Abstract
Emerging evidence indicates a link between inflammation and cancer metastasis, but the molecular mechanism(s) remains unclear. Uteroglobin (UG), a potent anti-inflammatory protein, is constitutively expressed in the lungs of virtually all mammals. UG-knock-out (UG-KO) mice, which are susceptible to pulmonary inflammation, and B16F10 melanoma cells, which preferentially metastasize to the lungs, provide the components of a model system to determine how inflammation and metastasis are linked. We report here that B16F10 cells, injected into the tail vein of UG-KO mice, form markedly elevated numbers of tumor colonies in the lungs compared with their wild type littermates. Remarkably, UG-KO mouse lungs overexpress two calcium-binding proteins, S100A8 and S100A9, whereas B16F10 cells express the receptor for advanced glycation end products (RAGE), which is a known receptor for these proteins. Moreover, S100A8 and S100A9 are potent chemoattractants for RAGE-expressing B16F10 cells, and pretreatment of these cells with a blocking antibody to RAGE suppressed migration and invasion. Interestingly, in UG-KO mice S100A8/S100A9 concentrations in blood are lowest in tail vein and highest in the lungs, which most likely guide B16F10 cells to migrate to the lungs. Further, B16F10 cells treated with S100A8 or S100A9 overexpress matrix metalloproteinases, which are known to promote tumor invasion. Most notably, the metastasized B16F10 cells in UG-KO mouse lungs express MMP-2, MMP-9, and MMP-14 as well as furin, a pro-protein convertase that activates MMPs. Taken together, our results suggest that a lack of an anti-inflammatory protein leads to increased pulmonary colonization of melanoma cells and identify RAGE as a potential anti-metastatic drug target.
Keywords: Cell Migration, Chemokines, Inflammation, Lung, Tumor Metastases, CC10, Tumor Colonization, Uteroglobin
Introduction
It is estimated that more than 90% of human cancer deaths result from metastasis (1–8), a complex process in which the cells from a primary tumor successfully invade and colonize a distant organ (6–9). It has long been suspected that a functional relationship exists between inflammation and cancer. Indeed, in 1863, Virchow observed that cancer develops at locations where chronic inflammation is present. This hypothesis is partly based on his assumption that some substances that cause tissue injury and inflammation also enhance cell proliferation (10). Although it is clear now that proliferation alone does not account for cancer, the cause and effect relationship between inflammation and cancer is widely accepted. Nonetheless, many aspects of the molecular and cellular mechanisms that link inflammation, migration, and establishment of tumor colonies in a distant organ remain unresolved (5). Recently, it has been reported that a tumor microenvironment actively contributes to cancer initiation, progression, and metastasis (11, 12). It has also been suggested that reductions in the rates of cancer morbidity and mortality may be achieved by gaining greater understanding of the molecular mechanism(s) of tumor cell migration, invasion, and establishment of colonies (13). In this regard, experimental animal models may provide valuable insight into the molecular mechanism(s) by which inflammation and pulmonary colonization of tumor cells may be linked and facilitate the identification of drug targets for the development of novel anti-metastatic therapies.
Uteroglobin (UG)3 (14), also known as CC10 protein (Clara cell 10-kDa protein) (15), is a steroid-inducible secreted protein with potent anti-inflammatory properties. It is constitutively expressed in the lungs of all mammals, and targeted disruption of the UG gene in mice (16) increases susceptibility to pulmonary inflammation (17). Moreover, we (18) and others (19) have previously demonstrated that overexpression of UG in cancer cells reverses their transformed phenotype and neoplastic potential. We also demonstrated that UG-knock-out (UG-KO) mice (16) are highly susceptible to lung tumorigenesis by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a carcinogen in cigarette smoke (20). Previously, we reported that the UG gene is constitutively expressed at a high level in the lung epithelia (21). However, UG expression in both malignant lung tumors originating from lung epithelia as well as in the cell lines derived from these tumors is either drastically suppressed or totally undetectable (22, 23). Further, we demonstrated that UG prevents migration and extracellular matrix invasion of cancer cells in vitro (24).
Pathogenesis of many diseases mediated via aging, infectious agents, inflammation, or genetic damage often leads to changes in gene expression (reviewed in Ref. 25). Recent reports indicate a link between inflammation and cancer metastasis (10, 12). Moreover, emerging evidence suggests that pre-existing inflammation in the tumor microenvironment stimulates angiogenesis and promotes cancer cell survival and metastasis (4, 12). The molecular mechanism by which inflammation is linked to metastasis is beginning to emerge. The receptor for advanced glycation end products (RAGE) is a multi-ligand, pattern-recognizing receptor of the immunoglobulin superfamily of proteins (26, 27). RAGE signaling has been reported to activate NF-κB, mitogen-activated protein kinases (MAPKs), and Src kinases leading to inflammation and cell proliferation. Among its various ligands, this receptor also interacts with the S100 family of Ca2+-binding proteins (28) including S100A8 (also known as MRP8, calgranulin A) (29) and S100A9 (also called MRP14, calgranulin B) (27) and plays critical roles in transducing inflammatory response (30, 31).
