Abstract
Angiogenin (ANG) is a secreted ribonuclease that cleaves tRNA to initiate a stress-response program in mammalian cells. Here we show that ANG inhibits protein synthesis and promotes arsenite- and pateamine A-induced assembly of stress granules (SGs). These effects are abrogated in cells transfected with the ANG inhibitor RNH1. Transfection of natural or synthetic 5′- but not 3′-tRNA fragments (tRNA-derived stress-induced RNAs; tiRNAs) induces the phospho-eukaryotic translation initiation factor 2α-independent assembly of SGs. Natural 5′-tiRNAs but not 3′-tiRNAs are capped with a 5′-monophosphate that is required for optimal SG assembly. These findings reveal that SG assembly is a component of the ANG- and tiRNA-induced stress response program.
Keywords: Diseases/Neurodegeneration, RNA/Metabolism, RNA/Modification, RNA/MicroRNA, RNA/Transfer RNA, RNA/Translation, Translation/Initiation Factors
Introduction
In response to environmental stress, eukaryotic cells activate stress response programs that down-regulate energy-expensive processes, such as transcription and translation. These regulatory programs reduce the expression of common housekeeping genes while increasing the expression of genes that repair stress-induced damage and promote cell survival. At the level of translation, this is achieved by exploiting the differential sensitivity of mRNAs to changes in the availability or activity of general initiation factors, such as eukaryotic translation initiation factor 2α (eIF2α)3 (1). Phosphorylation of eIF2α by one of several stress-activated kinases reduces the availability of the eIF2-GTP-tRNAiMet ternary complex to inhibit translation initiation. This reduces the translation of most transcripts but enhances the translation of transcripts possessing regulatory upstream open reading frames, such as transcription factor ATF4, a component of the integrated stress response program. Thus, phospho-eIF2α triggers a profound reprogramming of cellular protein synthesis that helps cells adapt to adverse environmental conditions.
Stress-induced cleavage of tRNA initiates a complementary stress response program found in both prokaryotes and eukaryotes (2). In mammals, stress-induced tRNA cleavage is mediated by angiogenin (ANG) (3, 4), a 14-kDa member of the pancreatic RNase superfamily. ANG is a secreted endoribonuclease that possesses both angiogenic (5) and cytoprotective activities (6, 7). Secreted ANG enters cells via receptor-mediated endocytosis (8, 9), translocates to the nucleus (10), and promotes ribosomal RNA transcription and cellular proliferation (11–13). ANG secretion is stimulated by hypoxia, suggesting that it may serve as a stress-induced paracrine factor that protects neighboring cells from deleterious effects of stress.
In a previous study, we showed that stress promotes ANG-mediated tRNA cleavage to produce tRNA-derived stress-induced RNAs (tiRNAs) (4). Cleavage occurs preferentially in the anticodon loop of mature tRNA to produce 5′- and 3′-fragments (5′- and 3′-tiRNAs, respectively). The addition of recombinant wild type but not RNase-inactive mutant ANG to cultured cells promotes tiRNA production and inhibition of protein synthesis. Thus, the ribonuclease activity of ANG is absolutely required for its angiogenic and cytoprotective properties as well as for tiRNA production and repression of translation. Moreover, transfection of 5′-tiRNAs but not 3′-tiRNAs inhibits translation in cultured cells. Importantly, tiRNAs inhibit protein translation in an eIF2α-independent manner (4), suggesting that ANG and tiRNAs may activate complementary components of an integrated stress response program.
Stress-induced phosphorylation of eIF2α inhibits translation and promotes the assembly of stress granules (SGs), cytoplasmic foci at which untranslated mRNPs are transiently concentrated (14). SGs contain stalled 48 S preinitiation complexes that remain after elongating ribosomes have run off their transcripts. SGs are assembled in a phospho-eIF2α-independent manner by the xenobiotic agents pateamine A and hippuristanol, compounds that inhibit the RNA helicase activity of initiation factor eIF4A to prevent 48 S scanning (15–19). The ability of tiRNAs to inhibit translation in a phospho-eIF2α independent manner (4) suggests that they might also inhibit 48 S scanning. If so, tiRNAs, like pateamine A and hippuristanol, should induce phospho-eIF2α independent assembly of SGs. Indeed, we find that transfection of natural 5′-tiRNAs but not 3′-tiRNAs induces phospho-eIF2α-independent SG assembly in cultured cell lines. Transfection of synthetic 5′-tiRNAAla also inhibits protein synthesis and induces SG assembly, processes that are enhanced by the presence of a 5′-monophosphate modification. Although recombinant ANG does not induce SG assembly on its own, it enhances the assembly of sodium arsenite and pateamine A-induced SGs. Taken together, our data implicate ANG, tiRNAs, and SGs in a phospho-eIF2α-independent stress response program.
EXPERIMENTAL PROCEDURES
Tissue Culture and Cell Treatments
Parental U2OS cells as well as stable transfectants expressing recombinant proteins were maintained at 37 °C in a CO2 incubator in minimal essential medium supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin/streptomycin (Sigma). Treatment of cells with the indicated doses of sodium arsenite (Sigma), pateamine A (desmethyldesamino-modified; a gift from Jun Liu (The Johns Hopkins University, Baltimore, MD)), or emetine (Sigma) was as described (20). U2OS cells were treated with recombinant wild type or mutant ANG (0.5 μg/ml) (21) for the indicated times.
Antibodies
Goat polyclonal antibody to the N terminus of eIF3b and to the C terminus of TIAR, rabbit polyclonal antibody to eIF4G or eIF4E, and mouse monoclonal antibodies to HuR or the C terminus of p70 S6 kinase (SK1-hedls) were purchased from Santa Cruz Biotechnology. Chicken polyclonal anti-G3BP was from Abcam, mouse monoclonal anti-HA was from Covance, mouse monoclonal antibodies to FLAG M2 and to actin were from Sigma, rabbit polyclonal anti-RCK was from Bethyl, and rabbit polyclonal phosphospecific anti-eIF2α (serine 52) was from Assay Designs. Anti-mouse and anti-rabbit secondary antibodies conjugated with horseradish peroxidase were from GE Healthcare. Cy2-, Cy3-, and Cy5-horseradish peroxidase or streptavidin-conjugated secondary antibodies were purchased from Jackson Immunoresearch Laboratories.
