Abstract
ComA of Streptococcus is a member of the bacteriocin-associated ATP-binding cassette transporter family and is postulated to be responsible for both the processing of the propeptide ComC and secretion of the mature quorum-sensing signal. The 150-amino acid peptidase domain (PEP) of ComA specifically recognizes an extended region of ComC that is 15 amino acids in length. It has been proposed that an amphipathic α-helix formed by the N-terminal leader region of ComC, as well as the Gly-Gly motif at the cleavage site, is critical for the PEP-ComC interaction. To elucidate the substrate recognition mechanism, we determined the three-dimensional crystal structure of Streptococcus mutans PEP and then constructed models for the PEP·ComC complexes. PEP had an overall structure similar to the papain-like cysteine proteases as has long been predicted. The active site was located at the bottom of a narrow cleft, which is suitable for binding the Gly-Gly motif. Together with the results from mutational experiments, a shallow hydrophobic concave surface of PEP was proposed as a site that accommodates the N-terminal helix of ComC. This dual mode of substrate recognition would provide the small PEP domain with an extremely high substrate specificity.
Keywords: ABC Transporter, Bacterial Signal Transduction, Cysteine Protease, Enzyme Mechanisms, Enzyme Structure, Biofilm, Quorum Sensing, Streptococcus
Introduction
The quorum-sensing system is a bacterial cell-cell signaling system mediated by an inherent signal molecule (pheromone) to properly respond to environmental changes and survive as a “community” (1). This signal pathway alters the gene expression profile of the target cells after a sufficient number of cells are accumulated to form the “quorum,” and thereby the concentration of a released pheromone reaches the threshold to bind to either the cell surface or intracellular receptors (2, 3). The quorum-sensing system is believed to regulate the diversities of the physiological functions in Gram-positive and Gram-negative bacteria, including bioluminescence (4), virulence factor expression (5), and competence for genetic transformation (6). In addition, many opportunistic pathogens form a biofilm in response to this system, which causes chronic infections. The biofilm complicates the clinical treatment because of its resistance to antibiotics (7). Oral streptococci such as Streptococcus mutans are not only cariogenic but also cause life-threatening infective endocarditis by forming a biofilm on the native or prosthetic heart valves. The biofilm formation has been observed to be regulated through quorum sensing in S. mutans (8), Streptococcus gordonii (9), and Streptococcus intermedius (10).
Although several quorum-sensing systems have already been reported, the most thoroughly described systems are the acyl-homoserine lactone-mediated system of Gram-negative bacteria and the peptide-based signaling system of Gram-positive bacteria (11). Signal-producing enzymes play a key role in the first step of this pathway. In Gram-negative bacteria, the cytosolic LuxI-type protein, which was initially discovered in Vibrio fisheri, synthesizes the acyl-homoserine lactone, and the pheromone molecule is thought to be excreted by passive diffusion or membrane transporters. Extensive studies have been made on these proteins (12), and now their three-dimensional structures, including those of Pantoea stewartii EsaI (13), Pseudomonas aeruginosa LasI (14), and Vibrio cholerae CqsA (15), are available. In contrast, signal-producing proteins of Gram-positive bacteria are membrane-embedded proteins that are responsible for both the production and secretion of the peptide-based pheromone molecules. Thus, we have fallen far behind in the biochemical and structural characterizations compared with the studies of their counterparts of Gram-negative bacteria.
The mechanism of Gram-positive Streptococcus quorum sensing has been well studied in S. pneumoniae. The competence-stimulating peptide, which functions as a quorum-sensing signal in this bacterium, is cleaved from the precursor peptide ComC and concomitantly exported to the extracellular milieu by ComA with help of the accessory protein, ComB. The accumulated competence-stimulating peptide binds to the extracellular domain of the ComD receptor, which subsequently phosphorylates the response regulator, ComE, by the histidine kinase activity of the cytosolic domain of ComD. Phosphorylated ComE up-regulates the transcriptions of the early genes such as comX and comW, leading the DNA uptake (competence) and recombination. Homologues of these com genes were found in other Streptococcus species (6, 16).
ComA of Streptococcus is a member of the bacteriocin-associated ATP-binding cassette (ABC)2 transporter family, which comprises the three following domains: the N-terminal peptidase domain, a transmembrane domain consisting of six membrane-spanning segments, and a C-terminal ATP-binding domain located on the cytoplasmic face of the membrane (17). The peptidase domains of this family are thought to cleave the propeptides after the consensus Gly-Gly motif. To date, peptidase domains of this family, such as Lag D, a transporter of lactococcin G in Lactococcus lactis (17), and CvaB, a transpofter of colicin V in Escherichia coli (18), have been demonstrated to have proteolytic activity. Recently, we succeeded in the heterologous overexpression of the 150-amino acid peptidase domain (PEP) of ComA and the propeptide ComC from several species of Streptococcus as soluble proteins in E. coli (19, 20). These advances enabled us to do a detailed biochemical analysis of the initial steps of the pheromone production in Streptococcus.
Based on sequence alignments, PEP is thought to belong to a papain-like cysteine protease family (17, 21). The analysis with purified PEP and ComC from S. pneumoniae (PPEP and PComC, respectively) identified the essential Cys residue (19). Using combinations of PEPs and ComCs from the species of Streptococcus, together with the mutagenesis analyses of ComC, it was found that four conserved hydrophobic residues in the N-terminal leader region of ComC extending from the −15 to −4 positions, as well as the Gly-Gly motif at the cleavage site, are critical for the interaction between PEP and ComC (20). It was also suggested that ComC undergoes a structural transition from the random coil to helix upon binding to PEP and that the four conserved residues of ComC could form a hydrophobic face of the helix (20). Based on these findings, we have hypothesized that there is a large hydrophobic patch on the surface of the PEP protein, which interacts with the hydrophobic face of the N-terminal helix of ComC, and there is a narrow region in the vicinity of the active center, which interacts with the Gly-Gly motif of ComC.
