Abstract
A group of phosphoinositide 3-kinase (PI3K) inhibitors, such as 3-methyladenine (3-MA) and wortmannin, have been widely used as autophagy inhibitors based on their inhibitory effect on class III PI3K activity, which is known to be essential for induction of autophagy. In this study, we systematically examined and compared the effects of these two inhibitors on autophagy under both nutrient-rich and deprivation conditions. To our surprise, 3-MA is found to promote autophagy flux when treated under nutrient-rich conditions with a prolonged period of treatment, whereas it is still capable of suppressing starvation-induced autophagy. We first observed that there are marked increases of the autophagic markers in cells treated with 3-MA in full medium for a prolonged period of time (up to 9 h). Second, we provide convincing evidence that the increase of autophagic markers is the result of enhanced autophagic flux, not due to suppression of maturation of autophagosomes or lysosomal function. More importantly, we found that the autophagy promotion activity of 3-MA is due to its differential temporal effects on class I and class III PI3K; 3-MA blocks class I PI3K persistently, whereas its suppressive effect on class III PI3K is transient. Because 3-MA has been widely used as an autophagy inhibitor in the literature, understanding the dual role of 3-MA in autophagy thus suggests that caution should be exercised in the application of 3-MA in autophagy study.
Keywords: Enzymes/Inhibitors, Subcellular Organelles/Lysosomes, Autophagy, Cell Death, mTOR, Phosphatidylinositol 3-Kinase, 3-Methyladenine, Autophagy-related Gene
Introduction
Autophagy refers to an evolutionarily conserved process in which intracellular proteins and organelles are sequestered in autophagosomes and subsequently degraded by lysosomal enzymes for the purpose of recycling cellular components to sustain metabolism during nutrient deprivation and to prevent accumulation of damaged proteins and organelles (1, 2). Autophagy is a dynamic process, consisting of several sequential stages (initiation, nucleation, elongation, and maturation) controlled by a group of autophagy-related genes (Atg genes). So far, more than 30 Atg genes have been identified in yeast, and many of them have homologues in mammalian cells (3). Upstream of ATG proteins, mammalian target of rapamycin (mTOR)4 has been well studied as the key regulatory molecule (4). mTOR is a serine/threonine protein kinase serving as the convergence point for many of the upstream stimuli and pathways to regulate cell growth, cell proliferation, cell motility, cell survival, protein synthesis, translation, and autophagy (5–7). Abundance of nutrients, including growth factors, glucose, and amino acids will activate mTOR and suppress autophagy, whereas nutrient deprivation will suppress mTOR, leading to activation of autophagy. At present, the molecular mechanisms downstream of mTOR responsible for its anti-autophagic function have not been fully understood. In yeast, TOR directly targets the ATG13-ATG1 complex and suppresses its function at the initiation stage of autophagy (8). In mammalian cells, the complex containing ULK1 (the ATG1 homologue), ATG13, and FIP200 is directly controlled by mTOR and is a critical part of the autophagy machinery in response to nutritional status (9, 10).
Among many signaling pathways controlling mTOR activation, phosphoinositide 3-kinase (PI3K) is the key element in response to growth factors, such as insulin (11). PI3K is a lipid kinase that phosphorylates phosphatidylinositol (PI) at the 3′-position of the inositol ring. In mammalian cells, there are three classes of PI3K: the class I PI3K mainly phosphorylates PI 4,5-bisphosphate to produce phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3), whereas the class III PI3K/hVps34 only phosphorylates PI to generate phosphatidylinositol 3-phosphate (PI3P). Little is currently known about the class II PI3K, which appears to catalyze PI3P and PI 3,4-bisphosphate from PI (12, 13). The class I PI3K is a heterodimer consisting of a p85 regulatory and a p110 catalytic subunit and is mainly activated via the insulin receptor, leading to activation of AKT by two kinases: PDK1 (phosphoinositide-dependent kinase-1) and mTORC2 (mTOR complex 2). The fully activated AKT then acts on the tuberous sclerosis complex (consisting of TSC1-TSC2) and Rheb, leading to activation of mTOR complex 1 and subsequently suppression of autophagy (6, 14). In contrast, the class III PI3K/hVps34 is known to be a positive regulator of autophagy, in addition to its function in vesicular trafficking in the endosomal/lysosomal system (12, 15). Recent studies have revealed that hVps34 mediates autophagy at both the initiation and maturation stage of autophagosomes by forming different protein complexes with various partners, including ATG6/Beclin 1, ATG14L, UVRAG, and Rubicon (16–20).
A group of PI3K inhibitors, including 3-methyladenine (3-MA), wortmannin, and LY294002, have been well established as autophagy inhibitors (21–23). Although all of these tested PI3K inhibitors target both class I and class III PI3K indiscriminately (24, 25), they have been proposed to suppress autophagy by inhibiting the class III PI3K to block the production of PI3P (26), which is essential for the initiation of autophagy via recruitment of other ATG proteins at the isolation membrane or phagophore (4, 27). Notably, most of the earlier studies proving the anti-autophagic function of these inhibitors were conducted in conditions where cells were isolated from starved animals or cultured in nutrient-deprived medium with relatively short periods of time (23, 26, 28). The effects of these inhibitors on autophagy induced by other stimuli in a nutrient-rich environment have not been evaluated systematically.
Here we investigated the effect of two commonly used PI3K inhibitors 3-MA and wortmannin on autophagy under both nutrient-rich and deprivation conditions. To our surprise, 3-MA is found to promote autophagy when treated in full medium for a prolonged period, whereas it is still capable of inhibiting starvation-induced autophagy. In contrast, wortmannin is able to suppress autophagy regardless of the nutrient status. The autophagy promotion activity of 3-MA is due to its differential temporal effects on class I and class III PI3K; 3-MA blocks class I PI3K persistently, whereas its suppressive effect on class III PI3K is transient. Because PI3K inhibitors, such as 3-MA, are widely used as autophagy inhibitors in the literature, understanding the dual role of 3-MA in autophagy thus suggests that great caution should be exercised in the application of 3-MA in autophagy study and in the data interpretation using this inhibitor. Our data also indicate that wortmannin is a more suitable autophagy inhibitor due to its more persistent inhibition on class III PI3K.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Anti-LC3, anti-ULK1, anti- FLAG, and anti-α-tubulin antibodies were purchased from Sigma. Anti-p62 and anti-ATG5-ATG12 antibodies were from Abnova and Nanotools, respectively. Anti-ATG7 antibody was from Prosci, and anti-GFP antibody was from Covance. All other antibodies were obtained from Cell Signaling. 3-MA, wortmannin, doxycyclin, chloroquine diphosphate (CQ), rapamycin, cycloheximide (CHX), actinomycin D, and Earles' balanced salt solution (EBSS) were purchased from Sigma. Cathepsin B substrate (Z-Arg-Arg-AMC), cathepsin L substrate (Ac-His-Arg-Tyr-Arg-ACC), E-64d, and pepstatin A were purchased from Calbiochem. Lysosensor green DND-189 was from Invitrogen.
Cell Culture
Mouse embryonic fibroblasts (MEFs), L929 cells, HEK293T cells, and HeLa cells were maintained in Dulbecco's modified Eagle's medium (Sigma) containing 10% fetal bovine serum (HyClone) and 1% penicillin/streptomycin (Invitrogen) (defined as full medium in this study) in a 5% CO2 atmosphere at 37 °C. The Atg5−/− MEFs and the Tet-off Atg5 MEFs were provided by Dr. N. Misushima (Tokyo Medical and Dental University). The Tsc2−/− MEFs were a generous gift from Dr. D. J. Kwiatkowski.