DNA Plasmids, siRNAs, and Synthetic tiRNAs
To construct FLAG-RNH1 and FLAG-LSM1 plasmids, open reading frames of RNH1 and LSM1 were amplified by PCR and subcloned into pCI-neo-FLAG vector using XhoI and NotI sites. Plasmid pET-HA, which expresses either wild type eIF2α or non-phosphorylatable eIF2α-S51A mutant construct, was described previously (22). siGENOME SMART pools for heme-regulated inhibitor kinase (HRI) and nonspecific control sequences were purchased from Fisher. siGENOME SMART pool for HRI (4) was purchased from Fisher and control siRNA (D0): 5′-GCATTCACTTGGATAGTAA-3′ was purchased from Applied Biosystems. All synthetic RNA oligonucleotides (synthetic tiRNAs) used in this study were purchased from Integrated DNA Technologies: P-ctiRNA, 5′-UGUGAGUCACGUGAGGGCAGAAUCUGCUC-3′; P-5′-tiRNAAla, 5′-GGGGGUGUAGCUCAGUGGUAGAGCGCGUG-3′; P-5′-tiRNAVal, 5′-GTTTCCGTAGTGTAGTGGTTATCACGTTCGCCT-3′; P-5′-tiRNAGly, 5′-GCGCCGCTGGTGTAGTGGTATCATGCAAGAT-3′; P-3′-tiRNAAla, 5′-CUUAGCAUGCACGAGGCCCCGGGUUCAAUCCCCGGCACCUCCA-3′. The 3′-end-biotinylated counterparts of these RNAs were synthesized by Integrated DNA Technologies.
Cell Transfection
Cells were transfected with DNA plasmids, siRNAs, or tiRNAs using Lipofectamine 2000 (Invitrogen). Before transfection, DNA or RNA complexes were preincubated in serum-free medium (Opti-MEM medium; Invitrogen) for 20 min at room temperature. For DNA plasmid transfections, U2OS cells were grown to ∼50% confluence in a 10-cm dish and then transfected with a total of 3 μg of each DNA plasmid complexed with 3 μl of Lipofectamine; 36 h post transfection, cells were trypsinized and replated 24 h prior to immunofluorescence experiments (see “Immunofluorescence Microscopy”). For siRNA transfections, U2OS cells (1.5 × 105/well) were plated in 6-well plates and grown to about 30% confluence, and 6 h later, they were transfected with 40 nm HRI siRNA or control siRNA using 2 μl of Lipofectamine. Two days after siRNA transfection, cells were trypsinized and replated for immunofluorescence experiments (see “Immunofluorescence Microscopy”). For tiRNA transfection, U2OS cells (0.9 × 105/well) were plated in 24-well plates for 24 h and then transfected with 750 nm synthetic or natural tiRNAs using 2.5 μl of Lipofectamine.
tiRNA Isolation
tiRNA extraction from ANG-treated cells was done as previously described (4).
Metabolic Labeling
U2OS cells (1 × 105/well) were plated in 24-well plates and 24 h later were incubated with metabolic labeling medium (DME without l-glutamine, sodium pyruvate, l-methionine, or l-cystine (Invitrogen), supplemented with 5% dialyzed fetal bovine serum (Fisher). After 30 min, fresh metabolic labeling medium containing ∼200 μCi of l-[35S]methionine (EasyTag Express 35S Protein Labeling Mix; PerkinElmer Life Sciences) and wild type or P112L mutant ANG (0.5 μg/ml) in the presence or absence of 70 μm sodium arsenite or 15 nm pateamine A was added to each well, and cells were then incubated at 37 °C for 1 h. To extract proteins, cells were washed three times with PBS, harvested in 200 μl of lysis buffer (2% SDS, 20 mm Hepes, pH 7.4), and then lysed with an ultrasonic sonicator (Branson). Proteins from clarified supernatants were precipitated by 60% acetone and resuspended in 70 μl of lysis buffer, and 10 μl of each sample was used to measure protein concentration or to count 35S incorporation. The amount of 35S incorporated into the whole cell protein was quantified with a scintillation counter (Beckman Coulter), and the percentage of incorporation was calculated for each sample by dividing the counts of 35S incorporation by the concentration of total protein and then multiplying this value by 100. Protein concentration was measured using the BCA protein assay reagent kit (Pierce) with a bovine serum albumin protein standard.
Testing of 5′-Monophosphate Modification of tiRNAs
Natural tiRNAs were purified as described (4). 500 pmol of native tiRNAs or synthetic control RNAs were treated with Terminator 5′-phosphate-dependent exonuclease (Epicenter Biotechnologies) for 30 min at 30 °C. The reaction was terminated by phenol extraction followed by ethanol precipitation of RNA. The resulting RNA was separated by 15% urea-PAGE and stained with SYBR Gold.
Western Blotting
Cells were solubilized in lysis buffer containing 5 mm MES (Sigma), pH 6.2, and 2% SDS. Proteins were separated on a 4–20% gradient SDS-polyacrylamide gel and transferred to nitrocellulose filter membranes (0.45-μm pore size; Invitrogen). The membranes were blocked with 5% normal horse serum in 1× TBS at room temperature for 1 h and incubated with rabbit polyclonal phospho-eIF2α (serine 52) diluted 1:1000 in TBS containing 5% normal horse serum overnight at 4 °C. The membranes were washed three times with 1× TBS containing 0.1% Tween 20 and incubated with anti-rabbit immunoglobulin G antibody conjugated with horseradish peroxidase (1:5000) (GE Healthcare). For the detection of actin, the membranes were incubated with the primary antibodies diluted 1:10,000 in TBS containing 5% normal horse serum, followed by incubation with anti-mouse conjugated with horseradish peroxidase (1:5000) (GE Healthcare). After washing, the specific proteins were detected using the Super Signal chemiluminescent detection system (Pierce) and autoradiography on X-Omat film (Eastman Kodak Co.).
Immunofluorescence Microscopy
Cells (9 × 104) were seeded onto coverslips (Fisher) and 24 h later were transfected with the indicated concentrations of natural or synthetic tiRNAs using Lipofectamine 2000 (Invitrogen) and/or treated with the indicated drugs. At various times, cells were fixed in 4% para-formaldehyde for 15 min and permeabilized using 100% chilled methanol for 10 min. The cells were rinsed several times with PBS and incubated overnight with blocking buffer (5% normal horse serum in PBS containing 0.02% sodium azide) at 4 °C. An appropriate primary antibody diluted in blocking buffer (1:200 for anti-eIF3b, anti-eIF4G, anti-eIF4E, anti-TIAR, and anti-HuR; 1:1000 for anti-SK1-hedls, anti-RCK, anti-HA, and anti-FLAG M2; and 1:2000 for anti-G3BP antibody) was then added to the cells and incubated overnight at 4 °C. The cells were washed three times with PBS and incubated with the appropriate secondary antibodies (Jackson Immunoresearch, ML grade) diluted 1:200 in blocking buffer containing 0.5 μg/ml Hoechst 33258 dye (Molecular Probes) for 1 h at room temperature. After washing with PBS, coverslips were mounted in polyvinyl mounting medium, and the cells were viewed and photographed with an Eclipse E800 fluorescence microscope (Nikon) equipped with a digital camera (CCD-SPOT RT, Diagnostic Instruments) using a ×60 oil immersion objective. The images were merged and analyzed using Adobe Photoshop (version 10).
