Abstract
Yersinia pestis has acquired a variety of complex strategies that enable the bacterium to overcome defenses in different hosts and ensure its survival and successful transmission. A full-genome microarray analysis on Caenorhabditis elegans infected with Y. pestis shows enrichment in genes that are markers of innate immune responses and regulated by a conserved PMK-1/p38 MAPK. Consistent with a role in regulating expression of immune effectors, inhibition of PMK-1/p38 by mutation or RNA interference enhances susceptibility to Y. pestis. Further studies of mosaic animals where PMK-1/p38 is exclusively inhibited or overexpressed in a tissue-specific manner indicate that PMK-1/p38 controls expression of a CUB-like family of immune genes at the cell-autonomous level. Given the conserved nature of PMK-1/p38 MAPK-mediated signaling and innate immune responses, PMK-1/p38 MAPK may play a role in the immune response against Y. pestis in natural hosts.
Keywords: C. elegans, Gene Expression, Gene Regulation, Immunology, Innate Immunity, MAP Kinases (MAPKs), p38 MAPK, Host Defense Response, Host-Pathogen Interactions, Infection
Introduction
Recently evolved from enteropathogenic Yersinia pseudotuberculosis, Yersinia pestis acts as a blood-borne pathogen capable of parasitizing insects and causing systemic disease in mammals (1, 2). Y. pestis has acquired a variety of complex strategies to overcome defense responses in different hosts to ensure its multiplication and survival (3–5). Although a number of virulence determinants contributing to Y. pestis persistence in mammals have been identified, interactions between Y. pestis and the host immune system remain poorly understood.
During its vector-mammal transmission cycle, Y. pestis must evade components of innate immunity in both insects and mammals. One of the initial immune responses against pathogens in vertebrates and invertebrates alike is the inducible humoral defense, including the production of antimicrobial peptides and reactive oxygen species (6, 7). These inducible effectors, possessing potent antimicrobial activity, are components of the phylogenetically ancient innate immune system that predates the origins of adaptive immunity. Evolutionary conservation among pathogen recognition receptors and signaling pathways contributing to the inducible immune response has permitted the use of alternative model hosts to study innate immunity.
In recent years, the nematode Caenorhabditis elegans has become an attractive alternative model host to study certain aspects of bacterial pathogenesis and innate immunity. Even though C. elegans lacks professional immune cells, it lives in soil environments where it is in contact with soil-borne microbes and has evolved physiological mechanisms to respond to different pathogens by activating the expression of innate immune response genes that are conserved across metazoans. Typically, C. elegans is grown in the laboratory by feeding them Escherichia coli. E. coli is effectively disrupted by the C. elegans pharyngeal grinder, and almost no intact bacterial cells can be found in the intestinal lumen. Once in the gut, however, pathogenic bacteria can overcome innate immune responses to proliferate and kill C. elegans. Infection of C. elegans with Y. pestis KIM5 leads to a persistent and lethal colonization of the nematode intestine (8). In addition, similar virulence factors are required for pathogenicity in nematodes and mice (8). Infectivity and persistence of Y. pestis KIM5 in the nematode makes C. elegans an attractive whole animal system for studying the host response to infection with the plague bacterium.
In this study, we examined the transcriptional response of C. elegans during infection with Y. pestis KIM5 to better understand conserved innate responses contributing to host defense during early stages of infection. Our results demonstrate a strong transcriptional response against Y. pestis highlighted by the induction of immune-related effectors that are predominantly regulated by PMK-1/p38. In C. elegans, as well as in other animals, PMK-1/p38 is likely expressed in a range of tissues where it can regulate immune responses at the cell-autonomous level or at the organismal level. Our studies indicate that PMK-1/p38 activity is required in the C. elegans intestine to regulate innate immunity at the cell-autonomous level.
EXPERIMENTAL PROCEDURES
Nematode and Bacterial Strains
C. elegans strains N2, KU25 pmk-1(km25), WM118 rde-1(ne300); neIs9[myo-3::HA::rde-1, rol-6], NR222 rde-1(ne219); kzIs9[pKK1260(plin-12::nls::gfp), pKK1253(plin-26::rde-1), rol-6] (9); and NR350 rde-1(ne219); kzIs20[pDM#715(phlh-1::rde-1), pTG95(psur-5::nls::GFP), rol-6] (9) were provided by the Caenorhabditis Genetics Center. Strain VP303 rde-1(ne219); kbIs7[pnhx-2::rde-1, rol-6] (10) was provided by Kevin Strange (Vanderbilt University). All strains were maintained at 20 °C on nematode growth medium (NGM)2 and fed with E. coli OP50. The following bacterial strains were used for experiments: E. coli OP50 (11) and Y. pestis KIM5 (12). E. coli was cultured overnight in LB broth at 37 °C, and Y. pestis was cultured in LB broth at 25 °C.
RNA Isolation
Gravid adult N2 nematodes were lysed using a solution of sodium hydroxide and bleach and washed, and eggs were synchronized overnight in S basal liquid medium at room temperature. Synchronized L1 larval animals were seeded onto modified NGM plates with 0.35% peptone containing E. coli OP50 and incubated at 25 °C until the nematodes had reached the L4 larval stage. Animals were collected and washed with M9 buffer before transferring to modified NGM plates containing Y. pestis KIM5 or E. coli OP50 for 24 h. After 24 h, animals were collected and washed with M9 buffer, and RNA was extracted using TRIzol reagent (Invitrogen). Residual genomic DNA was removed by DNase treatment (Ambion, Austin, TX). Three independent RNA isolations were performed with each pathogen for microarray analysis, and two additional RNA isolations were performed for samples used for quantitative PCR.
Microarray Analysis
For each experimental condition, RNA was isolated from three biological replicate samples. cRNA was synthesized from 10 μg of total RNA, and samples were hybridized to the C. elegans GeneChip (Affymetrix, Santa Clara, CA) by the Duke Microarray Facility. Microarray data were subjected to the robust multichip averaging algorithm using GeneSpring GX software (Agilent Technologies, Santa Clara, CA). Analysis of variance t test and fold-change calculations were also performed using GeneSpring software. Transcripts showing a corrected p value of <0.05 were considered differentially expressed between E. coli OP50 and Y. pestis KIM5 experimental treatments. The microarray data have been deposited in the Gene Expression Omnibus, accession number GSE20053.
Functional Enrichment Analysis
Genes showing a significant change in expression by microarray analysis (p < 0.05) were analyzed using FatiGO software (13). Genes were compared against a 21,249 C. elegans gene data base to identify over-represented Gene Ontology terms and Interpro motifs. Statistical analysis was performed by the FatiGO software using Fisher's exact test and corrected for false discovery rate using the methods of Benjamini and Hochberg. Significant functional terms were defined as p < 0.05.
