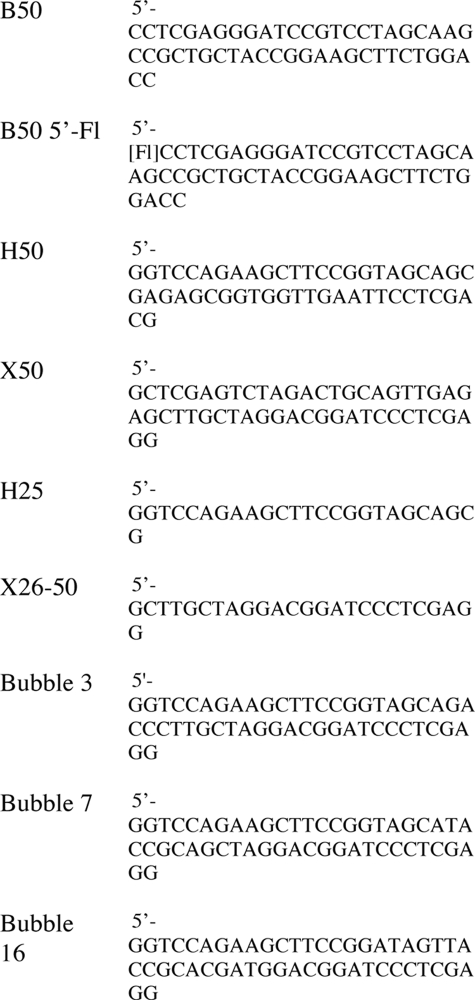Abstract
XPB helicase is an integral part of transcription factor TFIIH, required for both transcription initiation and nucleotide excision repair (NER). Along with the XPD helicase, XPB plays a crucial but only partly understood role in defining and extending the DNA repair bubble around lesions in NER. Archaea encode clear homologues of XPB and XPD, and structural studies of these proteins have yielded key insights relevant to the eukaryal system. Here we show that archaeal XPB functions with a structure-specific nuclease, Bax1, as a helicase-nuclease machine that unwinds and cleaves model NER substrates. DNA bubbles are extended by XPB and cleaved by Bax1 at a position equivalent to that cut by the XPG nuclease in eukaryal NER. The helicase activity of archaeal XPB is dependent on the conserved Thumb domain, which may act as the helix breaker. The N-terminal damage recognition domain of XPB is shown to be crucial for XPB-Bax1 activity and may be unique to the archaea. These findings have implications for the role of XPB in both archaeal and eukaryal NER and for the evolution of the NER pathway. XPB is shown to be a very limited helicase that can act on small DNA bubbles and open a defined region of the DNA duplex. The specialized functions of the accessory domains of XPB are now more clearly delineated. This is also the first direct demonstration of a repair function for archaeal XPB and suggests strongly that the role of XPB in transcription occurred later in evolution than that in repair.
Keywords: DNA Damage, DNA Helicase, DNA-Protein Interaction, DNA Repair, DNA Nucleotide Excision Repair, Nucleic Acid Enzymology, Archaea, Nuclease
Introduction
The superfamily 2 helicase XPB (Rad25 in Saccharomyces cerevisiae) is an essential component of transcription factor TFIIH. The ATPase activity of XPB is required for both nucleotide excision repair (NER)2 and transcription initiation from RNA polymerase II promoters (1, 2). The NER pathway is a highly flexible system that is required for the detection and removal of a wide variety of bulky and helix-distorting lesions, including photoproducts. Mutations in the xpb or xpd genes in humans can cause the serious genetic diseases xeroderma pigmentosum, trichothiodystrophy, and Cockayne's syndrome, due to defects in both transcription and repair (reviewed in Ref. 3). Examples of xpb mutations in humans are much more rare than those seen in the xpd gene, probably due to the crucial role of the XPB protein in basal transcription (4).
