Abstract
Germ line mutations of the ubiquitin ligase cullin 7 (CUL7) are linked to 3-M syndrome and Yakuts short stature syndrome, both of which are characterized by pre- and post-natal growth retardation. CUL7 knock-out mice show placental and embryonic defects similar to intrauterine growth retardation, suggesting a role of CUL7 in placentation. CUL7 was found in this study to be highly expressed in first trimester invasive human placental villi as well as in HTR8/SVneo and B6Tert cells, two cell lines derived from human first trimester trophoblast cells. However, CUL7 levels in term trophoblast cells or JEG-3 cells, which are derived from human choriocarcinoma but exhibit weak invasion capacity, were low or undetectable. Forced expression of CUL7 in JEG-3 cells induced cell morphological changes characteristic of epithelial-mesenchymal transition, which was accompanied by a complete loss of the epithelial markers E-cadherin and P-cadherin and a significant elevation of mesenchymal markers Vimentin and N-cadherin. JEG-3 cells expressing CUL7 exhibited enhanced cell migration and invasion. Conversely, CUL7-specific RNA interference in HTR8/SVneo cells resulted in increased E-cadherin expression and reduced cell migration and invasion. Furthermore, CUL7 expression down-regulated E-cadherin mRNA expression by up-regulating ZEB1 and Slug, two transcriptional repressors of E-cadherin. Finally, CUL7-induced loss of E-cadherin expression was partially reversed by treatment of CUL7-expressing cells with the proteasome inhibitor MG-132. These results suggest that the CUL7 E3 ligase is a key regulator in trophoblast cell epithelial-mesenchymal transition and placental development.
Keywords: Cell Differentiation, Cell Migration, Cell Motility, E3 Ubiquitin Ligase, Epithelial Cell, CUL7, E-cadherin, Epithelial-Mesenchymal Transition, Placenta, Trophoblast
Introduction
The cullin family proteins are scaffold proteins for the Ring finger type E3 ligases (1, 2). Humans express seven cullin proteins: CUL1–3, CUL4A, CUL4B, CUL5, and CUL7 (p185, p193, KIAA0076). Each cullin protein can form an E3 ligase similar to the prototype Ring-type E3 ligase Skp1-CUL1-F-box complex (3, 4). The Cullin-RING-finger type E3 ligases are important regulators in early embryonic development, as highlighted by genetic studies demonstrating that knock-out of CUL1, CUL3, or CUL4A in mice results in early embryonic lethality (5–7).
In contrast, CUL7 appears to be an important regulator of placental development. First, CUL7 knock-out in the mouse results in small and abnormal placentas, which are characterized by impaired formation of the spongiotrophoblast layer and vascular defects in the labyrinth layer (8). As a result of the placental defect, CUL7 knock-out mice exhibit smaller fetuses at late gestational stages and neonatal lethality. In support of the placental function of CUL7, mice lacking Fbxw8 (also known as Fbw6 or Fbx29), the only F-box protein identified as a CUL7 binding partner (9, 10), exhibit a phenotype almost identical to that of CUL7−/− mice; that is, small embryos but with abnormalities mainly restricted to the placenta (9, 11).
In addition to the genetic evidence in mice, CUL7 mutations have also been identified in 3-M syndrome and the Yakuts short stature syndrome, both of which are characterized by pre- and post-natal growth retardation but with relatively normal mental and endocrine functions, suggesting that CUL7 may also be crucial for human placental development (12, 13).
During human placental development, three main trophoblast populations can be identified: cytotrophoblast stem cells, syncytiotrophoblast, and extravillous cytotrophoblast (14). Villous trophoblast cells undergo terminal differentiation and fusion to form the multinucleated epithelial syncytiotrophoblast, whereas the extravillous cytotrophoblast undergoes an epithelial-mesenchymal transition (EMT),3 detaches from the distal cell columns, and infiltrates the endomyometrium to anchor the placenta. However, neither the external signals nor the major intracellular signaling pathways responsible for this EMT are known (14, 15).
Given the clear implication of CUL7 in placental development in both humans and mice, we investigated the possible roles of CUL7 in trophoblast cell differentiation and function. We first examined CUL7 expression in different types of trophoblast cells in human first trimester and term placentas as well as several well established cell models of trophoblast cell lineage. Through RNA interference and overexpression studies, we have demonstrated that CUL7 is a key factor in regulating the EMT of the trophoblast cell lineage.
EXPERIMENTAL PROCEDURES
Tissue Collection
Placental tissue collection was carried out in accordance with guidelines established by the Ethics Committee of the Xuan-Wu Hospital in Beijing. Informed consent was obtained from each woman donating her placenta. Ten normal human placentas at different gestational stages (seven at the first trimester and three at term) were collected from women undergoing legal elective abortion or normal full-term delivery. All placental tissues were washed with ice-cold phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 24 h at 4 °C followed by paraffin embedding.
Immunohistochemistry
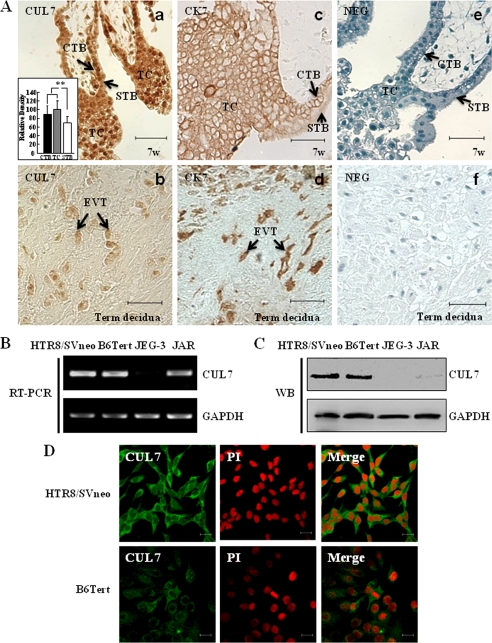
Immunohistochemical studies were performed using the Histostain-Plus kit and diaminobenzidine (Zhongshan Corp., Beijing, China) according to the manufacturer's instructions. Briefly, sections (5 μm) were deparaffinized and rehydrated in xylene and a graded series of ethyl alcohol. Antigen retrieval in 10 mm sodium citrate and 10 mm citric acid was performed by microwave irradiation. Tissue sections were then incubated with 3% H2O2 in methanol for 15 min to quench endogenous peroxidase followed by sequential incubation in normal goat serum for 30 min, primary antibodies against CUL7 (10 μg/ml, Bethyl Laboratories, Montgomery, TX) at 4 °C overnight, and biotinylated secondary antibody for 30 min. The diaminobenzidine solution was applied until a brown color developed. Sections were counterstained with hematoxylin and mounted with histomount (Zymed Laboratories Inc., San Francisco, CA). Negative controls were performed by replacing the primary antibody with control IgG. Semiquantitative determination of CUL7 expression in the various trophoblast cells (Fig. 1A, panel a, inset) was carried out using ImageJ Basics (National Institutes of Health, Version 1.42). At least 270 cells were counted for each cell type with the gray level of intercellular space taken as background.
