Abstract
Maspin (SERPINB5) is a tumor suppressor lost in breast and prostate cancer whose molecular function is unknown. It is a non-inhibitory member of the clade B serpins suggested to play a role in a plethora of intracellular and extracellular settings, yet its normal cellular distribution has never been clarified. Here we investigate the distribution of maspin in non-transformed human epithelial cells. By indirect immunofluorescence, maspin has a nucleocytoplasmic distribution in breast (MCF10A) and prostate (RWPE-1) cells and, by immunoblotting and pulse-chase analyses, is neither glycosylated nor secreted. Cell surface biotinylation studies also show that maspin is not present at the cell surface. Differentiation of MCF10A cells into three-dimensional acini results in the redistribution of maspin from the nucleus to the cytoplasm but does not result in secretion. Addition of an efficient conventional signal peptide to maspin directs it into the secretory pathway and results in glycosylation but not secretion. We further show that maspin in the cytoplasm of MCF10A cells is a soluble monomeric protein that is not detectably associated with the cytoskeleton or other extractable components. Taken together, these results suggest that maspin is restricted to an intracellular, possibly nuclear, role in which it influences cell-matrix interactions indirectly. It is probably released only as a consequence of cell damage or necrosis.
Keywords: Cancer/Breast, Cell/Differentiation, Protein/Intracellular Trafficking, Protein/Serpin, Protein/Targeting, Subcellular Organelles/Endoplasmic Reticulum
Introduction
Serpins comprise a large superfamily of intracellular and extracellular protease inhibitors with a small number of members that have evolved non-inhibitory functions (1). Inhibitory serpins inactivate target proteases through a “molecular mousetrap” mechanism that leads to the formation of a stable complex between serpin and protease. During this process, the serpin undergoes a dramatic conformational change that requires conserved “hinge” regions flanking the protease-binding site (reactive center loop). Most non-inhibitory serpins have non-conserved hinges.
Maspin (SERPINB5) is a 42-kDa non-inhibitory serpin first identified in a screen for potential tumor suppressors lost in human breast cancer cells (2). Reintroduction of maspin into tumor cells inhibits growth, cell migration and invasion, and angiogenesis and increases cell adhesion, all of which are hallmarks of a tumor suppressor. Multiple clinical studies have confirmed maspin loss in various carcinomas, particularly breast and prostate, and have demonstrated that retention of maspin expression is associated with a good prognosis.
Despite numerous studies implicating it in tumor suppression and striking evidence from a mouse model indicating its developmental importance (3), the normal function of maspin is unknown. It was originally suggested to act outside the cell by regulating cell-extracellular matrix adhesion in a mechanism dependent on the reactive center loop (4). For example, incubation of breast or prostate tumor cells with recombinant maspin or transfection with maspin cDNA inhibits invasion and motility (5), and yeast two-hybrid analysis demonstrated binding between maspin and type I and III collagens (6). It was also observed that maspin deficiency leads to mouse embryo death associated with failure to bind laminin (3), whereas maspin enhances the adhesion of cultured human corneal stromal cells to fibronectin, type I and IV collagens, and laminin (7). It has been suggested that maspin might modulate cell-matrix adhesion through an interaction with β1-integrin (8). Although maspin has features suggesting that it is non-inhibitory, it is reported to inhibit the extracellular urokinase-type plasminogen activator/urokinase-type plasminogen activator receptor system, but this is controversial (9–11).
More recently, intracellular functions of maspin have been suggested. It may remodel cell-extracellular matrix interactions inside-out via signaling through Rac and Cdc42 (12, 13). Other suggested roles include induction of apoptosis (14–17) as well as involvement in cellular stress responses (9, 18).
The above findings demand that maspin should have both an extracellular and an intracellular cytoplasmic distribution. Maspin belongs to clade B of the serpin superfamily, whose members lack a classical secretory signal peptide and are predominantly intracellular and nucleocytoplasmic (1, 19, 20). However, two clade B serpins, PAI-2 (SERPINB2) and SCCA-1 (SERPINB3), can be released from cells under certain circumstances. Furthermore, the prototype of the clade B group, chicken ovalbumin, is efficiently secreted. Analysis of ovalbumin and SERPINB2 shows that they carry unconventional signal peptides that are absent in other clade B members, including maspin (21).
On balance, therefore, it is likely that maspin is an intracellular protein with an intracellular role, but it remains possible that maspin resembles SERPINB2 and can be secreted as a glycoform. Here we unequivocally demonstrate, using non-transformed cells, that maspin is an intracellular nucleocytoplasmic protein that cannot be secreted.
EXPERIMENTAL PROCEDURES
Cell Culture
COS-1, MCF10A, and RWPE-1 cells were maintained as described (20, 22, 23). MCF10A acini were cultured as described (23). COS-1 cells were transfected using the DEAE-dextran/chloroquine method as described (24).
Antibodies
The mouse anti-maspin monoclonal antibody was purchased from BD Pharmingen. The mouse anti-maspin polyclonal and rabbit anti-maspin polyclonal antisera were raised against recombinant maspin produced in Escherichia coli (25). The anti-β1-integrin antibody P5D2, developed by E. A. Wayner, was obtained from the Developmental Studies Hybridoma Bank (Department of Biological Sciences, University of Iowa, Iowa City, IA). Alexa Fluor 594-conjugated concanavalin A (Molecular Probes, Eugene, OR) was used as a marker of the endoplasmic reticulum; a rabbit anti-giantin polyclonal antibody (Covance) was used as a marker of the Golgi apparatus; and rhodamine isothiocyanate (RITC)2-conjugated phalloidin (Invitrogen) was used as a marker of polymerized actin. The secondary antibody used in immunoblotting was sheep anti-mouse IgG conjugated to horseradish peroxidase (Chemicon), and the secondary antibodies used in indirect immunofluorescence were goat anti-mouse IgG conjugated to Alexa 488 (Invitrogen), goat anti-rabbit IgG conjugated to Alexa 488 (Invitrogen), goat anti-mouse IgG conjugated to Cy5 (Chemicon), and goat anti-rabbit conjugated to RITC (Sigma).