Plasmids and Transfection
The HA-ULK1 plasmid was a generous gift from Dr. N. Mizushima (Tokyo Medical and Dental University), which has been described elsewhere (9, 29). The mRFP-GFP tandem fluorescence-tagged LC3 construct (tfLC3) was provided by Dr. T. Yoshimori (Osaka University) (30). Transient transfection was performed using the Lipofectamine and Plus reagents (Invitrogen) according to the manufacturer's protocol. The tfLC3 stably transfected L929 cells have been described previously (31).
Co-immunoprecipitation and Immunoblotting
Immunoblotting analysis was performed following standard procedures. The cells were lysed in M2 lysis buffer: 20 mm Tris, pH 7, 0.5% Nonidet P-40, 250 mm NaCl, 3 mm EDTA, 3 mm EGTA, 2 mm dithiothreitol, phosphatase inhibitor mixture (Pierce), and the protease inhibitor mixture (Roche Applied Science). An equal amount of protein was resolved on SDS-PAGE and transferred onto polyvinylidene difluoride membrane (Bio-Rad). After it was blocked with 5% nonfat milk, the membrane was probed with the designated first and second antibodies and developed with the enhanced chemiluminescence method (Pierce) and visualized by a Kodak Image Station 440CF (Eastman Kodak Co.). The co-immunoprecipitation assay was performed following the established protocol with minor modifications (29). Briefly, cells after treatment were lysed for 1 h in the co-immunoprecipitation lysis buffer: 40 mm HEPES, pH 7.4, 2 mm EDTA, 0.3% CHAPS, phosphatase inhibitor mixture (Pierce), and protease inhibitor mixture. The lysate containing the same amount of protein was precleared by 20 μl of protein A/G-agarose beads (Pierce) for 1 h. The postcleared lysate was then incubated with 30 μl of protein A/G-agarose beads and 1 μg of antibody and mixed overnight at 4 °C. After incubation, the beads were extensively washed with lysis buffer five times, and the immunoprecipitated proteins were eluted by boiling for 5 min in sample buffer (Bio-Rad) for Western blotting.
Confocal Microscopy
Cells were seeded to a coverglass slide chamber (Lab-Tek®, NUNC), and after the designated treatments, the cells were examined under a confocal microscope (Olympus Fluoview 2000). The GFP-LC3 puncta were quantified by counting the number in cells as described elsewhere (32). Briefly, the GFP-LC3 puncta in a single cell were manually counted under a confocal microscope. For each group, 50 cells were randomly selected for the average number of GFP-LC3 puncta/cell. The data presented were from one representative experiment of at least three independent repeats.
Small Interfering RNA (siRNA)
The nonspecific siRNA oligonucleotides and siRNA oligonucleotides targeting mouse Atg7 (ON-TARGETplus SMARTpoolTM) were obtained from Dharmacon (Layfayette, CO). The siRNAs was transfected into the L929 cells with stable expression of tfLC3 using the DharmaFECT 4 transfection reagent according to the manufacturer's protocol.
Measurement of Cathepsin B/L Activity
Determination of cathepsin B/L activity in total cell extracts was conducted as described previously with modifications (33). Briefly, cells were lysed in M2 buffer, and the lysate (25 μg of cellular protein) was then incubated with a 50 μm concentration of the fluorogenic cathepsin B/L substrates, respectively, in 100 μl of cell-free system buffer (10 mm HEPES-NaOH, pH 7.4, 220 mm mannitol, 68 mm sucrose, 2 mm NaCl, 2.5 mm KH2PO4, 0.5 mm EGTA, 2 mm MgCl2, 5 mm pyruvate, 0.1 mm phenylmethylsulfonyl fluoride, and 1 mm dithiothreitol) in a 96-well plate for 1 h at 37 °C. The released AMC/ACC fluorophores were monitored by a fluorometer (Tecan SpectraFluor Plus) at an excitation wavelength of 409/380 nm and an emission wavelength of 505/460 nm. Data are presented as percentage of fluorescence intensity compared with the control group.
Measurement of Intralysosomal pH
The measurement of intralysosomal pH was performed based on an established method with modifications (34). Briefly, after designated treatments, cells were incubated with 1 μm Lysosensor probe (Invitrogen) for 1 h and then were trypsinized and collected and resuspended in phenol red-free medium. Ten thousand cells from each sample were analyzed using FACSCalibur flow cytometry (BD Biosciences) and CellQuest software.
Reverse Transcription-PCR
RNA was extracted with the RNeasy kit (Qiagen). 1 μg of total RNA from each sample was used as a template for cDNA synthesis with a QuantiTect reverse transcriptase kit (Qiagen). An equal volume of cDNA product was used in the PCR performed using the TopTaq Master Mix Kit (Qiagen). PCR amplification was performed using the following primers (purchased from Sigma): mouse p62 gene 5′-AGCTGCCCTCAGCCCTCTA-3′ (forward) and 5′-GGCTTCTCTTCCCTCCATGTT-3′ (reverse) and mouse glyceraldehyde-3-phosphate dehydrogenase 5′-GTGTTCCTACCCCCAATGT-3′ (forward) and 5′-TGTCATCATACTTGGCAGGTTTC-3′ (reverse). The PCR conditions were set according to the standard protocol. The PCR products were resolved on agarose gel containing GelRedTM nucleic acid gel stain (Biotium) and exposed using a Kodak Image Station 440CF.
Quantification of PI3P/PI(3,4,5)P3
Cells were seeded on 75-cm2 flasks and grown to 60% confluence and then treated as indicated in the legend to Fig. 5. The extraction and quantification of the PI3P/PI(3,4,5)P3 were determined by the PI3P/PI(3,4,5)P3 mass strip kits, respectively (Echelon Biosciences), following the instructions from the manufacturer. In addition, we also adopted another method for the quantification of PI(3,4,5)P3, following a method described previously (35). Briefly, cells were collected in 0.8 ml of ice-cold methanol, 12 n HCl (96:4), supplemented with 2 mm AlCl3. Lipids were then extracted by adding 0.4 ml of ice-cold chloroform and 0.4 ml of ice-cold water. The lower organic phase was collected, and remaining solution was re-extracted twice with 0.4 ml of ice-cold chloroform. The three lipid extracts were then combined and washed with 1 ml of ice-cold methanol, 2 mm oxalic acid in water (1:0.9). Lower organic phase was collected and dried using Speedvalc (Thermo Savant, Milford, MA) and deacylated by incubation with 0.5 ml of methylamine/water:n-butanol/MetOH (36:8:9:47) at 50 °C for 1 h. The aqueous phase was dried, resuspended in 0.5 ml of 1-butanol/petroleum ether/ethyl formate (20:40:1), and extracted twice with an equal volume of water. Aqueous extracts were dried, resuspended in water, and subjected to anion exchange high pressure liquid chromatography analysis using a Dionex Ion Chromatography 3000 system (Dionex, Sunnyvale, CA).
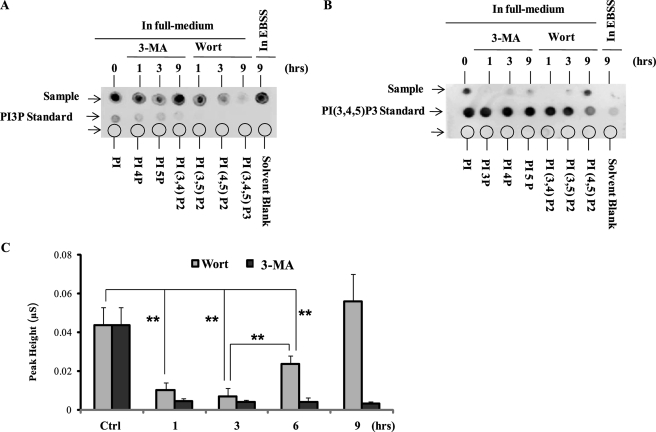
FIGURE 5.