Although the ATPase activities of both proteins are required for NER (5), the respective roles of the XPB and XPD helicase components of TFIIH are still a matter of debate. XPD is the more robust helicase (6), and it has been suggested to bind 5′ of the DNA lesion and translocate in a 5′ to 3′ direction toward the damage site, potentially acting as a sensor or proofreader of DNA damage for the NER pathway either by jamming directly on DNA lesions (7) or perhaps through damage sensing by its iron-sulfur cluster binding domain (8, 9). However, little direct evidence exists in support of these possibilities at present, and indeed it is not yet clear whether XPD or XPB binds first at repair sites or whether they bind the same or complementary strands in the repair bubble. The helicase activity of XPB is rather weak in vitro and is stimulated by its association with the TFIIH subunit p52 (10, 11). Mutations that target the helicase motifs of XPB do not disrupt the function of TFIIH in NER, leading to the suggestion that XPB should be considered as an ATP-dependent molecular switch, perhaps opening DNA structure locally (11). Recent studies confirm the essential requirement for XPB ATPase activity in the recruitment of TFIIH to DNA damage sites (12). In contrast, the ATPase activity of XPD is not required. These data suggest that XPB may bind first to repair sites, perhaps locally destabilizing the DNA duplex to allow subsequent XPD binding and extension of the repair bubble.
The archaea share many informational proteins in common with eukarya. Most archaea encode clear homologues of the eukaryal NER helicases XPB and XPD (13). XPD is an active 5′ to 3′ helicase with an essential iron-sulfur cluster (8). The crystal structure of archaeal XPD provided a molecular explanation for the effects of mutations causing xeroderma pigmentosum, trichothiodystrophy, and Cockayne's syndrome (9, 14, 15). Archaeal XPB on its own is an ssDNA-dependent ATPase in vitro, with weak helicase activity under some conditions (16, 17). A crystal structure of the core of XPB from Archaeoglobus fulgidus revealed the presence of two canonical motor domains and two accessory domains named the thumb (Thm) domain and damage recognition domain (DRD) with putative roles in DNA damage detection (17). The relevance of the archaeal XPB structure to an understanding of the eukaryal protein was emphasized by the finding that the Thm domain and a conserved RED motif identified in the archaeal enzyme are essential for the function of eukaryal XPB in NER (12).
Genes encoding archaeal XPB are usually found next to a gene encoding a protein of unknown function named Bax1 (16). Bax1 and XPB were shown previously to interact physically in vitro (16), and bioinformatic analyses have suggested that Bax1 might be a DNA endonuclease (18). This prediction was confirmed recently for Bax1 from Thermoplasma acidophilum (19).
Here we report the purification and characterization of a recombinant XPB-Bax1 complex from Sulfolobus solfataricus. We demonstrate that the complex functions as a helicase-nuclease partnership, unwinding and cleaving DNA substrates that are models for the early steps in NER. We show that the Thm domain of XPB is essential for DNA unwinding and Bax1-mediated cleavage, consistent with a role in DNA duplex unwinding. The DRD has a subtle but essential role in DNA processing by the XPB-Bax1 complex, and may be unique to archaeal XPBs. We conclude that archaeal XPB-Bax1 functions in archaeal NER, with Bax1 performing the role equivalent to XPG in eukaryal cells.
EXPERIMENTAL PROCEDURES
Cloning, Mutagenesis, Expression, and Purification
The xpb2 and bax1 genes (sso0473 and sso0475), which are organized as an operon in S. solfataricus (20), were amplified as a unit from S. solfataricus genomic DNA by PCR using the oligonucleotides 5′-CCATGGTAGGATTAGGATAC and 5′-CAGGATCCTTTAAACCTCTTTGATC and cloned into the pEHISTEV vector (21) using the BamHI/NcoI recognition sites for co-expression of recombinant XPB and Bax1 in E. coli. The N terminus of XPB carried a polyhistidine tag with a TEV protease cleavage site. The construct was sequenced fully to confirm the expected nucleotide sequences of both genes. Protein expression was carried out in C43 cells induced overnight for 16 h at 28 °C by the addition of isopropyl 1-thio-β-d-galactopyranoside (0.5 mm).