We have now determined the crystal structure of PEP from S. mutans (MuPEP1) and further constructed models of the PEP·ComC complexes. Based on these results, a series of mutations was then introduced into the putative substrate-binding site of PEP for the kinetic analysis and CD measurement. These experimental results supported the model, which proposes a unique substrate recognition mechanism of PEP. The present study provides a first glimpse into the signal production mechanism in the quorum-sensing pathway of Gram-positive bacteria.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
For crystallization of both the native and selenomethionine-labeled MuPEP1, a methionine auxotrophic E. coli strain, B834 (DE3), carrying an expression plasmid for MuPEP1, pSMuP1 (20), was used. The E. coli cells were grown by shaking at 37 °C in LB medium containing 50 μg/ml ampicillin. When the density of the cultures reached 1 × 108 cells/ml, 10 ml of this culture was transferred to 1 liter of LeMaster medium (22), containing 10 g of lactose and 50 μg/ml ampicillin, preincubated at 37 °C for 1 h. The 1-liter medium was supplemented with 25 mg of l-methionine or seleno-l-methionine for the native or selenomethionine-labeled MuPEP1, respectively. These cells were grown by shaking at 37 °C for 23 h and then harvested by centrifugation and resuspended in 20 ml of a buffer containing 20 mm Tris-HCl, 500 mm NaCl, and 5 mm imidazole, pH 7.9. The cell suspension was frozen and stored at −80 °C.
For the biochemical studies, the PEPs and ComCs were expressed and purified as previously described (20). Briefly, the wild-type and mutant PEPs were expressed in an E. coli strain, BL21 (DE3) pLysS, carrying each expression plasmid. The cells were grown and induced by 0.2 mm isopropyl-β-d-thiogalactopyranoside for 2 h at 37 °C for the MuPEP1 or for 5 h at 30 °C for the PPEPs. The PEPs were purified with His·Bind resin (Novagen) and dialyzed at 4 °C against a buffer containing 20 mm Tris-HCl, 200 mm ammonium sulfate, and 2 mm β-mercaptoethanol, pH 7.0.
For the expression of the ComCs, an E. coli strain, JM109 (DE3) pLysS, carrying each expression plasmid was grown and induced by 0.2 mm isopropyl-β-d-thiogalactopyranoside for 1 h at 37 °C for PComC or 2 h for at 37 °C for Streptococcus cristatus ComC (CComC). The ComCs were purified with the His·Bind resin followed by chromatography with a Mono Q column connected to an ÄKTA fast protein liquid chromatography system (Amersham Biosciences). The concentrations of the PEPs and ComCs were spectrophotometrically determined based on the number of the aromatic amino acid residues (19).
Protein Crystallization and Data Collection
Crystals of the native MuPEP1 were obtained by the sitting drop, vapor diffusion method at 20 °C (24-well Linbro plates were used). Drops were assembled with 2 μl of the 5.5 mg/ml protein, which was dissolved in 70 mm sodium phosphate and 2 mm β-mercaptoethanol, pH 7.0, mixed with an equal volume of the precipitant solution comprising 16% polyethylene glycol 3350 and 0.16 m di-ammonium hydrogen citrate (no buffer). Crystals were observed on day 5. The crystals were briefly soaked in the precipitant solution containing 24.5% polyethylene glycol 3350 and 0.12 m di-ammonium hydrogen citrate and then flash-frozen in a liquid nitrogen stream (−173 °C). Crystals of the selenomethionine-labeled MuPEP1 were obtained under conditions similar to the native MuPEP1 with a protein concentration of 4.9 mg/ml. multiple-wavelength anomalous dispersion data sets for the selenomethionine-labeled crystals and the single-wavelength data set for the native crystals were collected at the RIKEN Structural Genomics Beamline II BL26B2, SPring-8 (Hyogo, Japan) (23) by using a MarCCD 225 detector. The data sets were processed and scaled with the program package HKL2000 (24).
Structure Determination and Analysis
The initial phases were calculated by the program SHELX C/D/E implemented in the HKL2MAP interface (25) using the multiple-wavelength anomalous dispersion data sets. The space group of the crystal was determined to be P43212. The resulting phases were improved with the program PHENIX (26), followed by automatic model tracing with the program ARP/wARP (27). Model refinement was performed with the programs XtalView/X-fit (28) and CNS (29). The model was subsequently refined against the native data set at a 1.9-Å resolution. The final model was validated using the program PROCHECK in the CCP4 suite (30). The refinement statistics are summarized in supplemental Table S1. Figures were drawn using the programs PyMOL (31) and VMD (32). The atomic coordinates and structure factors have been deposited in Protein Data Bank (www.rcsb.org/) under the accession code 3K8U.
Molecular Modeling
The initial models for the subsequent molecular dynamics (MD) simulations were constructed using Xfit (28) (see text for details). These models were electrostatically checked using PDB 2PQR (33) with PROPKA (34). The proteins were placed in a box of water molecules, and sodium ions were randomly added to the system to maintain electrical neutrality using VMD (32). This resulted in a total atom count of 24,433 with the dimensions of 67 × 60 × 63 Å3 for the MuPEP1·MuComC model, 20,480 with the dimensions of 57 × 61 × 64 Å3 for the PPEP model, and 30,511 with the dimensions of 69 × 67 × 68 Å3 for the PPEP-PComC model.
MD simulations were carried out using NAMD2.6 (35) and the CHARMM22 force field for the protein and ligands (36), along with the TIP3P model for water (37). A cutoff of 12 Å (switching function starting at 10 Å) for the van der Waals interactions and real space electrostatic interactions was assumed. Periodic boundary conditions were used. The particle-mesh Ewald method (38) was used to compute the long range electrostatic forces. The SHAKE algorithm was used to constrain all bonds between the hydrogen atoms and heavy atoms (39). An integration time step of 2 fs was used, thus permitting a multiple time-stepping algorithm (40) to be employed in which the interactions involving covalent bonds and short range non-bonded interactions were computed for every time step, and long range electrostatic forces were computed every two time steps. Langevin dynamics was utilized to maintain a constant temperature of 300 K in all the simulations. Constant pressure simulations at 1 atm were conducted using the Nosé-Hoover Langevin piston method (41) with a decay period of 100 fs and a damping time scale of 50 fs. After an initial conjugate gradient minimization for 500 iterations, the equilibrated system was simulated for 1 ns. Analysis of the computed trajectories was performed by VMD.
Site-directed Mutagenesis
Mutagenesis was done using a QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All the single mutations of PPEP were introduced into pSPP1 (19). For a double mutant, L52A/V55A PPEP, the V55A mutation was introduced into L52A PPEP. The primer pairs used for the mutagenesis are listed in supplemental Table S2. The nucleotide sequences of the entire coding regions were verified.