3-MA inhibits class I and class III PI3K in different temporal patterns. A, the effect of 3-MA and wortmannin (Wort) on production of PI3P. WT MEFs were treated in EBSS or with 3-MA (5 mm), wortmannin (50 nm) in full medium as indicated. The levels of PI3P were measured using the PI3P mass strip kit as described under “Experimental Procedures.” B, the effect of 3-MA and wortmannin on production of PI(3,4,5)P3. WT MEFs were treated as in A, and the levels of PI(3,4,5)P3 were measured using the PI(3,4,5)P3 mass strip kit. C, WT MEFs were treated with 3-MA (5 mm) or wortmannin (50 mm) for 1, 3, 6, and 9 h in full medium, and the intracellular level of PI(3,4,5)P3 was measured using a chromatography-based method, as described under “Experimental Procedures.” The changes of PI(3,4,5)P3 are shown as relative peak height and are presented as means ± S.D. from three independent experiments and analyzed using Student's t test (**, p < 0.01).
Protein Degradation Assay
Protein degradation was measured as described previously (36). Briefly, HeLa cells were radiolabeled for 24 h with 0.05 mCi/ml l-[U-14C]valine. At the end of the labeling period, cells were rinsed three times with PBS. Cells were incubated for the designated times in either full medium or EBSS with or without the presence of 10 mm 3-MA.
Statistical Analysis
All data presented are representatives from at least three independent experiments. The numeric data from the protein degradation assay, cathepsin activity analysis, GFP-LC3 puncta counting, and quantification of PI(3,4,5)P3 using chromatography are presented as means ± S.D. from three independent experiments and analyzed using student t test. A p value less than 0.05 was considered as statistically significant.
RESULTS
Prolonged Treatment with 3-MA Induces Accumulation of Autophagic Markers under Nutrient-rich Conditions
PI3K inhibitors, such as 3-MA and wortmannin, have been widely used to inhibit autophagy (23, 26). Because most of the initial studies establishing the anti-autophagic function of these inhibitors were conducted under starvation condition, our initial intention was to examine the effects of these inhibitors on autophagy induced by other stimuli under nutrient-rich conditions (in full culture medium). To our surprise, it was found that prolonged treatment with 3-MA (up to 9 h) induced significant LC3 I to II conversion in wild type MEFs (Fig. 1A, left), whereas it was still effective in suppressing LC3 II conversation in the same cell type induced by starvation (by culturing in EBSS) (Fig. 1A, right). Moreover, similar results were also observed in L929 mouse fibrosarcoma cells (Fig. 1B) and human renal epithelial 293T cells (data not shown). These results thus suggest that the proautophagy effects of 3-MA are unlikely to be cell type-specific. The effectiveness of 3-MA as a PI3K inhibitor was confirmed by the suppression of S6 protein phosphorylation (Fig. 1, A and B, left), a surrogate marker for mTOR activity (37). In contrast, no similar results were observed in cells treated with wortmannin, another commonly used PI3K and autophagy inhibitor (26), in WT MEFs (Fig. 1C) and in L929 and 293T cells (data not shown). Interestingly, the inhibitory effect of wortmannin on S6 phosphorylation was found to be rather transient because the S6 protein phosphorylation level returned to normal after 6 h in cells cultured in full medium (Fig. 1C, left).
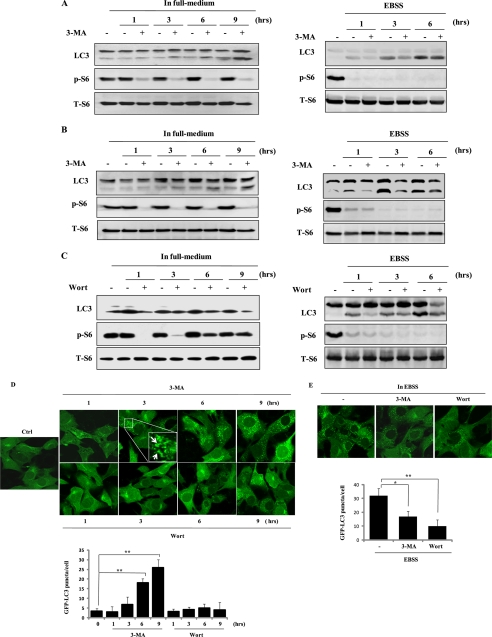
FIGURE 1.
Prolonged treatment with 3-MA in full medium leads to accumulation of autophagic markers. A, induction of LC3 I to II conversion by 3-MA in WT MEFs. WT MEFs were treated with 3-MA (5 mm) in full medium (left) or in EBSS (right; in the presence of 20 μm CQ) for the indicated periods of time. Cell lysate was collected and subject to immunoblotting. B, similar treatments were also performed in L929 cells. C, effect of wortmannin (Wort) on LC3 I to II conversion. WT MEFs were treated with wortmannin (50 nm) either in full medium (left) or in EBSS (right; in the presence of 20 μm CQ) for the indicated periods of time. D, induction of GFP-LC3 punctuation/aggregation in cells cultured in full medium by 3-MA but not by wortmannin. MEFs with stable expression of GFP-LC3 were treated with 3-MA (5 mm) or wortmannin (50 nm) as indicated and examined under a confocal microscope (×600, top). The enlarged area demonstrates the punctuated distribution of GFP-LC3 with a higher magnification. The number of GFP-LC3 puncta/cell were counted and presented (bottom; **, p < 0.01). E, effect of 3-MA and wortmannin on starvation-induced GFP-LC3 punctuation/aggregation. MEF cells with stable expression of GFP-LC3 as described in E were treated with 3-MA (5 mm) or wortmannin (50 nm) for 3 h under EBSS. The GFP-LC3 punctuation/aggregation was observed under a confocal microscope (×600, top). The GFP-LC3 puncta/cell were counted and presented (bottom; *, p < 0.05; **, p < 0.01).
In order to confirm the above results of LC3 detected by Western blot, we next tested the changes of the GFP-LC3 distribution pattern in MEFs with stable expression of the GFP-LC3 (38, 39). As shown in Fig. 1D, prolonged treatment with 3-MA, but not wortmannin, in full medium, markedly increased GFP-LC3 punctuation/aggregation. In contrast, both 3-MA and wortmannin were able to block starvation-induced GFP-LC3 punctuation/aggregation in cells cultured in EBSS (Fig. 1E). Taken together, these results demonstrate that prolonged treatment with 3-MA, but not wortmannin, under nutrient-rich conditions is capable of increasing LC3 II conversation and punctuate formation of GFP-LC3, indicating the possibility of enhanced autophagic flux.