Real Time Quantitative PCR
cDNA was synthesized from 5 μg of total RNA using random hexamers and SuperScript II reverse transcriptase (Invitrogen). Real time PCR was performed using SYBR Advantage quantitative PCR premix (Clontech) and gene-specific oligonucleotide primers (supplemental Table 4) on the LightCycler (Roche Applied Science). Relative fold-changes for transcripts were calculated using the comparative CT (2−ΔΔCT) method (14) and normalization to pan-actin (act-1, -3, -4) (15). Cycle thresholds of amplification were determined by LightCycler software (Roche Applied Science). All samples were run in triplicate.
RNA Interference
E. coli HT115(DE3) bacterial strains expressing double-stranded RNA (16) were grown in LB broth containing ampicillin (100 μg/ml) at 37 °C and plated onto NGM containing 100 μg/ml ampicillin and 3 mm isopropyl 1-thio-β-d-galactopyranoside. RNAi-expressing bacteria were allowed to grow overnight at 37 °C. L3 larval animals were placed on RNAi or vector control plates for 2 days at 20 °C until nematodes became gravid. Gravid adults were then transferred to fresh RNAi-expressing bacterial lawns and allowed to lay eggs for 4 h to synchronize a second generation RNAi population. Unc-22 RNAi was included as a positive control in all experiments to account for RNAi efficiency.
C. elegans Survival Analysis
C. elegans hermaphrodites treated with RNAi or a vector control were maintained at 20 °C until animals were young adults. Pathogen lawns for survival assays were prepared by inoculating modified NGM (in 6-cm Petri plates) with 60 μl of an overnight bacterial culture. Plates were incubated overnight at room temperature before animals were added. Young adult animals were transferred to modified NGM plates containing Y. pestis KIM5 bacterial lawns and incubated at 25 °C. Animals were scored every 12 or 24 h for survival and transferred to fresh pathogen lawns each of the first 5 days to avoid overgrowth by progeny. Animal survival was plotted using Kaplan-Meier survival curves and analyzed by log rank test using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). Survival curves resulting in p values of < 0.05 relative to control were considered significantly different. The time for 50% of the nematodes to die (TD50) was calculated using nonlinear regression analysis of survival proportions (GraphPad Prism) utilizing the following equation: Y = bottom + (top − bottom)/(1 + 10 (logEC50−X)·Hill slope), where Top is set at 100, Bottom is set at 0 ; X is the time, and Y is the percentage of nematodes alive at time X. In this instance, TD50 is equivalent to EC50.
Plasmid Constructs and Generation of Transgenic Lines
A GFP transcriptional reporter plasmid, pDB09.1, was constructed by cloning 1974 bp of the F35E12.5 promoter region (position −15 to −1989 upstream of ATG) into pPD95.79 (Fire Lab C. elegans Vector Kit, Addgene, Cambridge, MA). Wild-type N2 nematodes were microinjected with pDB09.1 and pRF4 to generate the extrachromosomal array acEx101. Integration of the extrachromosomal array acEx101 was achieved by UV irradiation, yielding strain AY101 acIs101[pDB09.1(pF35E12.5::gfp); pRF4(rol-6(su1006))]. Plasmid pDB09.2 was generated by cloning the cDNA sequence of pmk-1 into pPD95.77 (Addgene), creating a translational fusion between PMK-1 and GFP. The vha-6 promoter (17) was inserted upstream of the pmk-1::gfp coding sequence to confer intestine-restricted expression (18). pmk-1(km25) animals were microinjected with pDB09.2 and pRF4 to generate strain AY102 pmk-1(km25) acEx102[pDB09.2(pvha-6::pmk-1::gfp); pRF4(rol-6(su1006))].
Western Blot Analysis
Western blots were prepared by separating total nematode lysates from 50 adult animals on a 10% SDS-polyacrylamide gel and transferring separated proteins to Immuno-Blot polyvinylidene difluoride membranes (Bio-Rad). Blots were incubated with peroxidase-conjugated anti-GFP-horseradish peroxidase (1:5000; Rockland Immunochemicals) or anti-actin AC-40 (1:5000; Sigma) followed by horseradish peroxidase-conjugated anti-mouse antibody (1:1000; Bio-Rad). Blot were developed using SuperSignal chemiluminescence substrate (Pierce).
COPAS Biosorter GFP Analysis
Expression levels of the pF35E12.5::gfp reporter in AY101 transgenic animals were analyzed using the COPAS Biosort instrument for large particle flow cytometry (Union Biometrica, Holliston, MA). Synchronized animals treated with control vector or pmk-1 RNAi were exposed to E. coli or Y. pestis for 24 h and washed in M9 buffer prior to analysis. Fluorescence data were acquired for a minimum of 400 adult animals for each experimental sample. Plots were constructed using FlowJo flow cytometry analysis software (Tree Star, Inc., Ashland, OR).
RESULTS
Identification of Inducible Immune Responses to Y. pestis Infection
To investigate conserved innate immune mechanisms that play a role in early response to Y. pestis, we utilized Affymetrix GeneChip C. elegans Genome Arrays to find clusters of genes commonly up-regulated or down-regulated in response to Y. pestis infection. Infection of C. elegans was accomplished by transferring nematodes, propagated on E. coli OP50, onto bacterial lawns of Y. pestis KIM5. In previous studies, we have demonstrated that following ingestion Y. pestis establishes a persistent infection in the C. elegans intestine (8). Even though C. elegans is not known to be a natural host for Y. pestis, this persistent colonization of the nematode intestine by Y. pestis allows the host to properly recognize and respond to pathogen infection.
Host response to pathogen challenge was assessed by measuring transcriptional activity of a synchronized population of nematodes exposed to Y. pestis KIM5. RNA was collected from animals following 24 h of pathogen exposure allowing sufficient time for bacterial accumulation within the intestine, yet prior to extensive morbidity of infected animals. Transcript expression levels were compared between Y. pestis-treated animals and nematodes maintained on the relatively nonpathogenic E. coli OP50 to identify factors involved in host response to Y. pestis. Overall, analysis using the Affymetrix C. elegans genome array revealed a change in expression of 258 transcripts (supplemental Table 1). Of these, 99 genes were up-regulated greater than 2-fold, demonstrating a robust inducible response to Y. pestis (supplemental Table 1). Conversely, 27 genes were down-regulated greater than 2-fold, representing transcripts directly suppressed in response to infection or reduced as a consequence of host pathology.