FIGURE 1.
Expression of CUL7 in human placentas and trophoblast cell lines. A, shown is immunostaining of CUL7 in normal human placental villi at the seventh week of pregnancy (7w; panel a) or in the EVTs invaded into the maternal decidua at full term (panel b). Panels c and d show immunohistochemical staining with anti-cytokeratin 7 (CK7), a marker for cytotrophoblast cells (CTB), trophoblast column (TC), and EVT in the first trimester placental villi (c) or in the term decidua (d). Panels e and f show negative controls (NEG) on sections from first trimester placental villi (e) or term decidua (f) in which serum IgG from non-immunized rabbits was used in place of specific primary antibody. Relative staining intensities of CUL7 in different types of trophoblast cells are shown as an inset in panel a. **, p < 0.01. STB, syncytiotrophoblast. Bar = 50 μm. B and C, expression of CUL7 in different trophoblast cell lines was determined by RT-PCR (B) and Western blotting (WB; C), respectively. GAPDH was used as an internal control for RT-PCR or a loading control for Western blotting in this study. HTR8/SVneo, a human invasive EVT cell line derived from immortalized first trimester trophoblasts; B6Tert, immortalized cytotrophoblast cell line; JEG-3 and JAR, human choriocarcinoma cell lines. D, immunofluorescent staining of HTR8/SVneo or B6Tert cells with anti-CUL7 antibody is shown. Fluorescence signals specific to CUL7 antibody are visualized as green, and PI staining shows the nuclei (red). CUL7 was mainly detected in the cytoplasm of HTR8/SVneo and B6Tert cells. Bar = 20 μm.
Cell Lines and Cell Culture
All the experiments performed in this study were in accordance with guidelines established by the Ethics Committee, State Key Laboratory of Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences. The human extravillous trophoblast cell line HTR8/SVneo and choriocarcinoma cell line JAR were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The choriocarcinoma cell line JEG-3 was maintained in F-12/Dulbecco's modified Eagle's medium (1:1) containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. B6Tert cells are normal placental-origin cytotrophoblast cells stably transfected with human telomerase reverse transcriptase (hTERT) gene, which exhibit characteristics of normal extravillous cytotrophoblasts during the early weeks of gestation. B6Tert cells were cultured in F-12/Dulbecco's modified Eagle's medium with 1% bovine serum albumin, 200 mm l-glutamine, 1 mg/ml insulin, and 1 μg/ml epidermal growth factor. All cells were maintained at 37 °C with 5% CO2.
Stable Transfection
Full-length human CUL7 cDNA in pcDNA3 mammalian expression vector was a generous gift from Dr. Y. Xiong (Depart. of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC). JEG-3 cells were transfected with pcDNA3-CUL7 or empty vector (EV) with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. To establish cell lines stably expressing CUL7 or carrying the empty vectors, cells were trypsinized at 24 h after transfection and selected in medium in the presence of 0.75 mg/ml Geneticin (Invitrogen) for 7 days.
Transient Transfection of Wild-type and Mutant CUL7 Constructs
pcDNA3-CUL7 plasmids encoding full-length or truncated CUL7 mutants consisting of amino acid residues 1–1343 or 268–1698 were kindly provided by Dr. J. DeCaprio (Depart. of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA). In brief, JEG-3 cells were seeded onto coverslips in 60-mm dishes. Transfection was carried out when cells reached 50–60% confluence using Lipofectamine 2000 (Invitrogen) transfection. Four-microgram plasmid DNA/60-mm dish for full-length CUL7, mutant plasmid, or empty vector was used in this study. Cells transfected with pcDNA3-EV served as a negative control. Cells were incubated with Opti-MEM (Invitrogen) for 6 h and then with fresh F-12/ Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. At 48 h after transfection, cells grown on the coverslips were washed briefly with PBS and fixed with ice-cold methanol at −20 °C for 10 min. The coverslips were incubated with indicated primary antibody overnight at 4 °C, washed in PBS, and incubated for 1 h at 37 °C in PBS with fluorescein isothiocyanate-conjugated secondary antibody. Cells were washed in PBS and incubated for 5 min at room temperature with 4′,6-diamidino-2-phenylindole followed by mounting using 1,4-diazabicyclo(2.2.2)octane (DABCO) anti-fade mounting medium. Image acquisition was performed by confocal microscopy using a Carl Zeiss LSM 710 laser-scanning microscope.
Small RNA Interference
Stealth small interfering RNAs (siRNAs) were designed by Invitrogen. The sequences of siRNAs used are: CUL7, 5′-AUAAUCAGGACUACUCAACAUGUGC-3′; E-cadherin, 5′-AUAAUCAGGACUACUCAACAUGUGC-3′; ZEB1, 5′-AUAAGACCCAGAGUGUGAGAAGCGC-3′; Slug, 5′-UAAUGUGUCCUUGAAGCAACCAGGG-3′. As a nonspecific control, a universal control siRNA was used. siRNA duplex was transfected into cells with Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Final concentrations of siRNAs were 100 nm, and the cells were harvested 72 h after transfection.