3-MA Increases Autophagic Flux
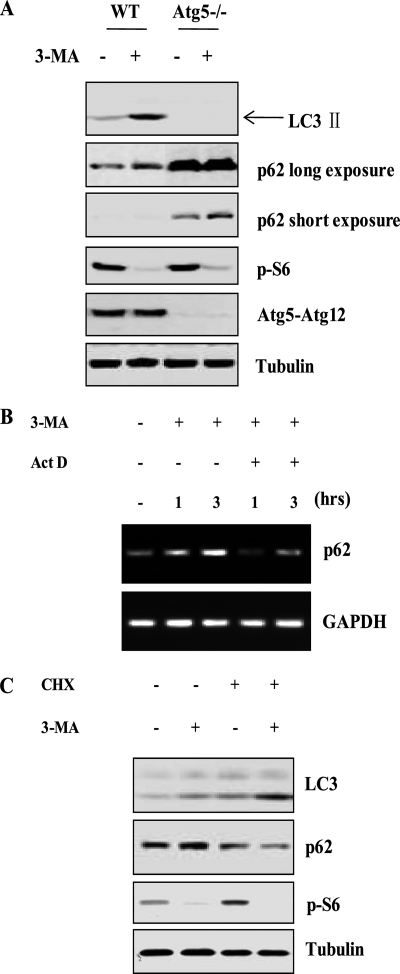
One important note in the evaluation of autophagy is that the increase of autophagic markers may represent the increased generation of autophagosomes and/or a blockage in autophagosomal maturation and degradation (33, 40–42). Moreover, it has been reported that LC3 I to II conversion could be caused by unidentified autophagy-independent mechanisms (29). Here we set out to examine whether the increased autophagic markers as observed above in cells treated with 3-MA are due to enhanced autophagic flux. First, we tested whether the induction of LC3 II conversion and punctuation/aggregation of GFP-LC3 upon 3-MA treatment are relevant to autophagy by using Atg5−/− MEFs in which the autophagy-dependent LC3 I to II conversion is known to be completely abolished (43). As shown in Fig. 2A, 3-MA failed to induce LC3 conversion in Atg5−/− MEF cells, suggesting that LC3 II accumulation is autophagy-dependent. Next, we utilized the Tet-off Atg5 MEFs in which the ATG5 and the GFP-LC3 were stably expressed (38). In this system, the Atg5 gene expression was abolished following treatment with doxycyclin (DOX) (Fig. 2B). Consistently, evident LC3 conversion was only observed in MEF cells with the presence of ATG5 (without treatment of DOX) but not in cells with the absence of ATG5 (pretreated with DOX) (Fig. 2B). Moreover, the punctuation/aggregation of GFP-LC3 was evidently induced in cells expressing ATG5 but not in cells without ATG5 (after treatment with DOX) (Fig. 2C). Previous studies have reported that LC3 II was present at both the outer and inner membrane of autophagosome, and the LC3 II will be degraded by lysosomal hydrolases after fusion with lysosome (44, 45). The GFP-tagged LC3 essentially follows the same process. Hence, the free GFP protein will be released after LC3 is degraded in autolysosome, which makes the free GFP another useful marker for autophagic flux (38, 46). As expected, in cells with ATG5, the free GFP protein increased upon 3-MA treatment, indicating the possible induction of autophagic flux, whereas no liberation of GFP was observed in Atg5-deficient MEFs (Fig. 2B).
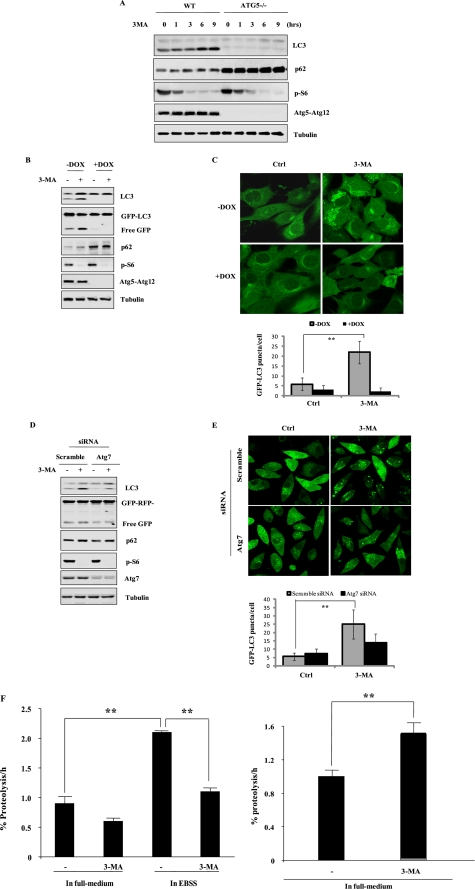
FIGURE 2.
3-MA increases autophagic flux. A, 3-MA-induced LC3 I to II conversion is ATG5-dependent. Both WT and Atg5−/− MEF cells were incubated with 3-MA (5 mm) for up to 9 h in full medium. Cell lysates were subject to immunoblotting. B, Tet-off Atg5 MEFs with stable expression of GFP-LC3 were pretreated with or without doxycyclin (DOX) for 4 days, and then cells were treated with 3-MA (5 mm) for 9 h in full medium, and cell lysate was subject to Western blotting. C, Tet-off Atg5 MEFs were prepared as described in B and treated with 3-MA (5 mm) for 9 h. Cells were examined with a confocal microscope for GFP-LC3 punctuation/aggregation (top). The GFP-LC3 puncta/cell were counted and presented (bottom; **, p < 0.01). D, L929 cells stably transfected with tfLC3 (tfLC3-L929) were transfected with Atg7 siRNA for 48 h. Cells were then treated with 3-MA (5 mm) for 9 h in full medium. Cell lysates were subject to immunoblotting. E, tfLC3-L929 as described in D were treated with 3-MA (5 mm) for 9 h in full medium, and the GFP-LC3 punctuation/aggregation was observed under a confocal microscope (×600, top). The GFP-LC3 puncta/cell were counted and presented (bottom; **, p < 0.01). F, 3-MA promotes long lived protein degradation in cells cultured in full medium. HeLa cells were radiolabeled for 24 h with 0.05 mCi/ml l-[U-14C]valine. At the end of the labeling period, cells were rinsed three times with PBS. Cells were then incubated in full medium or in EBSS with 10 mm valine in the presence or absence of 10 mm 3-MA for 4 h (top) or 8 h (bottom). The data in F are presented as means ± S.D. from three independent experiments and analyzed using Student's t test (*, p < 0.05; **, p < 0.01). Ctrl, control.
In order to further confirm the effect of 3-MA on autophagy, here we used L929 cells with stable expression of tfLC3. We silenced Atg7, another critical gene required for autophagy, in the above mentioned L929 cells and found that 3-MA-induced LC3 conversion and free GFP liberation were ATG7-dependent (Fig. 2D). Moreover, 3-MA-induced GFP-LC3 punctuation/aggregation was also diminished in L929 cells with Atg7 knockdown (Fig. 2E), further supporting the notion that 3-MA-induced accumulation of autophagic markers is dependent on autophagy and is indicative of increased autophagic flux.
The long lived protein degradation assay has been well established for assessing autophagy flux (36). In this study, the effect of 3-MA on long lived protein degradation was assessed in HeLa cells. As shown in Fig. 2F, treatment with 3-MA for a relatively short period of time (4 h) reduced the rate of protein degradation in cells cultured in EBSS (top). In contrast, longer treatment with 3-MA (8 h) in full medium significantly increased the rate of protein degradation (bottom), being consistent with the results obtained in both MEF and L929 cells. Taken together, data from this part of our study present clear evidence that prolonged treatment with 3-MA in full medium induces autophagy.