UG-KO mice that are highly susceptible to developing pulmonary inflammation and B16F10 melanoma cells, which preferentially metastasize to the lungs (6, 9), provide the components of a model system that can be utilized to explore whether: (i) the lack of UG promotes metastasis and if so, (ii) what might be the mechanism(s) that regulate cancer cell migration from a peripheral site of injection to a distant organ (e.g. lung) and finally establish metastatic tumors. Here we report that high level expression of S100A8 and S100A9 in the lungs of the UG-KO mice and the existence of a concentration gradient of these proteins from the peripheral circulation to the lungs provide a road map for the B16F10 cells to migrate to the lungs. We also discovered for the first time that B16F10 cells express RAGE. Thus, S100A8 and S100A9 provide the homing signal for RAGE-expressing B16F10 cells to migrate to this organ, which contains the highest concentration of these proteins. Most importantly, treatment of B16F10 cells with a blocking antibody to RAGE dramatically suppresses S100A8/S100A9-mediated chemotactic migration, suggesting that the migration of these cells is RAGE-specific. Taken together, our results show that the lack of an endogenous anti-inflammatory protein such as UG may lead to increased migration and colonization of melanoma cells in the lungs, identifying RAGE as a critical element in this process and a potential target for anti-metastatic drug development.
EXPERIMENTAL PROCEDURES
Animals
UG-KO mice were generated by targeted disruption of the UG gene in embryonic stem cells as described previously (18). Both UG-KO mice and their WT littermates were maintained under germ-free conditions, and all of the experiments were performed according to a protocol approved by the institutional Animal Care and Use Committee.
Cell Culture
B16-F10 cells were purchased from American Type Culture Collection and were cultured in DMEM containing 10% fetal bovine serum.
Tumor Metastasis
A suspension of 2 × 105 B16F10 cells in PBS was injected in the dorsal tail vain of UG-KO mice and their WT littermates. After 21 days of B16F10 cell injection, the animals were euthanized, and the lungs were perfused with PBS. The lung tissues were then fixed in 4% paraformaldehyde. The number of tumor colonies formed were counted and recorded.
Histological Analysis
The animals were sacrificed 21 days following injection of B16F10 cells into the dorsal tail vein. The lungs were then perfused with PBS and fixed by intratracheal administration of 4.0% neutral buffered formalin. After fixation, the tissues were embedded in paraffin and sectioned. The lung sections were stained with hematoxilin and eosin (American Histolab, Inc., Gaithersburg, MD). The stained tissue sections were examined using an Axioskop2 plus microscope (Carl Zeiss), and digital photomicrographs were recorded.
Generation of Conditioned Medium and Migration Assays
Perfused lungs from the UG-KO mice as well as those of their WT littermates were sectioned (2 mm2) using sterile instruments and cultured in DMEM for 90 min as described previously (32). The organ culture media were centrifuged, and the supernatants were stored at −80 °C until used as chemoattractants in the migration assay.
Suppression of RAGE Expression by siRNA
To suppress the RAGE expression, B16F10 cells were transfected with RAGE-specific siRNA (Ambion) as described earlier (33). As a negative control, the cells were transfected with a scrambled siRNA (Ambion). SiPORT lipid Transfection agent (Ambion) was used, and transfection was performed following the manufacturer's protocol. Total proteins were prepared from the cells 72 h after the transfection and expression of RAGE were checked by Western blotting to confirm RAGE siRNA-mediated down-regulation. The siRNA-transfected B16F10 cells were used after 72 h for in vitro invasion assay as well as injected to UG-KO mouse tail vein.
In Vitro Migration and Invasion Assays
The six-well chambers with polycarbonate filters with a pore size of 8.0 μm (BD Biosciences) were used to perform the migration assays. The lower compartment was filled with 2 ml of medium containing various chemoattractants (i.e. conditioned medium from culturing WT and UG-KO lung sections; DMEM containing 0.1% bovine serum albumin (BSA) with either recombinant S100A8 (100 pg/ml), S100A9 (1 ng/ml) (Abnova, Taiwan) or a combination of S100A8 and S100A9 (32). DMEM containing 0.1% BSA alone served as a control. B16F10 cells (1 × 105) were resuspended in 2 ml of DMEM and placed in the upper chamber. Cell migration through the porous filter was allowed to take place for 12 h in a humidified atmosphere of 5% CO2 at 37 °C. The upper surface of the filter was scraped with moist cotton swabs to remove nonmigrated cells, and then the cells attached to the lower surface of the filters (i.e. the migrated cells) were stained with HEMA3 (Fisher) according to the manufacturer's protocol. The migrated cells were photographed and counted in at least 12 different fields (magnification, ×200); the results are expressed as the means of at least three determinations ± S.D. In some experiments, B16F10 cells (1 × 105/ml) were pretreated with a blocking goat antibody to RAGE (R & D Systems) at a concentration of 10 μg/ml for 1 h in DMEM containing 0.1% BSA. The cells were washed three times to remove any unbound antibody before using these cells for the migration assay. Similarly, B16F10 cells were pretreated with a nonspecific goat IgG and used as a negative control. For in vitro invasion assay, similar steps were followed as for in vitro migration assay, but instead of chambers with only polycarbonate filters, chambers with Matrigel-coated filters (BD Biosciences) were used.
Stimulation of MMP Expressions in B16F10 Cells by S100A8/A9
B16-F10 cells were cultured in DMEM containing 10% fetal bovine serum with a seeding density 106/T75 flask. The cells were treated with various concentrations of S100A8 (0.2, 1.0, or 2.0 μg/ml) or S100A9 (0.2, 1.0, or 2.0 μg/ml) alone or in combination (0.2 or 1.0 μg/ml of each) for 24 h. B16-F10 cells without any treatment were kept as control. RNA and proteins were prepared from the treated as well as untreated control B16F10 cells and were used in real time RT-5PCR and Western blot analyses, respectively.
RNA Isolation and Quantitative Real Time PCR
Total RNA was isolated using a Qiagen RNeasy mini kit and treated with DNase (DNase I, 30 units/μg total RNA). Reverse transcribed and quantitative RT-PCR analysis was performed as described previously (33). The cDNA equivalent of 10 ng of total RNA was used as the template. The PCR primers used for real time RT-PCR are shown in supplemental Table S1. The data normalized to β-actin or glyceraldehyde-3-phosphate dehydrogenase were analyzed using ABI Prism Software version 1.01 (Applied Biosystems) and presented as fold change compared with controls. Quantitation was performed using at least three independent total RNA samples for each treatment groups, and the results are expressed as the means ± S.D.
