Abstract
The p53 pathway is disrupted in virtually every human tumor. In ∼50% of human cancers, the p53 gene is mutated, and in the remaining cancers, the pathway is dysregulated by genetic lesions in other genes that modulate the p53 pathway. One common mechanism for inactivation of the p53 pathway in tumors that express wild-type p53 is increased expression of MDM2 or MDMX. MDM2 and MDMX bind p53 and inhibit its function by distinct nonredundant mechanisms. Small molecule inhibitors and small peptides have been developed that bind MDM2 in the p53-binding pocket and displace the p53 protein, leading to p53-mediated cell cycle exit and apoptosis. To date, peptide inhibitors of MDMX have been developed, but no small molecule inhibitors have been reported. We have developed biochemical and cell-based assays for high throughput screening of chemical libraries to identify MDMX inhibitors and identified the first MDMX inhibitor SJ-172550. This compound binds reversibly to MDMX and effectively kills retinoblastoma cells in which the expression of MDMX is amplified. The effect of SJ-172550 is additive when combined with an MDM2 inhibitor. Results from a series of biochemical and structural modeling studies suggest that SJ-172550 binds the p53-binding pocket of MDMX, thereby displacing p53. This lead compound is a useful chemical scaffold for further optimization of MDMX inhibitors that may eventually be used to treat pediatric cancers and various adult tumors that overexpress MDMX or have similar genetic lesions. When combined with selective MDM2 inhibitors, SJ-172550 may also be useful for treating tumors that express wild-type p53.
Keywords: Cancer, Cell/Apoptosis, Cell/Cycle, Diseases/Cancer, Tumor/Suppressor/p53, Tumor/Therapy
Introduction
Tumorigenesis is a multistep process that involves dysregulation of several pathways that are crucial for cell growth and survival (1). The p53 pathway regulates cell survival in response to cellular stress (e.g. DNA damage) or oncogenic stress (e.g. Rb pathway dysregulation) (2, 3) and is suppressed in virtually every human cancer by genetic lesions in the p53 gene or other components of the pathway (4). Approximately half of all cancers express wild-type p53, and considerable research over the past decade has focused on inducing p53-mediated cell death in these tumors (4, 5). Most efforts to date have focused on inhibiting MDM2, a negative regulator of p53 (6–14).
Another key regulator of the p53 pathway is a protein related to MDM2 called MDMX (15–17). MDM2 and MDMX share homology in their p53-binding domains, but MDMX is believed to regulate p53 through distinct mechanisms. Specifically, MDM2 primarily regulates p53 stability and subcellular localization, whereas MDMX may directly regulate p53 transcription (17–21). MDMX is genetically amplified in 19% of breast carcinomas, 19% of colon carcinomas, 18% of lung carcinomas, and a smaller percentage of gliomas (17). One of the best characterized tumors with an MDMX amplification is retinoblastoma. Approximately 65% of human retinoblastomas have increased MDMX copy number, which correlates with increased MDMX mRNA and protein (22). Previous studies have demonstrated that the MDMX amplification suppresses p53-mediated cell death in Rb pathway-deficient retinoblasts (22).
A general consensus is emerging that to efficiently induce a p53 response in tumor cells that express wild-type p53, it may be necessary to inactivate both MDM2 and MDMX (18, 23, 24). To date, no screens to identify small molecule inhibitors of MDMX have been reported, and MDM2 inhibitors probably do not bind as efficiently to MDMX because of structural differences in the p53-binding pockets of the two proteins (25–27). Consistent with this theory, nutlin-3a binds MDMX with at least a 40-fold weaker equilibrium binding constant than for MDM2 (22).
Therefore, to identify small molecules that bind MDMX and prevent its interaction with p53, we developed biochemical and cell-based assays suitable for high throughput screening (HTS)3 of chemical libraries. Using this approach, we have identified the first MDMX inhibitor, SJ-172550, and demonstrated that it can efficiently kill MDMX-amplified retinoblastoma cells. SJ-172550 functions in an additive manner with the MDM2 inhibitor nutlin-3a, thereby confirming the importance of targeting both of these negative regulators of p53 in cancer cells. This validated MDMX inhibitor provides a valuable lead compound and chemical scaffold for further chemical modification to develop a high affinity MDMX inhibitor with good bioavailability, pharmacokinetics, and pharmacodynamics.
EXPERIMENTAL PROCEDURES
Plasmid Constructs and Protein Production
The p53-binding domain of mouse and human MDMX (amino acids 1–185) and human MDM2 (amino acids 1–188) were amplified by PCR and cloned into the pGEX-4T1 plasmid. Recombinant GST fusion proteins were prepared in BL21 (DE3) Escherichia coli cells. The lysates were cleared by spinning at 100,000 × g, and the supernatant was loaded onto a 5-ml GSTrap Fast-Flow column (GE Healthcare). Subsequent purification included a Mono Q column and an S200 gel filtration column. Peak fractions were combined and dialyzed against phosphate-buffered saline (pH 7.6) containing 2 mm phenylmethylsulfonyl fluoride.
Fluorescence Polarization Assays
Fluorescence polarization (FP) assays were conducted in assay buffer containing 10 mm Tris (pH 8.0), 42.5 mm NaCl, and 0.0125% Tween 20. The wild-type p53 peptide (amino acids 15–29) was GSGSSQETFSDLWKLLPEN, and the mutant AAA-p53 peptide was GSGSSQETFADLAKLAPEN. The FP assays were carried out using 2.5 nm FITC peptide (or 15 nm Texas Red) and 1 μm GST-MDMX or GST-MDM2. For MDM2-p53 or MDMX-p53 inhibitor assays, small molecules were preincubated with the recombinant protein for 30 min. The labeled peptide was then added and incubated for 45 min. FP assays were conducted in 384-well black microplates (Corning Glass). The FP FITC assays were analyzed using an EnVision multilabel plate reader with a 480-nm excitation filter, a 535-nm static and polarized filter, and an FP FITC dichroic mirror. The unlabeled competitor peptide and nutlin-3 were used as positive controls, and the alanine-substituted p53 peptide (AAA-p53) was used as a negative control. To minimize the possibility of false-positives caused by endogenous fluorescence from the compounds in the library, we also developed an FP assay with the Texas Red fluorophore. This assay was conducted as described above, except it required a 555-nm excitation filter, a 632-nm static and polarized filter, and a Texas Red FP dichroic mirror.