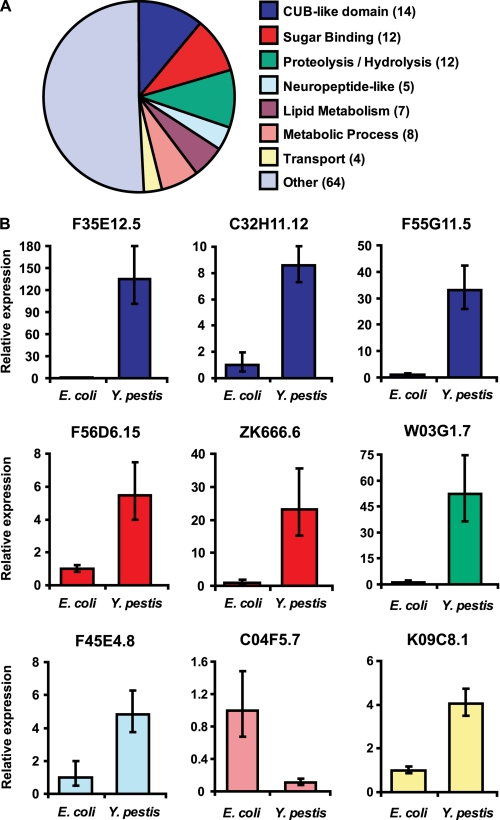
Several classes of transcripts that showed changes in expression in response to Y. pestis infection are markers of C. elegans immunity (Fig. 1A and Table 1). Members of gene families encoding CUB-like proteins, C-type lectins, and neuropeptide-like factors that are up-regulated in response to infection with several pathogens (15, 19–23) are similarly up-regulated in response to Y. pestis (Table 1). However, the overall transcriptional profile for genes belonging to these families was unique in comparison with the response to other pathogens. These results are in line with other studies that have reported shared and distinct inducible responses to pathogen exposure in C. elegans (22, 24).
FIGURE 1.
C. elegans immune effectors are differentially regulated in response to Y. pestis infection. A, pie chart of genes families showing >2-fold change during Y. pestis infection. The number of genes for each family represented by the chart is indicated in parentheses. B, expression levels of F35E12.5, C32H11.12, F55G11.5, F56D6.15, ZK666.6, W03G1.7, F45E4.8, C04F5.7, and K09C8.1 were determined using real time PCR as described under “Materials and Methods.” Relative expression levels of the indicated genes were determined using the comparative CT method (14) with normalization to pan-actin (act-1, -3, -4) (15). Bar graphs correspond to expression levels relative to average expression following control treatment (E. coli). Error bars represent standard deviation among independent biological samples.
TABLE 1.
Summary of C. elegans transcripts showing greater than 2-fold change in expression during Y. pestis infection
| Group | Gene | Fold-changea | p valueb | Group | Gene | Fold-changea | p valueb |
|---|---|---|---|---|---|---|---|
| CUB-like domain | F35E12.5 | 62.54 | 0.0062 | Proteolysis/hydrolysis | W03G1.7 | 13.55 | 0.0111 |
| C32H11.10 | 12.43 | 0.0355 | F53A9.1 | 9.37 | 0.0195 | ||
| C32H11.12 | 9.97 | 0.0217 | C05C10.4 | 5.48 | 0.0457 | ||
| F55G11.5 | 6.82 | 0.0178 | K10C2.3 | 5.12 | 0.0189 | ||
| C32H11.4 | 6.52 | 0.0217 | F21F8.4 | 4.45 | 0.0470 | ||
| F35E12.6 | 6.25 | 0.0217 | K11D2.2 | 3.36 | 0.0264 | ||
| F08G5.6 | 6.21 | 0.0195 | T10H4.12 | 2.57 | 0.0314 | ||
| C17H12.8 | 4.94 | 0.032 | F55F3.2 | 2.20 | 0.0195 | ||
| F55G11.8 | 4.68 | 0.0219 | F54F11.2 | 2.18 | 0.0256 | ||
| K08D8.5 | 3.59 | 0.0256 | F27E5.1 | 2.10 | 0.0195 | ||
| ZK896.5 | 2.57 | 0.0149 | Y16B4A.2 | 2.09 | 0.0460 | ||
| H20E11.1 | 2.03 | 0.0298 | C55B7.3 | −2.48 | 0.0249 | ||
| K10D11.5 | 2.00 | 0.0441 | Lipid metabolism | F54F3.3 | 11.53 | 0.0173 | |
| T05E12.6 | −4.52 | 0.0217 | Y65B4BR.1 | 10.01 | 0.0249 | ||
| Sugar binding | F56D6.15 | 10.09 | 0.0173 | K03H6.2 | 8.91 | 0.0062 | |
| F56D6.2 | 9.24 | 0.0233 | B0035.13 | 5.97 | 0.0227 | ||
| ZK666.6 | 6.88 | 0.0438 | F09C8.1 | 4.95 | 0.0382 | ||
| F35C5.5 | 3.77 | 0.0282 | F28H7.3 | 3.31 | 0.0217 | ||
| F35C5.9 | 3.59 | 0.0256 | W06D12.3 | −4.66 | 0.0216 | ||
| F40F4.6 | 3.42 | 0.0283 | Metabolic Process | F41E6.5 | 4.71 | 0.0460 | |
| Y54G2A.14 | 2.79 | 0.0282 | T25B9.7 | 2.03 | 0.0256 | ||
| F38A5.3 | 2.74 | 0.0322 | D1054.8 | −2.55 | 0.0460 | ||
| Y54G2A.8 | 2.58 | 0.0329 | C17C3.12 | −3.33 | 0.0139 | ||
| F21H7.4 | 2.47 | 0.0329 | F47C10.6 | −3.83 | 0.0305 | ||
| F35C5.8 | 2.00 | 0.0329 | C30G12.2 | −4.08 | 0.0382 | ||
| T09F5.9 | −4.19 | 0.0460 | C04F5.7 | −4.33 | 0.0322 | ||
| Neuropeptide-like | F45E4.8 | 7.78 | 0.0327 | C55B7.4 | −8.32 | 0.0441 | |
| Y45F10A.5 | 2.63 | 0.0295 | Transport | K09C8.1 | 4.26 | 0.0178 | |
| F37A8.4 | 2.58 | 0.0364 | C18H9.5 | −2.41 | 0.0012 | ||
| M01D7.5 | 2.54 | 0.0371 | C35A5.3 | −4.12 | 0.0219 | ||
| T23E7.4 | 2.26 | 0.0233 | T10H9.5 | −5.40 | 0.0280 |
a Fold change represents the ratio of expression in C. elegans exposed to Y. pestis relative to expression on E. coli. Average expression levels from three samples were used to calculate fold change.
b p values represent the corrected p value for multiple comparisons using the Benjamini-Hochberg method with a cutoff of 0.05.