Determination of Kinetic Parameters
The PPEP activity was assayed in a 100-μl reaction mixture containing 50 mm Tris-HCl, 150 mm ammonium sulfate, pH 7.0, and various concentrations of the substrate PComC. High-performance liquid chromatography analysis was performed as previously described (19). Briefly, the reaction mixtures were loaded onto a Waters μBondasphere C8 reversed-phase column (Waters) connected to a Beckman System Gold high-performance liquid chromatography system (Beckman-Coulter), and the peptides were separated on a linear gradient from 10 to 55% (v/v) acetonitrile containing 0.1% trifluoroacetic acid over 10 min at the flow rate of 1 ml/min at ambient temperature. Typically, <10% of the substrates were consumed in the reactions.
CD Measurements
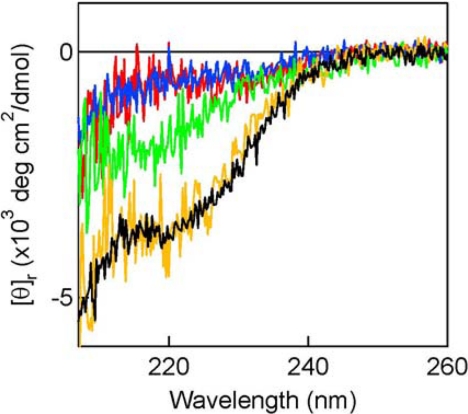
The CD spectra were recorded using a Jasco spectropolarimeter, model J-720WI (Jasco), equipped with a thermocontroller using a 0.1-cm light-path sample cell. The buffers and protein concentrations are described in the legend of Fig. 6.
FIGURE 6.
Structural transitions of CComC in the presence of PPEPs. The far-UV CD spectra of CComC in the region from 207 to 260 nm were measured in the absence or presence of the mutant PPEPs. The spectrum below 207 nm could not be obtained because of the large noise derived from the high UV absorption by the proteins. The ordinate is the mean residue ellipticity. The CD measurements were done using 20 μm CComC, and the mixture of 20 μm of CComC and 20 μm each of CAPEP, A51W CAPEP, or A67W CAPEP. The buffer solution contained 30 mm sodium phosphate, 150 mm ammonium sulfate, 15 mm sodium fluoride, and 0.025 mm dithiothreitol, pH 7.0. The spectra were measured at the scan speed of 50 nm/min and accumulated 30 times. The spectrum of CComC (0% TFE) is shown in blue. The spectrum of CComC in the same buffer containing 35% TFE is shown in black. The spectrum of each mutant PPEP was subtracted from that of the mixture of CComC and the corresponding PPEP, and the difference spectrum is shown: (CComC + CAPEP) − CAPEP (yellow); (CComC + A51W CAPEP) − A51W CAPEP (red); and (CComC + A67W CAPEP) − A67W CAPEP (green).
RESULTS
Overall Structure of MuPEP1
After an initial crystallization trial for PPEP yielded no diffraction-quality crystals, MuPEP1 was chosen for the subsequent crystallization, because it showed the highest thermostability among the PEPs from six species of Streptococcus that we obtained (20). The structure of MuPEP1 was determined to a near-atomic resolution of 1.9 Å with an Rwork of 21.2% (Rfree = 23.4%), and a root-mean square deviation (r.m.s.d.) from the ideal values in bond lengths and bond angles were 0.005 Å and 1.20°, respectively (supplemental Table S1). The final structure included residues 5–141 and 95 water molecules. No electron density was visible for the 4 N-terminal residues and the 15 C-terminal residues, including the His tag. The numbering of the MuPEP1 residues is based on the truncated form, in which the 44 N-terminal hydrophobic amino acid residues are removed from the native protein (20).
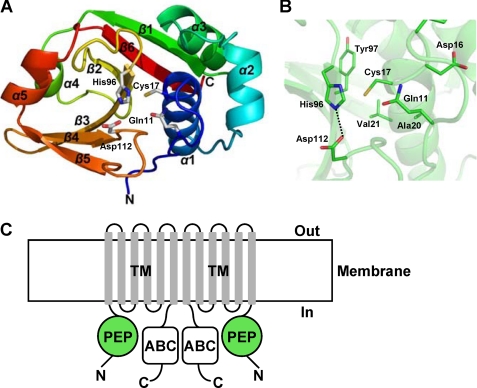
The structure of MuPEP1 has an α/β fold with six β-strands and five α-helices (Fig. 1A). The structure is organized around a central six-stranded antiparallel β-sheet with α-helices packing on both sides of the sheet to form two subdomains. The N-terminal subdomain, residues 5–62, consists of three α-helices, α1, α2, and α3. The C-terminal subdomain, residues 63–141, consists of six β-strands and two α-helices, and the order of these components is as follows: β1, α4, β2, β3, β4, β5, α5, and β6. The active site, containing the catalytic triad Cys-17, His-96, and Asp-112, is located in a narrow cleft at the interface of these two subdomains. Cys-17 and His-96 are at the N termini of α1 and β3, respectively, and Asp-112 is in a loop between β4 and β5 (Fig. 1A). The Sγ atom of Cys-17 is located 3.6 Å from the Nδ1 atom of His-96. The Oδ1 atom of Asp-112 forms a hydrogen bond with the Nϵ2 of His-96 at a distance of 2.7 Å (Fig. 1B).
FIGURE 1.
Structure of MuPEP1. A, schematic representation of the overall structure of MuPEP1. The secondary structure elements are rainbow colored with the N terminus in blue and the C terminus in red. The side chains of the catalytic triad residues, Cys-17, His-96, and Asp-112, and the oxyanion hole Gln-11 are shown as sticks. B, the active site of MuPEP1. Backbone atoms are indicated by the semitransparent schematic in green. The side chains of the residues in the vicinity of the active site are indicated by sticks. The oxygen, nitrogen, and sulfur atoms are colored in red, blue, and yellow, respectively. The dashed line indicates a hydrogen bond. C, schematic representation of putative ComA domain organization. PEP, peptidase domain; TM, transmembrane domain; ABC, ATP-binding domain. Bacteriocin-associated ABC transporters are presumed to function as homodimer (17).