Plasmids
For the expression of maspin in COS-1 cells, a maspin cDNA was modified by PCR amplification using the oligonucleotide primers 5′-GGG AGA TCT CAT GGA TGC CCT GCA ACT AGC-3′ (adds a BglII site to the 5′ end for cloning into pSHT) and 5′-CCC GCG GTT AAG GAG AAC AGA ATT TGC C-3′ and Taq DNA polymerase (New England Biolabs) for 25 cycles of 95 °C for 45 s, 55 °C for 45 s, and 72 °C for 90 s. The resulting 1.15-kb product was cloned into pCR-Blunt (Invitrogen) and then released and purified as a BglII-XbaI fragment. This was subcloned into pSHT (26) digested with BamHI and SpeI. To make pSVTf/maspin, maspin was released and purified from pCR-Blunt/maspin digested with EcoRI, and then subcloned into pSVTf also digested with EcoRI and dephosphorylated using calf intestinal alkaline phosphatase (New England Biolabs).
Pulse-Chase Experiments
Pulse-chase experiments were performed on transfected COS-1, MCF10A, and RWPE-1 cells as described (27), except that [35S]methionine was purchased from MP Biomedicals, and cells were labeled for 30 min. To immunoprecipitate β1-integrin, a rabbit polyclonal antiserum against mouse IgG (Dako) was used as a bridging antibody between protein A-Sepharose and the anti-β1-integrin antibody.
Endoglycosidase Treatments
Immunoprecipitates from transfected and labeled COS-1 cells or labeled MCF10A cells were treated with endoglycosidase H (Endo H) or peptide:N-glycosidase F (PNGase F), depending on the experiment, following the manufacturer's instructions (New England Biolabs). Briefly, immunoprecipitates were resuspended in 20 μl of water with 2% of the supplied denaturing buffer and boiled for 10 min. Immunoprecipitates were then incubated for 1 h at 37 °C with or without endoglycosidase. Samples were analyzed by SDS-PAGE and fluorography.
Cell Surface Biotinylation
MCF10A cells (4 × 106) were washed with ice-cold phosphate-buffered saline (PBS; 154 mm NaCl, 2.7 mm Na2HPO4, and 1.54 mm KH2PO4), pH 8.0, and then incubated on ice with EZ-Link sulfo-NHS-biotin at a final concentration of 0.5 mg/ml (Pierce) for 30 min and washed with ice-cold PBS with 100 mm glycine to quench excess biotin. From this point, all buffers were supplemented with 100 mm glycine. Equal amounts of cells were then lysed in either Nonidet P-40 lysis buffer (50 mm Tris, pH 8.0, 10 mm EDTA, and 1% (v/v) Nonidet P-40) or in 0.5% SDS in PBS and then diluted to a final concentration of 0.05% SDS with 150 mm NaCl, 5 mm EDTA, 50 mm Tris, pH 7.4, 0.05% (v/v) Nonidet P-40, 0.25% (w/v) gelatin, 60 Bloom, and 0.02% (w/v) NaN3. Nonidet P-40 lysates were used for immunoprecipitation of β1-integrin, and SDS lysates were used for the immunoprecipitation of maspin as described (27). A rabbit polyclonal antiserum raised against mouse IgG was used as a “bridging antibody” between protein A-Sepharose and the anti-β1-integrin monoclonal antibody. Immunoprecipitates were analyzed by nonreducing or by reducing SDS-PAGE, depending on the experiment, and immunoblotted for maspin.
Indirect Immunofluorescence and in Situ Cell Extractions
Cell monolayers grown on 10-well microscope slides were washed in PBS containing 0.1 mm CaCl2 and 1.0 mm MgCl2 (PBS+), fixed in 3.7% (w/v) formaldehyde in PBS+ for 20 min, quenched with 20 mm ammonium chloride, and where indicated permeabilized by incubation in 0.5% Triton X-100 in PBS+ for 5 min. Antigens were detected by incubation of the cells for 30 min with the appropriate dilution of primary antibody (1:1000 for mouse anti-maspin polyclonal antiserum, 1:500 for rabbit anti-maspin polyclonal antiserum, 1:50 for anti-β1-integrin, 1:800 for anti-giantin, and 1:50 for Alexa 594-conjugated concanavalin A). After being washed with PBS+, the cells were incubated with Alexa 488-conjugated secondary antibodies diluted 1:800 or RITC-conjugated secondary antibodies diluted 1:200. After 30 min, cells were washed in PBS+ and then mounted in Mowiol. Where double staining occurred, cells were first incubated with monoclonal and then with polyclonal primary antibodies, followed by incubation with the appropriate secondary antibodies. Cells were examined using either epifluorescence microscopy (Nikon TE-2000U Eclipse upright fluorescence microscope) or confocal laser scanning microscopy (Leica SP Multiphoton microscope, DMI6000 Invert).
For in situ extractions, 2.5 × 104 cells/well were grown on 10-well microscope slides, and acini were grown in 8-well chamber slides (23). Untreated cells were fixed, permeabilized, and stained as described above. Actin was marked with RITC-conjugated phalloidin (0.1 μg/ml final concentration). To remove proteins from the cytoplasm, cells were placed on ice, washed twice with ice-cold HMKE buffer (20 mm HEPES, pH 7.2, 5 mm MgCl2, 10 mm KCl, 1 mm EDTA, and 250 mm sucrose), and then exposed to 25 μg/ml digitonin in 50 μl of HMKE buffer containing protease inhibitors (1 μg/ml each aprotinin, leupeptin, and pepstatin, and 0.5 mm 4-(2-aminoethyl)benzenesulfonyl fluoride) for 10 min. Cells were then fixed and permeabilized for staining as described above.