Chemical Library and High Throughput Screening
The screening library consisted of 295,848 unique compounds from commercial sources (ChemDiv, ChemBridge, and Life Chemicals) arrayed individually at 10 mm in DMSO in 384-well polypropylene plates. The purity of compounds was reported by the vendor as ≥90%. HTS was carried out on a system developed by high resolution engineering with integrated plate incubators (Liconic). Plates were transferred from instrument to instrument by a Staübli T60 robot arm. Assay materials were dispensed in bulk by using Matrix Wellmates (Matrix Technologies). Compound plates were centrifuged in a Vspin plate centrifuge (Velocity11). All compound transfers were accomplished by using a 384-well pin tool with 10-nl hydrophobic surface-coated pins (V & P Scientific). These pins allowed for the delivery of 25 nl to achieve a final compound concentration of 10 μm. The fluorescent signal was measured using an EnVision multilabel plate reader.
RESULTS
Characterization of an MDMX-p53 Binding Assay for High Throughput Screening
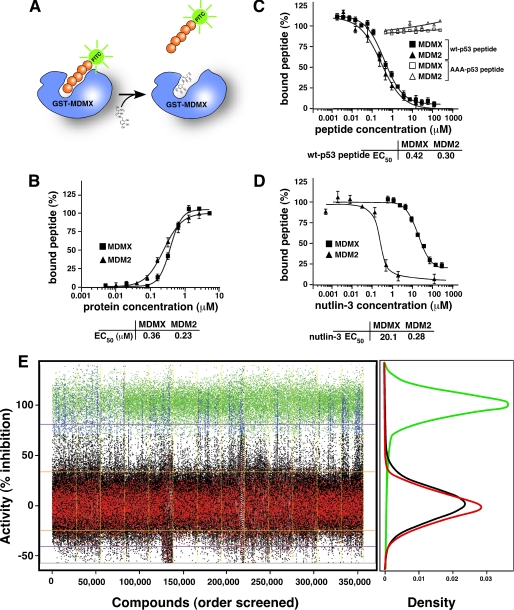
To identify MDMX inhibitors by HTS of chemical libraries, we developed an FP assay (28) to detect the binding of the p53 peptide to GST-MDMX-(1–185) in 384-well plates. This assay is based on the retention of polarization during fluorescence spectroscopy of the p53 peptide conjugated to a fluorophore such as FITC (Fig. 1A). FP is inversely proportional to the rotational diffusion of the fluorophore, and for our experiments, it was measured using an FP spectrometer. The polarization of free p53-FITC peptide (2.5 nm) was 100 mP, and with increasing concentrations of purified GST-MDMX-(1–185) protein (supplemental Fig. 1, A–D), the polarization peaked (100%) at 280 mP (Fig. 1B). Similar data were obtained using GST-MDM2-(1–188) (Fig. 1B and supplemental Fig. 1, A–D). From these curves, the EC50 for p53 peptide-MDMX was 0.36 μm and that for p53 peptide-MDM2 was 0.23 μm (Fig. 1B). Biacore experiments provided similar binding constants for GST-MDMX-(1–185) (Kd = 1.05 μm) and GST-MDM2-(1–188) (Kd = 1.03 μm) (supplemental Fig. 1E). Isothermal titration calorimetry measurements using a purified minimal p53-binding domain of MDMX-(23–111) confirmed these binding constants (supplemental Fig. 1, F–H).
FIGURE 1.
Biochemical assays for high throughput screening to identify MDMX inhibitors. A, schematic of the FP assay used to identify MDMX inhibitors. The protein used for the screen consisted of residues 1–185 of MDMX fused to GST. The peptide (orange) was conjugated to FITC (green) for the primary screen and to Texas Red for secondary assays. B, plot of the percentage of bound p53-FITC peptide (at a fixed concentration) with increasing concentrations of MDMX (squares) or MDM2 (triangles). C, plot of the percentage of p53-FITC peptide associated with indicated proteins in the presence of increasing concentrations of unlabeled wild-type p53 peptide or unlabeled alanine-substituted p53 peptide (AAA-p53) as a negative control. D, plot of the percentage of p53-FITC peptide associated with the indicated proteins in the presence of increasing concentrations of nutlin-3a. B–D, each data point is the mean ± S.D. of triplicate experiments. E, scatterplot of HTS of a chemical library for MdmX inhibitors. The blue data points indicate compounds that were selected for further analysis, and the black data points are compounds that did not exhibit activity in the HTS. DMSO was used as a negative control (red), and the unlabeled p53 peptide (green) was used as a positive control. The density plot illustrates the clear separation of the positive and negative control samples across the entire screen. Each day of screening is separated by a yellow line.
To test the specificity of our FP assay for MDMX/MDM2 binding to p53, we performed a competition experiment with unlabeled p53 peptide and a version of the AAA-p53 peptide that is defective for binding to MDM2/MDMX (29). The protein concentration in this assay was 1 μm, and the p53-FITC peptide concentration was 2.5 nm. The EC50 values from the competition experiments were 0.42 and 0.30 μm for GST-MDMX-(1–185) and GST-MDM2-(1–188), respectively (Fig. 1C), whereas AAA-p53 showed no evidence of competition at any concentration tested (0.5–200 μm) (Fig. 1C).
To test whether our FP assay was suitable for identifying small molecule inhibitors of MDMX-p53 binding, we performed a dose-response experiment using nutlin-3a at concentrations ranging from 0.5 nm to 300 μm (Fig. 1D). Nutlin-3a was originally identified as an MDM2 inhibitor (6), but it also binds to MDMX, albeit with a much weaker Kd value (22). The protein concentration was held constant at 1 μm, and the peptide concentration was 2.5 nm for each concentration of nutlin-3a tested. The EC50 value for binding of nutlin-3a to MDM2 was 0.28 μm and that to MDMX was 20.1 μm (Fig. 1D).