Western Blot Analyses
Total protein (30 μg) from each sample was resolved by electrophoresis using 4–12% NuPAGE Bis-Tris gels (Invitrogen) under reducing conditions and electrotransferred to nitrocellulose membrane (Invitrogen). Immunoblot analysis was performed using S100A8, S100A9 (1:1000; R & D Systems), RAGE (1:750; Abcam), β-actin (1:5000; US Biological), albumin (1:1000; Upstate Biotechnology), MMP-2 (1:1000; Abcam), MMP-9 (1:1000; Millipore), MMP-14 (1:1000; Abcam), and furin (1:500; Santa Cruz Biotechnology) antibodies. Horseradish peroxidase-conjugated anti-rabbit IgGs, anti-goat IgGs, and anti-mouse IgGs (Santa Cruz Biotechnology) were used as the second antibodies. Chemiluminescent detection was performed by using Supersignal west pico luminol/enhancer solution (Thermo Scientific) according to the manufacturer's protocol.
Confocal Microscopy
The B16F10 cells were incubated at 37 °C in an atmosphere of 5% of CO2 and 95% air for 24 h. The cells were washed three times with PBS, pH 7.6, and incubated in 4.0% formaldehyde solution for 15 min for fixation. The fixed B16F10 cells and the lung tissue sections of B16F10-injected UG-KO mice (see “Histological Analysis”) were incubated with antibodies to either RAGE (1:200), S100A8 (1:50), or S100A9 (1:50) overnight at 4 °C in PBS, pH 7.6, containing 2% BSA. Secondary antibodies were: goat anti-rabbit Alexafluor 488 (for RAGE; see Fig. 3B), goat anti-rabbit Alexafluor 594 (for RAGE; see Fig. 4), anti-goat Alexafluor 488 (for S100A8 or S100A9; Fig. 4 and supplemental Fig. S2A), or anti-goat Alexafluor 594 (for S100A8 or S100A9; see Fig. 3, C and D) conjugated secondary antibodies (Invitrogen) in PBS, pH 7.6, containing 2% BSA at room temperature for 1 h. The nuclei were stained with 4,6-diamidino-2-phenylindole dihydrochloride (Sigma). For the detection of MMPs in the tumor infiltrated lungs, sections of the lung tissues were preheated in citric acid buffer using microwaves as described previously (34). Briefly, the slides were placed in a plastic jar containing 500 ml of 0.1 m sodium citrate buffer with pH 6.0. The slides were heated in the microwave oven (Haier) for four 5-min cycles at 800 W with a break between the cycles to check the buffer levels. After heat treatment, the jar was allowed to cool for 30 min. The slices were then washed with distilled water once followed by two washes with PBS and immunostained as follows. The lung sections were first blocked with 5% BSA for 1 h and then overlaid with anti-MMP-2, anti-MMP-9, and anti-MMP-14 antibodies (1:100) and incubated overnight at 4 °C. The slides were washed three times with PBS and further incubated with anti-rabbit Alexafluor 594 (1:250) for 1 h. After three washes with PBS, fluorescence was visualized using LSM-510 inverted confocal microscope (Zeiss), and the images were processed with the LSM Image Browser version 4.2 (Zeiss).
FIGURE 3.
Expression of RAGE as well as S100A8 and S100A9 in B16F10 cells. A, left panel, products obtained from RT-PCR using B16F10 cDNA and primers specific for RAGE, S100A8, or S100A9 were run on an agarose gel to show the presence of RAGE mRNA and absence of any detectable amounts of S100A8 or S100A9 in B16F10 cells. Middle panel, Western blot analysis of B16F10 cell lysates using RAGE-antibody confirmed RAGE-production in these cells. Right panel, Western blot analysis of B16F10 cell lysates with S100A8 or S100A9 antibodies failed to show either S100A8 or S100A9 protein bands. UG-KO mice lung lysate was used as the positive control for S100A8/S100A9. B, immunocytochemistry of B16F10 cells with the antibody against RAGE clearly showed the presence of RAGE (green) in the B16F10 cells. C and D, immunocytochemical analysis of B16F10 cells using RAGE (green) and S100A8 (red) or RAGE (green) and S100A9 (red) antibodies showed B16F10 cells are positive for RAGE but do not produce S100A8 (C) and S100A9 (D). 4,6-Diamidino-2-phenylindole dihydrochloride (DAPI, blue) was used to stain the nuclei.
FIGURE 4.
Colocalization of RAGE and S100A8/A9 on the B16F10 tumors in UG-KO lungs. Confocal microscopy of lung tissues from UG-KO mice injected with B16F10 cells using antibodies against RAGE as well as S100A8 and S100A9 shows that RAGE (red) colocalized with both S100A8 (A, green) and S100A9 (B, green). 4,6-Diamidino-2-phenylindole dihydrochloride (DAPI, blue) was used to stain the nuclei.
Statistical Analysis
Statistical analyses of the data were performed by Student's t test using Excel Office 2000 (Microsoft), and p values of <0.05 were considered statistically significant.