Size-exclusion Chromatography
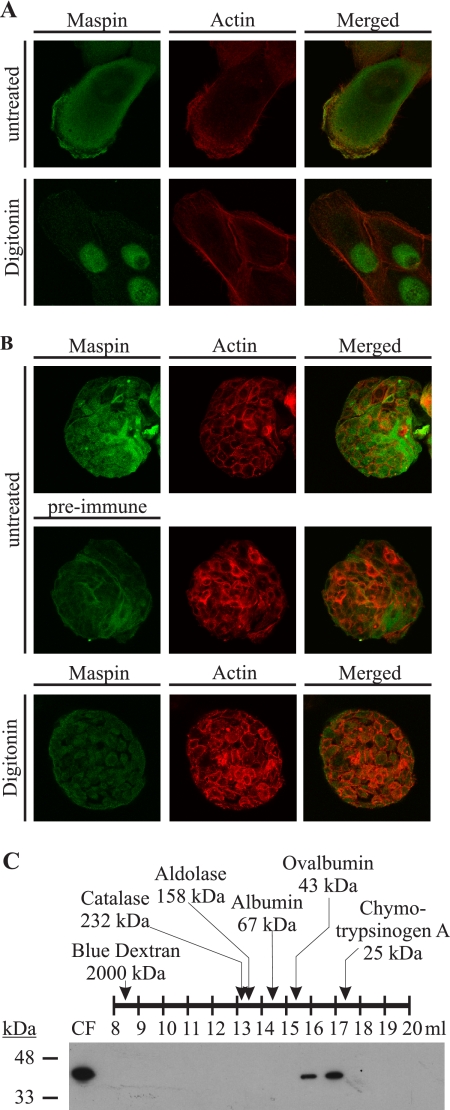
MCF10A cells resuspended at 107 cells/ml in ice-cold 50 mm Tris-HCl, pH 7.4, 25 mm KCl, and 5 mm MgCl2 were disrupted by homogenization, and only the supernatant was isolated to generate cytosol and non-nuclear membrane fractions as described (28). Size-exclusion chromatography was performed on a Superdex 200 10/300 GL high performance column (GE Healthcare) equilibrated with Tris-buffered saline (20 mm Tris, 150 mm NaCl). Sixteen fractions were resolved by SDS-PAGE and analyzed by immunoblotting for maspin. To determine the size of the eluted maspin, individual runs on the same column were performed for a set of protein standards (blue dextran, 2000 kDa; catalase, 232 kDa; aldolase, 158 kDa; albumin, 67 kDa; ovalbumin, 43 kDa; and chymotrypsinogen, 25 kDa), and the elution fraction for each protein was determined and then compared with that of maspin.
RESULTS
Maspin Has a Nucleocytoplasmic Localization and Is Not Present at the Cell Surface
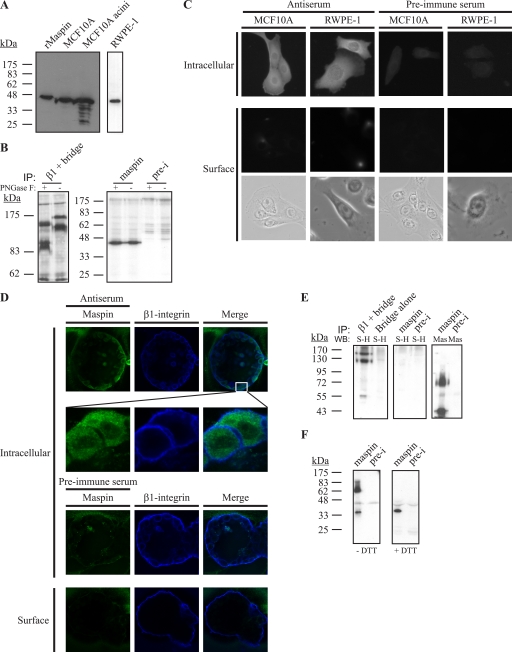
MCF10A is an immortalized non-transformed mammary epithelial cell line that exhibits features of normal breast epithelium (23) and in three-dimensional cultures has the ability to differentiate into acinus-like spheroids with a hollow lumen. RWPE-1 is an immortalized non- tumorigenic prostate epithelial cell line (22). We confirmed expression of endogenous maspin in these cell lines by immunoblotting, where it appears as a 42-kDa protein (Fig. 1A). Importantly, there is no evidence of larger (glyco)forms that would be expected if maspin enters the endoplasmic reticulum and one or more of the three potential asparagine-linked glycosylation sites on the molecule is used. This was confirmed using peptide PNGase F to probe the glycosylation state of maspin in MCF10A cells (Fig. 1B). PNGase F removes high mannose, hybrid, and complex oligosaccharides from N-linked glycoproteins. Although PNGase F successfully removed complex oligosaccharides from the control glycoprotein β1-integrin, resulting in a decrease in molecular weight (Fig. 1B, left panel), it had no effect on maspin (Fig. 1B, right panel), demonstrating that endogenous maspin in MCF10A cells is not glycosylated. We then looked at the distribution of maspin in MCF10A and RWPE-1 cells by indirect immunofluorescence. In permeabilized cells, maspin appears in the cytoplasm and nucleus, and no staining was observed in non-permeabilized cells, indicating that it is not found on the cell surface (Fig. 1C). We also examined a human breast myoepithelial cell line, Hs578Bst, for maspin, as it has been suggested that myoepithelial maspin may inhibit breast carcinoma cell invasion (29). However, we could not detect the presence of maspin in these cells by either immunoblotting or immunofluorescence (data not shown).
FIGURE 1.