RESULTS
ANG Promotes Arsenite-induced SG Assembly
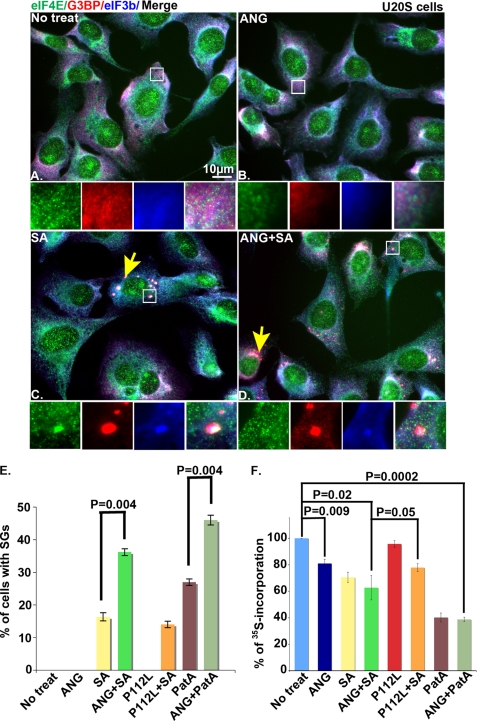
To determine whether ANG promotes SG assembly, U2OS cells were treated with recombinant ANG in the absence or presence of mild oxidative stress (70 μm sodium arsenite (SA), a dose that induces SGs in less than 20% of cells) for 45 min. The cells were fixed, stained for SG markers (eIF4E, G3BP, and eIF3b), and visualized using fluorescence microscopy. As shown in Fig. 1, neither wild type ANG nor the inactive P112L mutant (21) induced SG assembly in the absence of stress (Fig. 1, B and E). In cells treated with low dose arsenite, wild type (but not mutant) ANG significantly increased the number of cells with SGs (Fig. 1, D and E). Wild type but not mutant ANG similarly enhanced pateamine A-induced SG assembly (Fig. 1E), indicating that it potentiates phospho-eIF2α-dependent and independent inducers of SG assembly.
FIGURE 1.
Effect of ANG on SG assembly. U2OS cells were cultured in media alone (No treat) (A) or media containing ANG (0.5 μg/ml) (B), SA (70 μm) (C), or ANG plus sodium arsenite (ANG + SA) (D) for 45 min before processing for immunofluorescence microscopy using anti-eIF4E (green), anti-G3BP (red), and eIF3b (blue). Insets, enlarged views of the boxed region stained for eIF4E (green), G3BP (red), and eIF3b (blue) and merged channels (from left to right). The yellow arrows point out cells with SGs. E, quantification of the percentage of cells with SGs. Data report the average percentage of SGs from three independent experiments in which 200 cells were counted in each experiment. Error bars, S.D. (n = 3). F, quantification of [35S]methionine incorporation. Cells were incubated with metabolic labeling medium for 30 min, and then a fresh metabolic medium containing [35S]methionine and wild type ANG or mutant P112L (0.5 μg/ml) in the presence or absence of 70 μm sodium arsenite or 15 nm of pateamine A was added to the cells for 1 h. [35S]Methionine incorporation is reported as a percentage of that observed in untreated cells. The average percentage of [35S]methionine incorporation was calculated from three independent experiments and plotted for each experimental condition. The error bars indicate the S.D. (n = 3).
We confirmed that wild type but not mutant ANG significantly inhibited protein synthesis (Fig. 1F). The combination of ANG and sodium arsenite or pateamine A did not significantly inhibit protein synthesis more than either treatment alone (Fig. 1F). The finding that ANG inhibits protein synthesis but does not induce SG assembly suggests that a second signal is required for untranslated mRNPs to aggregate at SGs. We have found that stress-induced glycosylation of ribosomal proteins is required for the aggregation of untranslated mRNPs at SGs but not for stress-induced translational repression per se (23). This may be one of the stress-induced second signals playing a role in ANG-induced SG assembly.
Natural 5′-tiRNAs Induce SG Assembly
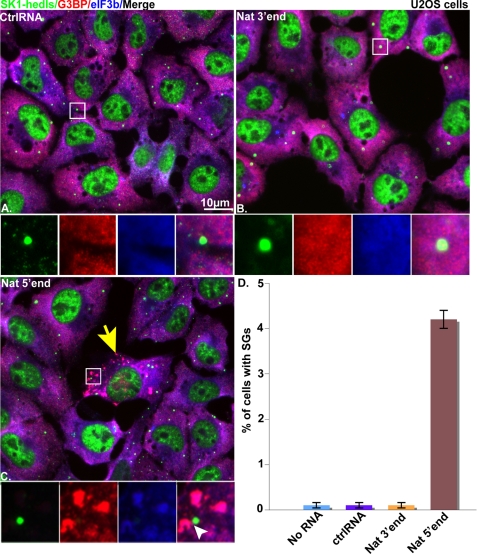
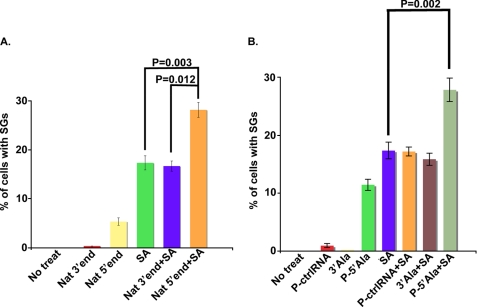
We previously reported that transfection of 5′- but not 3′-ANG-induced natural tiRNAs inhibits protein synthesis in U2OS cells (4). To determine whether this result correlates with the assembly of SGs, we transfected U2OS cells with natural 5′-tiRNAs (Fig. 2C) or 3′-tiRNAs (Fig. 2B) or control RNAs (Fig. 2A; 750 nm final concentrations) using Lipofectamine 2000. Time course analysis revealed that optimal induction of SGs occurred 7 h after transfection (data not shown). At this time point, immunofluorescence microscopy using antibodies reactive with SG markers (G3BP and eIF3b) identified very few non-transfected (Fig. 2D), control RNA-transfected (Fig. 2, A and D), or natural 3′-tiRNA-transfected (Fig. 2, B and D) cells with SGs. In contrast, SGs were observed in ∼5% of cells transfected with natural 5′-tiRNAs (Fig. 2C, yellow arrows and Fig. 2D). These data reveal that tiRNA-induced translational repression correlates with SG assembly in a small percentage of cells.
FIGURE 2.
Effect of natural tiRNAs on SG assembly in U2OS cells. Shown is immunofluorescence microscopy of U2OS cells transfected with 750 nm control RNA (ctrlRNA) (A), natural 3′-tRNA (Nat 3′end) (B), or natural 5′-tRNA (Nat 5′end) (C) and stained with SG markers (anti-G3BP (red) and eIF3b (blue)) and a PB marker (anti-SK1-hedls (green)) at 7 h post-transfection. Insets, enlarged views showing individual and merged channels. The yellow arrows indicate cells with SGs. The white arrowheads indicate PBs detected in juxtaposition interaction with SGs. D, quantification of the percentage of cells with SGs. Cells that showed SGs were counted in a total number of 600 cells/experiment, and the average percentage of cells that showed SGs was calculated from three independent experiments and plotted for each experimental condition. Error bars, S.D. (n = 3).