Although only a limited number of factors encoded by the aforementioned gene families have been directly associated with antimicrobial activity (25) or resistance to pathogen-mediated killing (15, 19, 26), strong induction during infection supports that these factors participate in the nematode immune response. Changes in host transcript levels for nine representative genes were confirmed using quantitative real time PCR (Fig. 1B). For each of the selected genes, the trend in gene expression revealed by real time PCR was similar to fold-change regulation as determined by microarray analysis (Fig. 1B and Table 1).
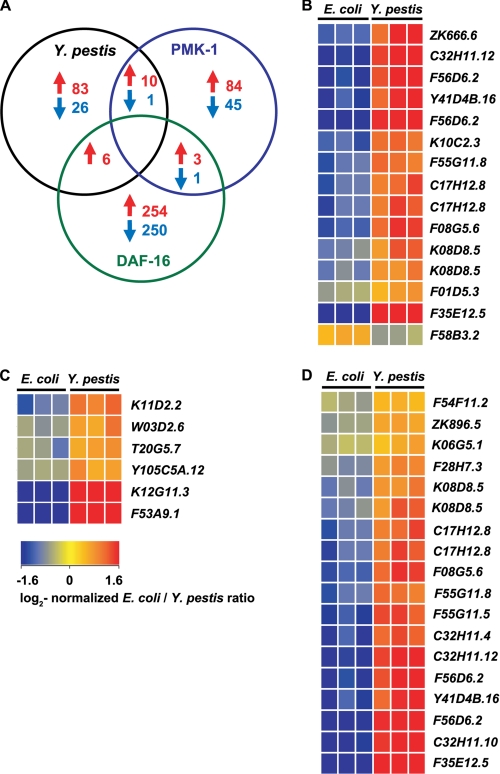
To identify immunity pathways regulating the transcriptional response to Y. pestis, we compared the 126 genes changed >2-fold by Y. pestis with transcripts regulated by PMK-1 and those regulated by the forkhead transcription factor DAF-16 (Fig. 2). Overall, there was significant overlap of genes up-regulated by Y. pestis infection with genes up-regulated by PMK-1 (Fig. 2, A and B, and Table 2). A lysozyme gene, F58B3.2 (lys-5), down-regulated by PMK-1 was similarly down-regulated in response to Y. pestis. Genes up-regulated by Y. pestis infection also showed overlap with genes that were up-regulated by DAF-16 (Fig. 2, A and C); however, greater overlap of genes up-regulated in response to Y. pestis was observed with genes down-regulated by DAF-16 (Fig. 2D). Taken together, these data suggest that PMK-1 plays an important role in the control of genes that are up-regulated in response to Y. pestis.
FIGURE 2.
Genes induced in response to Y. pestis overlap with factors regulated by PMK-1 MAPK. A, Venn diagram illustrating the number of transcripts up-regulated and down-regulated by Y. pestis (p < 0.05, 2-fold) that were similarly influenced by PMK-1 (20) and DAF-16 (44). B–D, heat maps reflecting relative levels of gene expression during Y. pestis infection for clusters of PMK-1-regulated genes (20) (B), genes up-regulated by DAF-16 (44) (C), and genes down-regulated by DAF-16 (44)(D).
TABLE 2.
Y. pestis induces expression of transcripts similar to those regulated by PMK-1
Genes showing a significant change in expression during Y. pestis infection (p < 0.05; see supplemental Table 1) were compared with genes regulated by the PMK-1 pathway (20). Those genes whose expression significantly changed in response to Y. pestis and that are dependent upon pmk-1 are shown.
| Description | Sequence ID | Y. pestis regulationa | pmk-1 regulationb |
|---|---|---|---|
| CUB-like | F35E12.5 | 62.54 | 3.6 |
| C32H11.12 | 9.97 | 5.0 | |
| F08G5.6 | 6.21 | 28.9 | |
| C17H12.8 | 5.28 | 8.3 | |
| F55G11.8 | 4.68 | 6.8 | |
| K08D8.5 | 4.07 | 3.5 | |
| C-type lectin | F56D6.2 | 9.24 | 5.0 |
| ZK666.6 | 6.88 | −3.3 | |
| DUF274 | Y41D4B.16 | 7.96 | 3.3 |
| Peptidase | K10C2.3 | 5.12 | 2.5 |
| Metridin-like Shk toxin | F01D5.3 | 2.48 | 3.3 |
| Lysozyme | F58B3.2 | −2.33 | −2.9 |
a Fold-change of transcript expression was following 24 h of Y. pestis infection in wild-type N2 animals.
b Fold-change of transcript expression in daf-2(e1368) animals was compared with daf-2(e1368); pmk-1(km25) animals, as described previously (20).
Genes That Are Markers of Response to Infection Contribute to C. elegans Resistance against Y. pestis-mediated Killing
The expression profiling studies revealed that genes encoding CUB-like domains were among the most highly induced genes in response to Y. pestis (Table 1). The CUB domain, named for its founding members C1r/C1s, Uegf, and Bmp1, contains 110 amino acids found in extracellular and plasma membrane-associated proteins involved in a variety of different functions, including complement activation, development, tissue repair, tumor suppression, and inflammation (27–29). Mounting evidence supports the role of proteins carrying CUB-like domains in C. elegans immunity. Microarray expression analysis examining the response of C. elegans to several pathogens, including Microbacterium nematophilum (19), Serratia marcescens (21, 22), and Pseudomonas aeruginosa (15, 20, 22), have previously demonstrated induction of different CUB-like factors during infection. Moreover, distinct subsets of CUB-like genes are induced in response to different classes of pathogens (22), suggesting that different regulatory mechanisms may tightly control their expression.
Of 50 C. elegans genes encoding CUB-like factors, 16 showed a significant change in expression in response to Y. pestis (Table 3), with expression of several of these genes being highly induced (Table 1). This over-representation of CUB-like genes in the inducible response to Y. pestis suggests that they may play a role in host defense. To address whether CUB-like factors contribute to host defense against Y. pestis, we used RNAi to inhibit the expression of individual CUB-like genes and determined animal susceptibility to infection. Of nine Y. pestis-induced CUB-like genes that were evaluated, RNAi inhibition of F08G5.6, C17H12.8, and C32H11.12 enhanced the susceptibility of the nematodes to Y. pestis-mediated killing (Table 4). The minor changes observed by inhibiting individual genes by RNAi could be attributed to incomplete RNAi or redundancy of function among members of the CUB-like family that are induced during infection. Consistent with our findings, RNAi of individual genes that are markers of inducible innate immunity has little effect on the resistance of C. elegans to different pathogens, maybe due to functional redundancy (15, 20, 21, 23).