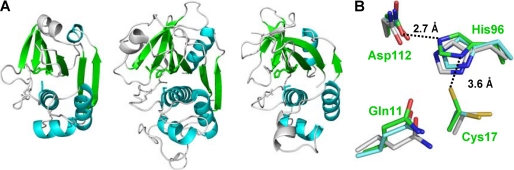
Despite the lack of a significant sequence similarity, the crystal structures show a common structural scaffold between MuPEP1 and the papain-like cysteine proteases (Fig. 2A). The secondary structure elements of MuPEP1 are analogous to those of the papain-like cysteine proteases, including three helices in the N-terminal subdomain and most of a central β-sheet and a helix (α4) in the C-terminal subdomain (Fig. 2A). MuPEP1 has a Cα r.m.s.d. of 3.0 Å for 137 selected residues and that of 2.7 Å for 133 selected residues when compared with papain and staphopain A, respectively, using the FATCAT server (42). Especially, the spatial arrangements of the catalytic triad and the oxyanion hole, Gln-11 of MuPEP1, are highly conserved. The overlaps of the Cα atoms of these four catalytic residues of MuPEP1 with those of papain or staphopain A equally yielded an r.m.s.d. value of 0.59 Å (Fig. 2B).
FIGURE 2.
Structural comparisons of MuPEP1 with cysteine proteases. A, schematic representation of MuPEP1 (left), papain (PDB ID code 9PAP) (center), and staphopain A (PDB ID code 1CV8) (right). The structures are aligned based on least-squares superimposition of the Cα atoms of the active-site residues such as Gln-11, Cys-17, His-96, and Asp-112. These residues are depicted by stick models. The structurally identical α-helices, α1–4, and β-strands, β1–6, are colored in cyan and green, respectively. B, active-site superimposition of MuPEP1 and papain-like cysteine proteases. Active-site residues of MuPEP1, Gln-11, Cys-17, His-96, and Asp-112, are shown by green sticks. The corresponding residues in papain (cyan) and staphopain A (gray) are shown by sticks after least-squares superimposition on the Cα atoms. The oxygen, nitrogen, and sulfur atoms are colored in red, blue, and yellow, respectively. Distances between atoms are shown with dashed lines.
Acyl-intermediate Model of MuPEP1·MuComC
To investigate the substrate recognition mechanism of PEP, we first attempted to express the recombinant ComC from S. mutans (MuComC), but could not get it accumulated in E. coli as a soluble protein. Next, MuComC was chemically synthesized, but was found to be almost insoluble in aqueous buffers. A fusion protein of MuPEP1 (C17A)–MuComC with a six-glycine linker was then constructed, expressed as a soluble protein, and purified, but it did not yield any crystals. We further tried to obtain the complex structure of MuPEP1 (C17A, C17S, or H96A) with ComCs from other Streptococcus species by co-crystallization and soaking methods. Some combinations yielded crystals, but the diffraction data revealed no ComC molecules.
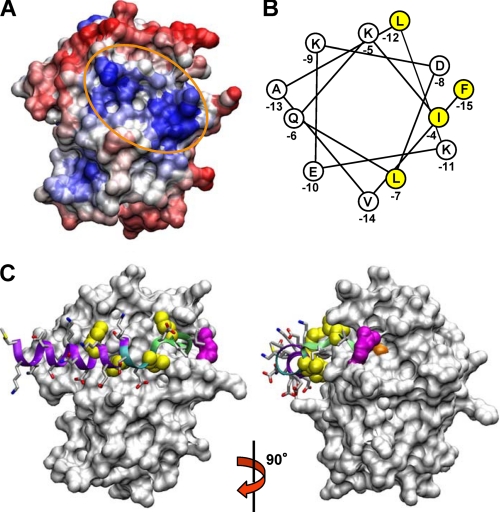
Thus, we set out to construct a computer model of the MuPEP1·MuComC complex. As the first step, the surface of the MuPEP1 protein was searched for a hydrophobic region, which is suitable for the interaction with the hydrophobic face of the putative N-terminal helix of ComC (20). The HotPatch program (43) was performed using the mode that predicts protein-protein interactions based on the hydrophobicity of a protein surface. Six major hydrophobic patches were identified. After taking into account the general consensus for the substrate orientation in other papain-like proteases, only one shallow concave surface adjacent to the active site remained as a candidate for the binding site for the N-terminal helix of ComC (Fig. 3A). This concave surface is located at the subdomain interface and comprises a cluster of hydrophobic residues, such as Ala-51, Leu-52, Val-55, Leu-94, and Leu-134.
FIGURE 3.
Acyl-intermediate model of MuPEP1·MuComC. A, surface representation of the result of the HotPatch program (43). Hydrophilic region is colored in red and hydrophobic region is in blue. The hydrophobic region adjacent to the active site is encircled by an orange oval line. B, α-helical wheel representation of the N-terminal region of PComC. Highly conserved hydrophobic amino acids, which are postulated to be critical in the interaction with PPEP (20), are shaded yellow. C, acyl-intermediate model of MuPEP1·MuComC. MuPEP1 is shown by a surface representation. The side chains of MuComC are indicated by sticks. The oxygen, nitrogen, and sulfur atoms are colored in red, blue, and yellow, respectively. The four highly conserved residues of MuComC, Phe-(−15), Ile-(−12), Leu-(−7), and Ile-(−4), are indicated by spheres and colored in yellow. The secondary structure elements, random coil, turn, and α-helix, of MuComC are colored in green, cyan, and purple, respectively. Cys-17 and Arg-93 (see text for detail) are colored in orange and magenta, respectively. The right structure is a view of the left one rotated by 90° along the y-axis.