Western Blotting
Total protein extracts were prepared in whole-cell lysis buffer (50 mm HEPES, 150 mm NaCl, 1 mm EGTA, 10 mm sodium pyrophosphate, 1.5 mm MgCl2, 100 mm sodium fluoride, 10% glycerol, and 1% Triton X-100) containing an inhibitor mixture (1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 1 mm sodium orthovanadate). Protein concentrations were determined using a standard Bradford assay, and 50 μg of total protein was subjected to SDS-PAGE followed by electrotransfer onto nitrocellulose membranes. Membranes were incubated overnight at 4 °C with primary antibodies against human CUL7 (1 μg/ml; Bethyl Laboratories), human Slug (0.25 μg/ml; Abgent, San Diego, CA), human ZEB1 (0.2 μg/ml; Santa Cruz Biotechnology Inc.), human E-cadherin (0.2 μg/ml, Santa Cruz), or human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (0.1 μg/ml; Abcam, Cambridge Science Park, UK) followed by incubation in secondary antibodies. Signals were developed using the Enhanced Chemiluminescence system (Pierce).
Semiquantitative Reverse Transcription (RT)-PCR
HTR8/SVneo, B6Tert, or JEG-3 cells were lysed and processed for total RNA extraction using Trizol reagent (Invitrogen) according to the manufacturer's instructions. The first cDNA strand was synthesized by Superscript reverse transcriptase. The sequences of the primers used for the PCR amplification are shown in Table 1. The cDNA of the housekeeping GAPDH gene was also amplified for normalization.
TABLE 1.
Primers used for RT-PCR experiments
| Gene | NCBI accession no. | Primer sequence (5′–3′) | |
|---|---|---|---|
| CUL7 | NM_014780 | Forward | CCATCTCAGAGTCCCAACAC |
| Reverse | TTCAGCACCACGGCATAG | ||
| E-cadherin | NM_004360 | Forward | ACGCATTGCCACATACA |
| Reverse | CGTTAGCCTCGTTCTCA | ||
| Vimentin | NM_003380 | Forward | ATGGCTCGTCACCTTCG |
| Reverse | AGTTTCGTTGATAACCTGTCC | ||
| N-cadherin | NM_001792 | Forward | GAAAGACCCATCCACG |
| Reverse | CCTGCTCACCACCACTA | ||
| P-cadherin | NM_001793 | Forward | GGTGGTTCTCCGCAATG |
| Reverse | GCCGCCTTCAGGTTCTC | ||
| ZEB1 | NM_030751 | Forward | AGAAAGCAGGCGAACC |
| Reverse | TCATCCTCCCAGCAGT | ||
| Snail | NM_005985 | Forward | AATCGGAAGCCTAACTACAGCG |
| Reverse | GTCCCAGATGAGCATTGGCA | ||
| Slug | AF042001 | Forward | AGCAGTTGCACTGTGATGCC |
| Reverse | ACACAGCAGCCAGATTCCTC | ||
| GAPDH | M33197 | Forward | AGCCACATCGCTCAGACA |
| Reverse | TGGACTCCACGACGTACT | ||
| Twist | NM_000474 | Forward | CAGCTACGCCTTCTCGG |
| Reverse | CACGCCCTGTTTCTTTGA | ||
| SIP1 | NM_014795 | Forward | AAATGATGAGGTGCTTCC |
| Reverse | TCAATGATGTAGGGCTTC | ||
Immunofluorescence
JEG-3, HTR8/SVneo, or JAR cells cultured on coverslips were fixed for 10 min in ice-cold methanol and permeabilized for 7 min with 0.01% Triton X-100 in PBS (PBST). After incubation with blocking buffer (3% bovine serum albumin), cells were incubated with primary antibodies against CUL7 (10 μg/ml, Bethyl Laboratories), E-cadherin (10 μg/ml, Santa Cruz), or Vimentin (10 μg/ml, Santa Cruz). Cells were washed with PBST and incubated with appropriate fluorescein isothiocyanate-conjugated secondary antibodies (10 μg/ml; Zhongshan Corp). For staining of filamentous actin, cells were incubated with fluorescein isothiocyanate-phalloidin (Molecular Probes, Inc.) at 3 units/ml dilution. Nuclei were counterstained with propidium iodide (PI). Slides were mounted with 1,4-diazabicyclo(2.2.2)octane anti-fade mounting medium and analyzed by confocal microscopy using a Carl Zeiss LSM 710 laser scanning microscope.
Matrigel Invasion Assay
Invasion of cells was measured in Matrigel (BD Biosciences)-coated Transwell inserts (6.5 mm, Costar, Cambridge) containing polycarbonate filters with 8-μm pores as detailed previously (16). The inserts were coated with 50 μl of 1 mg/ml Matrigel matrix according to the manufacturer's recommendations. After transfection with siRNAs for 24 h, HTR8/SVneo cells (2 × 105), B6Tert cells (2.5 × 105), or JEG-3 cells (2.5 × 105) in 200 μl of serum-free medium were plated in the upper chamber, whereas 600 μl of medium with 10% fetal bovine serum were added to the lower well. After incubation for 22 h at 37 °C, non-invaded cells on top of the Transwell were scraped off with a cotton swab. The filters attached with invaded cells on the other side were washed with PBS, fixed in methanol for 10 min, and stained with hematoxylin and eosin. The invaded cells were counted under a light microscope (Olympus IX51) in 10 randomly selected fields at a magnification of ×200. Each experiment was performed in triplicate. Invasion of cells under different treatments was normalized to the control and expressed as the means of invasion percentage (%) ±S.E.
Transwell Cell Migration Assay
Trophoblast migration was carried out using Transwell inserts (6.5 mm, Costar) containing polycarbonate filters with 8-μm pores. Methods used in cell migration assay were similar to Matrigel invasion assay except that the Transwell insert was not coated with Matrigel.