TABLE 3.
Over-represented functional terms in the response to Y. pestis
C. elegans transcripts showing a significant change in expression during Y. pestis infection (p < 0.05; see supplemental Table 1) were analyzed using FatiGO software for over-representation of functional terms. Over-represented Interpro terms are shown.
| Terma | Domain name | Genesb | p valuec |
|---|---|---|---|
| IPR003366 | CUB-like region | 16/50 | 1.61E-15 |
| IPR002035 | von Willebrand factor, type A | 8/52 | 5.62E-05 |
| IPR001461 | Peptidase A1 | 5/18 | 1.60E-03 |
| IPR001304 | C-type lectin | 15/260 | 4.86E-03 |
| IPR001969 | Peptidase aspartic, active site | 4/23 | 4.04E-02 |
| IPR009673 | Unknown function, DUF 1261 | 4/11 | 4.04E-02 |
a Interpro entry accession number is below.
b Number of genes that show a significant change in expression during Y. pestis infection relative to the total number of C. elegans genes for the indicated domain.
c p values were calculated by FatiGO software using Fisher's exact test and adjusted for multiple comparisons using the Benjamini and Hochberg method.
TABLE 4.
Effect of RNAi inhibition of CUB-like genes on susceptibility to Y. pestis infection
Wild-type (N2) nematodes were fed E. coli expressing double-stranded RNA to knock down expression of the indicated gene. Second generation RNAi animals were allowed to develop to the young adult stage and were then transferred to plates containing Y. pestis. Survival of animals on Y. pestis was monitored daily.
| RNAi treatment | Survival ratio (No. of animals)a |
p valueb | |
|---|---|---|---|
| Experiment 1 | Experiment 2 | ||
| Vector control | 1.00 (89) | 1.00 (91) | |
| F35E12.5 | 1.02 (82) | 0.98 (90) | 0.8363 |
| C32H11.10 | 0.90 (85) | 0.88 (82) | 0.0589 |
| C32H11.12 | 0.82 (82) | 0.98 (84) | 0.0018* |
| F55G11.5 | 0.99 (88) | 0.81 (84) | 0.4396 |
| C32H11.4 | 0.90 (84) | 1.00 (87) | 0.1850 |
| F35E12.6 | 0.93 (93) | 0.86 (86) | 0.1417 |
| F08G5.6 | 0.81 (86) | 0.88 (83) | 0.0017* |
| C17H12.8 | 0.86 (90) | 0.85 (92) | 0.0006* |
| K08D8.5 | 0.92 (94) | 0.89 (89) | 0.0806 |
a Represents the ratio of the TD50 for animals given the designated RNAi treatment relative to the TD50 of animals given control vector treatment. TD50 values were calculated using nonlinear regression analysis (GraphPad Prism). The number of animals scored as dead in each individual experiment are indicated in parentheses. Data from two independent experiments are shown.
b Statistical analyses were conducted on survival data pooled from two independent experiments. p values were determined using the log rank test. p values <0.05 were considered significant and are denoted by an asterisk.
PMK-1/p38 MAPK Plays a Key Role in the Regulation of CUB-like Genes in Response to Y. pestis Infection
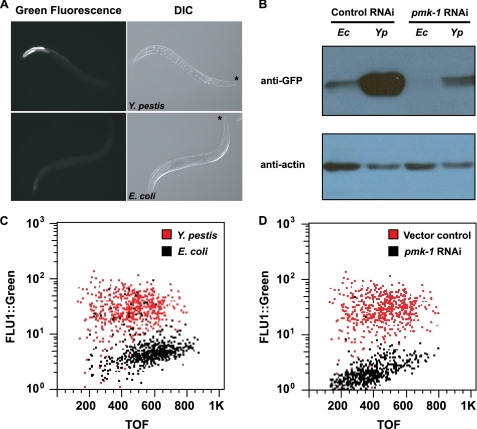
The microarray analysis indicated that at least six CUB-like genes that are up-regulated in response to Y. pestis infection may be positively regulated by PMK-1 (Table 2). Thus, we examined whether increased expression of the most highly induced CUB-like gene in response to Y. pestis infection, F35E12.5, was dependent upon PMK-1. Using AY101 transgenic animals, carrying a transcriptional reporter for F35E12.5 (pF35E12.5::gfp), the expression of gfp was examined following infection with Y. pestis. Although only minimal expression of pF35E12.5::gfp was observed in AY101 animals fed E. coli, pF35E12.5::gfp was highly expressed in the intestine of animals exposed to Y. pestis (Fig. 3A). Consistent with visual observation, Western blot analysis and large particle flow cytometry (COPAS Biosort instrument) confirmed strong expression of GFP protein levels in animals infected with Y. pestis (Fig. 3, B and C). RNAi inhibition of pmk-1 considerably reduced pF35E12.5::gfp expression in AY101 animals (Fig. 3, B and D), demonstrating that inducible expression of F35E12.5 in response to Y. pestis was largely dependent upon PMK-1.
FIGURE 3.
PMK-1 MAPK contributes to the induction of C. elegans genes in response to Y. pestis. A, AY101 acIs101[pDB09.1(pF35E12.5:: gfp);pRF4(rol-6(su1006))] animals containing a transcriptional reporter for F35E12.5 were exposed to Y. pestis (top panels) or E. coli (bottom panels) for 24 h and imaged using fluorescence microscopy. An asterisk indicates the head of the animals. DIC, differential interference contrast. B, Western blot analysis of GFP expression levels in AY101 animals. AY101 transgenic animals were treated with a control vector or pmk-1-specific RNAi. Total nematode lysates were collected from 50 adult animals following 24 h of exposure to E. coli (Ec) or Y. pestis (Yp). C and D, analysis of GFP fluorescence intensity in AY101 animals using the COPAS BIOSORT instrument (Union Biometrica, Holliston, MA) (45). C, AY101 animals treated with a control RNAi vector were exposed to Y. pestis or E. coli for 24 h. D, AY101 animals treated with control vector or pmk-1 RNAi were exposed to Y. pestis. GFP fluorescence intensity (FLU-1) was plotted against adult animal size, measured as time of flight (TOF). Each dot represents an individual nematode. All results are representative of three or more independent experiments.