High Throughput Screening of a Chemical Library to Identify Novel MDMX Inhibitors
To identify novel MDMX inhibitors, we performed an HTS of the St. Jude chemical library (295,848 unique compounds) by using the GST-MDMX-(1–185)/p53-FITC peptide FP assay (Fig. 1E). A total of 356,352 wells (compounds and controls) were screened over the course of 13 days by using 928 plates (chemical structures and screening data are available for download at the following url: www.stjuderesearch.org/guy/data/mdmx). We selected the mouse MDMX (MdmX) protein for the HTS, because expression of the recombinant protein in E. coli was more efficient than human MDMX. Compounds were screened at a final concentration of 10 μm. The scatterplot of activities demonstrates clear separation between the positive and negative controls (Fig. 1E). The average z-prime for the assays was positive but low (<0.4) because of variance in the control distributions (supplemental Fig. 2D). However, the reference peptide EC50 value was stable and within experiment error, and no major plate artifacts were detected (supplemental Fig. 2).
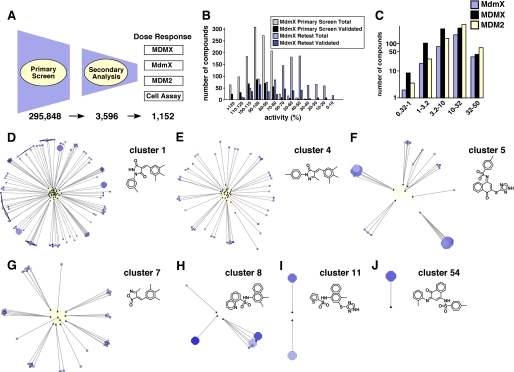
To reduce the probability of selecting false-positive compounds from the primary screen, we performed a receiver operating characteristic analysis using different activity thresholds for compound selection. This analysis led us to select the 70% activity threshold to obtain 3,596 compounds for subsequent validation. To eliminate compounds with intrinsic fluorescence emission spectra that overlapped with FITC, we performed a secondary FP assay using a Texas Red-conjugated p53 peptide. This analysis was performed on all 3,596 compounds that met the 70% activity cutoff (10 μm) in triplicate. The top 1,000 compounds were then selected from the Texas Red FP assay based on their percentage inhibition. An additional 152 structural analogs of these 1000 compounds were included (total 1,152) to provide sufficient coverage of chemical scaffolds to begin to establish structure-activity relationships for our candidate MDMX inhibitors (Fig. 2A).
FIGURE 2.
Identification of diverse chemotypes with candidate MDMX inhibitors. A, work flow schematic of the primary HTS, secondary analysis, and dose-response and cell-based assays. Numbers of compounds that were selected for each round of analysis are indicated. B, histogram of the distribution of MDMX activities for the 1,152 compounds in this study. Gray bars represent the number of compounds within the indicated range of activity from the FITC-FP primary screen; black bars represent the number of those compounds that were confirmed as true positives via dose response. Similarly, the light and dark blue bars represent the distributions from the Texas Red FP retest screen. The Texas Red FP assay is better than the FITC FP assay at discriminating true-positives from false-positives. C, distribution of EC50 values for MdmX, MDMX, and MDM2 for all 1,152 compounds calculated from the dose response in triplicate using the Texas Red FP assay. D–J, visual representation of seven chemotype clusters. The black triangles are Murcko scaffolds, and screened compounds are represented as nodes that are connected to their parent scaffold by gray lines. Nodes are colored by potency against MDMX (blue = high and gray = low) and sized according to selective cytotoxicity for retinoblastoma cells versus BJ cells (large = selective for retinoblastoma). Large dark blue circles have low binding constants for MDMX and selective cytotoxicity for retinoblastoma cells. Eleven compounds were selected from these chemotype clusters for further characterization.
Analysis of Active Compounds from MDMX Inhibitor High Throughput Screening
To further characterize the 1,152 active compounds, we measured the binding constant of each compound to MdmX (mouse), MDMX (human), and MDM2 (human) by performing a dose-response assay in triplicate (Fig. 2, A–C). Compounds that showed activity ≥70% in the primary FITC MdmX assay displayed a wider range of activities in the Texas Red retest assay (Fig. 2B, compare gray bars to light blue bars). Moreover, the median retest activity was a better predictor of validated compounds (compounds with a well behaved dose response) than the primary screen single point activity. Therefore, the to further characterize the 1,152 active compounds, we measured the binding constant of each compound to MdmX (mouse), MDMX (human), and MDM2 (human) by performing a dose-response assay in triplicate (Fig. 2, A–C). FP assay demonstrated better discriminatory power than the FITC FP assay.
To complement these biochemical studies, we carried out a cell-based assay to further characterize the activity of the 1,152 compounds on retinoblastoma cells that have an MDMX amplification (Weri1) or a cell line that is p53-deficient (SJmRbl-8) (22). CellTiter-Glo (Promega) assay was used to measure intracellular ATP levels as an indicator of viability. SJmRbl-8 cells and Weri1 cells showed a linear relationship between luminescence and cell number (supplemental Fig. 3, A and B). As a positive control for cytotoxicity, we used vincristine, which is a microtubule inhibitor that disrupts chromosome segregation during mitosis and kills both cell lines with similar LC50 values (supplemental Fig. 3C). As a positive control for p53-selective cytotoxicity, we used nutlin-3a, which selectively kills Weri1 cells with an MDMX amplification and is less cytotoxic against p53-deficient SJmRbl-8 cells (supplemental Fig. 3D). We also used BJ cells, an hTERT-immortalized human foreskin fibroblast cell line, as an additional control to estimate the general cytotoxicity of compounds in our lead compound collection. We carried out a dose-response cytotoxicity assay for each cell line on all 1,152 compounds in triplicate. Overall, the assay performed well in this HTS format, and we identified a subset of compounds from our active compounds set with significant selective cytotoxicity against the retinoblastoma cells (supplemental Fig. 3, E and F).