Natural 5′-tiRNAs Have 5′-Terminal Monophosphate Modifications
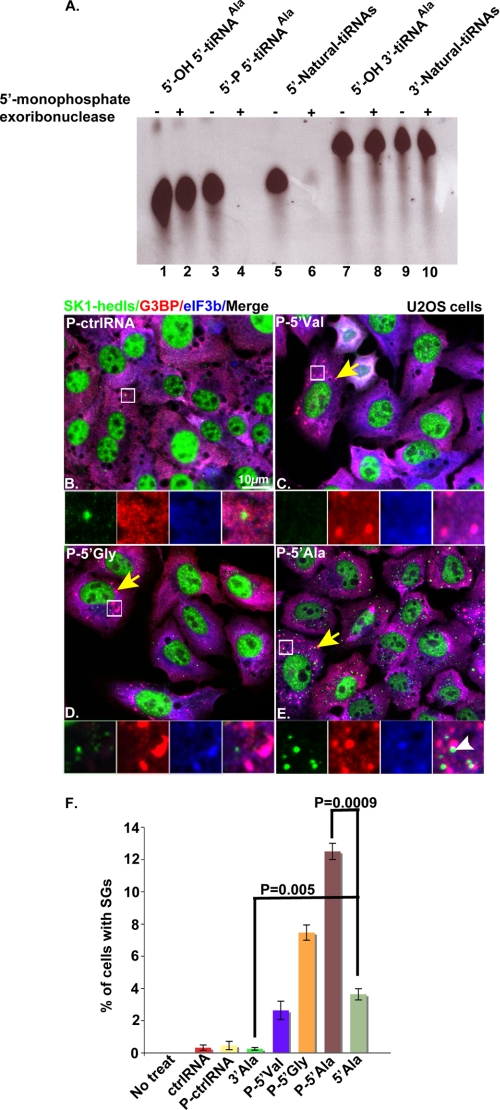
Because mature tRNAs are the precursor of tiRNAs, it is likely that 5′- but not 3′-tiRNAs possess a monophosphate modification at their 5′-ends. To test this prediction, we treated natural 5′- and 3′- tiRNAs or control RNAs with 5′-terminal monophosphate exoribonuclease. Although 5′-hydroxyl-containing synthetic RNAs corresponding to 5′- or 3′-halves of tRNAAla (5-OH 5′-tiRNAAla and 5′-OH 3′-tiRNAAla; Fig. 3A, lanes 1 and 2 and lanes 7 and 8, respectively) were not degraded, the 5′-monophosphate-modified 5′-tiRNAAla was degraded by this enzyme (Fig. 3A, compare lanes 3 and 4). Similarly, natural 5′-tiRNAs (Fig. 3A, lanes 5 and 6) but not 3′-tiRNAs (Fig. 3A, lanes 9 and 10) were degraded by this enzyme. Thus, 5′- but not 3′-tiRNAs possess 5′-terminal monophosphate modifications.
FIGURE 3.
Natural 5′-tiRNAs possess 5′-monophosphates that enhance SG assembly. A, synthetic 5′-OH-5′-tiRNAAla (lanes 1 and 2), 5′-P-5′-tiRNAAla (lanes 3 and 4), or 5′-OH-3′-tiRNAAla (lanes 7 and 8) were used as controls for the specificity of 5′-monophosphate exoribonuclease treatments. The control synthetic RNAs, natural 5′-tiRNAs (lanes 5 and 6) and natural 3′-tiRNAs (lanes 9 and 10) were treated with 5′-monophosphate exoribonuclease (lanes 2, 4, 6, 8, and 10) or a buffer control (lanes 1, 3, 5, 7, and 9) for 30 min prior to separation on a TBE-urea gel and visualization using CYBR Gold. U2OS cells were transfected with 750 nm synthetic phosphorylated control RNA (P-ctrlRNA) (B) or phosphorylated 5′-tiRNAs (P-5′Val, P-5′Gly, or P-5′Ala) (C–E) and then stained with anti-Sk1-hedls (green), anti-G3BP (red), and eIF3b (blue) antibodies at 7 h after transfection. Insets, enlarged views showing individual and merged channels. The yellow arrows point out representative cells with SGs. The white arrowheads indicate PBs in juxtaposition interaction with SGs. F, quantification of the percentage of cells with SGs. Cells that showed SGs were counted in a total number of 600 cells/experiment, and the average percentage of cells that showed SGs was calculated from three independent experiments and plotted for each experimental condition. The error bars indicate the S.D. (n = 3).
Synthetic tiRNAs Promote Stress Granule Assembly
Transfection of natural 5′-tiRNAs into U2OS cells represses translation and promotes the assembly of SGs. Although the majority of these natural RNAs correspond to tRNA halves (3, 4), minor non-tRNA-derived RNAs might contribute to the observed effects on translation and SG assembly. To determine whether 5′-tRNA halves are responsible for these effects, we tested the biological activities of synthetic 5′-tiRNAs. U2OS cells were transfected with synthetic 5′-monophosphate-modified RNA fragments corresponding to 5′-tiRNAVal (Fig. 3C), 5′-tiRNAGly (Fig. 3D), or 5′-tiRNAAla (Fig. 3E) (3, 4) before processing for immunofluorescence microscopy to quantify SGs. SGs were observed in ∼3, ∼7, and ∼12% of cells transfected with P-5′-tiRNAVal, P-5′-tiRNAGly, and P-5′-tiRNAAla, respectively (Fig. 3, C–E (yellow arrows) and F). In contrast, few if any cells displaying SGs were observed in untreated, unphosphorylated control (ctrlRNA), 5′-phosphorylated control (P-ctrlRNA), or 3′-tiRNAAla-transfected cells (Fig. 3F).
To determine whether the small percentage of cells with SGs was a consequence of inefficient transfection efficiency, U2OS cells were transfected with biotin-tagged control or tiRNAs before processing for immunofluorescence microscopy to visualize intracellular biotin (supplemental Fig. 1). Biotin RNA was visualized within endosomes in all cells, indicating that Lipofectamine-mediated RNA uptake is highly efficient. It is possible that the low percentage of tiRNA-induced SGs is a consequence of inefficient egress of tiRNAs from endosomes. A dose-response analysis showed that 75 nm is the threshold for tiRNAAla-induced SG assembly (supplemental Fig. 2). Although the percentage of SGs increased with increasing concentrations of tiRNAAla, at most, 12% of transfected cells assembled SGs. This result suggests that egress from endosomes may well contribute to the low percentage of SGs in tiRNAAla transfected cells. It is also possible the rare cells that assemble SGs are somehow “primed” for SG assembly. This might involve heterogeneous expression of proteins that are required for SG assembly. We tested this possibility by surveying tiRNA-induced SG assembly in U2OS cells expressing recombinant proteins that are known to promote SG assembly (e.g. G3BP (24), Tudor-SN (25), and ADAR (25); supplemental Fig. 3). The percentages of 5′-tiRNAAla-induced SGs in these cells are greater than that observed in the parental U2OS cells. Thus, the poor efficiency of tiRNA-induced SG assembly may reflect heterogeneity in the expression of key SG proteins. The ability of synthetic 5′-tiRNAs to induce the assembly of SGs confirms that the effects of natural 5′-tiRNAs are not due to non-tRNA contaminants.