Next, we constructed the acyl-intermediate formed between MuPEP1 and MuComC, in which the nucleophilic Cys-17 of MuPEP1 is acylated by the carbonyl carbon of Gly-(−1) of MuComC. The cleaved C-terminal region of MuComC is omitted in the present model. The backbone structure from Ile-(−3) to Gly-(−1) of MuComC was fitted into the active-site cleft based on the structure of the ubiquitin C-terminal hydrolase in a complex with its inhibitor ubiquitin aldehyde (PDB ID code 1CMX), which also has the Gly-Gly sequence at the cleavage site, and a covalent bond is formed between the carbonyl carbon of the C-terminal Gly and the Sγ atom of a nucleophilic Cys residue (44). The position of the backbone of Ile-(−4) was restricted by this fitting. Because the length of the putative helix of MuComC was unknown, the longest possible region from Met-(−25) to Ile-(−5) was initially forced to form a helix. This helix was then roughly docked on MuPEP1 with the angle that is in agreement with the assumption that the conserved hydrophobic residues (Fig. 3B), Phe-(−15), Ile-(−12), and Leu-(−7), interact with the hydrophobic concave shape of MuPEP1. To connect these two regions of MuComC, the backbone structure from Leu-(−7) to Ile-(−4) was manually coordinated to yield a suitable conformation with distance and dihedral angle constraints considered. Finally, the MD simulations were performed at 300 K. During the MD simulation, the r.m.s.d. of the MuPEP1·MuComC complex increased to ∼1.5 Å for the first 0.4 ns and reached a plateau phase continuing until 1.0 ns (supplemental Fig. S1). Hence, the structure obtained at 1.0 ns, after being subjected to the energy minimization, is shown in Fig. 3C. The r.m.s.d. positional fluctuations of each Cα atom of MuPEP1 during the MD simulation (0.4–1.0 ns) showed four prominent triangular signals (supplemental Fig. S2A). The peaks of the triangles correspond to the loop structures of MuPEP1: position 76 at the C terminus of α4, which itself is located in a large loop between β1 and β2; positions 91–95 in the loop between β2 and β3; position 105 in the turn between β3 and β4; and position 116 in the loop between β4 and β5. These positions are exposed to the solvent, implying that MuPEP1 did not show anomalous behaviors during the MD simulation. The r.m.s.d. positional fluctuations of MuComC relative to MuPEP1 gradually rose from Thr-(−22) to Met-(−25) (supplemental Fig. S2B), suggesting that the N-terminal residues of MuComC are being detached from MuPEP1. All the residues of MuComC are located in favored positions in the Ramachandran plot.
In the model of Fig. 3C, the region of MuComC from −25 to −10 retains the α-helix conformation. The side chains of the conserved residues, Phe-(−15) and Ile-(−12), in this helix and Leu-(−7) in the following turn structure, are stably accommodated by the hydrophobic surface of MuPEP1. The backbone of MuComC bends in the region from Leu-(−7) to Ile-(−4), which is located at the inlet to the active site. The side chain of Ile-(−4) is tightly locked by the side chains of Gln-(−47) and Thr-(−50). On the other hand, the hydrophilic residues of MuComC, such as Lys-(−14), Glu-(−13), Lys-(−11), and Thr-(−10), are fully exposed to the solvent.
The Gly-Gly motif of MuComC fits into the narrow cleft of MuPEP1 in this model (Fig. 4). This cleft is located at the interface of the two subdomains described above. The left side wall of the cleft (in the left and right views of Fig. 4) is mainly composed of Lys-92 through Gln-95 in the loop between β2 and β3, and the right side wall is Ile-12 through Arg-15 in the N-terminal chain and Lys-46 through Gly-48 in the loop between α2 and α3. The active-site residues are located at, and comprise a part of, the bottom of the cleft. The side chain of Arg-93 closed the active site like a lid (shown in magenta in Fig. 3C) after the MD simulations. The Nη1 and Nη2 atoms of Arg-93 form indirect hydrogen bonds with the backbone carbonyl oxygen of Lys-46 via two different water molecules. The aliphatic portion of the side chain of Arg-93 makes van der Waals contacts with the Gly-2 of MuComC. These interactions would partly suppress the fluctuation of the loop between β2 and β3 (a “dip” signal in supplemental Fig. S2A).
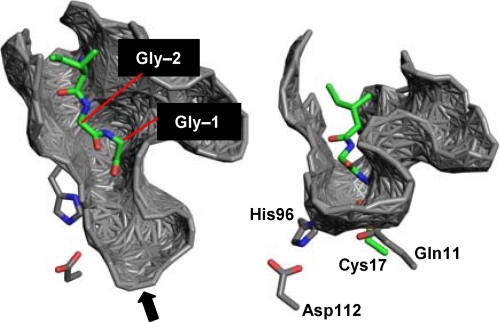
FIGURE 4.
Active-site cleft surrounding Gly-Gly. The acyl-intermediate model of MuPEP1·MuComC (green) obtained from molecular dynamics simulation using the program NAMD2.6 (35) was superimposed on the MuPEP1 crystal structure (gray) using the least-squares overlap of the Cα atoms of the active-site residues, Gln-11, Cys-17, His-96, and Asp-112. The surface of the cleft of the MuPEP1 structure, analyzed by PDBsum (47), is shown. Only Gly-(−1), Gly-(−2), and Ile-(−3) of MuComC are indicated here. The oxygen, nitrogen, and sulfur atoms are colored in red, blue, and yellow, respectively. The right structure is a view of the left one from the indicated direction (arrow).
Experimental Verification of the Model
To corroborate the present model, mutations were introduced into the putative substrate-binding site of PPEP. The PPEP-PComC pair was chosen, because it is the only native pair that can be experimentally examined (20).
For this purpose, an acyl-intermediate model of PPEP-PComC was newly constructed. First, the PPEP model was constructed using the MuPEP1 structure as a template. The 137 residues of the MuPEP1 structure were replaced with the corresponding residues of PPEP, which shows a 57% identity on the basis of the amino acid sequence alignment of these proteins (20), the MD simulations were completed, and then the energy minimization was carried out. The resultant PPEP model indicated that there is a hydrophobic concave shape analogous to that of MuPEP1 predicted by the HotPatch program (supplemental Fig. S3). The PPEP-PComC acyl-intermediate model was constructed according to the same procedures as described in the MuPEP1·MuComC modeling. Although the final model is almost comparable to the MuPEP1·MuComC model, the helix structure of PComC is partly unfolded during the MD simulation in the region between Leu-(−18) and Glu-(−17) (Fig. 5A). The r.m.s.d. positional fluctuations of the PComC residues relative to PPEP during the MD simulation suggest that the N-terminal region from Met-(−24) to Leu-(−18) of PComC is detached from PPEP (data not shown).
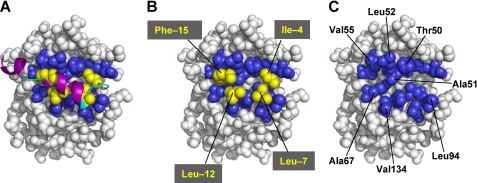
FIGURE 5.
Acyl-intermediate model of PPEP-PComC. A, Acyl-intermediate model of PPEP-PComC. A spherical representation of the PPEP complexed with PComC is shown. The secondary structure elements of PComC, random coil, turn, and α-helix, are colored in green, cyan, and purple, respectively. The side chains of the four highly conserved residues of PComC are colored in yellow. The residues of PPEP that are predicted to interact with these four PComC residues are colored in blue. B, The main chain of PComC is removed from A. Phe-(−15) of PComC interacts with Ala-51, Leu-52, Val-55, and Ala-67 of PPEP; Leu-(−12) interacts with Ala-67 and Val-134; Leu-(−7) interacts with Leu-94; Ile-(−4) interacts with Thr-50. C, The entire PComC is removed from A to show the hydrophobic concave surface of PPEP.