To integrate and visualize the dose-response data from the biochemical assays of MDMX and MDM2 and the cell-based data and chemical scaffolds represented in the 1,152 compounds, we overlaid biochemical and cell-based data onto a network graph constructed to represent related families of chemotypes within the lead compound set (supplemental Fig. 4). This approach allowed us to quickly identify scaffolds with the desired profiles showing strong binding to MDMX and cytotoxicity against an MDMX-amplified retinoblastoma cell line. On the basis of these data, we selected 11 representative compounds from clusters 1, 4, 5, 7, 8, 11, and 54 for further analysis (Fig. 2, D–J, supplemental Fig. 4, and supplemental Table 1). Several clusters that looked promising based on the aforementioned criteria were eliminated from further analysis because of the presence of potentially problematic chemical functionalities such as unsubstituted quinones, maleimides, Michael acceptors, and thioesters; potential redox activity; or other medicinal chemistry liabilities (supplemental Fig. 4).
SJ-134433 and SJ-044557 Covalently Modify the MDMX Protein
Among the 11 compounds selected for further analysis, SJ-134433 and SJ-044557 had excellent profiles (supplemental Fig. 6) with good binding constants for MDMX, some selectivity for MDMX over MDM2, and efficient killing of retinoblastoma cells with selectivity for the Weri1 line (supplemental Table 1). To begin characterizing these compounds, we performed an isothermal denaturation assay and a redox assay on SJ-134433, SJ-044557, and the other nine compounds (supplemental Fig. 5 and supplemental Table 1). The rationale underlying the isothermal denaturation assay is that binding of a small molecule to the p53-binding pocket of MDMX may stabilize the protein, and in the presence of the SYPRO orange hydrophobic dye, the temperature for denaturation and dye binding would shift (30, 31). Indeed, GST-MDMX-(1–185) showed a melting point shift in the presence of p53 peptide from 46.9 ± 0.6 °C for native GST-MDMX-(1–185) protein to 50.8 ± 0.6 °C for GST-MDMX-(1–185) protein bound to the p53 peptide (supplemental Fig. 5, A and B). However, neither SJ-044557 nor SJ-134433 exhibited a thermal shift; in fact, they appeared to destabilize the protein (supplemental Fig. 5).
To explore the possibility that these two compounds exhibited redox activity, we performed an assay to detect compounds capable of reducing resazurin to resorufin, a redox couple relevant to oxygen tension in mammalian cells (32). DMSO was used as a negative control, and a 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione-containing compound was used as the positive control. SJ-044557 and SJ-134433 showed some redox activity (supplemental Table 1). Next, we explored the stability of the compounds in our FP buffer to determine whether they were unstable and if any of the degradation products were reactive species that covalently modified the MDMX protein and blocked p53 binding. Both compounds were unstable after 24 h, and SJ-134433 degraded after 2 h in FP buffer (supplemental Fig. 6, B and E). To directly determine whether the purified MDMX-(23–111) protein was covalently modified by either compound, we performed high resolution mass spectrometry following incubation with SJ-044557 or SJ-134433. Both showed a shift in their mass consistent with covalent modifications (supplemental Fig. 6, H and I). Although these inhibitors might prove useful as tools for interrogating MDMX function, they are not suitable for further development and were thus abandoned.
SJ-172550 Is Stable and Reversibly Binds MDMX to Inhibit p53 Binding
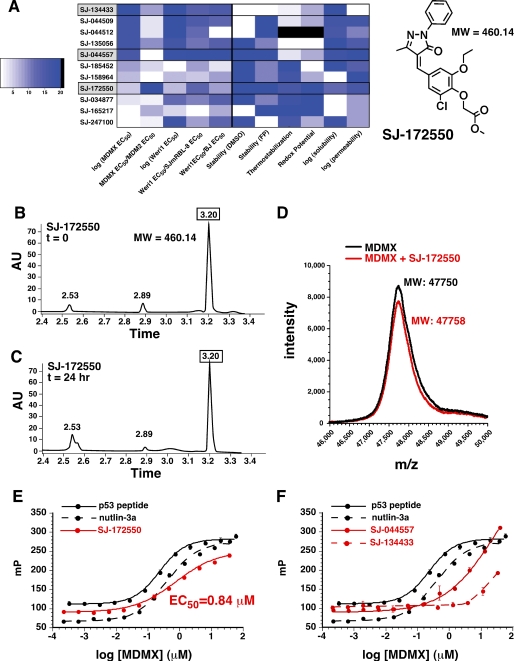
A more detailed analysis of the 11 selected compounds revealed that SJ-172550 had an excellent chemical profile, with respect to chemical stability, thermal stability, redox potential, and solubility. Unlike SJ-134433 and SJ-044557, SJ-172550 was stable in solution (Fig. 3, B and C) and did not modify the MDMX protein by covalent binding in our FP assay buffer (Fig. 3D). Moreover, the compound exhibited strong thermal stabilization (supplemental Fig. 5) and had undetectable redox activity (Fig. 3A and supplemental Table 1). To confirm that SJ-172550 binds MDMX reversibly, we incubated the compound with MDMX for 2 h in FP buffer and then removed the compound by dialysis. As a positive control, we used the p53 peptide and nutlin-3a. Our results showed that SJ-172550 bound MDMX reversibly (Fig. 3E), unlike SJ-044557 or SJ-172550 (Fig. 3F).
FIGURE 3.
SJ-172550 reversibly binds MDMX. A, heat map of normalized activity for the 11 compounds selected for follow-up characterization. Dark blue is more favorable for each measurement, and the compounds are listed in order of binding constant for MDMX from best (top) to worst (bottom). These data suggested that with subsequent biochemical analyses, SJ-134433 and SJ-044557 were less suitable for follow up, and SJ-172550 was preferred. High performance liquid chromatography of compound SJ-172550 showed that it is stable for 24 h in FP buffer (B and C), and MALDI mass spectrometry showed that it does not covalently modify the MDMX-(23–111) protein (D). The MDMX protein was incubated with SJ-172550 for 2 h in FP buffer and then dialyzed away in a large excess of dialysis buffer. The ability of the dialyzed protein to bind the p53-FITC peptide was then measured using the FP assay (red line). E, EC50 value for binding to p53 peptide after removal of SJ-172550 was indistinguishable from that of the untreated protein. A similar experiment with nutlin-3a also demonstrated that it binds reversibly to MDMX. F, SJ-044557 and SJ-134433, which covalently modified MDMX, did not reversibly bind MDMX (red lines). A.U., absorbance units.