RESULTS
Elevated Levels of Metastasis in the Lungs of Mice Lacking UG
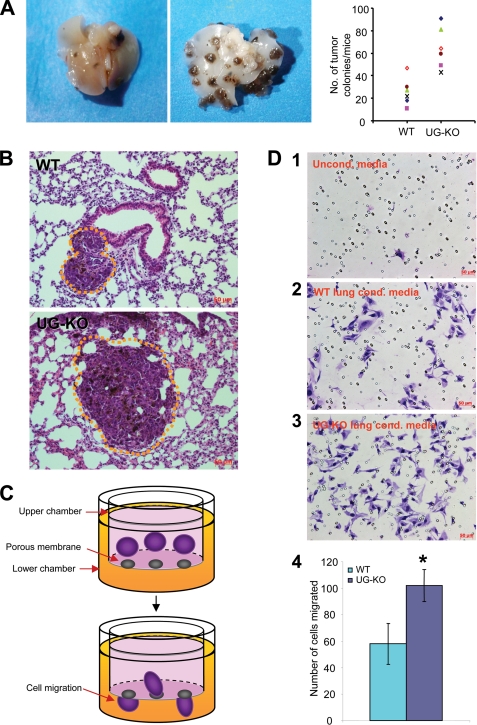
To determine whether UG deficiency predisposes mice to increased cancer cell metastasis, we injected B16F10 melanoma cells in the dorsal tail vein of the UG-KO mice and their WT littermates (see “Experimental Procedures”). We then quantitated the number of metastasized tumors in the lungs. We chose B16F10 cells because these cells are known to be preferentially invasive to the lungs (6, 9). Our results show that although there were only a few small tumors detectable in the lungs of the WT mice (Fig. 1A, left panel), numerous medium to large size tumor colonies were detectable in those of the UG-KO littermates (Fig. 1A, middle panel). The quantitation of the tumors in the lungs confirmed that metastasis of B16F10 cells occurred in the lungs of the UG-KO mice at a much higher level compared with that of their WT littermates (Fig. 1A, right panel). Histological analysis showed infrequent small colonies of tumors in the parenchyma of the lungs from WT mice (Fig. 1B, upper panel), whereas medium to large B16F10 tumors were readily found in those of the UG-KO littermates (Fig. 1B, lower panel).
FIGURE 1.
Lack of UG promotes B16F10 melanoma cell metastasis in the lungs. A, B16F10 melanoma cells were injected in the dorsal tail vein of UG-KO mice and their WT littermates. Although the lungs of the WT mice contained a small number of tumor colonies (left panel) those of the UG-KO littermates showed numerous moderate to large tumor colonies (middle panel). Right panel, quantitation of tumor colonies in WT and UG-KO lungs. B, histological analysis of the lung tissues shows a small colony of melanin producing B16F10 cells in the lung parenchyma of a WT mouse (upper panel), whereas the tumor size in the lungs of a UG-KO littermate (lower panel) appears considerably larger. Tumor colonies are outlined with dotted lines. C, graphic representation of the methods used for determining B16F10 cell migration in response to conditioned media. D, unconditioned medium, which was used as a control, caused virtually no cell migration (panel 1). The conditioned medium obtained from culturing WT lungs slices only modestly stimulated the migration of B16F10 cells (panel 2), whereas the conditioned medium from UG-KO lungs manifested a significantly higher level of migration of these cells (panel 3). These results were further confirmed by quantitation of the migrating cells (panel 4), suggesting that the conditioned medium from the UG-KO lungs contained potent chemoattractant(s) that may have facilitated the migration of the B16F10 cells in vitro.
UG-KO Mouse Lungs Contain Soluble Chemoattractants for B16F10 Cells
To determine whether the lungs of the UG-KO mice contain soluble factor(s) that promote homing of B16F10 cells to the lungs, we performed in vitro migration assays (Fig. 1C). In these experiments, we used conditioned media generated by culturing slices of lung tissues from either UG-KO mice or their WT littermates and used these media as chemoattractants (see “Experimental Procedures”). Nonconditioned medium was used as control. The results show that although nonconditioned medium failed to stimulate chemotactic migration (Fig. 1D, panel 1), the conditioned medium generated by culturing WT lung slices only slightly stimulated the migration of B16F10 cells from the upper to the lower chamber (Fig. 1D, panel 2). However, the conditioned media obtained from culturing lung slices from UG-KO mice dramatically stimulated the migration of these tumor cells (Fig. 1D, panel 3). These results were further confirmed by quantitation of the cells that completed migration across the filter separating the upper chamber from the lower chamber (Fig. 1D, panel 4), suggesting that the conditioned media from the UG-KO mouse lung cultures contain potent chemoattractant(s) that stimulate the migration of the B16F10 cells in vitro.
UG-KO Mouse Lungs Express High Levels of S100A8 and S100A9
We previously reported that UG-KO mice are susceptible to pulmonary inflammation (17), and recently it has been demonstrated that RAGE and its ligands may constitute a new pathway for pulmonary inflammation (35). The S100 protein family, especially S100A8 (29) and S100A9 (27), are cytoplasmic EF-hand Ca2+-binding proteins (36), which are some of the ligands that have been linked to human inflammatory and neoplastic diseases (37). Thus, we sought to determine the levels of expression of S100 proteins and RAGE in the lungs of UG-KO mice. Accordingly, using total RNA from the lung tissues of UG-KO mice and their WT littermates, we first determined the mRNA levels of several S100 genes that are associated with inflammation. The results showed that among six S100 genes surveyed (supplemental Fig. S1), the mRNA levels of only two genes, S100A8 and S100A9, were significantly higher in the lungs of the UG-KO mice compared with those of the WT littermates (Fig. 2A). Consistent with these results, the S100A8/S100A9 protein levels were also higher in the lungs of the UG-KO mice (Fig. 2B). We also performed immunohistochemical analysis to determine which cell type(s) in the UG-KO lungs produced S100A8 and S100A9. The results showed that the lung parenchymal cells are immunopositive for these proteins (supplemental Fig. S2A). Moreover, we found that the serum levels of these proteins in the UG-KO mice are also elevated (Fig. 2C). Interestingly, although the expression level of RAGE mRNA was slightly higher in the lungs of the UG-KO mice (supplemental Fig. S2B), a clear difference in RAGE protein levels between the WT and UG-KO lungs were not readily discernable (supplemental Fig. S2C).