Maspin is expressed by MCF10A and RWPE-1 cells but is absent from the cell surface. A, lysates of MCF10A cells and acini or RWPE-1 cells were separated via 12.5% SDS-PAGE and immunoblotted with recombinant maspin (rMaspin) as a control. The membrane was probed with mouse anti-maspin monoclonal antibody diluted 1:1000 and detected with horseradish peroxidase-conjugated secondary against mouse IgG. RWPE-1 lysates were analyzed on a separate gel. B, maspin in MCF10A cells is not glycosylated. Lysates of 30-min metabolically labeled MCF10A cells were immunoprecipitated (IP) using mouse anti-β1-integrin monoclonal antibody (left panel), rabbit anti-maspin polyclonal antiserum (right panel), or preimmune (pre-i) serum and then either treated with PNGase F or left untreated. Maspin samples were separated by 12.5% SDS-PAGE, and β1-integrin samples were separated by 7.5% SDS-PAGE. Gels were analyzed by fluorography. C, maspin is in the nucleus and cytoplasm and not on the cell surface. MCF10A and RWPE-1 cells were fixed and permeabilized or fixed alone to detect cell surface proteins and then probed with mouse anti-maspin polyclonal antiserum or with the preimmune serum. The primary antibody was detected using goat Alexa 488-conjugated secondary antibodies. Cells were examined by epifluorescence microscopy. Brightfield images of cells examined for cell surface staining are shown. D, maspin is intracellular in each luminal epithelial cell and does not co-localize with β1-integrin. MCF10A cells were grown on Matrigel, and acini were developed for 20 days. Acini were prepared as described for B and examined by confocal microscopy. Images shown are single optical sections. E, MCF10A cells were surface-biotinylated, and lysates were immunoprecipitated using rabbit anti-maspin polyclonal antiserum, preimmune serum, or mouse anti-β1-integrin monoclonal antibody. Immune complexes were collected and analyzed by 10% SDS-PAGE, and the immunoblot (WB) was probed with horseradish peroxidase-conjugated streptavidin (S-H) diluted 1:5000. The blot was stripped and re-probed with mouse anti-maspin monoclonal antibody (Mas) and detected with horseradish peroxidase-conjugated secondary against mouse IgG. Bridge refers to samples immunoprecipitated with bridging antibody alone. F, MCF10A cell lysates were immunoprecipitated, and immune complexes were left nonreduced (without dithiothreitol (−DTT)) or reduced (+DTT) and analyzed via 12.5% SDS-PAGE and immunoblotting with mouse anti-maspin monoclonal antibody followed by horseradish peroxidase-conjugated secondary against mouse IgG.
We investigated the localization of maspin in MCF10A acini. Because maspin is thought to be important for cell-extracellular matrix interactions, and such interactions are important for proper differentiation of individual cells into acini, we expected that maspin would localize to the cell surface upon differentiation. However, confocal analysis of MCF10A acini stained for maspin as well as for β1-integrin to mark the surface of these structures shows that in these differentiated cells, maspin remains intracellular and not on the cell surface (Fig. 1D). A close-up view of the luminal epithelial cell wall shows that maspin is clearly intracellular and that no co-localization with β1-integrin can be observed (Fig. 1D). Interestingly, acini lack maspin in the nucleus, suggesting that the protein redistributes during differentiation.
To confirm the absence of maspin on the cell surface, we biotinylated the MCF10A cell surface using cell-impermeable biotin and then immunoprecipitated total cell lysates using antibodies to maspin or β1-integrin. β1-Integrin serves as a positive control for surface biotinylation because MCF10A cells secrete a matrix of laminin-5 and adhere to this matrix via α3β1-integrin (30). Immunoprecipitated samples were then analyzed by SDS-PAGE and streptavidin-horseradish peroxidase to detect maspin or β1-integrin that had been biotinylated on the cell surface. As expected, β1-integrin was detected as a 130-kDa protein (Fig. 1E, left panel, first lane). By contrast, maspin was not detected (Fig. 1E, middle panel), demonstrating that it was not present at the cell surface during biotinylation. To ensure that maspin was present in the lysates, the same blot was stripped and re-probed with monoclonal antibody to maspin, which showed maspin species at 42 and 62 kDa (Fig. 1E, right panel). Fig. 1F shows that the 62-kDa species arises because of nonspecific oxidative dimerization of maspin during immunoprecipitation (which was carried out in the absence of reductant), as it can be reduced to 42 kDa, the expected size of maspin. Taken together, these results suggest that maspin is an intracellular serpin and is not found at the cell surface.
Maspin Is Not Released from MCF10A or RWPE-1 Cells
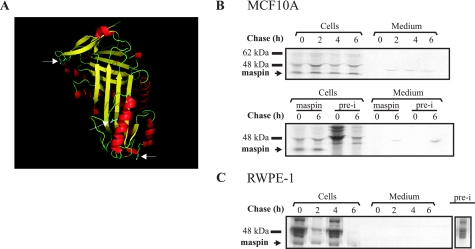
We next investigated whether maspin is released from MCF10A or RWPE-1 cells. Although maspin lacks a conventional secretory peptide, it is possible that it enters the classical secretory pathway using an unusual signal, as is the case for SERPINB2. If this were to occur, maspin should be glycosylated, increase in size, and be detected in medium conditioned by the cells. Maspin has three potential glycosylation sites (Fig. 2A), so increases in size of up to 10–12 kDa from 42 kDa can be expected. Alternately, if released via an unconventional pathway, maspin should be evident in conditioned medium as a 42-kDa species. To investigate this, we employed a pulse-chase approach. Fig. 2 (B and C) shows that maspin, migrating at a non-glycosylated size of 42 kDa, was synthesized by both cell lines. Maspin appears only in the cell lysates, and no release into the medium is evident. The immune complexes seen in the medium samples are not specific to maspin, as they also occur in samples immunoprecipitated using preimmune serum (Fig. 2B, lower panel). Taken together, these results show that maspin is not released from MCF10A or RWPE-1 cells by either the classical or unconventional pathways.
FIGURE 2.
