the consensus Link module and secondary structure elements within the immediate C-terminal region, features that are diagnostic of the extended HA-binding domain in CD44. Indeed, equivalents for all four additional beta strands of this extension, β0, β7, β8, and β9, were anticipated by secondary structure predictions and by molecular modeling of the Lyve-1 HABD using coordinates from the recently derived CD44 crystal structure. The close resemblance with CD44 was underscored by the conserved location and orientation of the putative HA-binding site, which was shown to occupy a complementary position below the hook-like β4/β5 loop region of the consensus Link fold in the Lyve-1 structural model. Notably, the binding site in Lyve-1 also contained the characteristic hydrophobic “pocket” that tethers the GlcNAc methyl group of HA in CD44 (22). Aside from these topographical features, however, the nature of the ligand-binding interaction was shown to differ markedly from that of the CD44 HABD. Specifically, the size of the HA-binding surface was considerably smaller than that of CD44. In addition, most of the residues (4/6) identified by site-directed mutagenesis were either charged or polar (Arg-99, Asn-103, Lys-105, and Lys-108), and the interaction was completely disrupted by conditions of ionic strength >200 mm. This stands in marked contrast to CD44 in which the interaction with HA is dominated by shape and hydrogen bonding complementarity and is mediated by hydrogen bonds and van der Waals forces that are unaffected by ionic strength (22). Interestingly, the mode of HA binding to Lyve-1 ectodomains resembles that of the selectins, which bind the prototype fucosylated tetrasaccharide sialyl Lewis-X through recognition of the negatively charged carboxylate group of NeuNAc by primarily lysine and arginine residues (39). Indeed the sensitivity of the Lyve-1 HA interaction to ionic strength (I50 150 mm) is almost identical to that of L-, E-, and P-selectins with sLeX (39, 40). However, preliminary experiments with Lyve-1 transfectants indicate this sensitivity to ionic strength is not apparent in intact cells (see supplemental Fig. S3). It may be that the HA-binding domain in the native receptor is shielded from the bulk solvent due to the nature of its local environment. Further experiments will be required to evaluate this possibility.
In addition to the smaller HA-binding surface and different character of HA binding in Lyve-1, we obtained evidence during Biacore analyses that Lyve-1 receptor function is influenced by elements even beyond the extended Link module. Whereas soluble CD44 constructs that contained only the extended Link module (mCD44Δ172) were fully functional as monomers, those containing the equivalent unit in Lyve-1 (mLyve-1Δ165) were functional only as dimers. Importantly, the mLyve-1Δ165 monomers appeared to be folded correctly as assessed by conformation-selective mAb binding. Moreover, HA binding to similar dimers has been previously documented as both specific and saturable as well as open to efficient competition by free HA in preference to other glycosaminoglycans such as chondroitin and heparan sulfates and selective obstruction by Lyve-1-blocking mAbs (13, 14, 29). As we did not observe the same functional requirement for dimerization in constructs encompassing the full-length Lyve-1 ectodomain (hLyve-1Δ232), our results therefore point to the involvement of additional regions downstream of the extended Link module in stabilizing the monomeric receptor, forming a larger regulatory unit in Lyve-1 than previously envisaged. Notably, the C-terminal membrane proximal region of the Lyve-1 ectodomain is highly O-glycosylated and carries a single unpaired cysteine residue (13, 14). Elements within this section may well interact with or otherwise stabilize the extended Link module. Experiments to define the nature of these elements and to delineate the boundaries of the stable monomer HA-binding unit using limited proteolysis are currently under way in our laboratory.
Finally, we explored the possibility that Lyve-1 might engage in receptor self-association or clustering at the cell surface, analogous to that observed for CD44 (see, for example, Refs. 32 and 33). However, physical measurement of self-association using BRET analyses showed no convincing evidence that the functional receptor on the surface of transfected 293T cells forms either constitutive or ligand-induced Lyve-1 homodimers. It remains a possibility that multivalent interactions between Lyve-1 and large HA polymers may induce longer range clustering of the receptor than that detectable by BRET (≤10 nm). It is tempting to speculate that such clustering by appropriate conformations of HA in the form of HA-protein complexes such as HA cables (Refs. 41–43; reviewed in Ref. 44) or serum IαI heavy chain-HA adducts including SHAP (serum hyaluronan-associated protein) (45, 46) might have the capacity to activate Lyve-1 just as they activate CD44 in vivo. Further studies will be required to evaluate these possibilities.