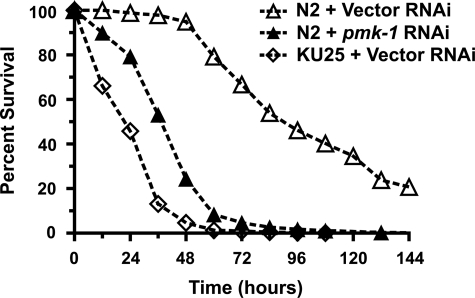
To determine the overall contribution of PMK-1-dependent effectors on host defense to Y. pestis, we measured susceptibility to infection in animals lacking expression of pmk-1. RNAi-mediated knockdown of pmk-1 in wild-type animals revealed an enhanced susceptibility to pathogen infection, indicating a crucial role for PMK-1 in host defense against Y. pestis infection (Fig. 4). Strain KU25, containing the pmk-1(km25) deletion allele, similarly exhibited enhanced susceptibility to Y. pestis (Fig. 4). Interestingly, KU25 animals were more susceptible to Y. pestis infection than pmk-1 RNAi animals. This difference in susceptibility to pathogen infection between KU25 and wild-type animals receiving pmk-1 RNAi suggests that RNAi may not be efficiently inhibiting pmk-1 in all the tissues involved in C. elegans immune responses.
FIGURE 4.
PMK-1 MAPK is necessary for host defense against Y. pestis in the nematode C. elegans. Kaplan-Meier survival analysis of C. elegans strains challenged with Y. pestis KIM5. N2 wild-type animals were treated with either a control vector or pmk-1 RNAi. Strain KU25 contains the pmk-1(km25) deletion allele. Survival assays were performed as described under “Materials and Methods,” and animal survival was monitored every 12–24 h. Log rank analysis confirmed a significant decrease in survival following RNAi knockdown of pmk-1 (p < 0.0001) and a significant decrease in survival in strain KU25 (p < 0.0001) when compared with N2 animals.
Intestinal PMK-1/p38 MAPK Is Critical for Immunity toward Y. pestis
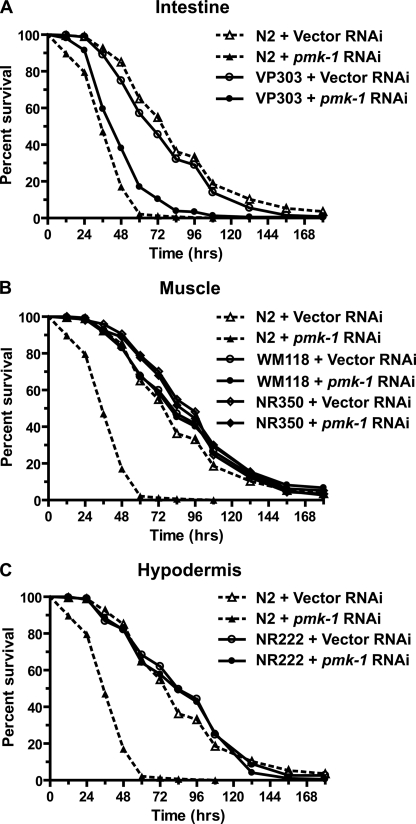
To evaluate tissue-specific contributions of PMK-1 in response to Y. pestis infection, we utilized C. elegans strains that enrich RNAi activity to the intestine (strain VP303), muscle (strains WM118 and NR350), or hypodermis (strain NR222). Enriched RNAi knockdown of pmk-1 in the intestine of VP303 animals resulted in an enhanced susceptibility to Y. pestis, similar to pmk-1 RNAi in wild-type N2 animals (Fig. 5A). In contrast, RNAi of pmk-1 in muscle or hypodermis had only minimal effects on the susceptibility to Y. pestis infection (Fig. 5 and supplemental Table 2). These findings indicate that the immune response to Y. pestis requires intestinal activity of PMK-1.
FIGURE 5.
Intestinal PMK-1 is required for survival on Y. pestis. Kaplan-Meier survival analysis of C. elegans strains challenged with Y. pestis KIM5 following RNAi knockdown of pmk-1 in the intestine (A), muscle (B), and hypodermis (C). Survival assays were performed as described under “Materials and Methods,” and animal survival was monitored every 12–24 h. Each plot represents the combined data of two or more experiments and a minimum of 90 animals (supplemental Table 2). Log rank analysis confirmed a significant decrease in survival following RNAi knockdown of pmk-1 in wild-type strain N2 (p < 0.0001) and strain VP303 (p < 0.0001) when compared with control RNAi treatment.
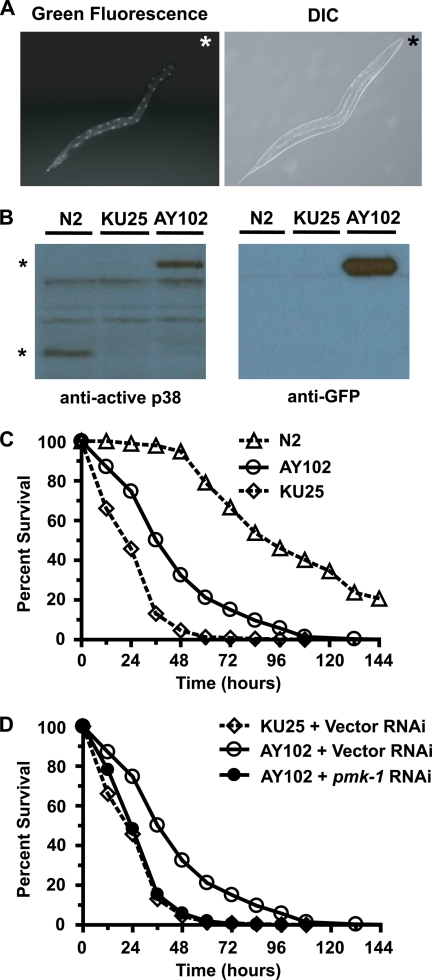
As knockdown of intestinal PMK-1 increased susceptibility to Y. pestis in wild-type animals, we hypothesized that expression of intestinal PMK-1 would conversely increase resistance to Y. pestis in C. elegans strain KU25, which contains the pmk-1(km25) deletion allele and rapidly succumbs to infection (Fig. 4). AY102 transgenic animals were developed by introducing the acEx102[pvha-6::pmk-1::gfp; rol-6(su1006)] extrachromosomal array into KU25 animals, resulting in expression of PMK-1::GFP exclusively within the nematode intestine. In AY102 animals, PMK-1::GFP fluorescent fusion protein was observed in nuclei and the cytosol of intestinal cells (Fig. 6A). Western blot analysis using an antibody that recognizes phosphorylated, active PMK-1 recognized a phosphorylated form of PMK-1::GFP (Fig. 6B), indicating that PMK-1::GFP may be functional. Consistent with the observation supporting that intestinal PMK-1::GFP is active, AY102 animals are significantly more resistant to Y. pestis infection than KU25 animals (Fig. 6C and supplemental Table 3). Interestingly, intestinal expression of PMK-1::GFP did not fully rescue the enhanced susceptibility to Y. pestis infection of KU25 animals (Fig. 6C). This indicates that even though PMK-1 activity in the intestine is crucial in defense against Y. pestis, it may also be required in other tissues. Alternatively, despite phosphorylation of PMK-1::GFP, the fusion protein might not retain complete functionality in comparison with wild-type PMK-1. Importantly, the increased resistance to infection in AY102 animals was specific to expression of intestinal PMK-1::GFP as the enhanced resistance to Y. pestis was abolished following treatment with pmk-1 RNAi (Fig. 6D).