Statistical Analysis
Each experiment was performed in triplicate. Quantification of the bands from RT-PCR and Western blotting were determined by MetaView Image Analyzing System (Version 4.50; Universal Imaging Corp., Downingtown, PA). Results were presented as the mean ± S.E. Statistical analyses were performed using the Statistical Package for Social Science (SPSS for Windows package release 10.0; SPSS Inc., Chicago, IL) and are indicated under “Results” and in the legends to Figs. 1, 4, and 5. *, p < 0.05. **, p < 0.01.
FIGURE 4.
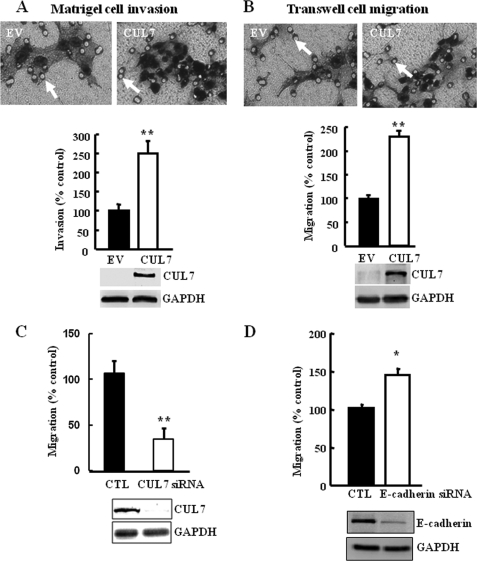
Overexpression of CUL7 in JEG-3 cells enhanced cell invasion and migration (A and B). Representative images of filters containing invaded cells in the Matrigel invasion assay (A) or Transwell cell migration assay (B) are shown. White arrows indicate pores on the filter of the Transwell. Bar graphs show the statistical analysis of three independent experiments (t test; **, p < 0.01). C, silencing CUL7 by siRNA in JEG-3 cells stably transfected with CUL7 decreased the migration capacity of JEG-3 cells based on Transwell cell migration assay (t test; **, p < 0.01). D, silencing of E-cadherin by siRNA resulted in increased migration capacity of JEG-3 cells, based on Transwell cell migration assay (t test; *, p < 0.05). Below the graphs are representative immunoblots showing the efficiency of overexpression or RNAi.
FIGURE 5.
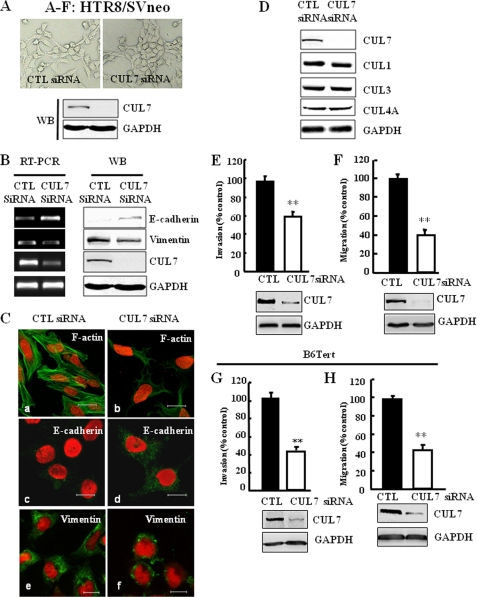
CUL7 siRNA interference increased E-cadherin expression in HTR8/SVneo cells. A, shown are representative bright field images of HTR8/SVneo cells transfected with scrambled siRNA (CTL siRNA) or CUL7 siRNA. Efficiency of siRNA interference was assessed as described below. WB, Western blot. B, silencing CUL7 increased E-cadherin but decreased Vimentin expression in HTR8/SVneo cells. C, HTR8/SVneo cells transfected with CTL siRNA or CUL7 siRNA for 48 h were immunostained for filamentous actin (F-actin) using fluorescein isothiocyanate-conjugated phalloidin, E-cadherin, or Vimentin and were photographed with confocal laser scanning microscopy. Nuclei were shown by PI staining. Bar = 20 μm. D, specificity of CUL7 siRNA was tested by examining the expression of several other cullins in HTR8/SVneo cells transfected with CUL7 siRNA. E and F, bar graphs show that HTR8/SVneo cells transfected with CUL7 siRNA exhibit significantly reduced levels of cell invasion (E) and cell migration (F). Immunoblotting of a typical transfection experiment is shown to indicate siRNA interference efficiency (t test; **, p < 0.01). G and H, bar graphs show that B6Tert cells transfected with CUL7 siRNA exhibit significantly reduced levels of cell invasion (G) and cell migration (H). Immunoblotting of a typical transfection experiment is shown below the bar graphs to indicate siRNA interference efficiency. (t test; **, p < 0.01).
RESULTS
CUL7 Expression in First Trimester Human Placental Tissues and Invasive Extravillous Trophoblast Cells
We first carried out immunohistochemical studies to examine the expression pattern of CUL7 in different placental compartments of women at the first trimester or at term. As shown in Fig. 1A, CUL7 expression levels were higher in cytotrophoblast cells (CTB) and the trophoblast column (TC) than those in syncytiotrophoblasts (STB) at first trimester (panel a). Extravillous trophoblast cells (EVT) invading into the maternal decidua at term had a lower level of CUL7 (panel b). Cytokeratin 7 (CK7) is a marker to show positive staining in cytotrophoblast cells and the trophoblast column in first trimester placental villi (panel c) and EVT in term decidua (panel d). Control IgG exhibited no specific staining in either first trimester placental villi (panel e) or term decidua (panel f). Expression of CUL7 in several human trophoblast cell lines was also examined by RT-PCR, Western blotting, and immunofluorescence microscopy. CUL7 mRNA could be detected in HTR8/SVneo, B6Tert, and choriocarcinoma JAR cells but was very low in JEG-3 cells (Fig. 1B). Similarly, CUL7 protein was present in HTR8/SVneo cells and B6Tert cells but was undetectable in JEG-3 cells (Fig. 1C). JAR cells expressed low but detectable levels of CUL7 proteins (Fig. 1C). Immunofluorescence microscopy confirmed these findings and further revealed that CUL7 proteins were predominantly present in the cytoplasm of HTR8/SVneo and B6Tert cells (Fig. 1D).