To determine whether monophosphate modifications on 5′- but not 3′-tiRNAs contribute to bioactivity, U2OS cells were transfected with synthetic 5′-tiRNAAla with or without the 5′-phosphate modification. The 5′-monophosphate-modified version induced 4-fold more SG-positive cells than the unmodified version (data not shown) (Fig. 3F). Thus, 5′-end phosphorylation of 5′-tiRNAAla enhances its ability to induce SG assembly.
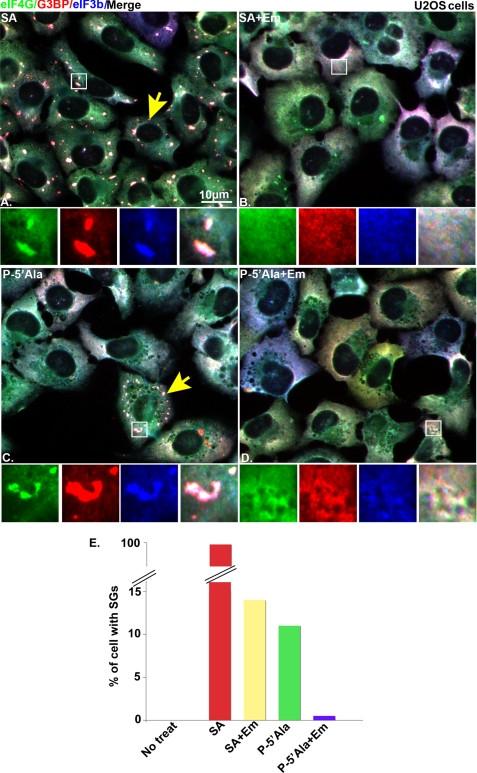
To confirm that the granules induced by 5′-tiRNAs are bona fide SGs, we determined the effect of emetine, a drug that inhibits SG assembly by preventing polysome disassembly (26), on 5′-tiRNAAla-induced SG assembly. U2OS cells were mock-transfected or transfected with synthetic P-5′-tiRNAAla (Fig. 4, C and D) and then treated (Fig. 4, B and D) or not treated (Fig. 4, A and C) with 50 μg/ml emetine for 60 min. SGs (yellow arrows) were detected using eIF4G (green), G3BP (red), and eIF3b (blue). Mock-transfected cells treated with SA served as a positive control (Fig. 4A). Emetine inhibits the formation of SGs in cells transfected with P-5′-tiRNAAla (Fig. 4, D and E) to a similar extent as cells treated with SA (Fig. 4, B and E). The assembly of SGs induced by P-5′-tiRNAAla was confirmed using three additional SG markers, HuR, RCK, and TIAR (data not shown). No SGs were detected in cells transfected with either control RNA (P-ctrlRNA) or 3′-tiRNAAla (data not shown). Because SGs are found in physical association with processing bodies (PBs), an RNA granule linked to mRNA decay (27), we used PBs as another marker to confirm that tiRNAs induce the assembly of bona fide SGs. SGs induced by natural 5′-tiRNA or synthetic 5′-tiRNAAla interact with PBs (Figs. 2C and 3E, insets, white arrowheads, respectively). These results indicate that 5′-tiRNAAla promotes canonical SG assembly.
FIGURE 4.
Emetine inhibits P-5′-tiRNAAla-induced SGs. A–D, immunofluorescence microscopy of non-transfected cells treated with 200 μm SA for 60 min (A), 200 μm SA for 60 min followed by 50 μg/ml emetine (SA + Em) treatment for another 60 min (B), or cells transfected with 750 nm P-5′-tiRNAAla (C) and then treated with 50 μg/ml emetine (P-5′Ala + Em) (D) for 60 min. Insets, enlarged views showing individual and merged channels. The yellow arrows point out representative cells with SGs. E, quantification of the percentage of cells with SGs. The percentage of cells that showed SGs were quantified by counting 200 cells.
We next determined whether transfection of tiRNAs promotes SG assembly in cells treated with low doses of sodium arsenite. U2OS cells were transfected with natural 5′- or 3′-tiRNAs, synthetic tiRNAs (P-5′-tiRNAAla, or 3′-tiRNAAla), or control RNA (P-ctrlRNA) and then treated with low dose arsenite (70 μm, 60 min). Natural 5′-tiRNAs (Fig. 5A) and synthetic P-5′-tiRNAAla (Fig. 5B) significantly increased the arsenite-induced assembly of SGs. In contrast, natural 3′-tiRNAs (Fig. 5A), synthetic control RNA (Fig. 5B), and synthetic 3′-tiRNAAla (Fig. 5B) failed to promote arsenite-induced SG assembly. The finding that ANG treatment alone induces tiRNA production but not SG assembly suggests that Lipofectamine-mediated transfection imparts a mild stress that is required for tiRNA-induced SG assembly. Thus, ANG appears to lower the threshold for SG assembly.
FIGURE 5.
5′-tiRNAs enhance arsenite-induced SG assembly. A and B, quantification of the percentage of cells with SGs in control U2OS cells cultured in the absence (No treat) or presence (SA) of 70 μm SA or cells transfected with 750 nm natural 5′-tiRNAs (Nat 5′end) or 3′-tiRNAs (Nat 3′end) or synthetic tiRNAs (P-ctrlRNA, P-3′tiRNAAla, P-5′tiRNAAla) for 7 h and then treated with 70 μm SA (3′end + SA, 5′end + SA, P-ctrlRNA (P-ctrlRNA + SA), P-3′tiRNAAla (P-3′Ala + SA), or P-5′-tiRNAAla (P-5′-Ala + SA)) for 60 min. Cells were stained with SG markers (anti-eIF3b, anti-G3BP, and anti-eIF4G). The percentage of cells with SGs was quantified by counting 200 cells/experiment. Data show the average percentage of SGs from three independent experiments. Error bars, S.D. (n = 3).