FIGURE 2.
Expression of S100A8 and S100A9 in WT and UG-KO mice and expression of RAGE in B16F10 cells. S100A8 and S100A9 mRNA levels are dramatically elevated in the lungs of UG-KO mice compared with those of the WT littermates (A). The S100A8 and S100A9 protein levels were also significantly higher in the lung tissues (B) as well as in the serum (C) of the UG-KO mice compared with those of the WT littermates.
B16F10 Cells Express RAGE but Not S100A8 and S100A9
Because RAGE acts as a receptor for the S100A8 and S100A9 proteins, we sought to determine whether B16F10 cells express both of these ligands and RAGE. Accordingly, we determined the mRNA and protein levels of S100A8/S100A9 as well as their receptor, RAGE, in B16F10 cells. The results show that although the expression of RAGE mRNA (Fig. 3A, left panel) and RAGE protein (Fig. 3A, middle panel) are readily detectable in B16F10 cells, the expression of S100A8 and S100A9 was undetectable (Fig. 3A, right panel). Consistent with these results, RAGE immunofluoresce analysis also showed high level of expression of this S100 receptor on B16F10 cells (Fig. 3B). Remarkably, the expression of S100A8 and S100A9 in B16F10 cells was virtually undetectable (Fig. 3, C and D). We then determined RAGE expression in metastasized B16F10 cells in UG-KO lungs by immunofluorescence. The results show high levels of RAGE-specific immunofluorescence colocalizing with S100A8 and S100A9 over the tumors (Fig. 4). Taken together, these results demonstrate that although the lung tissues of the UG-KO mice express high levels of S100A8/S100A9, the B16F10 cells express their binding partner, RAGE, at an elevated level. This raises the possibility that circulating S100A8/S100A9 provides the homing signal for the RAGE-expressing B16F10 cells to the UG-KO mouse lungs, which contain the highest levels of these proteins.
Chemotactic Migration of B16F10 Cells Mediated by S100A8/S100A9 Requires RAGE
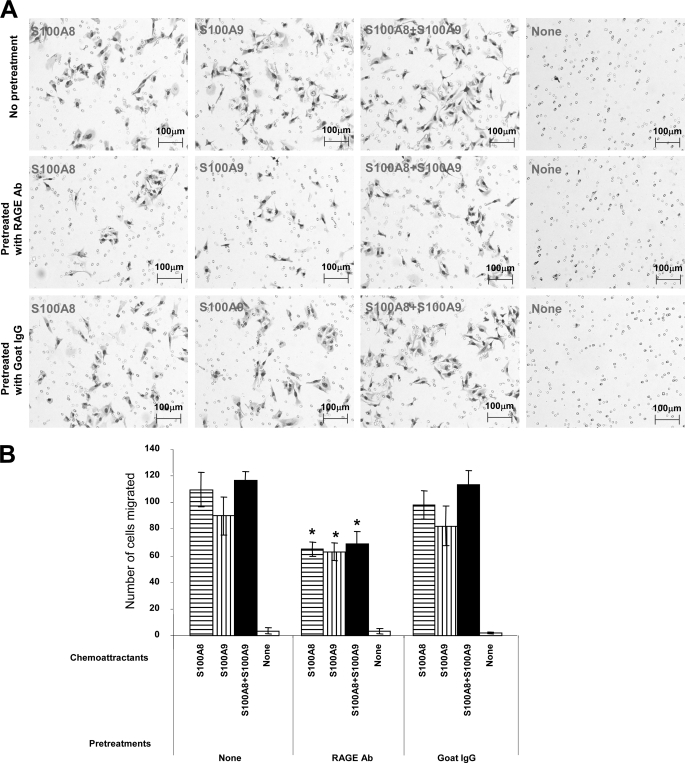
To determine whether S100A8 and S100A9 in the lungs of the UG-KO mice are indeed the chemoattractants for RAGE-positive B16F10 cells, we performed in vitro migration assays using medium containing either recombinant S100A8 or S100A9 or a combination of the two proteins. Medium without these proteins served as control. Our results show that compared with the control, both S100A8- and S100A9-containing media stimulated significantly higher levels of migration of the B16F10 cells, and the combination of both proteins appears to have a mild additive effect (Fig. 5A, top row). We then sought to delineate the role of RAGE in S100A8/S100A9-mediated chemotactic migration of B16F10 cells. Accordingly, we pretreated the B16F10 cells with a blocking antibody to RAGE and performed the chemotactic migration assays using media containing recombinant S100A8, S100A9, or a combination of the two proteins. The results showed that although the B16F10 cells migrated toward the media containing recombinant S100A8 or S100A9, such migration was markedly inhibited when the cells were pretreated with blocking antibody to RAGE (Fig. 5A, middle row). Notably, pretreatment of the cells with a nonspecific IgG did not inhibit S100A8- or S100A9-induced chemotactic migration (Fig. 5A, bottom row). The quantitation of the migrated cells confirmed these findings (Fig. 5B). We also performed in vitro invasion assays using nontransfected, scrambled siRNA-transfected and RAGE siRNA-transfected B16F10 cells using S100A8 and S100A9 as chemoattractants. The results showed that only RAGE-specific siRNA transfection, which inhibited the level of RAGE protein (supplemental Fig. S3A), markedly suppressed B16F10 invasion in vitro (supplemental Fig. S3B). We then injected the RAGE siRNA-transfected and scrambled siRNA-transfected B16F10 cells in the tail vein of the UG-KO mice to determine whether suppression of RAGE inhibited B16F10 colonization in the lungs. The results showed that only RAGE-specific siRNA-transfection suppressed lung colonization (supplemental Fig. S3C). Taken together, these results demonstrate that the migration and invasion of the B16F10 cells in vitro as well as in vivo requires the S100A8/S100A9 and RAGE.