FIGURE 6.
Expression of intestine-restricted PMK-1 enhances resistance to Y. pestis. A, AY102 transgenic animals expressing intestinal PMK-1::GFP were imaged using fluorescence microscopy. An asterisk indicates the head of the animal. B, Western blot analysis of total nematode lysates from wild-type (N2) animals, KU25 animals containing the pmk-1(km25) deletion allele, and AY102 animals. An asterisk indicates PMK-1 phosphoprotein (lower band) and PMK-1::GFP phosphoprotein (upper band). C and D, Kaplan-Meier survival analysis of N2, KU25 pmk-1(km25), and AY102 pmk-1(km25) acEx102[pDB09.2(pvha-6::pmk-1::gfp); pRF4(rol-6(su1006))] animals following treatment with a control vector (C) or comparison of control vector with pmk-1 RNAi (D). Each plot represents the combined data of four or more experiments and a minimum of 500 animals (supplemental Table 3). Log rank analysis confirmed a significant increase in resistance in strain AY102 (p < 0.0001), when compared with strain KU25. Additionally, knockdown of pmk-1 abolished the increased resistance in strain AY102 (p < 0.0001), relative to control treatment of AY102 animals.
DISCUSSION
Successful transmission of the vector-borne pathogen Y. pestis requires that the bacterium be able to rapidly adapt to diverse host environments. Following transmission to a mammalian host, plague bacteria that withstand early innate defenses can persist in the extracellular environment through expression of factors that enable increased resistance to serum and phagocytosis, induce apoptosis of immune cells, and suppress the inflammatory response (30–38). Thus, for the host, efficient recognition and elimination of the pathogen by the innate immune response during early stage infection is critical in defense against Y. pestis. Using the invertebrate host C. elegans to model early stage infection with Y. pestis, we demonstrate a robust inducible response during infection and identify the PMK-1/p38 MAPK pathway as a central component in protective immunity.
PMK-1/p38 MAPK regulates the expression of several classes of genes induced during pathogen infection such as those encoding CUB-like domains, C-type lectins, and ShK toxins (20). Factors including the CUB-like genes F08G5.6, F20G2.5, and F35E12.7 (15, 26) and the C-type lectin genes ZK666.6, E03H4.10, and C54D1.2 (19) have been demonstrated to affect the host response during infection with either P. aeruginosa or M. nematophilum, supporting a function in immunity for these factors. Consistent with these observations, a significant number of PMK-1-regulated factors were over-represented in the inducible response to Y. pestis (Fig. 2 and Table 2). Our findings also reveal a role for the CUB-like genes F08G5.6, C17H12.8, and C32H11.12 in host defense to Y. pestis. Although knockdown of several CUB-like factors induced in response to Y. pestis failed to alter pathogen susceptibility, possibly due to redundancy, enhanced susceptibility of animals lacking pmk-1 demonstrates a convincing role overall for PMK-1-/p38-dependent factors in the host response to Y. pestis. Similar studies show a lack of a phenotype when candidate immune effector genes are inhibited by RNAi but show strong phenotypes when upstream regulators of their expression are altered (15, 20, 23).
In C. elegans, PMK-1/p38 MAPK signaling is involved in the response to diverse physiological stimuli and environmental stresses. Cell-autonomous activity of PMK-1/p38 MAPK signaling in C. elegans has been described during oxidative stress in the intestine (39) and in the localized response to Drechmeria coniospora infection in the epidermis (40). Our findings highlight a cell-autonomous role of PMK-1/p38 MAPK in the intestinal response to pathogen infection. This response of PMK-1/p38 MAPK in the intestine is consistent with the localization of Y. pestis colonization (8) and is also the major site of expression for markers of C. elegans immunity (Fig. 3) (22).
In summary, our findings reveal that C. elegans mounts a striking inducible response to Y. pestis consisting of several factors that are prominent markers of nematode immunity. This inducible response is largely regulated by the cell-autonomous activity of intestinal PMK-1/p38. Although cell-autonomous function of PMK-1/p38 MAPK is shown to regulate intestinal host defense in the present study, recent observations from our laboratory indicate that the nematode nervous system can also influence the p38 MAPK intestinal response to pathogens (41). Additional examples of non-cell-autonomous regulation of inducible immunity have been demonstrated for the DAF-16 signaling in the intestine (42) and the transforming growth factor-β-mediated response in the epidermis (43). Together, these observations indicate that mechanisms underlying the regulation of PMK-1/p38 MAPK immunity can be quite complex and likely involve both a localized response to pathogen infection while jointly integrating cues from the rest of the organism.
Supplementary Material
Acknowledgments
We thank the Duke Microarray Facility, Dr. Timothy Hammond (Durham Veterans Affairs Medical Center), for use of the COPAS Biosort instrument, and members of the Aballay laboratory for helpful discussions. Some strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota), which is funded by the National Institutes of Health National Center for Research Resources. Strain VP303 was kindly provided by Kevin Strange (Vanderbilt University).
This work was supported, in whole or in part, by National Institutes of Health Grants GM070977 (to A. A.) and AI075844 (to D. D. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–4.
- NGM
- nematode growth medium
- RNAi
- RNA interference
- MAPK
- mitogen-activated protein kinase
- GFP
- green fluorescent protein.