From the inspection of this model, Thr-50, Ala-51, Leu-52, Val-55, Ala-67, Leu-94, and Val-134 were picked for the mutational analysis, because each side chain of these residues was expected to make contact with one or two of the four conserved hydrophobic residues of PComC (Fig. 5). Also, these residues were predicted to be exposed to the solvent, and therefore, mutagenesis on these residues would minimally affect the overall structure of PPEP. Table 1 shows the kinetic parameters of the mutant PPEPs for PComC. The catalytic efficiencies decreased by 1.8- to 16-fold upon the mutations of T50S, L52A, V55A, L94A, and V134A. A double mutant, L52A/V55A PPEP, showed a significant decrease in the affinity of PPEP for PComC (16-fold increase in Km), whereas the catalytic rate for this mutant only slightly decreased. In contrast, the mutations of A51W and A67W, which are expected to introduce steric hindrance into the putative substrate-binding site of PPEP, severely impair the catalytic efficiency (1400- and 110-fold, respectively) by reducing both the affinity and the catalytic rate for ComC.
TABLE 1.
Kinetic parameters of wild-type and mutant PPEPs for PComC
The experiments were carried out at 25 °C in 50 mm Tris-HCl, 150 mm ammonium sulfate, and 0.2 mm dithiothreitol, pH 7.0, as described under “Experimental Procedures.” The concentrations of PComC were 50–750 μm, and those of the mutant PPEPs were 1.0–7.5 μm for the PPEPs with kcat/Km values of <20 m−1s−1. For the other mutant PPEPs, the concentrations of PComC were 12.5–500 μm, and that of the mutant PPEPs was 0.25 μm.
| PPEPs | kcat | Km | kcat/Km |
|---|---|---|---|
| min−1 | μm | m−1s−1 | |
| Wild typea | 1.4 | 86 | 270 |
| T50S | 0.46 ± 0.03 | 94 ± 14 | 82 |
| L52A | 0.64 ± 0.04 | 110 ± 20 | 97 |
| V55A | 0.47 ± 0.05 | 450 ± 100 | 17 |
| L52A/V55A | 0.88 ± 0.03 | 1400 ± 100 | 10 |
| L94A | 0.43 ± 0.01 | 130 ± 10 | 55 |
| V134A | 0.80 ± 0.03 | 89 ± 9 | 150 |
| A51W | 0.0064 ± 0.0003 | 550 ± 50 | 0.19 |
| A67W | 0.071 ± 0.007 | 490 ± 90 | 2.4 |
a Data from Kotake et al. (20).
We have demonstrated that the structural transition of ComC upon the interaction with PEP can be observed by CD measurements (20). To examine how the steric hindrance caused by the A51W and A67W mutations affect the binding and helix formation of ComC, the CD spectra of ComC were measured in the absence and presence of PEP bearing the A51W or A67W mutation. These mutations were introduced into C17A PPEP (CAPEP), because CAPEP has lost its catalytic activity and cannot cleave PComC (19). CComC was chosen because of its lowest Km value for PPEP among the examined ComCs (20). The CD spectrum of 20 μm CComC in the aqueous buffer (Fig. 6, blue line) and that in the same buffer containing 35% trifluoroethanol (TFE) (black line) exhibit typical random coil and α-helix structures, respectively, as previously shown (20). The structural transitions of CComC in the presence of CAPEPs were measured by subtracting the spectrum of each CAPEP (20 μm) from that of the mixture of each CAPEP (20 μm) and CComC (20 μm). The resultant difference spectrum of CComC in the presence of CAPEP (yellow line) is almost identical to the spectrum of CComC measured in 35% TFE. On the other hand, the difference spectrum of CComC in the presence of A51W CAPEP (red line) overlaps with the spectrum of CComC taken in the aqueous buffer. The difference spectrum of CComC in the presence of A67W CAPEP (green line) is intermediate between that in the presence of CAPEP and that in the presence of A51W CAPEP.
DISCUSSION
The assumption based on primary structures that the peptidase domain of the bacteriocin-associated ABC transporters is a papain-like cysteine protease (17) has been fully supported by the three-dimensional structure of MuPEP1 in the present study. C17S, C17A, and H96A PPEPs (19) and C17A MuPEP1 exhibited complete loss of activity, indicating the essential role of the Cys residue in the catalysis. The Sγ atom of Cys-17 is located 3.6 Å from Nδ1 of His-96, and the distance is slightly longer than the ideal distance of a hydrogen bond, 2.6–3.5 Å. The distances between the corresponding atoms of Cys and His of papain and staphopain A were also reported to be 3.7 Å and 4.0 Å, respectively (Fig. 2B). The pKa values of Cys-17, His-96, and Asp-112 of MuPEP1 were predicted to be 3.5, 12, and 0.7, respectively, by the PROPKA program (34). Therefore, as proposed in other papain-like cysteine proteases (45, 46), Cys-17 and His-96 would form a thiolate-imidazolium ion pair under physiological conditions. Asp-112, which is in a hydrogen-bonding distance to His-96, would function to stabilize the spatial orientation and protonated form of His-96. Thus, MuPEP1 has all the structural components required for the catalytic mechanism of a cysteine protease.
Despite the overall structural resemblance to other protein-degrading cysteine proteases (Fig. 2A), the PEPs exhibit a peculiar substrate specificity, which strictly discriminates ComCs against other proteins (19, 20). Our model of the PEP·ComC complexes would provide the structural basis for the substrate recognition of the PEPs. Consistent with our prediction, two distinct regions were found on the surface of MuPEP1 to interact with the ComCs. One of these regions is the active-site cleft, which accommodates the consensus Gly-Gly motif of the ComCs (Fig. 4). When each Gly-(−1) and Gly-(−2) of MuComC of our model is mutated to Ala, the side-chain methyl group clashes with the side walls of the cleft: For example, Ala-(−1) is against the main-chain atoms of Thr-14 and Ala-(−2) is against that of Gln-95. Previous studies showed that even a minimum change (Gly to Ala) at these two sites of PComC resulted in an 800-fold decrease in the catalytic efficiency of PPEP (19). In addition, PPEP was not sensitive to the typical peptide-mimetic inhibitors, such as antipain, leupeptin, and E-64, which broadly inhibit the cysteine proteases (19). These findings support the restricted geometry at the S1 and S2 sites of PEPs that only the Gly-Gly motif can fit.