FIGURE 5.
Suppression of chemotactic migration of B16F10 cells by blocking antibody to RAGE. B16F10 cells were preincubated with a blocking goat antibody (Ab) to RAGE or with a nonspecific goat IgG for 1 h prior to performing the in vitro migration assay using medium containing either S100A8 or S100A9 or a combination of S100A8 and S100A9 as chemoattractants. Medium containing no chemoattractants served as control. The results showed that treatment of the cells with the blocking antibody to RAGE significantly suppressed the migration of the B16F10 cells. Representative photomicrographs for each treatment are shown in A. These results were further confirmed by quantitation of the migrating cells under the microscope (B).
Concentrations of S100A8/S100A9 from the Tail Vein and in the Lungs of UG-KO Mice
How might B16F10 cells, introduced in the tail vein, migrate and colonize the lungs of the UG-KO mice? We entertained the hypothesis that there is a low to high gradient of S100A8 and S100A9 proteins from the tail vein (where the cancer cells are injected) to the UG-KO mouse lungs, which produce the highest levels of S100A8 and S100A9. Accordingly, we obtained blood samples by canulating the tail vein as well as the pulmonary vein, and the sera were analyzed for S100A8 and S100A9 levels by Western blot analysis. The results showed that the levels of both of these proteins were appreciably lower in the blood samples from the tail vein compared with those in the blood samples obtained from the pulmonary vein (supplemental Fig. S4). Interestingly, we found that treatment of B16F10 cells with recombinant S100A8 or S100A9 augments the level of RAGE expression (supplemental Fig. S5). This may indicate that after encountering increasing concentrations of S100A8/A9, these cells elevate the RAGE levels to bind more ligand. Taken together, these results suggested the existence of a low to high S100A8/S100A9 gradient, which most likely provides a road map for the RAGE-positive B16F10 cells, injected in the tail vein, to migrate to the lungs, where the concentration of these proteins was the highest.
B16F10 Cells Acquire Invasiveness by Expressing MMPs Stimulated by S100A8/S100A9
How might B16F10 cells, upon arrival at the lungs, acquire the invasiveness required for metastasis to the lung parenchyma? It is a general consensus that the extracellular matrices of cancer cells as well as those of the noncancerous cells within the tumors have profound impact on tumor progression and metastasis (38). Interestingly, MMPs, a family of proteolytic enzymes (especially MMP-2, MMP-9, and MMP-14), have been shown to regulate the nature of the tumor microenvironment, and increased expression as well as activation of these enzymes are found in virtually all metastasizing human cancers (39–41). Thus, we sought to determine whether RAGE-positive B16F10 cells, upon encountering high levels of S100A8/S100A9 in the UG-KO mouse lungs, overexpress MMPs that confer the invasiveness essential for metastasis on these cells. Accordingly, we treated B16F10 cells with recombinant S100A8 or S100A9 and determined the expression of mRNAs for MMP-2, MMP-9, and MMP-14 by real time RT-PCR. The results showed elevated mRNA levels for all three MMPs in B16F10 cells treated with S100A8 or S100A9 (supplemental Fig. S5). Because it has been reported that the S100A8 and S100A9 heterodimerize to yield maximum the stimulation of gene expression, we treated B16F10 cells with a combination of recombinant S100A8 (1 μg/ml) and S100A9 (1 μg/ml) and determined the mRNA and protein levels of MMP-2, MMP-9, and MMP-14. We chose to study these three MMPs because it has been previously reported that these proteinases confer invasiveness to tumor cells promoting metastasis (42, 43). The results of real time RT-PCR revealed that the mRNA levels of MMP-2, MMP-9, and MMP-14 in B16F10 cells treated with a combination of S100A8 and S100A9 were markedly elevated (Fig. 6A). To determine whether these MMPs are enzymatically active, we analyzed the levels of mature (active) MMP-2, MMP-9, and MMP-14 by Western blot analysis of homogenates of untreated and S100A8/S100A9-treated B16F10 cells. The results showed elevated levels of mature MMP-2, MMP-9, and MMP-14 proteins in S100A8/S100A9-treated B16F10 cells (Fig. 6B). Interestingly, treatment of B16F10 cells with S100A8/S100A9 also stimulated the expression of furin (Fig. 6, A and B), a pro-protein convertase that regulates the enzymatic activity of some MMPs such as MMP-14 (MT1-MMP) (44). Thus, it is likely that furin activated MMP-14, which in turn activated MMP-2. This notion is supported by the demonstration that heterodimerization of activated MMP-14 with MMP-2 activates MMP-2 (44). The mechanism of activation of MMP-9 remains unclear. To determine whether B16F10 cells after arriving at the S100A8/S100A9-laden lungs of UG-KO mice produce MMPs in vivo, we conducted immunofluorescence studies using antibodies against MMP-2, MMP-9, and MMP-14. The results showed that all three MMPs were expressed by the metastasized B16F10 tumors in the UG-KO lungs at a high level (Fig. 6C). Taken together, our results suggest that S100A8 and S100A9 not only stimulate the expression of MMPs associated with cancer metastasis but also increase the levels of mature (active) MMP-2, MMP-9, and MMP-14 in B16F10 cells, which are essential for metastasis of these cancer cells to the UG-KO mouse lungs.