Supplementary Material
This work was supported by grants from the UK Medical Research Council (to D. G. J.) and by a Medical Research Council D.Phil. studentship (to B. R. S. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- Lyve-1
- lymphatic vessel endothelial receptor-1
- BRET
- bioluminescent resonance energy transfer
- BRETeff
- BRET efficiency
- HA
- hyaluronan
- HABD
- hyaluronan-binding domain
- Fl-HA
- fluorescein-conjugated hyaluronan
- TEV
- tobacco etch virus
- mAb
- monoclonal antibody
- FACS
- fluorescence-activated cell sorting
- Luc
- luciferase
- GFP
- green fluorescent protein
- bHA
- biotinylated hyaluronan.
REFERENCES
- 1.Swartz M. A. (2001) Adv. Drug Deliv. Rev. 50, 3–20 [DOI] [PubMed] [Google Scholar]
- 2.Oliver G., Alitalo K. (2005) Annu. Rev. Cell Dev. Biol. 21, 457–483 [DOI] [PubMed] [Google Scholar]
- 3.Zhou B., Weigel J. A., Saxena A., Weigel P. H. (2002) Mol. Biol. Cell 13, 2853–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris E. N., Weigel J. A., Weigel P. H. (2008) J. Biol. Chem. 283, 17341–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser J. R., Kimpton W. G., Laurent T. C., Cahill R. N., Vakakis N. (1988) Biochem. J. 256, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson D. G. (2003) Trends Cardiovasc. Med. 13, 1–7 [DOI] [PubMed] [Google Scholar]
- 7.von Andrian U. H., Mempel T. R. (2003) Nat. Rev. Immunol. 3, 867–878 [DOI] [PubMed] [Google Scholar]
- 8.Randolph G. J., Angeli V., Swartz M. A. (2005) Nat. Rev. Immunol. 5, 617–628 [DOI] [PubMed] [Google Scholar]
- 9.Johnson L. A., Jackson D. G. (2008) Ann. N. Y. Acad. Sci. 1131, 119–133 [DOI] [PubMed] [Google Scholar]
- 10.Clark R. A., Alon R., Springer T. A. (1996) J. Cell Biol. 134, 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pure E., Cuff C. A. (2001) Trends Mol. Med. 7, 213–221 [DOI] [PubMed] [Google Scholar]
- 12.Ponta H., Sherman L., Herrlich P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 33–45 [DOI] [PubMed] [Google Scholar]
- 13.Banerji S., Ni J., Wang S. X., Clasper S., Su J., Tammi R., Jones M., Jackson D. G. (1999) J. Cell Biol. 144, 789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prevo R., Banerji S., Ferguson D. J., Clasper S., Jackson D. G. (2001) J. Biol. Chem. 276, 19420–19430 [DOI] [PubMed] [Google Scholar]
- 15.Jackson D. G. (2004) APMIS 112, 526–538 [DOI] [PubMed] [Google Scholar]
- 16.Brissett N. C., Perkins S. J. (1996) FEBS Lett. 388, 211–216 [DOI] [PubMed] [Google Scholar]
- 17.Hardingham T. E., Fosang A. J. (1992) FASEB J. 6, 861–870 [PubMed] [Google Scholar]
- 18.Kohda D., Morton C. J., Parkar A. A., Hatanaka H., Inagaki F. M., Campbell I. D., Day A. J. (1996) Cell 86, 767–775 [DOI] [PubMed] [Google Scholar]
- 19.Day A. J., Prestwich G. D. (2002) J. Biol. Chem. 277, 4585–4588 [DOI] [PubMed] [Google Scholar]
- 20.Banerji S., Day A. J., Kahmann J. D., Jackson D. G. (1998) Protein Expr. Purif. 14, 371–381 [DOI] [PubMed] [Google Scholar]
- 21.Teriete P., Banerji S., Noble M., Blundell C. D., Wright A. J., Pickford A. R., Lowe E., Mahoney D. J., Tammi M. I., Kahmann J. D., Campbell I. D., Day A. J., Jackson D. G. (2004) Mol. Cell. 13, 483–496 [DOI] [PubMed] [Google Scholar]
- 22.Banerji S., Wright A. J., Noble M., Mahoney D. J., Campbell I. D., Day A. J., Jackson D. G. (2007) Nat. Struct. Mol. Biol. 14, 234–239 [DOI] [PubMed] [Google Scholar]
- 23.Jackson D. G. (2004) Glycoforum, http://www.glycoforum.gr.jp/science/hyaluronan/HA28/HA28E.html