The other region, a hydrophobic surface adjacent to the active-site cleft (Fig. 3A and supplemental Fig. S3), would interact with the conserved hydrophobic residues of ComCs at the positions of −15, −12, −7, and −4. Although the entire region of this N-terminal leader sequence of ComC was postulated to form an α-helix (Fig. 3B) in the starting structure, the present model of the PEP·ComC complexes showed that the region from −7 to −4 is in a random coil or turn structure (Figs. 3C or 5A, respectively). The hydrophobic surface is slightly concave in shape, and the residues comprising the surface, such as Thr at position 50, Ala at 51, Leu at 52, Val at 55, His at 87, Leu at 94, Thr at 132, Leu/Val at 134, and Ile/Leu at 136, are highly conserved among the Streptococcus PEPs (20). The importance of this concave surface was examined by analyzing the effects of the mutations introduced into this region of PPEP on its catalytic activity (Table 1). The decreases in the catalytic efficiencies by the mutations at the positions of 50, 52, 55, 94, and 134 were rather moderate (1.8- to 16-fold) compared with those (31- to 180-fold) observed in the reactions of PPEP with the mutant PComCs, Phe-(−15) → Ala, Leu-(−12) → Ala, Leu-(−7) → Ala, and Ile-(−4) → Ala (20). This was expected because each of the four conserved residues of ComC would interact with several residues of PEP. For example, the side chain of Phe-(−15) of PComC is supposed to make contacts with Ala-51, Leu-52, Val-55, and Ala-67 of PPEP (Fig. 5B), and although the Phe-(−15) to Ala mutation of ComC removes all these interactions, the Leu-52 to Ala mutation of PEP removes only part of them. Indeed, the affinity of PPEP for PComC was reduced in an additive manner by the double mutations at positions 52 and 55. The kcat/Km value of PEPP for Phe-(−15) → Ala PComC decreased by 110-fold (20), and that of L52A/V55A PPEP for PComC decreased by 27-fold. Thus, the contribution of Leu-52 and Val-55 of PPEP to the stabilization of the transition state is estimated to be 68% (RT ln 27/RT ln 110) of the total effect attributed to the interaction of Phe-(−15) of PComC and residues of PPEP. To more significantly impair the activity by the least number of mutations, thus to more clearly show that this hydrophobic region is the substrate-binding site, steric hindrance was introduced into this region. As expected, the A51W and A67W mutations severely decreased the catalytic efficiency, and the effect of the mutation of Ala-51, which is located at the center of this putative binding site, was larger than that of Ala-67 located at the periphery. The A51W mutation was also introduced into MuPEP1, resulting in almost complete loss of the activity, which imply the conserved importance of this region in the Streptococcus PEPs.
These results are well correlated with the impairment in the helical transition of ComC in the presence of A51W CAPEP and A67W CAPEP (Fig. 6). These findings further support the idea that the N-terminal leader region of ComC undergoes a structural transition from the random coil to helix upon binding to the hydrophobic region of PEP and that this is a prerequisite step for the PEP·ComC complex to take the productive form. The A51W and A67W mutations resulted in large decreases in the kcat value, not only increases in the Km value. Together with the fact that PEPs do not cleave proteins with only the Gly-Gly sequence, it is thought that the substrate backbone flanking Gly-Gly bears a strain around the cleavage site in the ground state (the E·S complex). This strain would be released in the transition state, thereby lowering the activation energy and increasing the kcat. Apparently, a large binding energy between the hydrophobic regions of PEP and ComC described above would be necessary to cause the strain. However, this interaction is not enough by itself, and the C-terminal region must also be tightly held by binding to PEP and/or other parts of ComA to realize the strain around the cleavage site. To clarify this, the models for the ground state and the tetrahedral intermediate, a metastable structure near the transition state, are required. The acyl-intermediate structure shown here should be different from the structures of both the ground state and the tetrahedral intermediate. In fact, the acyl carbon oxygen does not point to the putative oxyanion hole Gln-11 in the models. Additionally, because the cleaved C-terminal region, the mature signal molecule, must be transferred to the transmembrane domain to be excreted without leaking into the cytoplasm, exploring the binding site for the C-terminal region of ComC would be very important for the biological function of ComA.
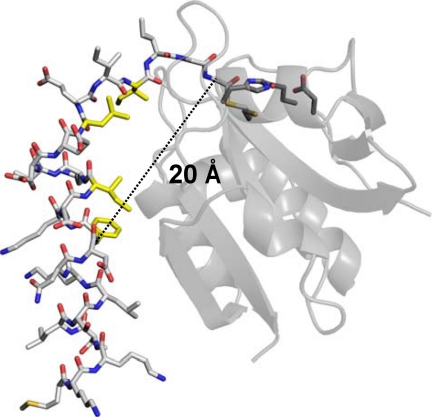
The present study also proposes a model that explains how a small protease like PEP can specifically recognize an amino acid residue of the substrate that is 15 residues apart from the cleavage site. In the MuPEP1·MuComC model, the Cα atom of Phe-(−15) of ComC is ∼20 Å away from that of Gly-(−1) (Fig. 7). To the best of our knowledge, PEP is a protease that specifically recognizes the most extended region of the substrate. The residues comprising the hydrophobic concave of the PEPs, except for the residue at position 55, are also conserved in the family of the bacteriocin-associated ABC transporters, implying that these transporters share the common substrate recognition mechanism with ComA. Therefore, the present results would provide a prototypical model for studying the peptidase domains of the other members of bacteriocin-associated ABC transporters. Based on the fact that the family of the bacteriocin-associated ABC transporters has so far been found only in prokaryotes, the PEP domains of ComAs would be an ideal target for, and the structure of MuPEP1 might help, the development of drugs that inhibit the biofilm formation of Streptococcus.
FIGURE 7.
The distance between the Cα atom of Phe-(−15) and that of Gly-(−1). The overall structure of MuPEP1 is indicated by the semitransparent schematic in gray. The active-site residues of MuPEP1, Gln-11, Cys-17, His-96, and Asp-112, are indicated by sticks. MuComC is represented by white sticks. The side chains of the four highly conserved residues of MuComC are colored in yellow. The oxygen, nitrogen, and sulfur atoms are colored in red, blue, and yellow, respectively. The dashed line indicates the distance between the Cα atom of Phe-(−15) and that of Gly-(−1).