3-MA-induced Accumulation of Autophagic Markers Is Not Due to Blockage of Lysosomal Functions
Here we attempted to further exclude the possibility that 3-MA-induced accumulation of autophagic markers is due to suppression of autophagosome maturation and lysosomal degradation. The tfLC3 has been reported to be a valuable tool for examining autophagosome maturation and autolysosome formation (30). Essentially, this assay is based on the difference in the nature of the two fluorescent proteins (GFP and RFP) (i.e. RFP is much more resistant to lysosomal quenching than GFP in the acidic environment). Therefore, if the autolysosome maturation proceeds normally, it would give rise to more red-only puncta. Conversely, if the autophagosome does not fuse with lysosome or the lysosome function is impaired, most of the puncta should exhibit both red and green signals and appear to be yellow (30). Here we used the L929 cells with stable transfection of tfLC3 to analyze the effect of 3-MA on this process. As shown in Fig. 3A, treatment with 3-MA (in full medium for 9 h) led to more mRFP punctuation and accumulation than that with GFP, similar to the effect of rapamycin that was used as a positive control. More importantly, CQ, an agent capable of impairing lysosomal acidification and suppressing the protease activity, can obviously enhance both GFP and mRFP puncta formation and colocalization induced by 3-MA. The similar effects of CQ on rapamycin were also observed. These results thus provide further evidence that 3-MA is unlikely to inhibit autophagosome maturation and autolysosome formation. We further confirmed the above findings in MEF cells with stable expression of GFP-LC3 (the same cell system in Fig. 1E). As shown in Fig. 3B, CQ is able to further enhance punctuation/aggregation of GFP-LC3 in cells treated with 3-MA. Consistently, the presence of CQ also further increased the LC3 II protein level induced by 3-MA (Fig. 3C).
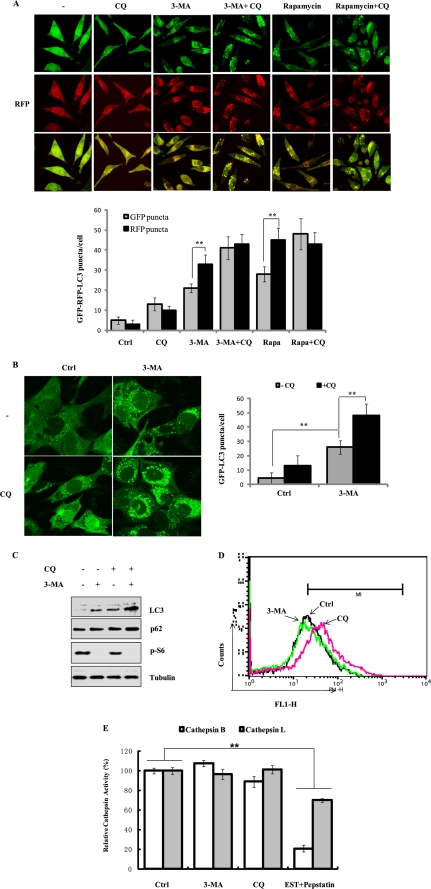
FIGURE 3.
3-MA does not affect lysosomal functions. A, effect of 3-MA on autolysosome maturation and lysosomal degradation. The L929 cells with stable expression of the tfLC3 construct (tfLC3-L929) were treated with CQ (20 μm), 3-MA (5 mm), 3-MA + CQ and rapamycin (10 nm), or rapamycin + CQ for 9 h in full medium. The cells were examined using a confocal microscope (×600, top). The RFP- and GFP-LC3 puncta/cell were counted and presented (bottom; **, p < 0.01). B, effect of CQ on GFP-LC3 punctuation/aggregation induced by 3-MA. WT MEFs with stable expression of GFP-LC3 (same as in Fig. 2B without DOX treatment) were treated with 3-MA (5 mm), CQ (20 μm), or 3-MA + CQ for 9 h in full medium. The cells were examined by confocal microscopy (×600, left). The GFP-LC3 puncta/cell were counted and presented (right; **, p < 0.01). C, MEFs were treated as described in B, and the cell lysate were subjected to immunoblotting. D, effect of 3-MA on intralysosomal pH. WT MEF cells were treated with 3-MA (5 mm) or CQ (20 μm) for 9 h in full medium, and the intralysosomal pH values were determined using a Lysosensor probe coupled with flow cytometry, as described under “Experimental Procedures.” E, effect of 3-MA on cathepsin B/L activity. WT MEFs were treated with 3-MA (5 mm), CQ (20 μm), or cathepsin inhibitors (E64d + pepstatin A, 20 μg/ml each) for 9 h, and the lysate was subjected to the cathepsin B/L activity assay, as described under “Experimental Procedures.” The data in E are presented as means ± S.D. from three independent experiments and analyzed using Student's t test (**, p < 0.01). Ctrl, control.
On the other hand, because the intralysosomal pH is a critical factor in determining the cathepsin enzymatic activity and lysosomal functions, we examined the effect of 3-MA on intralysosomal pH by using the Lysosensor (34). As shown in Fig. 3D, 3-MA treatment (in full medium for 9 h) did not affect intralysosomal pH, whereas treatment with CQ evidently enhanced the intralysosomal pH value. Finally, we tested whether 3-MA can directly affect the enzymatic activity of cathepsins measured with cell lysates on known cathepsin substrates. Treatment with 3-MA did not have any effect on cathepsin B and L activities (Fig. 3E). In contrast, evident inhibitory effect was found with the two known cathepsin inhibitors (E64d and pepstatin A). The negative effect of CQ on cathepsin enzymatic activity is understandable because CQ acts through neutralization of lysosomal pH. Taken together, these results collectively demonstrate that 3-MA has no adverse effect on lysosomal function, thus further strengthening our earlier observation that 3-MA-induced accumulation of autophagic markers is due to an increase of autophagic flux.
3-MA Up-regulates p62/SQSTM1 Gene Transcription
One intriguing observation from our study is that 3-MA treatment led to evident increase of p62 protein level (Figs. 2 (A, B, and D) and 3C). p62/SQSTM1 is a multifunctional protein that binds to LC3 and can be destructed within the autolysosome (47, 48). Thus, reduction of p62 has been regarded as a marker for increase of autophagic flux (49, 50). In this study, the p62 protein level in Atg5−/− MEFs (Fig. 2A) and in the Tet-off Atg5 MEFs pretreated with DOX (Fig. 2B) was much higher than that in the control cells, being consistent with the common understanding that a defect in autophagy could result in accumulation of p62 (49, 50). Thus, an intriguing question arises. How does 3-MA result in a higher level of p62 with enhanced autophagic flux? When we carefully examined the change of p62 upon 3-MA treatment, it was found that 3-MA increased the p62 level even in Atg5−/− MEFs (p62 short exposure in Fig. 4A) as well as in cells with DOX-mediated deletion of ATG5 (Fig. 2B), thus suggesting that such a change of p62 is independent of autophagy and is not the result of autophagy suppression.
FIGURE 4.
3-MA up-regulates p62/SQSTM1 gene transcription. A, effect of 3-MA on p62 protein level in Atg5−/− MEFs. WT and Atg5−/− MEFs were treated with 3-MA (5 mm) for 9 h in full medium, and then cell lysate was collected and subjected to immunoblotting. B, effect of 3-MA on p62 mRNA level. WT MEFs were treated with 3-MA (5 mm), actinomycin D (Act D) (10 μm), or 3-MA + ActD for 1 or 3 h in full medium. The p62 mRNA level was determined by reverse transcription-PCR. C, CHX blocks 3-MA-induced increase of p62 protein level. WT MEFs were treated with 3-MA (5 mm), CHX (10 μm), or 3-MA + CHX for 9 h in full medium, and cell lysate was subjected to immunoblotting. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To determine whether the increase of p62 protein level by 3-MA is caused by augmented gene transcription, we tested the effect of 3-MA on p62 mRNA level. As shown in Fig. 4B, it is clear that treatment with 3-MA markedly enhanced the mRNA level of p62, with a temporal pattern consistent with the change of the p62 protein level (Fig. 2A). As expected, 3-MA-induced p62 mRNA induction was abolished by Act D, a de novo mRNA synthesis inhibitor (Fig. 4B). We next investigated the effect of 3-MA on p62 protein level when de novo protein synthesis was blocked by CHX. As shown in Fig. 4C, in the presence of CHX, 3-MA could no longer enhance the p62 protein level. Importantly, 3-MA still induced LC3 conversion in the presence of CHX. These data thus suggest that the increased p62 protein level observed in cells treated with 3-MA is mediated by p62 gene expression and is not the result of autophagy suppression.