SJ-172550 Inhibits MDMX-p53 Binding in Cultured Cells
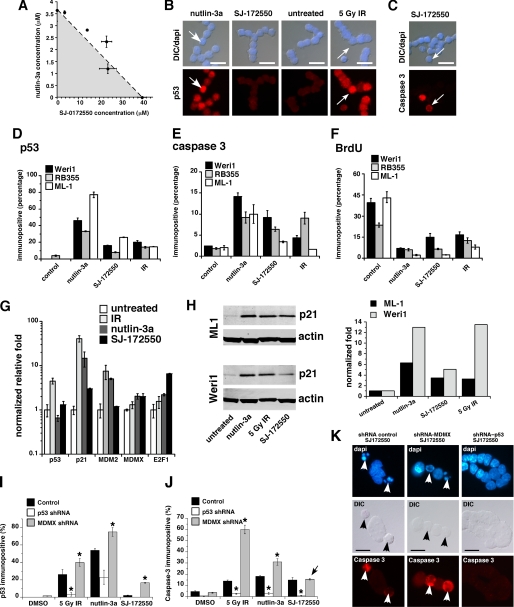
To test whether SJ-172550 and nutlin-3a have additive or synergistic effects on retinoblastoma cells, we performed an isobologram experiment with these two compounds. The data suggested that nutlin-3a and SJ-172550 act in an additive manner to kill MDMX-amplified human retinoblastoma cells (Fig. 4A). Next, we exposed Weri1 and RB355 retinoblastoma cells and ML-1 leukemia cells (with wild-type p53) to SJ-172550 (20 μm) for 20 h and analyzed the p53 and activated caspase-3 levels by immunofluorescence to study the mechanism of cell death. As positive controls, we exposed Weri1 cells to nutlin-3a (5 μm) for 20 h or 5 gray ionizing radiation (IR). DMSO was used as the negative control. As expected, the Weri1 cells exposed to nutlin-3a or IR showed a robust accumulation of p53 (Fig. 4, B and D). In contrast, the cells exposed to SJ-172550 did not exhibit the same level of accumulation of p53 (Fig. 4, B and D). This is consistent with the role of MDMX in regulating transcriptional activation of p53-responsive promoters but not p53 protein levels. Apoptosis was robustly induced after exposure to SJ-172550 (Fig. 4, C and E), and cells exited the cell cycle (Fig. 4F). Real time RT-PCR and immunoblotting analysis of these cells revealed that there was also an induction of p53 target genes, but it was not as robust as that observed with nutlin-3a or IR (Fig. 4, G and H). More importantly, the cell death mediated by SJ-172550 was p53-dependent (Fig. 4, I–K). In addition, HCT116 cells were sensitive to SJ-172550, but p53-deficient HCT-116 cells were not (supplemental Fig. 8). SJSA-X cells expressing high levels of MDMX were also sensitive to SJ-172550 (supplemental Fig. 8).
FIGURE 4.
SJ-172550 disrupts the MDMX-p53 interaction in cells maintained in culture. A, isobologram shows the additive inhibition of Weri1 cell growth when the combination of SJ-172550 and nutlin-3a was used to treat cells. Multiple ratios were tested, and the plot shows that the LC50 value (dashed line) was additive but not synergistic (shaded area) or adverse (area above the dashed line). The error bars represent standard deviation from two independent experiments. B and C, immunostaining of compound-treated Weri1 cells. The top panels are merged images of differential interference contrast (DIC) and 4′,6-diamidino-2-phenylindole (DAPI) staining, and the lower panels show p53 levels (B) or caspase-3 activation (C). Nutlin-3a and IR were used as positive controls, and DMSO was used as a negative control. D–F, quantification of the percentage of immunopositive cells shown in B and C and bromodeoxyuridine (BrdU). G, real time PCR quantification of p53 target genes p21, MDM2, and E2F1 activated by drug or IR treatments. MDMX levels were also tested. H, immunoblot and quantification for p21 protein levels activated by drug or IR treatment. Gy, gray. I and J, histogram of the proportion of p53 or activated caspase-3 immunopositive cells following treatment with 5 gray IR, nutlin-3a or SJ-172550. In parallel samples, p53 was knocked down using a p53 shRNA, and MDMX was knocked down using an MDMX shRNA (22), or the samples were treated with a control (scrambled) shRNA. Each bar represents the mean ± S.D. scoring in triplicate of 100 cells for each condition and each shRNA. K, representative images of Weri1 retinoblastoma cells following treatment with SJ-172550 when they lacked p53 (p53 shRNA) or MDMX (MDMX shRNA) as compared with a control shRNA. Arrows indicate activated caspase-3 immunopositive cells in the red (Cy3) channel. shRNA, small hairpin RNA. Scale bars, 10 μm.
To determine whether SJ-172550 disrupts the MDMX-p53 interaction of cells in culture, we performed co-immunoprecipitation experiments in the presence of the compound. Reciprocal co-immunoprecipitation experiments with antibodies against MDMX and p53 in C33A (human cervical carcinoma) cells demonstrated partial inhibition of MDMX-p53 binding in cells (supplemental Fig. 7). Similar data were obtained using human embryonic retina cells and Weri1 retinoblastoma cells (supplemental Fig. 7). Together, these results suggest that the MDMX-p53 interaction was at least partially inhibited by SJ-172550, despite its relatively low cell permeability (see supplemental Table 1).