- 24.Nightingale T. D., Frayne M. E., Clasper S., Banerji S., Jackson D. G. (2009) J. Biol. Chem. 284, 3935–3945 [DOI] [PubMed] [Google Scholar]
- 25.Katoh S., Zheng Z., Oritani K., Shimozato T., Kincade P. W. (1995) J. Exp. Med. 182, 419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh S., Miyagi T., Taniguchi H., Matsubara Y., Kadota J., Tominaga A., Kincade P. W., Matsukura S., Kohno S. (1999) J. Immunol. 162, 5058–5061 [PubMed] [Google Scholar]
- 27.Gee K., Kozlowski M., Kumar A. (2003) J. Biol. Chem. 278, 37275–37287 [DOI] [PubMed] [Google Scholar]
- 28.Banerji S., Noble M., Teriete P., Wright A. J., Blundell C. D., Campbell I. D., Day A. J., Jackson D. G. (eds) (2005) in Structure of the CD44 Hyaluronan-binding Domain and Insight into Its Regulation by N-Glycosylation, Vol. II, pp. 625–630, Matrix Biology Institute, Edgewater, NJ [Google Scholar]
- 29.Johnson L. A., Prevo R., Clasper S., Jackson D. G. (2007) J. Biol. Chem. 282, 33671–33680 [DOI] [PubMed] [Google Scholar]
- 30.Dougherty W. G., Carrington J. C., Cary S. M., Parks T. D. (1988) EMBO J. 7, 1281–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James J. R., Oliveira M. I., Carmo A. M., Iaboni A., Davis S. J. (2006) Nat. Methods 3, 1001–1006 [DOI] [PubMed] [Google Scholar]
- 32.Sleeman J., Rudy W., Hofmann M., Moll J., Herrlich P., Ponta H. (1996) J. Cell Biol. 135, 1139–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesley J., Hascall V. C., Tammi M., Hyman R. (2000) J. Biol. Chem. 275, 26967–26975 [DOI] [PubMed] [Google Scholar]
- 34.Mohamadzadeh M., DeGrendele H., Arizpe H., Estess P., Siegelman M. (1998) J. Clin. Invest. 101, 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siegelman M. H., DeGrendele H. C., Estess P. (1999) J. Leukoc. Biol. 66, 315–321 [DOI] [PubMed] [Google Scholar]
- 36.Nandi A., Estess P., Siegelman M. H. (2000) J. Biol. Chem. 275, 14939–14948 [DOI] [PubMed] [Google Scholar]
- 37.English N. M., Lesley J. F., Hyman R. (1998) Cancer Res. 58, 3736–3742 [PubMed] [Google Scholar]
- 38.Jackson D. G., Prevo R., Clasper S., Banerji S. (2001) Trends Immunol. 22, 317–321 [DOI] [PubMed] [Google Scholar]
- 39.Bouyain S., Rushton S., Drickamer K. (2001) Glycobiology 11, 989–996 [DOI] [PubMed] [Google Scholar]
- 40.Torgersen D., Mullin N. P., Drickamer K. (1998) J. Biol. Chem. 273, 6254–6261 [DOI] [PubMed] [Google Scholar]
- 41.de La Motte C. A., Hascall V. C., Calabro A., Yen-Lieberman B., Strong S. A. (1999) J. Biol. Chem. 274, 30747–30755 [DOI] [PubMed] [Google Scholar]
- 42.de la Motte C. A., Hascall V. C., Drazba J., Bandyopadhyay S. K., Strong S. A. (2003) Am. J. Pathol. 163, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Majors A. K., Austin R. C., de la Motte C. A., Pyeritz R. E., Hascall V. C., Kessler S. P., Sen G., Strong S. A. (2003) J. Biol. Chem. 278, 47223–47231 [DOI] [PubMed] [Google Scholar]
- 44.Day A. J., de la Motte C. A. (2005) Trends Immunol. 26, 637–643 [DOI] [PubMed] [Google Scholar]
- 45.Huang L., Yoneda M., Kimata K. (1993) J. Biol. Chem. 268, 26725–26730 [PubMed] [Google Scholar]
- 46.Zhuo L., Kanamori A., Kannagi R., Itano N., Wu J., Hamaguchi M., Ishiguro N., Kimata K. (2006) J. Biol. Chem. 281, 20303–20314 [DOI] [PubMed] [Google Scholar]
- 47.Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meiler J., Baker D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12105–12110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.