Maspin is not secreted from MCF10A or RWPE-1 cells. A, ribbon structure of maspin. White arrows denote asparagine residues that may be glycosylated. B and C, pulse-chase experiments were performed on MCF10A cells (B) and the primary prostate cell line, RWPE-1 (C), as indicated under “Experimental Procedures.” Cell lysates and media were collected at each time point and immunoprecipitated with rabbit anti-maspin polyclonal antiserum. Immune complexes were collected, reduced, and analyzed by 10% SDS-PAGE and fluorography. Comparison of antiserum and preimmune (pre-i) serum complexes for MCF10A immunoprecipitation is shown in the middle panel. The preimmune complex from RWPE-1 immunoprecipitation shown in C is from a separate autoradiograph.
Maspin Is Not Released from Cells on Addition of a Conventional Signal Sequence
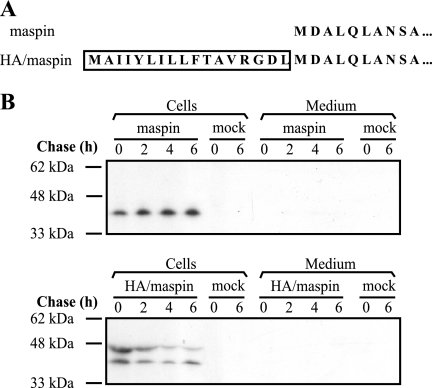
The above results do not rule out the secretion of maspin in response to a specific but as yet undetermined stimulus. Our inability to detect secreted maspin could also be explained by the absence of a specific cofactor required for an unconventional or inefficient signal sequence to function in these cells. However, if maspin has evolved to be released (like SERPINB2), it should be glycosylated and move efficiently through the secretory pathway once directed there by its signal sequence. To mimic this scenario and avoid the need for a specific stimulus (Fig. 3A), we provided maspin with the efficient influenza virus hemagglutinin (HA) signal sequence, which functions as a signal peptide that facilitates endoplasmic reticulum (ER) association and subsequent transmembrane movement (31). As a control, cDNA encoding wild-type maspin was cloned into the expression vector pSVTf.
FIGURE 3.
Maspin directed into the classical secretory pathway is not released into the extracellular milieu. A, comparison of the N terminus of HA/maspin and maspin. Boxed residues comprise the HA signal peptide. B, COS cells were transfected with maspin (upper panel) and HA/maspin (lower panel) DNA. Forty-eight hours after transfection, cells were used in a pulse-chase experiment as described under “Experimental Procedures.” COS cells transfected without DNA (mock) were included as a negative control.
To investigate whether the addition of the HA signal sequence could induce the secretion of maspin, HA/maspin and maspin expression plasmids were transfected into COS-1 cells, and a pulse-chase analysis was performed. A mock transfection without the addition of expression plasmids was also performed as a control. In lysates of cells producing maspin, a 42-kDa protein was detected, and there was no evidence of release into the medium (Fig. 3B, upper panel). This was consistent with the results observed in the MCF10A and RWPE-1 cells, showing that native maspin is not glycosylated or secreted. By contrast, in lysates of cells producing HA/maspin, proteins migrating at 42 and 47 kDa were evident, with the larger form probably resulting from glycosylation at two of the three potential sites, indicating that maspin had entered the ER (Fig. 3B, lower panel). However, HA/maspin was not detected in the medium (Fig. 3B, lower panel), suggesting that it cannot exit the secretory pathway.
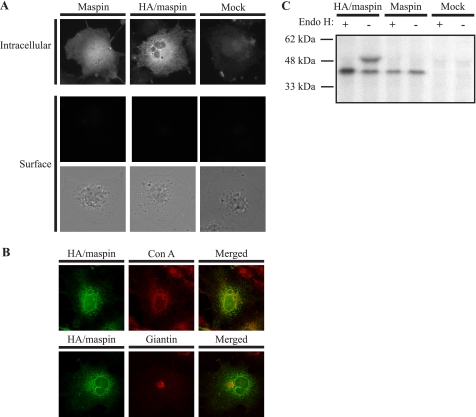
The observation that HA/maspin could not be detected in the medium suggests that it is sequestered somewhere along the secretory pathway. By indirect immunofluorescence, it was determined that maspin in COS-1 cells has a nucleocytoplasmic distribution and is not at the cell surface, whereas HA/maspin shows a reticular pattern of staining consistent with localization in the endoplasmic reticulum (Fig. 4A). Surface staining of cells showed that HA/maspin is not on the cell surface, suggesting that it is sequestered inside the cell (Fig. 4A). To confirm that HA/maspin is trapped in the ER, co-localization studies with concanavalin A, a marker of the ER, as well as giantin, a marker of the Golgi, were performed. HA/maspin co-localized with concanavalin A but not with giantin (Fig. 4B), suggesting that HA/maspin is indeed sequestered in the ER and cannot move through to the Golgi.
FIGURE 4.
Glycosylated maspin is retained in the ER. A, COS-1 cells transfected with HA/maspin or maspin were prepared as described in the legend to Fig. 1 and probed with mouse anti-maspin polyclonal antiserum. As a control, COS-1 cells were mock-transfected. The primary antibody was detected with goat anti-mouse IgG conjugated to Alexa 488, and cells were examined by epifluorescence microscopy. The lower panel shows brightfield images of cells examined for cell surface staining. B, COS-1 cells transfected with HA/maspin were fixed, permeabilized, and probed with mouse anti-maspin monoclonal antibody. The primary antibody was detected with goat anti-mouse IgG conjugated to Alexa 488. The ER was marked with Alexa Fluor 594-conjugated concanavalin A (Con A), and the Golgi was marked with rabbit anti-giantin polyclonal antibody detected with goat anti-rabbit IgG conjugated to RITC. Cells were examined by confocal microscopy. Images show single optical sections. C, COS-1 cells were transfected with HA/maspin or maspin DNA or without DNA (Mock). Forty-eight hours after transfection, cells were labeled for 30 min in medium lacking methionine containing 100 μCi of [35S]methionine and then incubated in complete medium for 6 h. Cell lysates and medium were then collected and immunoprecipitated with rabbit anti-maspin polyclonal antiserum. Immune complexes were either treated (+) or left untreated (−) with Endo H. Samples were then reduced and analyzed by 10% SDS-PAGE and fluorography.