Computational Model of SJ-172550 Binding to MDMX
X-ray crystallographic studies have provided high resolution structures of MDM2 and MDMX bound to p53 (Fig. 5 and supplemental Fig. 9) (26, 27, 33, 34). We overlaid these two structures and determined that the Cα root mean square deviation was 3.9 Å, suggesting that although the overall fold was well conserved, the tertiary structures of MDM2 or MDMX bound to p53 are significantly different. This can be more readily visualized using a space-filling representation of the overlaid structures. In particular, the structure of the p53-binding pocket of MDMX was smaller than that of MDM2 (supplemental Fig. 9B). When nutlin-3a was bound to MDM2, the tertiary structure of the pocket underwent a small change (Cα root mean square deviation = 0.82 Å) (supplemental Fig. 9C). When the structure of MDM2 bound to nutlin-3a was overlaid with that of MDMX bound to p53, the smaller binding pocket of MDMX may explain the lower affinity binding of nutlin-3a to MDMX, as compared with that of MDM2 (supplemental Fig. 9, D and E).
FIGURE 5.
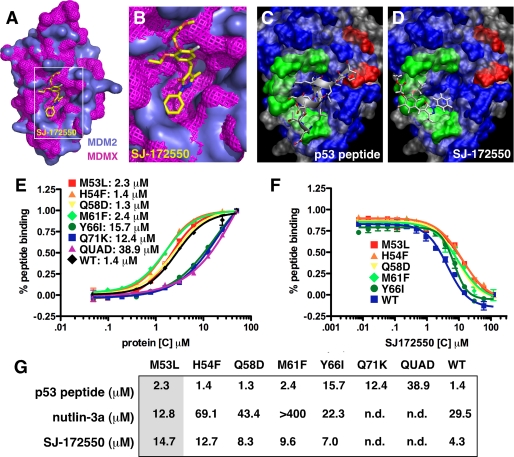
A and B, space-filling model of the overlaid MDM2-nutlin-3a (teal) with MDMX-p53 (pink) showing SJ-172550 bound to the p53-binding pocket of MDM2/MDMX in the secondary docking pose. C, position of p53 peptide based on the MDM2/peptide crystal structure. D, primary docking pose of SJ-172550 to MDMX superimposed on the crystal structure of p53 peptide bound to MDM2. C and D, the gray is the solvent-excluded surface of MDM2 (Protein Data Bank code 2Z5T), and the blue is the surface for MDMX. Green residues represent single mutations, and residues shown in red formed a quadruple mutant. Mutations designed to displace SJ-172550 based on the models of the most energetically favorable docking poses (Fig. 5, B and D) are Q58D, M61F, Y66I, and Q71D. A second series of residues were changed to make the MDMX-binding pocket more like MDM2 to determine whether SJ-172550 was binding in the p53-binding pocket. These mutations include M53L, H54F, and a quadruple mutant (QUAD) with P95H/S96R/P97K/R103Y. From top to bottom, the green residues are Met-53, His-54, Gln-58, Met-61, Tyr-66, and Gln-71 with the red residues being Arg-103, Pro-97, Ser-96, and Pro-95. E, plot of direct binding of each MDMX protein to Texas Red-labeled p53 peptide exactly as described for the HTS. Each point is the mean ± S.D. of triplicate assays. The EC50 values for each protein are indicated. F, competition experiments with each MDMX mutant and increasing concentrations of SJ-172550. Each data point is the mean ± S.D. of triplicate assays. The proteins that did not show direct binding of p53 peptide could not be analyzed in this competition experiment. G, summary of the EC50 values for direct binding of p53 peptide and competition by nutlin-3a and SJ-172550. The shaded column (M53L) is one of the mutants that was predicted to make MDMX more like MDM2 and increase nutlin-3a binding while inhibiting SJ-172550 binding without affecting peptide binding. n.d., not determined; WT, wild type.
We used both AutoDock 4.2 (35) and the fastdock algorithm in Scigress Explorer version 7.7 to model the binding of nutlin-3a to MDM2 and found excellent agreement between the computational model and the native conformation reported in previous co-crystallization studies (supplemental Fig. 9F) (6). Using the same computational approach, the binding of SJ-172550 to the p53-binding pocket of MDMX was modeled, yielding two important structures (Fig. 5, B and D, and supplemental Fig. 9G). Together, these results provide a plausible mechanism of action for SJ-172550. SJ-172550 may occlude the p53-binding pocket of MDMX, thereby inhibiting p53 binding. To test this directly, we generated a series of seven mutants in the MDMX-binding pocket (Fig. 5, C and D, supplemental Fig. 9, F and G, and supplemental material) and purified the protein for binding studies (Fig. 5, E–G, supplemental Fig. 9, H and I and supplemental information). Some of the mutants (e.g. H54F) were predicted to displace SJ-172550 without affecting peptide binding, and other mutants (e.g. M53L) were predicted to make the binding pocket of MDMX more like MDM2 and thereby reduce binding of SJ-172550 and increase binding of nutlin-3a without affecting peptide binding. These data provide additional validation for our proposed mechanism of SJ-172550 inhibition of the MDMX-p53 interaction.
DISCUSSION
Although several small molecule inhibitors of MDM2 have been identified (6, 14), this is the first report to identify a small molecule inhibitor of MDMX. There is growing evidence that MDM2 and MDMX inhibit p53 through distinct mechanisms and that simultaneous inhibition of these two proteins in tumor cells that express wild-type p53 may be more effective at killing the cells than the inhibition of MDM2 alone. Our results complement important previous studies on high affinity peptide inhibitors of MDMX and MDM2 (9, 34). MDMX inhibitors alone (peptide or small molecule) may be useful for treating tumors such as retinoblastoma that show increased MDMX expression (22). In addition, they may be effective when combined with MDM2 inhibitors to induce a robust p53 response in cancer cells that express wild-type p53 (36).
We have developed and optimized a biochemical assay for HTS of MDMX inhibitors. We screened a diverse chemical library and identified compound SJ-172550 as the first small molecule inhibitor of MDMX with a low micromolar binding constant. SJ-172550 reduced p53 binding in vitro and had little or no redox activity. It did not covalently modify MDMX but rather thermostabilized the protein and reversibly bound it, which was consistent with our modeling of SJ-172550 binding to the p53-binding pocket of MDMX. When retinoblastoma cells expressing wild-type p53 and high levels of MDMX were exposed to SJ-172550 in vitro, they showed evidence of p53-mediated cytotoxicity. More importantly, the death was p53-dependent because an shRNA to p53 prevented SJ-172550-mediated cell death (Fig. 4, J and K). Although these data point to a p53-dependent cell death mechanism, they do not rule out the possibility of off-target binding of SJ-172550. Indeed, many lead compounds and drugs show off-target effects.