For purification, cells were lysed by sonication in lysis buffer (20 mm Tris (pH 8.0), 500 mm NaCl, 0.1% Triton X-100, and 1 mm EDTA) containing a mixture of protease inhibitors (Roche Applied Science). The lysate was centrifuged (48,000 × g, 30 min, 4 °C), and filtered through a 0.45-μm syringe filter before being applied to a HisTrap column (HisTrap HP 5 ml, GE Healthcare), previously charged with NiCl2 and equilibrated with loading buffer (20 mm Tris (pH 8.0), 500 mm NaCl, 30 mm imidazole, and 10% glycerol). Proteins bound to the column were eluted with a linear gradient of imidazole (0.03–0.5 m) in loading buffer. Fractions containing the XPB-Bax1 complex were identified by SDS-PAGE, pooled, and purified to homogeneity by using a HiLoad 26/60 Superdex 200 size exclusion column (GE Healthcare) equilibrated with gel filtration buffer (25 mm Tris, pH 8.5, 500 mm NaCl, 1 mm EDTA, 0.5 mm dithiothreitol, and 10% glycerol). Fractions containing the XPB-Bax1 complex were identified by SDS-PAGE, pooled, and concentrated, and the polyhistidine tag was removed by cleavage overnight at 4 °C using 0.2 mg/ml TEV protease in the same buffer. Following cleavage, the protein was applied to the HisTrap column in loading buffer to separate tagged and untagged protein. The untagged complex was collected from the flow-through, pooled, and concentrated to 5 mg/ml in storage buffer (20 mm Tris, pH 8.0, 500 mm NaCl, 10% glycerol) and stored aliquoted at −80 °C until needed.
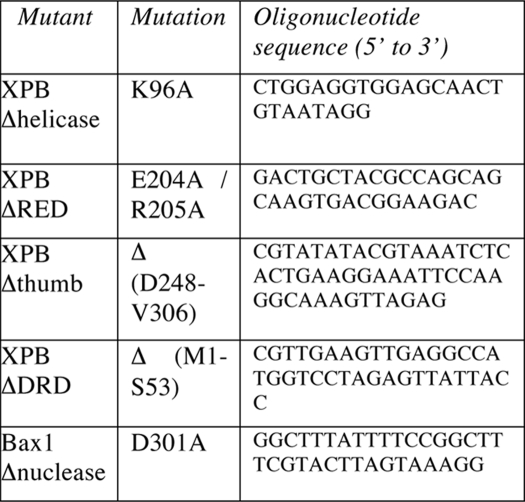
The Δnuclease, Δhelicase, ΔRED, and ΔThumb mutants were generated using a XL QuikChange mutagenesis kit (Stratagene). The ΔDRD mutant was generated by introducing an NcoI site before the codon corresponding to Val54 by mutagenesis. The plasmid was then digested by NcoI to remove the DNA corresponding to residues 1–53, which constitute the DRD, and the vector circularized by ligation and transformed into E. coli. Mutations and deletions were confirmed by DNA sequencing and mass spectrometry of the pure proteins. Oligonucleotides for mutagenesis are listed in Table 1.
TABLE 1.
Oligonucleotides for mutagenesis
DNA Substrate Preparation
Oligonucleotides for DNA substrates were gel-purified and ethanol-precipitated as described previously (22). The oligonucleotides in Table 2 were purchased from Operon Biotechnologies GmbH (Cologne, Germany) and annealed to make the substrates as follows. Bubble 3, Bubble 7, and Bubble 16 were assembled by annealing oligonucleotide B50 with the appropriate Bubble oligonucleotide. For the fluorescent DNA substrates, oligonucleotide B50 5′-Fl was annealed with oligonucleotide H50 for the 5′-labeled splayed duplex, oligonucleotide X50 for the 3′-splayed duplex, oligonucleotide H25 for the 5′-overhang, and oligonucleotide X26–50 for the 3′-overhang, respectively.
TABLE 2.