Forced Expression of CUL7 in JEG-3 Cells Induced Changes Characteristic of EMT
We noticed that JEG-3 cells, which lacked CUL7 expression, exhibited cobblestone-like morphology (not shown but similar to JEG-3-EV as in Fig. 2A, middle panel), characteristic of epithelial cells. In contrast, HTR8/ SVneo cells, which expressed CUL7, were of more elongated, fibroblast-like mesenchymal appearance (Fig. 1D). To investigate whether the lack of CUL7 expression in JEG-3 cells contributed to their epithelial cell morphology, we generated JEG-3 cells stably expressing CUL7. Overexpression of CUL7 in JEG-3 cells was confirmed by RT-PCR and Western blotting (Fig. 2A, left panel). JEG-3 cells expressing CUL7 lost the epithelial cell morphology and instead exhibited elongated mesenchymal cell shape (Fig. 2A, right panel)). In contrast, JEG-3 cells harboring the EV maintained the cobblestone-like epithelial morphology (Fig. 2A, middle panel).
FIGURE 2.
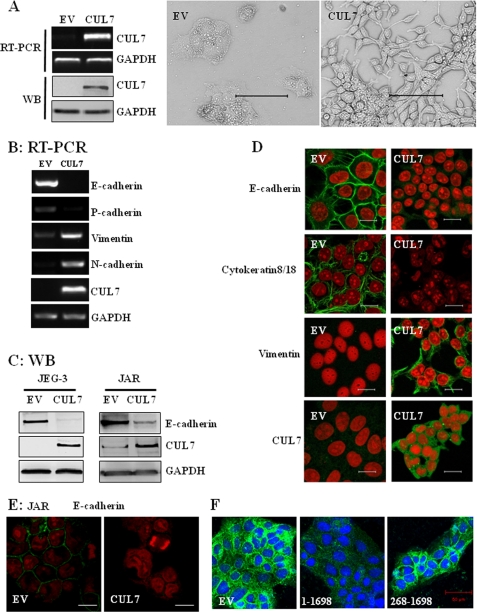
Overexpression of CUL7 in JEG-3 cells induced morphological changes characteristic of EMT. A, JEG-3 cells stably transfected with EV or CUL7 were analyzed by RT-PCR or immunoblotting (WB; left panel). Shown in the middle and right panels are typical phase contrast micrographs of the two types of cells grown to subconfluence. Bar = 100 μm. B, expression levels of the epithelial markers (E-cadherin and P-cadherin) and mesenchymal markers (N-cadherin and Vimentin) were determined by RT-PCR in the two types of cells as in A. C, expression of E-cadherin and CUL7 was analyzed by immunoblotting of whole cell lysates of JEG-3 or JAR cells transfected with EV or CUL7. D and E, typical immunofluorescence images are shown of JEG-3 (D) and JAR cells (E) transfected with EV or CUL7 with the indicated primary antibodies. The nuclei were stained with PI. Bar = 20 μm. F, JEG-3 cells were transiently transfected with EV, full-length CUL7 (1–1698) or a truncated mutant (268–1698) for 48 h and were fixed and immunostained with anti-E-cadherin antibodies (green) and counter-stained with 4′,6-diamidino-2-phenylindole (blue). Bar = 50 μm.
To confirm that CUL7 expression caused EMT in JEG-3 cells, we examined the expression of both epithelial markers (E-cadherin and P-cadherin) (17) and mesenchymal markers (Vimentin and N-cadherin) (17, 18). Overexpression of CUL7 in JEG-3 cells resulted in a substantial loss of E-cadherin and P-cadherin as well as a markedly increased expression of Vimentin and N-cadherin (Fig. 2, B and C). Immunofluorescence microscopy revealed that JEG-3 cells (not shown) and JEG-3 cells transfected with a control vector exhibited E-cadherin staining typical of epithelial cells (Fig. 2D, EV), whereas CUL7 expression in these cells caused a complete loss of E-cadherin in the cell junctions (Fig. 2D, CUL7). Analyzing another epithelial marker, cytokeratin 8/18, revealed a similar result, with prominent expression in JEG-3 cells transfected with EV but an absence in CUL7-expressing JEG-3 cells (Fig. 2D). In contrast, Vimentin was undetectable in vector-transfected JEG-3 cells but was prominently expressed in CUL7-expressing JEG-3 cells (Fig. 2D). The effect of forced expression of CUL7 was also examined in JAR cells, another cell line derived from human choriocarcinoma that exhibits low endogenous CUL7 (Fig. 1, B and C). Significantly, overexpression of CUL7 also diminished E-cadherin expression (Fig. 2, C and E).
To explore whether CUL7-mediated EMT in JEG-3 cells required the associated E3 ligase activity, a CUL7 N-terminal-truncated mutant, CUL7 (268–1698), lacking the ability to bind FBXW8 was utilized (8). As shown in Fig. 2F, transient expression of full-length CUL7 (1–1698) eliminated E-cadherin expression, similar to the results obtained in the stable line (Fig. 2D). In contrast, the FBXW8 binding-deficient mutant, CUL7 (268–1698) was ineffective in inhibiting E-cadherin expression. The two CUL7 constructs were expressed at similar levels in these transient expression experiments (not shown). These results strongly suggest that the ability of CUL7 to bind FBXW8 is necessary for its ability to inhibit E-cadherin expression and to cause EMT in JEG-3 cells.