In combination with the MDM2 inhibitor nutlin-3a, SJ-172550 showed additive cytotoxicity in cells that expressed wild-type p53. Thus, we propose that SJ-172550 binds the p53-binding pocket of MDMX, thereby freeing p53 to induce apoptosis. This compound represents a bona fide lead molecule and, together with the other promising lead scaffolds from this study, can now be used for further medicinal chemical analyses, including optimization of affinity, specificity, and cell permeability and assessment of pharmacokinetics and toxicity.
It has been shown that MDM2 and MDMX can form a heterodimer through their Ring domains, and this may regulate MDM2-mediated degradation of p53 (37–39). We found that the overall p53 protein levels are not dramatically altered following exposure of cultured cells to SJ-172550. The identification of the first small molecule inhibitor will allow researchers to probe this mechanism further by comparing the effect of MDMX protein loss to inhibition of MDMX-p53 binding on p53 stability.
One of our key findings from the analysis of biochemical and cell-based assay data was that the compounds that had the best binding constants for MDMX were not necessarily the ones that were most suitable for follow-up. For example, SJ-134433 had a very good binding constant for MDMX, but further analysis showed it was unstable in FP buffer, did not thermostabilize MDMX, had significant redox activity, and covalently modified the protein. These data clearly emphasize the importance of performing comprehensive characterization of active compounds to rule out nonspecific mechanisms of action that make compounds unsuitable for further development. They also emphasize the need to select candidates for further work based on a balance of chemical and biological properties, rather than purely on potency or biochemical mechanism of action.
We have not yet fully validated the mechanism of action of SJ-172550, but it seems probable based upon our modeling and data that it binds the p53-binding pocket of MDMX and frees p53 to activate its target genes leading to cell cycle exit and apoptosis. Consistent with this model, we observed moderate p53 pathway activation in MDMX-amplified retinoblastoma cells and partial disruption of the MDMX-p53 interaction in cell lysates from retinoblastoma cells and other cell lines. We do not believe that the mechanism of action is through p53 protein stabilization based on immunoblotting and single cell immunostaining. A structural alignment of the binding pockets of MDMX and MDM2 was produced using the backbone atoms and provides some clues about where SJ-172550 may bind MDMX to induce p53 pathway activation. Mutations generated in the p53-binding pocket of MDMX provided additional support for this proposed mechanism. Additional x-ray crystallography and other structural studies are required to definitively show that SJ-172550 binds to the p53-binding pocket of MDMX. Nonetheless, this is the first small molecule MDMX inhibitor that has been identified with a low micromolar binding constant. It is important to note that SJ-172550 also binds MDM2, although less effectively. Our compound is clearly less effective against MDM2 than nutlin-3a (6) or other MDM2 inhibitors (14), and it is difficult to predict whether further refinement of MDMX binding will similarly improve MDM2 binding or lead to a high affinity MDMX-selective inhibitor.
Supplementary Material
Acknowledgments
We thank Taosheng Chen, Fu-Yue Zeng, Wenwei Lin, Jimmy Cui, and David Bouck for assistance with HTS at the St. Jude High Throughput Screening Center. We thank Aaron Kosinski and David Miller for assistance with protein preparation. We thank Erin H. Seeley, Jamie L. Allen, and Richard M. Caprioli (all of the Mass Spectrometry Research Center, Vanderbilt Medical Center, Nashville, TN) for assistance with high resolution MALDI mass spectrometry analyses. We also thank Brett Waddell for assistance with Biacore analysis and Angie McArthur for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health grants from NCI, Cancer Center Support. This work was also supported by Howard Hughes Medical Institute, American Cancer Society, Research to Prevent Blindness, Pearle Vision Foundation, International Retinal Research Foundation, Pew Charitable Trust, and American Lebanese Syrian Associated Charities (to M. A. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Figs. 1–9, Table 1, and additional references.
- HTS
- high throughput screening
- FP
- fluorescence polarization
- FITC
- fluorescein isothiocyanate
- MALDI
- matrix-assisted laser desorption ionization
- IR
- ionizing radiation
- GST
- glutathione S-transferase
- shRNA
- small hairpin RNA.