Supplementary Material
Acknowledgments
We thank Seiki Kuramitsu and Noriko Nakagawa for help with the structural analysis and Takeshi Murakawa for help with the graphics work.
This work was supported by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (Category C Grant 19570141 to S. I.) and a grant from the Osaka Medical Research Foundation for Incurable Diseases (to T. Y.)
The atomic coordinates and structure factors (code 3K8U) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
- ABC
- ATP-binding cassette
- PEP
- peptidase domain of strepcococcal ComA
- PPEP
- S. pneumoniae PEP
- CAPEP
- C17A PPEP
- PComC
- S. pneumoniae ComC
- r.m.s.d.
- root mean square deviation
- MD
- molecular dynamics
- TFE
- trifluoroethanol
- CComC
- S. cristatus ComC
- MuPEP1
- S. mutans PEP
- MuComC
- S. mutans ComC.
REFERENCES
- 1.Fuqua W. C., Winans S. C., Greenberg E. P. (1994) J. Bacteriol. 176, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuqua C., Parsek M. R., Greenberg E. P. (2001) Annu. Rev. Genet. 35, 439–468 [DOI] [PubMed] [Google Scholar]
- 3.Miller M. B., Bassler B. L. (2001) Annu. Rev. Microbiol. 55, 165–199 [DOI] [PubMed] [Google Scholar]
- 4.Greenberg E. P. (2003) J. Clin. Invest. 112, 1288–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleerebezem M., Quadri L. E. (2001) Peptides 22, 1579–1596 [DOI] [PubMed] [Google Scholar]
- 6.Cvitkovitch D. G., Li Y. H., Ellen R. P. (2003) J. Clin. Invest. 112, 1626–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costerton J. W., Stewart P. S., Greenberg E. P. (1999) Science 284, 1318–1322 [DOI] [PubMed] [Google Scholar]
- 8.Li Y. H., Tang N., Aspiras M. B., Lau P. C., Lee J. H., Ellen R. P., Cvitkovitch D. G. (2002) J. Bacteriol. 184, 2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilmore K. S., Srinivas P., Akins D. R., Hatter K. L., Gilmore M. S. (2003) Infect. Immun. 71, 4759–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen F. C., Pecharki D., Scheie A. A. (2004) J. Bacteriol. 186, 6327–6331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raina S., De Vizio D., Odell M., Clements M., Vanhulle S., Keshavarz T. (2009) Biotechnol. Appl. Biochem. 54, 65–84 [DOI] [PubMed] [Google Scholar]
- 12.Williams P. (2007) Microbiology 153, 3923–3938 [DOI] [PubMed] [Google Scholar]
- 13.Watson W. T., Minogue T. D., Val D. L., von Bodman S. B., Churchill M. E. (2002) Mol. Cell 9, 685–694 [DOI] [PubMed] [Google Scholar]
- 14.Gould T. A., Schweizer H. P., Churchill M. E. (2004) Mol. Microbiol. 53, 1135–1146 [DOI] [PubMed] [Google Scholar]
- 15.Jahan N., Potter J. A., Sheikh M. A., Botting C. H., Shirran S. L., Westwood N. J., Taylor G. L. (2009) J. Mol. Biol. 392, 763–773 [DOI] [PubMed] [Google Scholar]
- 16.Luo P., Li H., Morrison D. A. (2004) Mol. Microbiol. 54, 172–183 [DOI] [PubMed] [Google Scholar]
- 17.Håvarstein L. S., Diep D. B., Nes I. F. (1995) Mol. Microbiol. 16, 229–240 [DOI] [PubMed] [Google Scholar]
- 18.Wu K. H., Tai P. C. (2004) J. Biol. Chem. 279, 901–909 [DOI] [PubMed] [Google Scholar]
- 19.Ishii S., Yano T., Hayashi H. (2006) J. Biol. Chem. 281, 4726–4731 [DOI] [PubMed] [Google Scholar]
- 20.Kotake Y., Ishii S., Yano T., Katsuoka Y., Hayashi H. (2008) Biochemistry 47, 2531–2538 [DOI] [PubMed] [Google Scholar]
- 21.Rawlings N. D., Morton F. R., Kok C. Y., Kong J., Barrett A. J. (2008) Nucleic Acids Res. 36, D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrickson W. A., Horton J. R., LeMaster D. M. (1990) EMBO J. 9, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueno G., Kanda H., Hirose R., Ida K., Kumasaka T., Yamamoto M. (2006) J. Struct. Funct. Genomics 7, 15–22 [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25.Pape T., Schneider T. R. (2004) J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 26.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 27.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 28.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 29.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project Number 4 (CCP4) (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 31.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 32.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 33.Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Robertson A. D., Jensen J. H. (2005) Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 35.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacKerell A. D., Bashford D., Bellot M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., et al. (1998) J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen W. L., Chandrasekhar J., Madura J. D., Imprey R. W., Klein M. L. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 38.Darden T. A., York D. M., Pedersen L. G. (1993) J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 39.Ryckaert J., Ciccotti G., Berendsen H. (1977) J. Comput. Phys. 23, 327–341 [Google Scholar]
- 40.Schlick T., Skeel R. D., Brunger A. T., Kale L. V., Hermans J., Schulten K., Board J. A. (1999) J. Comput. Phys. 151, 9–48 [Google Scholar]
- 41.Zhang Y., Feller S. E., Brooks B. R., Pastor R. W. (1995) J. Chem. Phys. 103, 10252–10266 [Google Scholar]
- 42.Ye Y., Godzik A. (2004) Nucleic Acids Res. 32, W582–W585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettit F. K., Bare E., Tsai A., Bowie J. U. (2007) J. Mol. Biol. 369, 863–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnston S. C., Riddle S. M., Cohen R. E., Hill C. P. (1999) EMBO J. 18, 3877–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicastro G., Menon R. P., Masino L., Knowles P. P., McDonald N. Q., Pastore A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10493–10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polgár L. (1974) FEBS Lett. 47, 15–18 [DOI] [PubMed] [Google Scholar]
- 47.Laskowski R. A., Chistyakov V. V., Thornton J. M. (2005) Nucleic Acids Res. 33, D266–D268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.