FIGURE 6.
Stimulation of MMP expression in B16F10 cells by S100A8 and S100A9. A, the levels of MMP-2, MMP-9, MMP-14, and furin mRNAs were significantly up-regulated upon treatment with S100A8 and S100A9 at a concentration of 1 μg/ml for 24 h. Cells without any treatment were used as control. Consistent with the mRNA levels, the protein levels of MMP-2, MMP-9, MMP-14, and furin were also up-regulated. B, the precursor (P), intermediate (I), and matured (active) forms (M) of the MMPs and furin are indicated by arrows. A splice variant (SV) of Furin is also shown. β-Actin is used as the loading control. C, immunohistochemistry of lung tissues from UG-KO mice injected with B16F10 cells using antibodies against MMP-2, MMP-9, and MMP-14 (right panels). Anti-rabbit Alexafluor 594 (red) was used as a secondary antibody. H&E-stained tissue sections of same regions are shown at the left panels. H&E, hemotoxylin and eosin.
DISCUSSION
Cancer metastasis is a multistep process, which requires coordinated actions of several genes (6–9, 45, 46). We previously reported that the lungs of the UG-KO mice express genes that are known to mediate inflammation (17). In the present study, we found that the expression of several other genes related to inflammation (e.g. CARMA1, Maspin, RANK, and tumor necrosis factor α) is also appreciably elevated in the UG-KO mouse lungs (supplemental Fig. S6). Thus, the lack of UG in the lungs of these mice creates a pro-inflammatory microenvironment conducive for B16F10 cell invasion and metastasis. Indeed, consistent with our findings, recent evidence indicates a critical role of tumor microenvironment on cancer development and metastasis (11, 12, 33, 47). Moreover, it has been shown that primary tumors secrete soluble factors such as transforming growth factor β and tumor necrosis factor α that stimulate the production of proteins S100A8 and S100A9 in epithelial and myeloid cells within the lung prior to tumor metastasis (32, 48). Thus, it is possible that S100A8/S100A9-mediated inflammatory response in the UG-KO mouse lungs provides a microenvironment conducive for B16F10 cells to anchor, invade, and establish metastasis.
The results of our experiments identify three important parameters that coordinately link inflammatory microenvironment in UG-KO mouse lungs with the progression of metastasis of B16F10 cells. These are: (i) UG causing elevated level of expression of S100A8/S100A9 proteins in the lungs; (ii) the presence of RAGE on B16F10 cells and the existence of a circulating S100A8/S100A9 gradient in the UG-KO mice, which serves as a homing signal for the cancer cells to arrive at the destination (lungs), which contains the highest level of these proteins; and (iii) elevated expression as well as activation of MMP-2, MMP-9, and MMP-14 in B16F10 cells in response to S100A8/S100A9. Most importantly, the suppression of S100A8/S100A9-mediated chemotactic migration of B16F10 cells by a blocking antibody to RAGE identifies RAGE as a potential drug target.
Supplementary Material
Acknowledgments
We thank I. Owens, J. Y. Chou, and S. W. Levin for critical review of the manuscript and helpful suggestions. We are also grateful to V. Schram (Microscopy and Imaging Core, NICHD, National Institutes of Health) for help with confocal microscopy.
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver NICHD, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
- UG
- uteroglobin
- UG-KO
- uteroglobin knock-out
- WT
- wild type
- RAGE
- receptor for advanced glycation end products
- MMP
- matrix metalloproteinase
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- siRNA
- small interfering RNA
- BSA
- bovine serum albumin
- RT
- reverse transcription.
REFERENCES
- 1.Marx J. (2004) Science 306, 966–968 [DOI] [PubMed] [Google Scholar]
- 2.Fidler I. J. (2003) Nat. Rev. Cancer 3, 453–458 [DOI] [PubMed] [Google Scholar]
- 3.Luo J. L., Tan W., Ricono J. M., Korchynskyi O., Zhang M., Gonias S. L., Cheresh D. A., Karin M. (2007) Nature 446, 690–694 [DOI] [PubMed] [Google Scholar]
- 4.Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 5.Coussens L. M., Werb Z. (2002) Nature 420, 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers A. F., Groom A. C., MacDonald I. C. (2002) Nat. Rev. Cancer 2, 563–572 [DOI] [PubMed] [Google Scholar]
- 7.Mehlen P., Puisieux A. (2006) Nat. Rev Cancer 6, 449–458 [DOI] [PubMed] [Google Scholar]
- 8.Liotta L. A., Kohn E. C. (2003) Nat. Genet. 33, 10–11 [DOI] [PubMed] [Google Scholar]
- 9.Fidler I. J. (1975) Cancer Res. 35, 218–224 [PubMed] [Google Scholar]
- 10.Balkwill F., Mantovani A. (2001) Lancet 357, 539–545 [DOI] [PubMed] [Google Scholar]
- 11.Joyce J. A., Pollard J. W. (2009) Nat. Rev. Cancer 9, 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu M., Polyak K. (2008) Curr. Opin. Genet. Dev. 18, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedley B. D., Chambers A. F. (2009) Adv. Cancer Res. 102, 67–101 [DOI] [PubMed] [Google Scholar]