5′-tiRNAAla Induces SG Formation Independently of eIF2α Phosphorylation
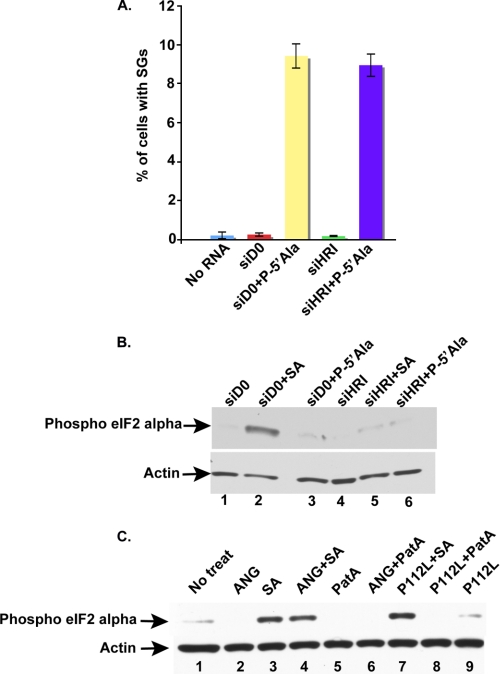
HRI phosphorylates eIF2α in response to SA treatment (28). We previously showed that phosphorylation of eIF2α is not required for the production of tiRNAs or for tiRNA-induced translational repression (4). Because phosphorylation of eIF2α inhibits translation initiation and promotes SG assembly (29), we asked whether tiRNAs require phospho-eIF2α to promote SG assembly. U2OS cells transfected with control (D0) or HRI-directed siRNAs were subsequently retransfected with P-5′-tiRNAAla. Cells treated with SA (200 μm) were used as a positive control for phospho-eIF2α-dependent assembly of SGs, whereas cells treated with pateamine A (50 nm) were used as a control for phospho-eIF2α-independent assembly of SGs. As expected, HRI knockdown effectively prevented arsenite-induced SG assembly (supplemental Fig. 4). In contrast, SGs were assembled in both control and HRI knockdown cultures treated with pateamine A (supplemental Fig. 4) or transfected with P-5′-tiRNAAla (Fig. 6A and supplemental Fig. 4). P-5′-tiRNAAla-induced SG assembly was similar in control and HRI knockdown cells (Fig. 6A and supplemental Fig. 4). Western blot analysis using a polyclonal antibody specific for the phosphorylated form of eIF2α detected high levels of eIF2α phosphorylation only in the control siRNA knockdown cells treated with SA (Fig. 6B, lane 2). In agreement with the indispensable role for HRI in eIF2α phosphorylation, HRI knockdown cells treated with SA showed background levels of eIF2α phosphorylation (Fig. 6B, lane 5). No increase in eIF2α phosphorylation was observed in cells transfected with P-5′-tiRNAAla (Fig. 6B, lane 3; compare with lanes 1, 4, and 6). Similar results were obtained using U2OS cells transfected with plasmids encoding HA-tagged wild-type or non-phosphorylatable eIF2α (S51A) prior to transfection with P-5′-tiRNAAla (supplemental Fig. 5).
FIGURE 6.
Effect of phospho-eIF2α on ANG- and/or P-5′-tiRNAAla. A, quantification of the percentage of cells with SGs in untransfected U2OS cells (No RNA) or cells transfected with control siRNA (siD0) or with HRI siRNA (siHRI) and then mock-transfected or transfected with P-5′-tiRNAAla (P-5′Ala; 750 nm) for 7 h. The percentage of cells with SGs was quantified by counting 500 cells/experiment. Data show the average percentage of SGs from three independent experiments. Error bars, S.D. (n = 3). B, Western blot analysis of phospho-eIF2α in U2OS cells transfected with either D0 or HRI siRNAs and then treated or not treated with 200 μm SA for 60 min. Lane 1, control knockdown cells (siD0); lane 2, control knockdown cells treated with 200 μm SA (siD0 + SA); lane 3, control knockdown cells transfected with P-5′-tiRNAAla (siD0 + P-5′Ala); lane 4, HRI knockdown cells (siHRI); lane 5, HRI knockdown cells treated with 200 μm SA (siHRI + SA); lane 6, HRI knockdown cells transfected with P-5′-tiRNAAla (siHRI + P-5′Ala). C, Western blot analysis of phospho-eIF2α in untreated cells (No treat; lane 1) or in U2OS cells treated with either ANG (0.5 μg/ml; lane 2), SA (70 μm; lane 3), ANG plus sodium arsenite (ANG + SA; lane 4), pateamine A (PatA; 15 nm; lane 5), ANG plus pateamine A (ANG + PatA; lane 6), ANG mutant + SA (P112L + SA; lane 7), P112L + PatA (lane 8), or P112L (lane 9). Phospho-eIF2α was detected with polyclonal antibodies specific for eIF2α phosphorylated on serine 52 (upper panels). Actin (lower panels) was used as a loading control.
We also quantified eIF2α phosphorylation in cells treated with ANG alone or in combination with either sodium arsenite or pateamine A. No change in eIF2α phosphorylation was observed upon treatment with ANG alone (Fig. 6C, lane 2) or in combination with pateamine A (Fig. 6C, lane 6). When ANG or its mutant P112L was combined with SA, eIF2α phosphorylation levels were similar to SA alone (Fig. 6C, compare lane 3 with lanes 4 and 7). These results indicate that neither ANG nor P-5′-tiRNAAla require eIF2α phosphorylation to induce SG assembly in U2OS cells.
RNH1 Interferes with ANG-induced SG Assembly in U2OS Cells
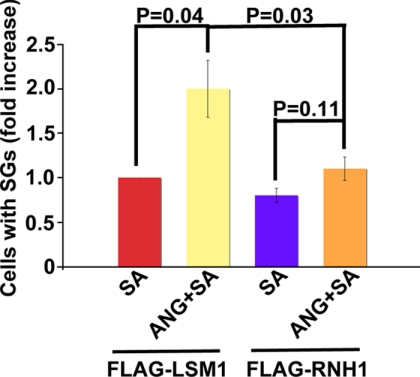
RNH1 (ribonuclease/ANG inhibitor 1) inhibits production of tiRNAs in U2OS cells (4). To determine whether RNH1 antagonizes the ANG-mediated assembly of SGs in cells treated with low doses of sodium arsenite, cells were transfected with either control FLAG-LSM1 (a protein whose overexpression does not induce SG assembly) or FLAG-RNH1 constructs prior to exposure to SA or SA plus ANG. RNH1 or LSM1 overexpression was detected using anti-FLAG antibody, and SGs were detected using anti-G3BP and anti-eIF3b antibodies. ANG significantly enhanced SG assembly in LSM1-overexpressing cells but not in RNH1-overexpressing cells (Fig. 7). These results indicate that RNH1 abrogates the effects of ANG on arsenite-induced SG assembly.
FIGURE 7.
Effect of RNH1 on ANG-mediated promotion of SG assembly. Quantification of the percentage of cells with SGs in U2OS cells transfected with either FLAG-LSM1 or FLAG-RNH1 and then treated with 60 μm SA in the presence or absence of ANG for 45 min. Cells were stained with SG markers (anti-FLAG, anti-G3BP, and anti-eIF3b). SGs were counted only in 40–50 cells overexpressing either protein. The average percentage of cells with SG was calculated from three independent experiments, expressed as -fold increase over ANG alone, and plotted for each experimental condition. The error bars indicate the S.D. (n = 3).