3-MA Inhibits Class I and Class III PI3K in Different Temporal Patterns
It has been well accepted that 3-MA inhibits autophagy through suppression of class III PI3K activity (26). Both 3-MA and wortmannin are known to inhibit both class I and III PI3K unselectively (24, 25), whereas they show different effects on autophagy with prolonged treatment under nutrient-rich conditions (Fig. 1). Thus, it would be of interest to examine and compare the effects of these two inhibitors on class I and class III PI3K activity in defined treatment conditions. As shown in Fig. 5A, treatment with 3-MA in full medium significantly reduced the level of PI3P, the product of class III PI3K, at early time points (1 and 3 h), whereas the PI3P level was restored to the control level at 9 h. In contrast, treatment with wortmannin resulted in sustained reduction of PI3P. Thus, data from this experiment demonstrate that 3-MA and wortmannin possess different temporal patterns on class III PI3K; the effect of 3-MA is transient, whereas that of wortmannin is more persistent. Interestingly, there was no evident reduction of PI3P level in cells cultured in EBSS for 9 h, consistent with earlier reports that the class III PI3K activation is largely constitutive and is not adversely affected by starvation (51, 52).
Next, we measured the level of PI(3,4,5)P3, the product of class I PI3K (Fig. 5B). It is indeed interesting to note that 3-MA almost completely blocked the product of PI(3,4,5)P3 up to 9 h, whereas the effect of wortmannin was transient because there was a recovery of PI(3,4,5)P3 at 9 h. As expected, culturing cells in EBSS completely eliminated PI(3,4,5)P3 production. In order to confirm the above findings, we utilized a chromatography-based method for quantification of PI(3,4,5)P3. As shown in Fig. 5C, the PI(3,4,5)P3 level was dramatically reduced for up to 9 h upon 3-MA treatment, whereas there was a recovery from 6 h onward in cells treated with wortmannin. The level of PI3P could not be measured using this method due to the technical difficulty in separating PI3P and PI4P (data not shown). Collectively, these data demonstrate that under nutrient-rich conditions, 3-MA and wortmannin inhibit both class I and III PI3K but in opposite temporal patterns. It is thus possible that the induction of autophagy by prolonged treatment with 3-MA in nutrient-rich conditions is due to suppression of class I PI3K while sparing the class III PI3K.
3-MA Disrupts the Anti-autophagic Function of mTOR Complex 1
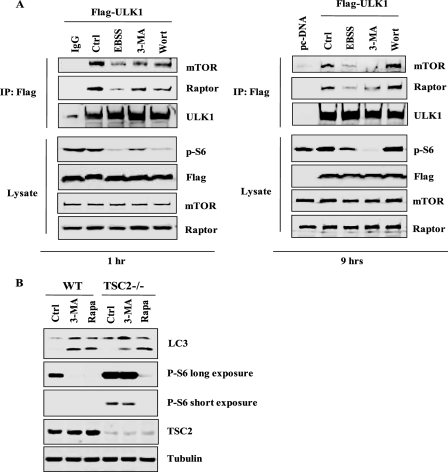
It has been well established that activation of class I PI3K will lead to activation of the Akt-mTOR signaling cascade, which in turn suppresses autophagy (14, 15). Recently, a complex consisting of ULK1 (mammalian homologue of ATG1), ATG13, and FIP200 has been identified as the downstream target molecule of mTORC1 (mTOR complex I) for autophagy regulation (9, 10, 29). Under nutrient-rich conditions, the ULK1-ATG13-FIP200 complex is binding to mTORC1 through the interaction between ULK1 and raptor, whereby mTORC1 phosphorylates ULK1 and ATG13 to inhibit autophagy. When the class I PI3K-Akt-mTOR pathway is inhibited, such as under starvation conditions, the ULK1-ATG13-FIP200 complex will disassociate from mTORC1 and become activated to promote autophagy. We thus utilized this model to determine whether 3-MA can trigger autophagy via modulation of mTORC1-ULK1 interaction. First, we found that treatment with 3-MA, wortmannin, and EBSS at an early time point (1 h) led to disruption of the interaction between ULK1 and mTORC1, evidenced by diminished mTOR and raptor that co-immunoprecipitated with ectopically expressed FLAG-ULK1 (Fig. 6A, left). The inhibitory effects of these treatments on the PI3K class I-Akt-mTORC1 pathway were confirmed by the reduction of p-S6 (Fig. 6A, left). Moreover, disruption of mTORC1 and ULK1 interaction and suppression of p-S6 were maintained in cells treated with 3-MA or EBSS up to 9 h, whereas they were restored in cells treated with wortmannin at 9 h (Fig. 6A, right). Such observations are indeed consistent with their different inhibitory capability on class I PI3K as shown earlier (Fig. 5).
FIGURE 6.
3-MA disrupts the function of mTOR complex I. A, 3-MA suppresses the interaction between ULK1 and mTOR complex 1. HEK293T cells were transfected with FLAG-ULK1 for 36 h, and cells were then treated in EBSS or with 3-MA (5 mm), wortmannin (50 nm) in full medium for 1 h (left) or 9 h (right). The cell lysate was subjected to immunoprecipitation (IP) using an antibody against FLAG, followed by immunoblotting. A fraction of the cell lysate was used for immunoblotting as indicated. B, the effect of 3-MA on autophagy in Tsc2−/− MEFs. WT and Tsc2−/− MEFs were treated with 3-MA (5 mm) or rapamycin (10 nm) for 9 h, and the cell lysate was subject to immunoblotting.
Finally, we utilized the Tsc2−/− MEFs to further confirm that sustained suppression of class I PI3K transforms 3-MA into an autophagy inducer. The deletion of TSC2 leads to constitutive activation of mTORC1 (53). As expected, there was a high level of p-S6 due to hyperactivation of mTOR in Tsc2−/− MEFs (Fig. 6B). Notably, 3-MA was unable to effectively inhibit p-S6 in Tsc2−/− cells, and as a result, 3-MA-induced LC3 I to LC3 II conversion was dramatically compromised in Tsc2−/− cells compared with wild type cells (Fig. 6B). Such observations thus support the notion that 3-MA suppresses the PI3K-Akt-mTOR signaling pathway upstream of mTOR to induce autophagy. It is also noteworthy that the autophagy machinery is still competent in Tsc2−/− MEFs, based on the observation that rapamycin was able to block mTORC1 activity and subsequently induce LC3 conversion (Fig. 6B).
DISCUSSION
3-MA as an autophagy inhibitor was first discovered via screening of purine-related substances using isolated hepatocytes from starved rats (28). Subsequent studies confirmed that 3-MA, together with wortmannin and LY294002, suppresses autophagy via inhibition of class III PI3K (23, 26). However, there are several concerns regarding the role of 3-MA as an autophagy inhibitor. For instance, 3-MA, which is usually used at very high concentrations to inhibit autophagy, can target other kinases and affect other cellular processes, such as glycogen metabolism, lysosomal acidification, endocytosis, and mitochondrial permeability transition (54–56). Moreover, it has been reported earlier that 3-MA can suppress proteolysis even in Atg5-deficient cells, suggesting that its effects on protein degradation extend beyond its role in autophagy inhibition (57). In this study, we present a surprising finding that 3-MA has a dual role in modulation of autophagy; although it is capable of suppressing autophagy induced by starvation, its prolonged treatment in full medium induces autophagy. We first observed that there were marked increases of the autophagic markers in cells treated with 3-MA in full medium for a prolonged period of time (up to 9 h) (Fig. 1). Second, we provided convincing evidence that the increase of autophagic markers was the result of enhanced autophagic flux (Fig. 2), not due to suppression of maturation of autophagosomes or lysosomal function (Fig. 3). Therefore, our study, for the first time, demonstrates the unique dual role of 3-MA in modulation of autophagy.