This was further supported by use of Endo H to probe the glycosylation state of HA/maspin. Endo H can remove “high mannose” oligosaccharides present on ER resident proteins or on nascent proteins that have not left the ER. However, proteins that enter the Golgi become resistant to Endo H. COS-1 cells transfected with HA/maspin and maspin were metabolically labeled, chased for 6 h, lysed, and immunoprecipitated using anti-maspin polyclonal antiserum. The immune complexes were then treated with or without Endo H. As shown in Fig. 4C, Endo H treatment of HA/maspin completely removed the larger glycoforms, further supporting the observations that HA/maspin is sequestered in the ER. Maspin, even when supplied with an efficient signal sequence, cannot move through the secretory pathway to the cell surface or be secreted, strongly suggesting that it has an intracellular role.
Maspin Is a Soluble Monomeric Protein in the Cytoplasm
With the observation that maspin is an intracellular serpin and has a diffuse nucleocytoplasmic distribution, we next questioned whether maspin is specifically associated with a cellular structure such as the cytoskeleton because it is thought to oppose cell migration and invasion. We investigated this through indirect immunofluorescence of MCF10A monolayers treated with digitonin, which permeabilizes the plasma membrane and allows the selective extraction of cytosolic but not nuclear proteins (20, 32). Maspin was fully extractable from the cytoplasm, suggesting that it is a soluble protein that is not associated with the cytoskeleton or any other cytoplasmic structure, and as expected, a fraction remained associated with the nucleus (Fig. 5A). Actin staining showed that the structure of the cells remained intact following digitonin treatment. In acini, maspin was also evident as a soluble cytoplasmic protein and was absent from the nucleus (Fig. 5B).
FIGURE 5.
Maspin is not stably associated with the cytoskeleton or other cellular proteins and exists as a monomeric serpin. A and B, maspin is a soluble cytoplasmic protein. MCF10A monolayers (A) growing on microscope slides and MCF10A acini (B) were extracted with HMKE buffer containing digitonin and then fixed, permeabilized, and probed with mouse anti-maspin polyclonal antiserum. The primary antibody was detected with goat anti-mouse IgG conjugated to Alexa 488. The actin cytoskeleton was marked by RITC-conjugated phalloidin. Cells were examined by confocal microscopy. Images show single optical sections. C, MCF10A cells were harvested in hypotonic buffer, and gel filtration was performed on the cytosolic fraction (CF) of lysate. Fractions were separated by 12.5% SDS-PAGE, analyzed by immunoblotting for maspin using mouse anti-maspin monoclonal antibody, and detected with horseradish peroxidase-conjugated secondary against mouse IgG. A series of molecular mass standards ranging between 25 and 2000 kDa was assessed, and the fractions in which they appeared are shown. The molecular mass of the eluted maspin was within the predicted monomeric range.
Because maspin does not bind to detergent-resistant fractions of the cell, we next investigated whether maspin could bind to an extractable cellular protein or whether it exists in the cytoplasm in an oligomeric form, as serpins have a propensity to oligomerize (33, 34). MCF10A cells were subjected to hypotonic lysis, and cytosolic fractions were evaluated by gel filtration through a Superdex 200 column. Based on gel filtration of protein standards from 25 to 2000 kDa through the Superdex 200 column, maspin eluted within the monomeric range of 22–67 kDa (Fig. 5C). Together, these results suggest that maspin in MCF10A cell cytoplasm is monomeric and is not stably associated with other cellular proteins.
DISCUSSION
We have shown here that maspin is an obligate intracellular serpin, which is neither secreted nor found at the plasma membrane of immortalized non-transformed mammary or prostate epithelial cells. Earlier research on maspin and its function as a tumor suppressor employed transformed breast and prostate cancer cell lines that do not express maspin and depended on the addition of purified recombinant maspin to the exterior of these cells or the re-introduction of a functional gene via transfection. This led to the general view that maspin functions extracellularly as a tumor suppressor by inhibiting cell motility and invasion and increasing cell adhesion but without evidence that endogenous maspin actually reaches the cell surface.
Although clade B serpins do not possess conventional signal peptides, there are examples of secreted and glycosylated clade B serpins, such as SERPINB2 and ovalbumin. However, the N termini of these two serpins have characteristics of uncleaved signal peptides that are absent from other clade B serpins, including maspin. In other words, maspin is predicted to be a cytoplasmic non-secreted protein (19, 21), which is confirmed by our data. It is important to note that SERPINB2 and ovalbumin are efficiently folded, glycosylated, and released from the secretory pathway and, unless mutated (35), show no evidence of retention in secretory pathway organelles. By contrast, we have shown that although maspin can be artificially targeted to the ER, it cannot travel through the secretory pathway. The HA signal sequence used here has been previously employed to successfully translocate nuclear or cytosolic proteins into the ER (27, 36, 37). Although such proteins enter the ER, where they can be glycosylated, they fail to move along the secretory pathway because of misfolding and degradation (38). However, the addition of the HA signal sequence to a protein that is normally secreted only enhances its trafficking and does not impair its usual processing and secretion (27). Based on these studies, the fact that maspin cannot be secreted even when provided with a signal sequence suggests that it is restricted to a nucleocytoplasmic role.