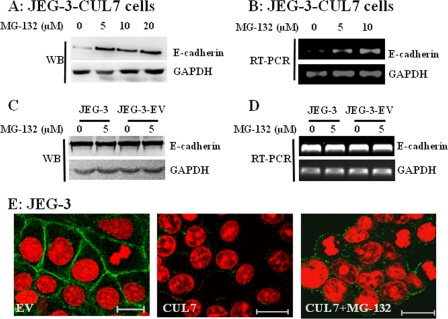
To further confirm the involvement of E3 ligase/proteasome in CUL7-mediated EMT in JEG-3 cells, we treated CUL7-expressing JEG-3 cells with the proteasome inhibitor MG-132. MG-132 significantly restored E-cadherin expression, as indicated by both immunoblotting and RT-PCR (Fig. 3, A and B). As a control, treating JEG-3 cells or JEG-3 cells transfected with EV with the same concentration of MG-132 did not alter E-cadherin expression (Fig. 3, C and D). Furthermore, the proteasome inhibitor partially restored E-cadherin at the cell junctions (Fig. 3E). These results clearly suggested that CUL7-mediated EMT in JEG-3 cells was dependent on its function to activate the E3/proteasome pathway.
FIGURE 3.
MG-132 partially restored E-cadherin expression in JEG-3 cells stably transfected with CUL7. JEG-3 cells stably expressing CUL7 (A and B), parental JEG-3 cells, and JEG-3 cells stably transfected with empty vector (C and D) were treated with the indicated concentrations of MG-132 for 6 h (RT-PCR) and 24 h (WB) and subjected to RT-PCR (B and D) and Western blotting (A and C) analysis of E-cadherin expression, respectively. E is an immunofluorescence micrograph showing the expression of E-cadherin (green) in JEG-3 cells transfected with EV (left panel), JEG-3 cells transfected with CUL7 (middle panel), or JEG-3-CUL7 cells treated with 10 μm MG-132 for 24 h. Nuclei were shown by PI staining. Bar = 20 μm.
Forced CUL7 Expression in JEG-3 Cells Enhanced Migration and Invasion
Epithelial cells undergoing EMT are characterized by increased motility. We, thus, attempted to determine whether overexpression of CUL7 increased cell motility by using the Matrigel invasion assay and Transwell migration assay. Cell motility was markedly increased in JEG-3 cells transfected with CUL7, as compared with control cells, which were transfected with the EV (Fig. 4, A and B). To test the specificity of the effect of CUL7 in this overexpression system, CUL7 siRNA was introduced into JEG-3 cells stably transfected with CUL7. Migration of CUL7-expressing JEG-3 cells was significantly decreased by CUL7 siRNA (Fig. 4C). As expected, transfection of JEG-3 cells with E-cadherin siRNA, but not with a scrambled siRNA (CTL), inhibited E-cadherin expression and increased cell motility (Fig. 4D).
CUL7-specific RNA Interference Increased E-cadherin Expression and Reduced Cell Motility
In contrast to JEG-3 cells, which lacked CUL7 and exhibited an epithelial cell phenotype, HTR8/SVneo and B6Tert cells expressed CUL7 and exhibited a fibroblast-like phenotype. To determine whether CUL7 expression is necessary for the fibroblast-like phenotype, we transfected HTR8/SVneo cells with siRNA targeted to CUL7. CUL7 siRNA significantly reduced CUL7 expression but did not affect the expression of other cullins tested (Fig. 5D). HTR8/SVneo cells transfected with CUL7 siRNA, but not control siRNA, exhibited a partial phenotypic change such that in some area the cells were clustered and appeared cobblestone-like morphology (Fig. 5A, upper right panel). Furthermore, CUL7 siRNA caused a significant increase in E-cadherin expression and a corresponding decrease in Vimentin expression (Fig. 5, B and C). Furthermore, CUL7 siRNA expression diminished the levels of stress fibers in these cells (Fig. 5C, panels a and b). Consistent with the notion that inhibition of CUL7 expression reduced the fibroblastic nature of HTR8/SVneo cells, the CUL7 siRNA-transfected cells also exhibited significantly decreased cell invasion (Fig. 5E) and migration (Fig. 5F). Similarly, interference with CUL7 expression in B6Tert cells also significantly reduced cell mobility (Fig. 5, G and H), although no significant impact was observed on cell morphology in this case.
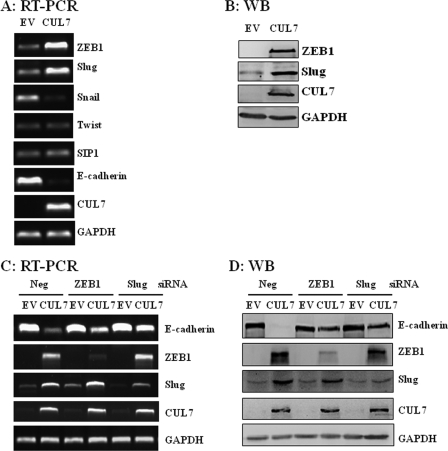
CUL7 Inhibition of E-cadherin Expression Required Two Known E-cadherin Transcription Repressors
Our results indicated that CUL7 overexpression in JEG-3 cells eliminated E-cadherin mRNA (Fig. 2B), suggesting that CUL7 E3 ligase might somehow inhibit E-cadherin transcription. To test this notion, we first determined whether CUL7 had any effect on the expression of various transcription factors that are known to repress E-cadherin transcription (17, 19). We showed that ZEB1 and Slug, but not Snail, Twist, or SIP1, were significantly up-regulated in JEG-3 cells expressing CUL7 (Fig. 6, A and B). To show whether ZEB1 and Slug were necessary for CUL7-mediated E-cadherin down-regulation, ZEB1 or Slug siRNA was introduced into JEG-3 cells overexpressing CUL7. We found that siRNA-mediated knockdown of ZEB1 or Slug significantly attenuated down-regulation of E-cadherin in these cells (Fig. 6, C and D), suggesting that down-regulation of E-cadherin by CUL7 in JEG-3 cells was mediated at least in part by increased expression of ZEB1 and Slug. In line with this observation, we found that the increased expression of E-cadherin in CUL7 siRNA-transfected HTR8/SVneo cells was accompanied by down-regulation of Slug (data not shown).
FIGURE 6.