The autophagy induction function by 3-MA is indeed intriguing. In order to understand the underlying mechanisms for the proautophagic effect of 3-MA, we compared the kinetics of 3-MA and wortmannin on class I and III PI3K with a prolonged treatment in full medium. Based on our data, it is believed that the induction of autophagy by 3-MA is due to its persistent inhibition on class I PI3K, whereas the class III PI3K is largely spared toward the end of the treatment (Fig. 5). It appears that under such specific treatment conditions, 3-MA acts similarly as rapamycin, a well established autophagy inducer via suppression of mTOR function (58). Here there are several important questions that remain to be investigated further. First, the molecular basis for a different temporal pattern by 3-MA on class I and class III PI3K is not known. Because the class I PI3K is mainly located within the plasma membrane (11), one can speculate that 3-MA may accumulate or associate with membrane to make the persistent suppression of class I PI3K activity possible. Second, it is not clear whether 3-MA has a similar temporal effect on class III PI3K under starvation conditions. In this study, we were logistically constrained to examine the effect of 3-MA on cells cultured in EBSS for more than 6 h, due to the fact that prolonged 3-MA treatment in EBSS in combination with CQ dramatically affected cell viability (data not shown). There is one important observation in this study that starvation (culturing cells in EBSS for 9 h) has little effect on class III PI3K activity (Fig. 5). Although at present it is still controversial whether class III PI3K activity is subject to modulation by nutrients (12), data from this study are indeed consistent with a number of earlier reports that the class III PI3K has been found to be nutrient-independent (26, 51, 52). For instance, simultaneous measurement of these two kinase activities showed that the presence of IL-13 only affects class I and not class III PI3K (26). Therefore, future studies examining the temporal effect of 3-MA on class III PI3K under starvation conditions will give rise to important information for understanding the effect of 3-MA as an autophagy inhibitor.
On the other hand, the inhibitory effects of wortmannin are just opposite to that of 3-MA: persistent on class III PI3K and transient on class I PI3K. The initial report showing that the inhibitory effect of wortmannin on PI3K was assessed based on class I PI3K found that wortmannin achieves its inhibitory effect via covalent binding with class I PI3K, and such binding is irreversible (22). Interestingly, the inhibition by wortmannin of class I PI3K is known to be transient (22, 26), thus consistent with the data from this study. Similar to 3-MA, the molecular basis for the different temporal pattern by wortmannin on class I and class III PI3K is not known and remains an important topic for future study. At present, the so-called class-specific PI3K inhibitors are not available; understanding the different temporal patterns of 3-MA and wortmannin on class I and III PI3K thus suggests a type of conditional selectivity by these two commonly used PI3K inhibitors. It is also obvious that data from this study tend to suggest that wortmannin is a more suitable autophagy inhibitor than 3-MA, due to its persistent inhibition on class III PI3K activity.
Another interesting finding from this study is that 3-MA is capable of up-regulating p62 protein expression at the mRNA level, a process independent of the autophagy machinery (Figs. 2 and 4). p62 is a ubiquitin-binding scaffold protein that can target ubiquitinated protein aggregates for selective degradation by autophagy (47, 59) It is now believed that p62 plays a critical role in the control of autophagy, apoptosis, and cancer development (60). On the other hand, the transcriptional regulation of p62 is relatively less understood. We have some preliminary data to show that 3-MA activates ERK and ERK inhibitors abolish the up-regulation of p62 (data not shown), indicating the possibility that 3-MA up-regulates p62 gene transcription via an ERK-dependent pathway. Further studies are required to examine the molecular mechanisms and the biological significance of p62 up-regulation in the course of autophagy induction.
Understanding the dual role of 3-MA in autophagy may have important implications in autophagy study because 3-MA is the first specific autophagy inhibitor identified and widely used even since 1982 (28, 61). One particularly important and yet highly controversial issue in autophagy study is the role of autophagy in cell death. Although autophagy is well established as a prosurvival mechanism under starvation and several other cell stress conditions, there are numerous reports demonstrating the prodeath function of autophagy (62–64). It is now acknowledged that part of the confusion is due to the misconception that the level of autophagosomes equals the autophagic flux (58). Now, with the recognition of the autophagy induction function of 3-MA with prolonged treatment under nutrient-rich conditions, it is possible that the use of 3-MA as an autophagy inhibitor may also contribute to such confusion regarding the role of autophagy in cell death. A careful examination of the literature shows that there are many reports using 3-MA for prolonged treatment (more than 24 h) in full medium to inhibit autophagy induced by stimuli, such as cancer therapeutics, DNA damage agents, and radiation (65–70), without providing direct evidence showing the effect of 3-MA on class III PI3K activity. Therefore, great cautions need to be exercised when 3-MA is used as an autophagy inhibitor with a prolonged period of treatment under nutrient-rich conditions.
Acknowledgments
We thank Dr. N. Mizushima, T. Yoshimori, and D. J. Kwiatkowski for providing the reagents.
The work was supported in part by research grants from the National University of Singapore (NUS) University Research Council and Singapore Biomedical Research Council and the Toxicology Program of the Life Science Institute, NUS.