A formal possibility remains that maspin is released from cells following an unconventional pathway in response to an unknown signal (39). In a recent report, maspin was identified through a mass spectrometry-based proteomics approach as a protein potentially secreted through an unconventional pathway regulated by activation of caspase-1, but the authors were unable to verify its secretion (40). In another study, proteomics analysis of conditioned medium from MCF10A, BT474, and MDA-MB-468 cells showed maspin in the medium, but it was not in the top 100 secreted proteins based on abundance. Furthermore, over 50% of the proteins released were identified as intracellular, suggesting that significant nonspecific release resulted from cell damage or death (41). The confounding effect of nonspecific release is also illustrated by an analysis of neoplastic (maspin-negative) prostate epithelium, which similarly identified over 50% intracellular proteins in the conditioned medium (42). Finally, maspin is released from H460 cancer cells in response to irradiation, possibly packaged in exosomes (43). Whether other cells can release maspin in the same way in response to stress remains to be discovered. Together, these results suggest that extracellular maspin arises as a consequence of cell damage or lysis and not via active secretion. Interestingly, the release of the related intracellular serpins SERPINB3 and SERPINB4 from tumor cells has also been explained as a consequence of cell damage or lysis (44).
What then is the molecular function of maspin within cells? Several lines of evidence suggest that maspin modulates the cell-matrix interaction (4, 6, 13, 45), and an inside-out signaling mechanism for regulating such interactions is not without precedent (46). Maspin has also been suggested to interact with the apoptotic machinery (14–17). However, as shown here by imaging and fractionation or by affinity-based approaches,3 we have been unable to detect association of maspin with cytoskeletal or other cellular structures or components, including β1-integrin, as previously reported (8). Alternatively, it is possible that maspin exerts its role in the nucleus at the level of gene or chromatin regulation and thus indirectly affects the cell-matrix interaction or differentiation state. This would be consistent with previous reports of nuclear maspin (47–54) and the redistribution of maspin from the nucleus to the cytoplasm on MCF10A differentiation shown in this study. It is also consistent with observations that maspin interacts with the transcription factor IRF6 (18) and that a related clade B serpin has chromatin remodeling capability (55).
This work was supported by National Health and Medical Research Council of Australia Program Grant 490900.
S. S. Y. Teoh, J. C. Whisstock, and P. I. Bird, unpublished data.
- RITC
- rhodamine isothiocyanate
- Endo H
- endoglycosidase H
- PNGase F
- peptide:N-glycosidase F
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Silverman G. A., Whisstock J. C., Askew D. J., Pak S. C., Luke C. J., Cataltepe S., Irving J. A., Bird P. I. (2004) Cell. Mol. Life Sci. 61, 301–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou Z., Anisowicz A., Hendrix M. J., Thor A., Neveu M., Sheng S., Rafidi K., Seftor E., Sager R. (1994) Science 263, 526–529 [DOI] [PubMed] [Google Scholar]
- 3.Gao F., Shi H. Y., Daughty C., Cella N., Zhang M. (2004) Development 131, 1479–1489 [DOI] [PubMed] [Google Scholar]
- 4.Ngamkitidechakul C., Warejcka D. J., Burke J. M., O'Brien W. J., Twining S. S. (2003) J. Biol. Chem. 278, 31796–31806 [DOI] [PubMed] [Google Scholar]
- 5.Sheng S., Carey J., Seftor E. A., Dias L., Hendrix M. J., Sager R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11669–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blacque O. E., Worrall D. M. (2002) J. Biol. Chem. 277, 10783–10788 [DOI] [PubMed] [Google Scholar]
- 7.Ngamkitidechakul C., Burke J. M., O'Brien W. J., Twining S. S. (2001) Invest. Ophthalmol. Vis. Sci. 42, 3135–3141 [PubMed] [Google Scholar]
- 8.Cella N., Contreras A., Latha K., Rosen J. M., Zhang M. (2006) FASEB J. 20, 1510–1512 [DOI] [PubMed] [Google Scholar]
- 9.Yin S., Lockett J., Meng Y., Biliran H., Jr., Blouse G. E., Li X., Reddy N., Zhao Z., Lin X., Anagli J., Cher M. L., Sheng S. (2006) Cancer Res. 66, 4173–4181 [DOI] [PubMed] [Google Scholar]
- 10.Biliran H., Jr., Sheng S. (2001) Cancer Res. 61, 8676–8682 [PubMed] [Google Scholar]
- 11.McGowen R., Biliran H., Jr., Sager R., Sheng S. (2000) Cancer Res. 60, 4771–4778 [PubMed] [Google Scholar]
- 12.Odero-Marah V. A., Khalkhali-Ellis Z., Chunthapong J., Amir S., Seftor R. E., Seftor E. A., Hendrix M. J. (2003) Cancer Biol. Ther. 2, 398–403 [DOI] [PubMed] [Google Scholar]
- 13.Shi H. Y., Stafford L. J., Liu Z., Liu M., Zhang M. (2007) Cell Motil. Cytoskeleton 64, 338–346 [DOI] [PubMed] [Google Scholar]
- 14.Liu J., Yin S., Reddy N., Spencer C., Sheng S. (2004) Cancer Res. 64, 1703–1711 [DOI] [PubMed] [Google Scholar]