CUL7 overexpression in JEG-3 cells increased expression of ZEB1 and Slug, two transcriptional repressors of E-cadherin. A, the mRNA levels of ZEB1, Slug, Snail, Twist, SIP1, E-cadherin, CUL7, and GAPDH in stable CUL7-expressing JEG-3 cells and the mock-transfected cells (EV) were determined by RT-PCR. B, immunoblots show protein levels of ZEB1 and Slug were significantly higher in CUL7-expressing JEG-3 cells. WB, Western blot. C and D, siRNA interference of ZEB1 or Slug partially restored E-cadherin expression in JEG-3/CUL7 cells. The RT-PCR (C) and immunoblot (D) results shown were representatives of three independent experiments. Neg, negative controls.
DISCUSSION
We have presented evidence here in support of a novel function of CUL7 in promoting EMT in trophoblast cell lineage. First, first-trimester trophoblast cell lines HTR8/SVneo and B6Tert expressed a high level of CUL7 and exhibited fibroblast-like morphology, whereas choriocarcinoma JEG-3 cells that lacked CUL7 expression exhibited epithelial cell phenotype. Second, forced expression of CUL7 in JEG-3 cells resulted in dramatic cell morphological changes characteristic of EMT, accompanied by a complete loss of three well established epithelial markers, E-cadherin, P-cadherin, and cytokeratin 8/18, and elevation of two mesenchymal markers, Vimentin and N-cadherin. Third, JEG-3 cells expressing CUL7 exhibited increased cell motility, as measured by two different assays (Matrigel cell invasion and Transwell cell migration assays). Fourth, CUL7-specific RNA interference in HTR8/SVneo cells restored E-cadherin expression, decreased Vimentin expression, and diminished stress fibers, which were accompanied by decreased cell motility.
EMT of trophoblast cells is critical for human placentation. In the course of human placental development, EMT takes place where a large subpopulation of trophoblast cells escapes from the anchoring columns, and single cells elongate and detach to infiltrate the maternal decidual interstitium and arteries (14). Aberrant trophoblast invasion leads to trophoblast-associated gestational diseases; aggressive and uncontrolled invasion leads to choriocarcinoma, a highly metastatic cancer (20), whereas insufficient trophoblast EMT and invasion lead to intrauterine growth restriction or pre-eclampsia (21–23). JEG-3 cells are derived from human choriocarcinoma and are of cytotrophoblast origin (24). JEG-3 cells exhibit epithelial cell morphology and are, therefore, considered trophoblast cells that have undergone an EMT during tumor metastasis and, later, a mesenchymal to epithelial transition after metastatic invasion of maternal tissues (25). In this sense, forced expression of CUL7 in JEG-3 cells may recapitulate cytotrophoblast cell EMT during embryo implantation and placental development. The fact that CUL7 expression alone elicited a complete EMT in JEG-3 cells suggested that CUL7 is a key regulator of EMT in the trophoblast cell lineage.
CUL7 is a scaffolding protein in a Skp1-CUL7-F-box-ROC1-like E3 ligase responsible for assembling an E3 complex consisting of Skp1, CUL7, FBXW8, and ROC1 (26). It has been established that the N-terminal region of CUL7 binds the E3 substrate recognition subunit, FBXW8 (26), and therefore, the N-terminal-truncated mutant, CUL7 (268–1698), should be unable to assemble a functional E3 ligase. The fact that this mutant, unlike full-length CUL7, failed to suppress E-cadherin expression (Fig. 2F) is supportive of the notion that suppression of E-cadherin expression and induction of EMT in JEG-3 cells are dependent on the ability of CUL7 to assemble a Skp1-CUL7-F-box RING E3 complex and the recruitment of the ubiquitin-proteasome pathway. This notion was further supported by the demonstration of a significant recovery of E-cadherin expression after MG-132 treatment of JEG-3-CUL7 cells (Fig. 3). In contrast, the same MG-132 treatment did not affect E-cadherin expression in either JEG-3 cells or JEG-3-EV cells, both of which lacked detectable CUL7 expression. Therefore, the effect of MG-132 described herein appeared to be specific for the CUL7-based E3 ligase/proteasome pathway in so far as E-cadherin expression was concerned. Interestingly, other E3 ligases have also been shown to be able to induce EMT, for example, the Smad ubiquitin regulatory factor 1 (Smurf1) E3 ligase, which has been implicated as a key regulator of mammary gland epithelial cell EMT (27). In that case, Smurf1 functions downstream of transforming growth factor β in promoting the loss of E-cadherin expression in mammary epithelial cells. It remains to be determined possible upstream regulators of CUL7 in promoting EMT of trophoblast cells, which undoubtedly will help to better understand the mechanism of trophoblast EMT under physiological or pathological conditions.
Our data indicated that two transcription repressors of E-cadherin, ZEB1 and Slug, were significantly up-regulated in JEG-3 cells overexpressing CUL7. Moreover, knocking-down ZEB1 or Slug attenuated CUL7-mediated E-cadherin repression. Taken together, these data reveal that CUL7 represses E-cadherin expression at least in part by activating E-cadherin repressors ZEB1 and Slug via a currently unknown mechanism. It remains to be determined how CUL7, an important component of RING E3 ligase, triggers the activation of the two transcriptional factors. One might speculate that it targets some negative regulators upstream of ZEB1 and Slug for degradation. Identification of the potential targets should be the focus of future studies.
CUL7 has been previously implicated in cellular transformation (28, 29). It has also been reported that CUL7, as an anti-apoptotic oncogene, cooperates with Myc to promote transformation (30). Because CUL7 is overexpressed in human lung cancers and is correlated with poor patient outcome, it is suggested that CUL7 may play an important role in transformation of tumor cells (30). Our results suggest another role for CUL7 in promoting EMT in the trophoblast cell lineage. Given the indispensable role of EMT in promoting progression of many carcinomas and the fact that trophoblast invasion shares many similarities with the invasion of tumor cells, our data raise the interesting question of whether CUL7 is also involved in EMT in tumor or other cell types.
In summary, our work has revealed a required role for CUL7 in EMT of trophoblast cell lineage and in promoting trophoblast migration and invasion. Our studies also suggest a possible pathological mechanism of CUL7-linked growth retardation syndromes.