- mTOR
- mammalian target of rapamycin
- PI3K
- phosphoinositide 3-kinase
- PI
- phosphatidylinositol
- PI(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- PI3P
- phosphatidylinositol 3-phosphate
- 3-MA
- 3-methyladenine
- CQ
- chloroquine diphosphate
- CHX
- cycloheximide
- EBSS
- Earles' balanced salt solution
- MEF
- mouse embryonic fibroblast
- GFP
- green fluorescent protein
- RFP
- red fluorescent protein
- mRFP
- monomeric RFP
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- siRNA
- small interfering RNA
- WT
- wild type
- DOX
- doxycyclin
- tfLC3
- tandem fluorescence-tagged LC3 construct
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Mizushima N. (2007) Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 2.Shintani T., Klionsky D. J. (2004) Science 306, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Z., Klionsky D. J. (2007) Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 4.Pattingre S., Espert L., Biard-Piechaczyk M., Codogno P. (2008) Biochimie 90, 313–323 [DOI] [PubMed] [Google Scholar]
- 5.Inoki K., Guan K. L. (2006) Trends Cell Biol. 16, 206–212 [DOI] [PubMed] [Google Scholar]
- 6.Reiling J. H., Sabatini D. M. (2006) Oncogene 25, 6373–6383 [DOI] [PubMed] [Google Scholar]
- 7.Sarbassov D. D., Ali S. M., Sabatini D. M. (2005) Curr. Opin. Cell Biol. 17, 596–603 [DOI] [PubMed] [Google Scholar]
- 8.Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000) J. Cell Biol. 150, 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. (2008) J. Cell Biol. 181, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., Kim D. H. (2009) Mol. Biol. Cell 20, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantley L. C. (2002) Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 12.Backer J. M. (2008) Biochem. J. 410, 1–17 [DOI] [PubMed] [Google Scholar]
- 13.Engelman J. A., Luo J., Cantley L. C. (2006) Nat. Rev. Genet. 7, 606–619 [DOI] [PubMed] [Google Scholar]
- 14.Corradetti M. N., Guan K. L. (2006) Oncogene 25, 6347–6360 [DOI] [PubMed] [Google Scholar]
- 15.Chang Y. Y., Juhász G., Goraksha-Hicks P., Arsham A. M., Mallin D. R., Muller L. K., Neufeld T. P. (2009) Biochem. Soc. Trans. 37, 232–236 [DOI] [PubMed] [Google Scholar]
- 16.Itakura E., Kishi C., Inoue K., Mizushima N. (2008) Mol. Biol. Cell 19, 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., Akira S., Noda T., Yoshimori T. (2009) Nat. Cell Biol. 11, 385–396 [DOI] [PubMed] [Google Scholar]
- 18.Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. (2009) Nat. Cell Biol. 11, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuya N., Yu J., Byfield M., Pattingre S., Levine B. (2005) Autophagy 1, 46–52 [DOI] [PubMed] [Google Scholar]
- 20.Liang C., Lee J. S., Inn K. S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J. U. (2008) Nat. Cell Biol. 10, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vlahos C. J., Matter W. F., Hui K. Y., Brown R. F. (1994) J. Biol. Chem. 269, 5241–5248 [PubMed] [Google Scholar]
- 22.Powis G., Bonjouklian R., Berggren M. M., Gallegos A., Abraham R., Ashendel C., Zalkow L., Matter W. F., Dodge J., Grindey G. (1994) Cancer Res. 54, 2419–2423 [PubMed] [Google Scholar]
- 23.Blommaart E. F., Krause U., Schellens J. P., Vreeling-Sindelárová H., Meijer A. J. (1997) Eur. J. Biochem. 243, 240–246 [DOI] [PubMed] [Google Scholar]
- 24.Kong D., Yamori T. (2008) Cancer Sci. 99, 1734–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight Z. A., Shokat K. M. (2007) Biochem. Soc. Trans. 35, 245–249 [DOI] [PubMed] [Google Scholar]
- 26.Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P. (2000) J. Biol. Chem. 275, 992–998 [DOI] [PubMed] [Google Scholar]
- 27.Zeng X., Overmeyer J. H., Maltese W. A. (2006) J. Cell Sci. 119, 259–270 [DOI] [PubMed] [Google Scholar]
- 28.Seglen P. O., Gordon P. B. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J. L., Oshiro N., Mizushima N. (2009) Mol. Biol. Cell 20, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura S., Noda T., Yoshimori T. (2007) Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 31.Wu Y. T., Tan H. L., Huang Q., Ong C. N., Shen H. M. (2009) Autophagy 5, 824–834 [DOI] [PubMed] [Google Scholar]
- 32.Vergne I., Roberts E., Elmaoued R. A., Tosch V., Delgado M. A., Proikas-Cezanne T., Laporte J., Deretic V. (2009) EMBO J. 28, 2244–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y. T., Tan H. L., Huang Q., Kim Y. S., Pan N., Ong W. Y., Liu Z. G., Ong C. N., Shen H. M. (2008) Autophagy 4, 457–466 [DOI] [PubMed] [Google Scholar]
- 34.Bains M., Heidenreich K. A. (2009) Methods Enzymol. 453, 145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nasuhoglu C., Feng S., Mao J., Yamamoto M., Yin H. L., Earnest S., Barylko B., Albanesi J. P., Hilgemann D. W. (2002) Anal. Biochem. 301, 243–254 [DOI] [PubMed] [Google Scholar]
- 36.Bauvy C., Meijer A. J., Codogno P. (2009) Methods Enzymol. 452, 47–61 [DOI] [PubMed] [Google Scholar]
- 37.Dann S. G., Selvaraj A., Thomas G. (2007) Trends Mol. Med. 13, 252–259 [DOI] [PubMed] [Google Scholar]
- 38.Hosokawa N., Hara Y., Mizushima N. (2006) FEBS Lett. 580, 2623–2629 [DOI] [PubMed] [Google Scholar]
- 39.Mizushima N. (2009) Methods Enzymol. 452, 13–23 [DOI] [PubMed] [Google Scholar]
- 40.Mizushima N. (2004) Int. J. Biochem. Cell Biol. 36, 2491–2502 [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N., Yoshimori T. (2007) Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 42.Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 44.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. (2005) Autophagy 1, 84–91 [DOI] [PubMed] [Google Scholar]
- 46.Ding W. X., Ni H. M., Gao W., Chen X., Kang J. H., Stolz D. B., Liu J., Yin X. M. (2009) Mol. Cancer Ther. 8, 2036–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 48.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bjørkøy G., Lamark T., Pankiv S., Øvervatn A., Brech A., Johansen T. (2009) Methods Enzymol. 452, 181–197 [DOI] [PubMed] [Google Scholar]
- 51.Tassa A., Roux M. P., Attaix D., Bechet D. M. (2003) Biochem. J. 376, 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi Y., Coppola D., Matsushita N., Cualing H. D., Sun M., Sato Y., Liang C., Jung J. U., Cheng J. Q., Mulé J. J., Pledger W. J., Wang H. G. (2007) Nat. Cell Biol. 9, 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C., Blenis J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caro L. H., Plomp P. J., Wolvetang E. J., Kerkhof C., Meijer A. J. (1988) Eur. J. Biochem. 175, 325–329 [DOI] [PubMed] [Google Scholar]
- 55.Punnonen E. L., Marjomäki V. S., Reunanen H. (1994) Eur. J. Cell Biol. 65, 14–25 [PubMed] [Google Scholar]
- 56.Xue L., Borutaite V., Tolkovsky A. M. (2002) Biochem. Pharmacol. 64, 441–449 [DOI] [PubMed] [Google Scholar]
- 57.Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. (2001) J. Cell Biol. 152, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meijer A. J., Codogno P. (2009) Crit. Rev. Clin. Lab. Sci. 46, 210–240 [DOI] [PubMed] [Google Scholar]
- 59.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 60.Moscat J., Diaz-Meco M. T. (2009) Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scarlatti F., Granata R., Meijer A. J., Codogno P. (2009) Cell Death Differ. 16, 12–20 [DOI] [PubMed] [Google Scholar]
- 63.Galluzzi L., Vicencio J. M., Kepp O., Tasdemir E., Maiuri M. C., Kroemer G. (2008) Curr. Mol. Med. 8, 78–91 [DOI] [PubMed] [Google Scholar]
- 64.Gozuacik D., Kimchi A. (2007) Curr. Top. Dev. Biol. 78, 217–245 [DOI] [PubMed] [Google Scholar]
- 65.Xiong H. Y., Guo X. L., Bu X. X., Zhang S. S., Ma N. N., Song J. R., Hu F., Tao S. F., Sun K., Li R., Wu M. C., Wei L. X. (2009) Cancer Lett. 288, 68–74 [DOI] [PubMed] [Google Scholar]
- 66.Gao M., Yeh P. Y., Lu Y. S., Hsu C. H., Chen K. F., Lee W. C., Feng W. C., Chen C. S., Kuo M. L., Cheng A. L. (2008) Cancer Res. 68, 9348–9357 [DOI] [PubMed] [Google Scholar]
- 67.Longo L., Platini F., Scardino A., Alabiso O., Vasapollo G., Tessitore L. (2008) Mol. Cancer Ther. 7, 2476–2485 [DOI] [PubMed] [Google Scholar]
- 68.Kanzawa T., Germano I. M., Komata T., Ito H., Kondo Y., Kondo S. (2004) Cell Death Differ. 11, 448–457 [DOI] [PubMed] [Google Scholar]
- 69.Ito H., Daido S., Kanzawa T., Kondo S., Kondo Y. (2005) Int. J. Oncol. 26, 1401–1410 [PubMed] [Google Scholar]
- 70.Li J., Hou N., Faried A., Tsutsumi S., Takeuchi T., Kuwano H. (2009) Ann. Surg. Oncol. 16, 761–771 [DOI] [PubMed] [Google Scholar]