- 15.Romani A. A., Soliani P., Desenzani S., Borghetti A. F., Crafa P. (2006) BMC Cancer 6, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W., Shi H. Y., Zhang M. (2005) BMC Cancer 5, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latha K., Zhang W., Cella N., Shi H. Y., Zhang M. (2005) Mol. Cell. Biol. 25, 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey C. M., Khalkhali-Ellis Z., Kondo S., Margaryan N. V., Seftor R. E., Wheaton W. W., Amir S., Pins M. R., Schutte B. C., Hendrix M. J. (2005) J. Biol. Chem. 280, 34210–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remold-O'Donnell E. (1993) FEBS Lett. 315, 105–108 [DOI] [PubMed] [Google Scholar]
- 20.Bird C. H., Blink E. J., Hirst C. E., Buzza M. S., Steele P. M., Sun J., Jans D. A., Bird P. I. (2001) Mol. Cell. Biol. 21, 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belin D., Guzman L. M., Bost S., Konakova M., Silva F., Beckwith J. (2004) J. Mol. Biol. 335, 437–453 [DOI] [PubMed] [Google Scholar]
- 22.Bello D., Webber M. M., Kleinman H. K., Wartinger D. D., Rhim J. S. (1997) Carcinogenesis 18, 1215–1223 [DOI] [PubMed] [Google Scholar]
- 23.Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Methods 30, 256–268 [DOI] [PubMed] [Google Scholar]
- 24.Chu M., Bird C. H., Teasdale M., Bird P. I. (1998) Thromb. Haemost. 80, 119–127 [PubMed] [Google Scholar]
- 25.Law R. H., Irving J. A., Buckle A. M., Ruzyla K., Buzza M., Bashtannyk-Puhalovich T. A., Beddoe T. C., Nguyen K., Worrall D. M., Bottomley S. P., Bird P. I., Rossjohn J., Whisstock J. C. (2005) J. Biol. Chem. 280, 22356–22364 [DOI] [PubMed] [Google Scholar]
- 26.Madison E. L., Bird P. (1992) Gene 121, 179–180 [DOI] [PubMed] [Google Scholar]
- 27.Scott F. L., Coughlin P. B., Bird C., Cerruti L., Hayman J. A., Bird P. (1996) J. Biol. Chem. 271, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 28.Brock T. G., Paine R., 3rd, Peters-Golden M. (1994) J. Biol. Chem. 269, 22059–22066 [PubMed] [Google Scholar]
- 29.Sternlicht M. D., Kedeshian P., Shao Z. M., Safarians S., Barsky S. H. (1997) Clin. Cancer Res. 3, 1949–1958 [PubMed] [Google Scholar]
- 30.Plopper G. E., Domanico S. Z., Cirulli V., Kiosses W. B., Quaranta V. (1998) Breast Cancer Res. Treat. 51, 57–69 [DOI] [PubMed] [Google Scholar]
- 31.Chao C. C., Bird P., Gething M. J., Sambrook J. (1987) Mol. Cell. Biol. 7, 3842–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulmasov B., Bruno J., Woost P. G., Edwards J. C. (2007) BMC Cell Biol. 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomas D. A., Evans D. L., Finch J. T., Carrell R. W. (1992) Nature 357, 605–607 [DOI] [PubMed] [Google Scholar]
- 34.Lomas D. A., Carrell R. W. (2002) Nat. Rev. Genet. 3, 759–768 [DOI] [PubMed] [Google Scholar]
- 35.Mikus P., Ny T. (1996) J. Biol. Chem. 271, 10048–10053 [DOI] [PubMed] [Google Scholar]
- 36.Bird P., Gething M. J., Sambrook J. (1987) J. Cell Biol. 105, 2905–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wirth D. F., Lodish H. F., Robbins P. W. (1979) J. Cell Biol. 81, 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 39.Nickel W., Seedorf M. (2008) Annu. Rev. Cell Dev. Biol. 24, 287–308 [DOI] [PubMed] [Google Scholar]
- 40.Keller M., Rüegg A., Werner S., Beer H. D. (2008) Cell 132, 818–831 [DOI] [PubMed] [Google Scholar]
- 41.Kulasingam V., Diamandis E. P. (2007) Mol. Cell. Proteomics 6, 1997–2011 [DOI] [PubMed] [Google Scholar]
- 42.Martin D. B., Gifford D. R., Wright M. E., Keller A., Yi E., Goodlett D. R., Aebersold R., Nelson P. S. (2004) Cancer Res. 64, 347–355 [DOI] [PubMed] [Google Scholar]
- 43.Yu X., Harris S. L., Levine A. J. (2006) Cancer Res. 66, 4795–4801 [DOI] [PubMed] [Google Scholar]
- 44.Uemura Y., Pak S. C., Luke C., Cataltepe S., Tsu C., Schick C., Kamachi Y., Pomeroy S. L., Perlmutter D. H., Silverman G. A. (2000) Int. J. Cancer 89, 368–377 [DOI] [PubMed] [Google Scholar]
- 45.Abraham S., Zhang W., Greenberg N., Zhang M. (2003) J. Urol. 169, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 46.Petrich B. G. (2009) Thromb. Haemost. 101, 1020–1024 [PubMed] [Google Scholar]
- 47.Bettstetter M., Woenckhaus M., Wild P. J., Rümmele P., Blaszyk H., Hartmann A., Hofstädter F., Dietmaier W. (2005) J. Pathol. 205, 606–614 [DOI] [PubMed] [Google Scholar]
- 48.Chua R., Setzer S., Govindarajan B., Sexton D., Cohen C., Arbiser J. L. (2009) J. Am. Acad. Dermatol. 60, 758–766 [DOI] [PubMed] [Google Scholar]
- 49.Lonardo F., Li X., Siddiq F., Singh R., Al-Abbadi M., Pass H. I., Sheng S. (2006) Lung Cancer 51, 31–39 [DOI] [PubMed] [Google Scholar]
- 50.Maass N., Hojo T., Ueding M., Lüttges J., Klöppel G., Jonat W., Nagasaki K. (2001) Clin. Cancer Res. 7, 812–817 [PubMed] [Google Scholar]
- 51.Machtens S., Serth J., Bokemeyer C., Bathke W., Minssen A., Kollmannsberger C., Hartmann J., Knüchel R., Kondo M., Jonas U., Kuczyk M. (2001) Int. J. Cancer 95, 337–342 [DOI] [PubMed] [Google Scholar]
- 52.Mohsin S. K., Zhang M., Clark G. M., Craig Allred D. (2003) J. Pathol. 199, 432–435 [DOI] [PubMed] [Google Scholar]
- 53.Murai S., Maesawa C., Masuda T., Sugiyama T. (2006) Cancer Sci. 97, 883–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu M., Zheng H., Tsuneyama K., Takahashi H., Nomoto K., Xu H., Takano Y. (2007) Hum. Pathol. 38, 1248–1255 [DOI] [PubMed] [Google Scholar]
- 55.Grigoryev S. A., Bednar J., Woodcock C. L. (1999) J. Biol. Chem. 274, 5626–5636 [DOI] [PubMed] [Google Scholar]