DISCUSSION
In response to environmental stress, mammalian cells reprogram global translation to down-regulate expression of housekeeping genes and enhance expression of genes involved in the adaptation to stress. This reprogramming is initiated by phospho-eIF2α-induced translational repression and facilitated by active sequestration of untranslated mRNAs into SGs. We recently described a phospho-eIF2α-independent stress response program that is initiated by ANG-induced cleavage of tRNAs. ANG contributes to stress-induced translational repression, and transfection of 5′- but not 3′-tiRNAs inhibits protein synthesis (4). This strongly implicates both ANG and tiRNAs as effectors of stress-induced translational repression. In the present study, we further investigate the role of ANG and tiRNAs in the regulation of a phospho-eIF2α-independent stress response program.
Whereas environmental stresses (e.g. heat, UV irradiation, oxidative conditions, and hyperosmolarity) activate eIF2α kinases to promote phospho-eIF2α-mediated translational repression, selected xenobiotic compounds mediate phospho-eIF2α-independent translational repression. The natural products pateamine A (extracted from marine sponge) and hippuristanol (extracted from coral) and the anti-inflammatory lipid mediator 15d-PGJ2 inhibit translation by inactivating eIF4A, the RNA helicase required for the 48 S scanning reaction (30). In yeast (31), trypanosomes (32), and Drosophila (33), heat shock triggers the phospho-eIF2α-independent assembly of SGs by an uncharacterized mechanism. Here we show that ANG-induced tiRNAs also trigger the phospho-eIF2α-independent assembly of SGs (Fig. 6 and supplemental Figs. 2 and 3). This is the first example of SG assembly triggered by small non-coding RNA. The mechanism by which ANG-generated 5′-tiRNAs induce SG formation is still under investigation.
ANG is a neuroprotective factor required for the survival of motor neurons subjected to hypoxic conditions (34). Hypoxia is one of the environmental stresses that induces the assembly of SGs (14). The finding that ANG potentiates SG assembly in cells subjected to mild oxidative stress (Fig. 1) suggests that SG formation contributes to the protective effects of ANG. Importantly, this ANG-specific modulation of SG assembly is dependent on the ribonuclease activity of ANG because ANG mutants lacking RNase activity (P112L) do not promote SG assembly (Fig. 1). This is in accord with studies showing that ALS-associated mutants of ANG that lack ribonuclease activity also lack neuroprotective activity (7, 35). It is tempting to speculate that the secretion of ANG from stressed cells serves to prime adjacent cells for SG assembly in response to a mild second stress, thus acting as a signaling factor similar to interferon.
Cells rapidly repress translation in response to stress. This translational repression, specifically at the step of translation initiation, correlates with the assembly of SGs (14). In the present study, we show that ANG-generated 5′-tiRNAs or synthetic 5′-monophosphorylated 5′-tiRNAAla (Figs. 3 and 4), but not ANG-generated 3′-tiRNAs or 3′-synthetic tiRNAAla, induce SG assembly in U2OS cells. These findings suggest that tiRNA-induced translational silencing may inhibit general translation initiation and thus induce SGs. Because ANG enhances SG assembly induced by mild doses of sodium arsenite or pateamine A (Fig. 1) and transfection of tiRNAs promotes SG assembly (Figs. 3 and 4), it is logical to suggest that tiRNAs are a downstream component of an ANG-dependent stress response pathway. A previous study showed that silencing of the translation initiation factor eIF3 inhibits apoptosis in tumor cells (36), suggesting a strong link between translation initiation and apoptosis. This hypothesis was recently supported by data showing that SGs, which contain stalled 48 S preinitiation complexes, inhibit apoptosis via down-regulation of the stress-activated p38 and Jun N-terminal kinase (JNK) mitogen-activated protein kinase (MAPK) (SAPK) pathways (6). In line with this, the ability of ANG-generated 5′-tiRNAs to induce both translational repression and SG assembly strongly suggests that tiRNAs may affect cell survival via SG formation. Further studies are needed to clarify the mechanism by which ANG-produced 5′-tiRNAs regulate cell survival.
5′-tiRNAs but not 3′-tiRNAs have a distinct and potent inhibitory effect on translation (4). In addition, in our present study, 5′-tiRNAs but not 3′-tiRNAs induce SG formation in U2OS cells (Fig. 3). Collectively, these results suggest that some structural feature of 5′-tiRNAs confers these functional effects. Different modifications of small ncRNAs have been previously reported to contribute to their biological activity, stability, and interaction with partner proteins (37, 38). Terminal monophosphate modifications found at the 5′-ends of siRNAs, microRNAs, and piwi-interacting RNAs is a biochemical feature of these ncRNAs that is required for their functions (39–41). We found that 5′- but not 3′-tiRNAs have 5′-terminal monophosphates (Fig. 3A). In agreement with this finding, transfection of synthetic 5′-tiRNAAla, lacking a 5′-monophosphate, induces 4-fold less SG formation than 5′-tiRNAAla possessing the 5′-monophosphate modification (Fig. 3F). In addition to the 5′-monophosphate modifications that are essential for the function of various ncRNAs, tiRNAs might possess additional structural features that are required for translational repression and SG assembly. Because small ncRNAs associate with PIWI/Argonaute proteins (42), it is possible that tiRNAs may be a part of and/or be analogous to miRNA/Argonaute complexes comprising RISC (RNA-induced silencing complex).
Previously, we showed that knockdown of RNH1, a cytoplasmic protein that complexes with ANG, leads to the production of tiRNAs (4). This tiRNA production, however, leads to a significant repression of cellular translation in the presence, but not in the absence, of oxidative stress. In our present study, we show that ANG enhances assembly of SGs in cells subjected to mild oxidative stress. This enhancement is reversed by RNH1 overexpression, suggesting that the RNH1/ANG ratio regulates both tiRNA formation and translation repression/stress granule promotion. Interestingly, cysteine-rich RNH1 has been shown to defend against oxidative stress by modulating intracellular redox homeostasis (43). Oxidation of RNH1 thiol groups inactivates this activity and promotes its degradation (44). It is thus possible that oxidative stress-induced inactivation of RNH1 explains the stress dependence of ANG-induced enhancement of SG assembly.
Altogether, our data support the hypothesis that ANG and tiRNAs are components of an alternative stress response pathway. This stress response program acts via formation of SGs and independently of global eIF2α phosphorylation-dependent translation control. Further studies are needed to understand how this novel pathway contributes to the survival of stressed cells.
Supplementary Material
Acknowledgment
We thank the Anderson laboratory for helpful discussions and advice.