REFERENCES
- 1.Achtman M., Zurth K., Morelli G., Torrea G., Guiyoule A., Carniel E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14043–14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wren B. W. (2003) Nat. Rev. Microbiol. 1, 55–64 [DOI] [PubMed] [Google Scholar]
- 3.Titball R. W., Hill J., Lawton D. G., Brown K. A. (2003) Biochem. Soc. Trans. 31, 104–107 [DOI] [PubMed] [Google Scholar]
- 4.Marceau M. (2005) Curr. Issues Mol. Biol. 7, 151–177 [PubMed] [Google Scholar]
- 5.Pujol C., Bliska J. B. (2005) Clin. Immunol. 114, 216–226 [DOI] [PubMed] [Google Scholar]
- 6.Boman H. G., Hultmark D. (1987) Annu. Rev. Microbiol. 41, 103–126 [DOI] [PubMed] [Google Scholar]
- 7.Vallet-Gely I., Lemaitre B., Boccard F. (2008) Nat. Rev. Microbiol. 6, 302–313 [DOI] [PubMed] [Google Scholar]
- 8.Styer K. L., Hopkins G. W., Bartra S. S., Plano G. V., Frothingham R., Aballay A. (2005) EMBO Rep. 6, 992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qadota H., Inoue M., Hikita T., Köppen M., Hardin J. D., Amano M., Moerman D. G., Kaibuchi K. (2007) Gene 400, 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espelt M. V., Estevez A. Y., Yin X., Strange K. (2005) J. Gen. Physiol. 126, 379–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner S. (1974) Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Une T., Brubaker R. R. (1984) Infect. Immun. 43, 895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Shahrour F., Minguez P., Vaquerizas J. M., Conde L., Dopazo J. (2005) Nucleic Acids Res. 33, W460–W464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 15.Shapira M., Hamlin B. J., Rong J., Chen K., Ronen M., Tan M. W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14086–14091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Welchman D. P., Zipperlen P., Ahringer J. (2003) Nature 421, 231–237 [DOI] [PubMed] [Google Scholar]
- 17.Wang J., Tokarz R., Savage-Dunn C. (2002) Development 129, 4989–4998 [DOI] [PubMed] [Google Scholar]
- 18.Oka T., Toyomura T., Honjo K., Wada Y., Futai M. (2001) J. Biol. Chem. 276, 33079–33085 [DOI] [PubMed] [Google Scholar]
- 19.O'Rourke D., Baban D., Demidova M., Mott R., Hodgkin J. (2006) Genome Res. 16, 1005–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troemel E. R., Chu S. W., Reinke V., Lee S. S., Ausubel F. M., Kim D. H. (2006) PLoS Genet. 2 (11):e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallo G. V., Kurz C. L., Couillault C., Pujol N., Granjeaud S., Kohara Y., Ewbank J. J. (2002) Curr. Biol. 12, 1209–1214 [DOI] [PubMed] [Google Scholar]
- 22.Alper S., McBride S. J., Lackford B., Freedman J. H., Schwartz D. A. (2007) Mol. Cell. Biol. 27, 5544–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerry S., TeKippe M., Gaddis N. C., Aballay A. (2006) PLoS ONE 1, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong D., Bazopoulou D., Pujol N., Tavernarakis N., Ewbank J. J. (2007) Genome Biol. 8, R194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couillault C., Pujol N., Reboul J., Sabatier L., Guichou J. F., Kohara Y., Ewbank J. J. (2004) Nat. Immunol. 5, 488–494 [DOI] [PubMed] [Google Scholar]
- 26.Nandakumar M., Tan M. W. (2008) PLoS Genet. 4, e1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bork P., Beckmann G. (1993) J. Mol. Biol. 231, 539–545 [DOI] [PubMed] [Google Scholar]
- 28.Abdul Ajees A., Gunasekaran K., Volanakis J. E., Narayana S. V., Kotwal G. J., Murthy H. M. (2006) Nature 444, 221–225 [DOI] [PubMed] [Google Scholar]
- 29.Mollenhauer J., Herbertz S., Holmskov U., Tolnay M., Krebs I., Merlo A., Schrøder H. D., Maier D., Breitling F., Wiemann S., Gröne H. J., Poustka A. (2000) Cancer Res. 60, 1704–1710 [PubMed] [Google Scholar]
- 30.Bartra S. S., Styer K. L., O'Bryant D. M., Nilles M. L., Hinnebusch B. J., Aballay A., Plano G. V. (2008) Infect. Immun. 76, 612–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolodziejek A. M., Sinclair D. J., Seo K. S., Schnider D. R., Deobald C. F., Rohde H. N., Viall A. K., Minnich S. S., Hovde C. J., Minnich S. A., Bohach G. A. (2007) Microbiology 153, 2941–2951 [DOI] [PubMed] [Google Scholar]
- 32.Anisimov A. P., Dentovskaya S. V., Titareva G. M., Bakhteeva I. V., Shaikhutdinova R. Z., Balakhonov S. V., Lindner B., Kocharova N. A., Senchenkova S. N., Holst O., Pier G. B., Knirel Y. A. (2005) Infect. Immun. 73, 7324–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Y., Rosqvist R., Forsberg A. (2002) Infect. Immun. 70, 1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang X. Z., Lindler L. E. (2004) Infect. Immun. 72, 7212–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viboud G. I., Bliska J. B. (2005) Annu. Rev. Microbiol. 59, 69–89 [DOI] [PubMed] [Google Scholar]
- 36.Cornelis G. R. (2002) J. Cell Biol. 158, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welkos S., Friedlander A., McDowell D., Weeks J., Tobery S. (1998) Microb. Pathog. 24, 185–196 [DOI] [PubMed] [Google Scholar]
- 38.Montminy S. W., Khan N., McGrath S., Walkowicz M. J., Sharp F., Conlon J. E., Fukase K., Kusumoto S., Sweet C., Miyake K., Akira S., Cotter R. J., Goguen J. D., Lien E. (2006) Nat. Immunol. 7, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 39.An J. H., Vranas K., Lucke M., Inoue H., Hisamoto N., Matsumoto K., Blackwell T. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16275–16280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pujol N., Zugasti O., Wong D., Couillault C., Kurz C. L., Schulenburg H., Ewbank J. J. (2008) PLoS Pathog. 4, e1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Styer K. L., Singh V., Macosko E., Steele S. E., Bargmann C. I., Aballay A. (2008) Science 322, 460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawli T., Tan M. W. (2008) Nat. Immunol. 9, 1415–1424 [DOI] [PubMed] [Google Scholar]
- 43.Zugasti O., Ewbank J. J. (2009) Nat. Immunol. 10, 249–256 [DOI] [PubMed] [Google Scholar]
- 44.Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C. (2003) Nature 424, 277–283 [DOI] [PubMed] [Google Scholar]
- 45.Pulak R. (2006) Methods Mol. Biol. 351, 275–286 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.