- 14.Mukherjee A. B., Zhang Z., Chilton B. S. (2007) Endocr. Rev. 28, 707–725 [DOI] [PubMed] [Google Scholar]
- 15.Ray M. K., Magdaleno S., O'Malley B. W., DeMayo F. J. (1993) Biochem. Biophys. Res. Commun. 197, 163–171 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z., Kundu G. C., Yuan C. J., Ward J. M., Lee E. J., DeMayo F., Westphal H., Mukherjee A. B. (1997) Science 276, 1408–1412 [DOI] [PubMed] [Google Scholar]
- 17.Mandal A. K., Zhang Z., Ray R., Choi M. S., Chowdhury B., Pattabiraman N., Mukherjee A. B. (2004) J. Exp. Med. 199, 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z., Kundu G. C., Panda D., Mandal A. K., Mantile-Selvaggi G., Peri A., Yuan C. J., Mukherjee A. B. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3963–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabo E., Goheer A., Witschi H., Linnoila R. I. (1998) Cell Growth Differ. 9, 475–485 [PubMed] [Google Scholar]
- 20.Yang Y., Zhang Z., Mukherjee A. B., Linnoila R. I. (2004) J. Biol. Chem. 279, 29336–29340 [DOI] [PubMed] [Google Scholar]
- 21.Peri A., Cordella-Miele E., Miele L., Mukherjee A. B. (1993) J. Clin. Invest. 92, 2099–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linnoila R. I., Jensen S. M., Steinberg S. M., Mulshine J. L., Eggleston J. C., Gazdar A. F. (1992) Am. J. Clin. Pathol. 97, 233–243 [DOI] [PubMed] [Google Scholar]
- 23.Broers J. L., Jensen S. M., Travis W. D., Pass H., Whitsett J. A., Singh G., Katyal S. L., Gazdar A. F., Minna J. D., Linnoila R. I. (1992) Lab. Invest. 66, 337–346 [PubMed] [Google Scholar]
- 24.Kundu G. C., Mandal A. K., Zhang Z., Mantile-Selvaggi G., Mukherjee A. B. (1998) J. Biol. Chem. 273, 22819–22824 [DOI] [PubMed] [Google Scholar]
- 25.Shames D. S., Minna J. D., Gazdar A. F. (2007) Curr. Mol. Med. 7, 85–102 [DOI] [PubMed] [Google Scholar]
- 26.Neeper M., Schmidt A. M., Brett J., Yan S. D., Wang F., Pan Y. C., Elliston K., Stern D., Shaw A. (1992) J. Biol. Chem. 267, 14998–15004 [PubMed] [Google Scholar]
- 27.Boyd J. H., Kan B., Roberts H., Wang Y., Walley K. R. (2008) Circ. Res. 102, 1239–1246 [DOI] [PubMed] [Google Scholar]
- 28.Donato R. (1986) Cell Calcium 7, 123–145 [DOI] [PubMed] [Google Scholar]
- 29.Gebhardt C., Németh J., Angel P., Hess J. (2006) Biochem. Pharmacol 72, 1622–1631 [DOI] [PubMed] [Google Scholar]
- 30.Hofmann M. A., Drury S., Fu C., Qu W., Taguchi A., Lu Y., Avila C., Kambham N., Bierhaus A., Nawroth P., Neurath M. F., Slattery T., Beach D., McClary J., Nagashima M., Morser J., Stern D., Schmidt A. M. (1999) Cell 97, 889–901 [DOI] [PubMed] [Google Scholar]
- 31.Heizmann C. W., Ackermann G. E., Galichet A. (2007) Subcell. Biochem. 45, 93–138 [DOI] [PubMed] [Google Scholar]
- 32.Hiratsuka S., Watanabe A., Sakurai Y., Akashi-Takamura S., Ishibashi S., Miyake K., Shibuya M., Akira S., Aburatani H., Maru Y. (2008) Nat. Cell Biol. 10, 1349–1355 [DOI] [PubMed] [Google Scholar]
- 33.Saha A., Kim S. J., Zhang Z., Lee Y. C., Sarkar C., Tsai P. C., Mukherjee A. B. (2008) FEBS Lett. 582, 3823–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warembourg M., Leroy D. (2000) J. NeuroSci. Methods 104, 27–34 [DOI] [PubMed] [Google Scholar]
- 35.Morbini P., Villa C., Campo I., Zorzetto M., Inghilleri S., Luisetti M. (2006) Mod. Pathol. 19, 1437–1445 [DOI] [PubMed] [Google Scholar]
- 36.Schäfer B. W., Heizmann C. W. (1996) Trends Biochem. Sci 21, 134–140 [DOI] [PubMed] [Google Scholar]
- 37.Nacken W., Roth J., Sorg C., Kerkhoff C. (2003) Microsc Res Tech 60, 569–580 [DOI] [PubMed] [Google Scholar]
- 38.Bissell M. J., Radisky D. (2001) Nat. Rev. Cancer 1, 46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werb Z. (1997) Cell 91, 439–442 [DOI] [PubMed] [Google Scholar]
- 40.Liotta L. A., Steeg P. S., Stetler-Stevenson W. G. (1991) Cell 64, 327–336 [DOI] [PubMed] [Google Scholar]
- 41.Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., Seiki M. (1994) Nature 370, 61–65 [DOI] [PubMed] [Google Scholar]
- 42.Hofmann U. B., Westphal J. R., Van Muijen G. N., Ruiter D. J. (2000) J. Invest. Dermatol. 115, 337–344 [DOI] [PubMed] [Google Scholar]
- 43.Moss N. M., Wu Y. I., Liu Y., Munshi H. G., Stack M. S. (2009) J. Biol. Chem. 284, 19791–19799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ra H. J., Parks W. C. (2007) Matrix Biol. 26, 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Couzin J. (2003) Science 299, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 46.Kang Y., Massagué J. (2004) Cell 118, 277–279 [DOI] [PubMed] [Google Scholar]
- 47.Bierie B., Moses H. L. (2006) Nat. Rev. Cancer 6, 506–520 [DOI] [PubMed] [Google Scholar]
- 48.Rafii S., Lyden D. (2006) Nat. Cell Biol. 8, 1321–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.