This work was supported, in whole or in part, by National Institutes of Health Grants AI065858, AI033600, and AR0514732.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- eIF2α
- eukaryotic translation initiation factor 2α
- ANG
- angiogenin
- tiRNA
- tRNA-derived stress-induced RNA
- SG
- stress granule
- PB
- processing body
- P-3′-tiRNA and P-5′-tiRNA
- phosphorylated 3′ and 5′-tiRNA, respectively
- HRI
- heme-regulated inhibitor kinase
- MES
- 4-morpholineethanesulfonic acid
- PBS
- phosphate-buffered saline
- SA
- sodium arsenite
- ncRNA
- non-coding RNA.
REFERENCES
- 1.Holcik M., Sonenberg N. (2005) Nat. Rev. Mol. Cell Biol. 6, 318–327 [DOI] [PubMed] [Google Scholar]
- 2.Thompson D. M., Parker R. (2009) J. Cell Biol. 185, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. (2009) FEBS Lett. 583, 437–442 [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki S., Ivanov P., Hu G. F., Anderson P. (2009) J. Cell Biol. 185, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fett J. W., Strydom D. J., Lobb R. R., Alderman E. M., Bethune J. L., Riordan J. F., Vallee B. L. (1985) Biochemistry 24, 5480–5486 [DOI] [PubMed] [Google Scholar]
- 6.Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. (2008) Nat. Cell Biol. 10, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 7.Sebastia J., Kieran D., Breen B., King M. A., Netteland D. F., Joyce D., Fitzpatrick S. F., Taylor C. T., Prehn J. H. (2009) Cell Death Differ. 16, 1238–1247 [DOI] [PubMed] [Google Scholar]
- 8.Hu G. F., Riordan J. F., Vallee B. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2204–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatzi E., Badet J. (1999) Eur. J. Biochem. 260, 825–832 [DOI] [PubMed] [Google Scholar]
- 10.Moroianu J., Riordan J. F. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1677–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibaragi S., Yoshioka N., Kishikawa H., Hu J. K., Sadow P. M., Li M., Hu G. F. (2009) Mol Cancer Res. 7, 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan Y., Wang F., Liu X. H., Gong D. J., Cheng H. Z., Huang S. D. (2009) Lung Cancer 66, 28–36 [DOI] [PubMed] [Google Scholar]
- 13.Tsuji T., Sun Y., Kishimoto K., Olson K. A., Liu S., Hirukawa S., Hu G. F. (2005) Cancer Res. 65, 1352–1360 [DOI] [PubMed] [Google Scholar]
- 14.Anderson P., Kedersha N. (2009) Curr. Biol. 19, R397–R398 [DOI] [PubMed] [Google Scholar]
- 15.Bordeleau M. E., Matthews J., Wojnar J. M., Lindqvist L., Novac O., Jankowsky E., Sonenberg N., Northcote P., Teesdale-Spittle P., Pelletier J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10460–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazroui R., Sukarieh R., Bordeleau M. E., Kaufman R. J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. (2006) Mol. Biol. Cell 17, 4212–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Low W. K., Dang Y., Schneider-Poetsch T., Shi Z., Choi N. S., Merrick W. C., Romo D., Liu J. O. (2005) Mol. Cell 20, 709–722 [DOI] [PubMed] [Google Scholar]
- 18.Low W. K., Dang Y., Bhat S., Romo D., Liu J. O. (2007) Chem. Biol. 14, 715–727 [DOI] [PubMed] [Google Scholar]
- 19.Dang Y., Kedersha N., Low W. K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J. O. (2006) J. Biol. Chem. 281, 32870–32878 [DOI] [PubMed] [Google Scholar]
- 20.Kedersha N., Tisdale S., Hickman T., Anderson P. (2008) Methods Enzymol. 448, 521–552 [DOI] [PubMed] [Google Scholar]
- 21.Wu D., Yu W., Kishikawa H., Folkerth R. D., Iafrate A. J., Shen Y., Xin W., Sims K., Hu G. F. (2007) Ann Neurol 62, 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kedersha N., Chen S., Gilks N., Li W., Miller I. J., Stahl J., Anderson P. (2002) Mol. Biol. Cell 13, 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. (2008) Nat. Cell Biol. 10, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., Tazi J. (2003) J. Cell Biol. 160, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Scadden A. D. (2007) Mol. Cell 28, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kedersha N., Cho M. R., Li W., Yacono P. W., Chen S., Gilks N., Golan D. E., Anderson P. (2000) J. Cell Biol. 151, 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. (2005) J. Cell Biol. 169, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J. J., Anderson P., Kaufman R. J. (2005) J. Biol. Chem. 280, 16925–16933 [DOI] [PubMed] [Google Scholar]
- 29.Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. (1999) J. Cell Biol. 147, 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim W. J., Kim J. H., Jang S. K. (2007) EMBO J. 26, 5020–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grousl T., Ivanov P., Frýdlová I., Vasicová P., Janda F., Vojtová J., Malínská K., Malcová I., Nováková L., Janosková D., Valásek L., Hasek J. (2009) J. Cell Sci. 122, 2078–2088 [DOI] [PubMed] [Google Scholar]
- 32.Kramer S., Queiroz R., Ellis L., Webb H., Hoheisel J. D., Clayton C., Carrington M. (2008) J. Cell Sci. 121, 3002–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farny N. G., Kedersha N. L., Silver P. A. (2009) RNA 15, 1814–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian V., Crabtree B., Acharya K. R. (2008) Hum. Mol. Genet. 17, 130–149 [DOI] [PubMed] [Google Scholar]
- 35.Kieran D., Sebastia J., Greenway M. J., King M. A., Connaughton D., Concannon C. G., Fenner B., Hardiman O., Prehn J. H. (2008) J. Neurosci. 28, 14056–14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J., Kahle A., Hershey J. W., Honchak B. M., Warneke J. A., Leong S. P., Nelson M. A. (2006) Oncogene 25, 4923–4936 [DOI] [PubMed] [Google Scholar]
- 37.Ghildiyal M., Zamore P. D. (2009) Nat. Rev. Genet. 10, 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim V. N., Han J., Siomi M. C. (2009) Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 39.Aravin A. A., Hannon G. J., Brennecke J. (2007) Science 318, 761–764 [DOI] [PubMed] [Google Scholar]
- 40.Nykänen A., Haley B., Zamore P. D. (2001) Cell 107, 309–321 [DOI] [PubMed] [Google Scholar]
- 41.Schwarz D. S., Hutvágner G., Haley B., Zamore P. D. (2002) Mol. Cell 10, 537–548 [DOI] [PubMed] [Google Scholar]
- 42.Höck J., Meister G. (2008) Genome Biol. 9, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monti D. M., Montesano Gesualdi N., Matousek J., Esposito F., D'Alessio G. (2007) FEBS Lett. 581, 930–934 [DOI] [PubMed] [Google Scholar]
- 44.Blázquez M., Fominaya J. M., Hofsteenge J. (1996) J. Biol. Chem. 271, 18638–18642 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.