Acknowledgments
We thank Drs. X. Johné Liu and Jay Baltz for helpful discussions and language assistance, Dr. Shengping Chen for clinical sample collection, and Dr. J. DeCaprio and Dr. Y. Xiong for kindly providing CUL7 plasmids.
This work was supported by 973 Program Grants 2006CB504006 and 2006CB944008 from the Ministry of Science and Technology of the People's Republic of China (to H. W.), Chinese Academy of Sciences Knowledge Innovation Program Grant KSCX2-YW-R-080 (to H. W.), and National Natural Science Foundation of China Grants 30971088 and 30770323 (to H. W. and H. Lin).
- EMT
- epithelial-mesenchymal transition
- PBS
- phosphate-buffered saline
- siRNA
- small interfering RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- RT
- reverse transcription
- EVT
- extravillous trophoblast cells
- EV
- empty vector
- PI
- propidium iodide.
REFERENCES
- 1.Nakayama K. I., Nakayama K. (2006) Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 2.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 3.Skaar J. R., Arai T., DeCaprio J. A. (2005) Mol. Cell. Biol. 25, 5579–5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardozo T., Pagano M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 5.Dealy M. J., Nguyen K. V., Lo J., Gstaiger M., Krek W., Elson D., Arbeit J., Kipreos E. T., Johnson R. S. (1999) Nat. Genet. 23, 245–248 [DOI] [PubMed] [Google Scholar]
- 6.Singer J. D., Gurian-West M., Clurman B., Roberts J. M. (1999) Genes Dev. 13, 2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li B., Ruiz J. C., Chun K. T. (2002) Mol. Cell. Biol. 22, 4997–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arai T., Kasper J. S., Skaar J. R., Ali S. H., Takahashi C., DeCaprio J. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9855–9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsutsumi T., Kuwabara H., Arai T., Xiao Y., Decaprio J. A. (2008) Mol. Cell. Biol. 28, 743–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarikas A., Xu X., Field L. J., Pan Z. Q. (2008) Cell Cycle 7, 3154–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsunematsu R., Nishiyama M., Kotoshiba S., Saiga T., Kamura T., Nakayama K. I. (2006) Mol. Cell. Biol. 26, 6157–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber C., Dias-Santagata D., Glaser A., O'Sullivan J., Brauner R., Wu K., Xu X., Pearce K., Wang R., Uzielli M. L., Dagoneau N., Chemaitilly W., Superti-Furga A., Dos Santos H., Mégarbané A., Morin G., Gillessen-Kaesbach G., Hennekam R., Van der Burgt I., Black G. C., Clayton P. E., Read A., Le Merrer M., Scambler P. J., Munnich A., Pan Z. Q., Winter R., Cormier-Daire V. (2005) Nat. Genet. 37, 1119–1124 [DOI] [PubMed] [Google Scholar]
- 13.Maksimova N., Hara K., Miyashia A., Nikolaeva I., Shiga A., Nogovicina A., Sukhomyasova A., Argunov V., Shvedova A., Ikeuchi T., Nishizawa M., Kuwano R., Onodera O. (2007) J. Med. Genet. 44, 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vićovac L., Aplin J. D. (1996) Acta. Anat. 156, 202–216 [DOI] [PubMed] [Google Scholar]
- 15.Blechschmidt K., Mylonas I., Mayr D., Schiessl B., Schulze S., Becker K. F., Jeschke U. (2007) Virchows Arch. 450, 195–202 [DOI] [PubMed] [Google Scholar]
- 16.Fu J. J., Lin P., Lv X. Y., Yan X. J., Wang H. X., Zhu C., Tsang B. K., Yu X. G., Wang H. (2009) Placenta. 30, 305–312 [DOI] [PubMed] [Google Scholar]
- 17.Thiery J. P. (2002) Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 18.Arias A. M. (2001) Cell 105, 425–431 [DOI] [PubMed] [Google Scholar]
- 19.Kang Y., Massagué J. (2004) Cell 118, 277–279 [DOI] [PubMed] [Google Scholar]
- 20.Knoeller S., Lim E., Aleta L., Hertwig K., Dudenhausen J. W., Arck P. C. (2003) Am. J. Reprod. Immunol. 50, 41–47 [DOI] [PubMed] [Google Scholar]
- 21.Kaufmann P., Black S., Huppertz B. (2003) Biol. Reprod. 69, 1–7 [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y., Damsky C. H., Chiu K., Roberts J. M., Fisher S. J. (1993) J. Clin. Invest. 91, 950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmieri C., Fisher R. A., Sebire N. J., Lindsay I., Smith J. R., McCluggage W. G., Savage P., Seckl M. J. (2005) Lancet 366, 688. [DOI] [PubMed] [Google Scholar]
- 24.Karmakar S., Dhar R., Das C. (2004) J. Biol. Chem. 279, 55297–55307 [DOI] [PubMed] [Google Scholar]
- 25.Yang J., Weinberg R. A. (2008) Dev. Cell 14, 818–829 [DOI] [PubMed] [Google Scholar]
- 26.Dias D. C., Dolios G., Wang R., Pan Z. Q. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16601–16606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozdamar B., Bose R., Barrios-Rodiles M., Wang H. R., Zhang Y., Wrana J. L. (2005) Science 307, 1603–1609 [DOI] [PubMed] [Google Scholar]
- 28.Ali S. H., Kasper J. S., Arai T., DeCaprio J. A. (2004) J. Virol. 78, 2749–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasper J. S., Kuwabara H., Arai T., Ali S. H., DeCaprio J. A. (2005) J. Virol. 79, 11685–11692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S. S., Shago M., Kaustov L., Boutros P. C., Clendening J. W., Sheng Y., Trentin G. A., Barsyte-Lovejoy D., Mao D. Y., Kay R., Jurisica I., Arrowsmith C. H., Penn L. Z. (2007) Cancer Res. 67, 9616–9622 [DOI] [PubMed] [Google Scholar]