REFERENCES
- 1.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 2.Dang J., Kuo M. L., Eischen C. M., Stepanova L., Sherr C. J., Roussel M. F. (2002) Cancer Res. 62, 1222–1230 [PubMed] [Google Scholar]
- 3.Vogelstein B., Lane D., Levine A. J. (2000) Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 4.Vousden K. H., Lu X. (2002) Nat. Rev. Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 5.Levine E. M., Passini M., Hitchcock P. F., Glasgow E., Schechter N. (1997) J. Comp. Neurol. 387, 439–448 [PubMed] [Google Scholar]
- 6.Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 7.Marine J. E. (2006) Heart Rhythm 3, 342–344 [DOI] [PubMed] [Google Scholar]
- 8.Anderson J. J., Challen C., Atkins H., Suaeyun R., Crosier S., Lunec J. (2007) Int. J. Oncol. 31, 545–555 [PubMed] [Google Scholar]
- 9.Hu B., Gilkes D. M., Chen J. (2007) Cancer Res. 67, 8810–8817 [DOI] [PubMed] [Google Scholar]
- 10.Udayakumar T. S., Hachem P., Ahmed M. M., Agrawal S., Pollack A. (2008) Mol. Cancer Res. 6, 1742–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tovar C., Rosinski J., Filipovic Z., Higgins B., Kolinsky K., Hilton H., Zhao X., Vu B. T., Qing W., Packman K., Myklebost O., Heimbrook D. C., Vassilev L. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1888–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coll-Mulet L., Iglesias-Serret D., Santidrián A. F., Cosialls A. M., de Frias M., Castaño E., Campàs C., Barragán M., de Sevilla A. F., Domingo A., Vassilev L. T., Pons G., Gil J. (2006) Blood 107, 4109–4114 [DOI] [PubMed] [Google Scholar]
- 13.Kojima K., Konopleva M., Samudio I. J., Shikami M., Cabreira-Hansen M., McQueen T., Ruvolo V., Tsao T., Zeng Z., Vassilev L. T., Andreeff M. (2005) Blood 106, 3150–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shangary S., Qin D., McEachern D., Liu M., Miller R. S., Qiu S., Nikolovska-Coleska Z., Ding K., Wang G., Chen J., Bernard D., Zhang J., Lu Y., Gu Q., Shah R. B., Pienta K. J., Ling X., Kang S., Guo M., Sun Y., Yang D., Wang S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3933–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel F., Schulz J., Böhnke A., Blümke K., Kappler M., Bache M., Schmidt H., Würl P., Taubert H., Hauptmann S. (2005) Int. J. Cancer 117, 469–475 [DOI] [PubMed] [Google Scholar]
- 16.Shvarts A., Bazuine M., Dekker P., Ramos Y. F., Steegenga W. T., Merckx G., van Ham R. C., van der Houven van Oordt W., van der Eb A. J., Jochemsen A. G. (1997) Genomics 43, 34–42 [DOI] [PubMed] [Google Scholar]
- 17.Danovi D., Meulmeester E., Pasini D., Migliorini D., Capra M., Frenk R., de Graaf P., Francoz S., Gasparini P., Gobbi A., Helin K., Pelicci P. G., Jochemsen A. G., Marine J. C. (2004) Mol. Cell. Biol. 24, 5835–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toledo F., Krummel K. A., Lee C. J., Liu C. W., Rodewald L. W., Tang M., Wahl G. M. (2006) Cancer Cell 9, 273–285 [DOI] [PubMed] [Google Scholar]
- 19.Marine J. C., Dyer M. A., Jochemsen A. G. (2007) J. Cell Sci. 120, 371–378 [DOI] [PubMed] [Google Scholar]
- 20.Marine J. C., Jochemsen A. G. (2004) Cell Cycle 3, 900–904 [PubMed] [Google Scholar]
- 21.Migliorini D., Lazzerini Denchi E., Danovi D., Jochemsen A., Capillo M., Gobbi A., Helin K., Pelicci P. G., Marine J. C. (2002) Mol. Cell. Biol. 22, 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurie N. A., Donovan S. L., Shih C. S., Zhang J., Mills N., Fuller C., Teunisse A., Lam S., Ramos Y., Mohan A., Johnson D., Wilson M., Rodriguez-Galindo C., Quarto M., Francoz S., Mendrysa S. M., Guy R. K., Marine J. C., Jochemsen A. G., Dyer M. A. (2006) Nature 444, 61–66 [DOI] [PubMed] [Google Scholar]
- 23.Hu B., Gilkes D. M., Farooqi B., Sebti S. M., Chen J. (2006) J. Biol. Chem. 281, 33030–33035 [DOI] [PubMed] [Google Scholar]
- 24.Gu J., Kawai H., Nie L., Kitao H., Wiederschain D., Jochemsen A. G., Parant J., Lozano G., Yuan Z. M. (2002) J. Biol. Chem. 277, 19251–19254 [DOI] [PubMed] [Google Scholar]
- 25.Böttger V., Böttger A., Garcia-Echeverria C., Ramos Y. F., van der Eb A. J., Jochemsen A. G., Lane D. P. (1999) Oncogene 18, 189–199 [DOI] [PubMed] [Google Scholar]
- 26.Popowicz G. M., Czarna A., Rothweiler U., Szwagierczak A., Krajewski M., Weber L., Holak T. A. (2007) Cell Cycle 6, 2386–2392 [DOI] [PubMed] [Google Scholar]
- 27.Popowicz G. M., Czarna A., Holak T. A. (2008) Cell Cycle 7, 2441–2443 [DOI] [PubMed] [Google Scholar]
- 28.Stricher F., Martin L., Barthe P., Pogenberg V., Mechulam A., Menez A., Roumestand C., Veas F., Royer C., Vita C. (2005) Biochem. J. 390, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu F., Chi S. W., Kim D. H., Han K. H., Kuntz I. D., Guy R. K. (2006) J. Comb. Chem. 8, 315–325 [DOI] [PubMed] [Google Scholar]
- 30.Senisterra G. A., Finerty P. J., Jr. (2009) Mol. Biosyst. 5, 217–223 [DOI] [PubMed] [Google Scholar]
- 31.Senisterra G. A., Soo Hong B., Park H. W., Vedadi M. (2008) J. Biomol. Screen. 13, 337–342 [DOI] [PubMed] [Google Scholar]
- 32.Lor L. A., Schneck J., McNulty D. E., Diaz E., Brandt M., Thrall S. H., Schwartz B. (2007) J. Biomol. Screen. 12, 881–890 [DOI] [PubMed] [Google Scholar]
- 33.Kallen J., Goepfert A., Blechschmidt A., Izaac A., Geiser M., Tavares G., Ramage P., Furet P., Masuya K., Lisztwan J. (2009) J. Biol. Chem. 284, 8812–8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pazgier M., Liu M., Zou G., Yuan W., Li C., Li C., Li J., Monbo J., Zella D., Tarasov S. G., Lu W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4665–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. (2009) J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurie N. A., Shih C. S., Schin-Shih C., Dyer M. A. (2007) Curr. Cancer Drug Targets 7, 689–695 [DOI] [PubMed] [Google Scholar]
- 37.Stad R., Little N. A., Xirodimas D. P., Frenk R., van der Eb A. J., Lane D. P., Saville M. K., Jochemsen A. G. (2001) EMBO Rep. 2, 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Cell Death Differ. 15, 841–848 [DOI] [PubMed] [Google Scholar]
- 39.Jackson M. W., Berberich S. J. (2000) Mol. Cell. Biol. 20, 1001–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.