Oligonucleotides for DNA substrates
XPB-Bax1 Activity Assays
For DNA cleavage assays with radioactive substrates, 300 nm XPB-Bax1 (wild type or mutant) was incubated with 200 nm DNA substrate in reaction buffer (20 mm Tris, pH 8.5, 100 mm glutamate, 0.1 mg/ml bovine serum albumin) in a total volume of 10 μl for 20 min at 50 °C. Divalent metal ions (10 mm) and nucleotide (ATP or ATPγS, 1 mm) were added as indicated. Reactions were stopped by the addition of loading dye with EDTA, and products were analyzed on denaturing polyacrylamide TBE gels as described previously (23).
For experiments using fluorescent DNA, 1.2 μm XPB-Bax1 was incubated with 1 μm DNA at 45 °C, with all other conditions as described above. Size markers (A and G) were prepared from labeled substrates using standard protocols. Gels were scanned in a Fuji FLA5000 imager and analyzed using Fuji ImageGauge software.
For XPB-Bax1 assays analyzed by native gel electrophoresis, assays were performed in reaction buffer containing 750 nm XPB-Bax1 and 500 nm fluorescent DNA in a total volume of 10 μl for 5 or 20 min at 45 °C. Manganese chloride (10 mm) and nucleotide (ATP or ATPγS, 1 mm) were added as indicated. Reactions were stopped by the addition of 20 μl of chilled stop solution (10 mm Tris, pH 8, 5 mm EDTA, 0.5% SDS, 1 mg/ml proteinase K, 5 μm unlabeled oligonucleotide B50) and incubated for 30 min at room temperature. Samples were electrophoresed on a 10% native acrylamide TBE gel for 2 h and imaged using a Fuji FLA5000 imager as described above.
Electrophoretic Mobility Shift Assay
The radiolabeled DNA (10 nm) was incubated with an increasing amount of XPB-Bax1 in binding buffer (20 mm Hepes, pH 7.05, 2 mm dithiothreitol, 50 mm NaCl, 0.002% Triton X-100, and 0.1 mg/ml bovine serum albumin) in a total volume of 10 μl at 50 °C for 15 min. Samples were electrophoresed on a 10% native acrylamide TBE gel for 90 min and imaged using a Fuji FLA5000 imager as described above.
RESULTS
Cloning, Expression, and Purification of the XPB-Bax1 Complex
The genes encoding S. solfataricus XPB (sso0473) and Bax1 (sso0475) were amplified together by PCR and cloned into the vector pEHISTEV. This allowed expression of both genes as an operon, with the XPB protein expressed with an N-terminal polyhistidine tag that is cleavable by Tev protease (21). The proteins were purified by immobilized metal affinity chromatography and gel filtration as described under “Experimental Procedures.” The N-terminal polyhistidine tag was removed from the XPB protein by cleavage with TEV protease prior to gel filtration. XPB and Bax1 co-purified on each column as a complex with an apparent stoichiometry of 1:1, as shown previously (16, 19). Mutated variants with inactivated XPB or Bax1 were prepared as described under “Experimental Procedures” and purified as for the wild type protein complex.
The XPB-Bax1 Complex Catalyzes the Metal-dependent Cleavage of a Model NER Substrate
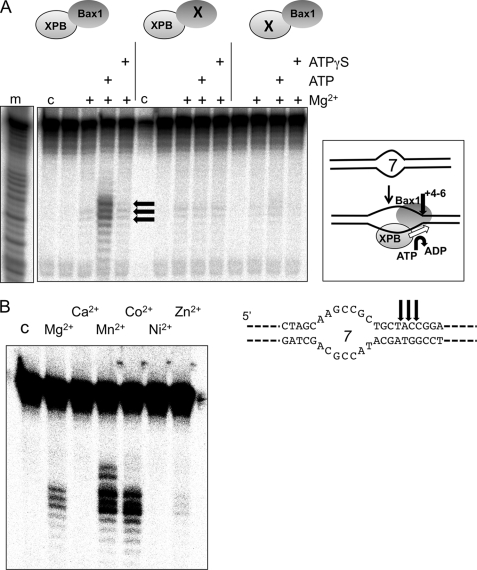
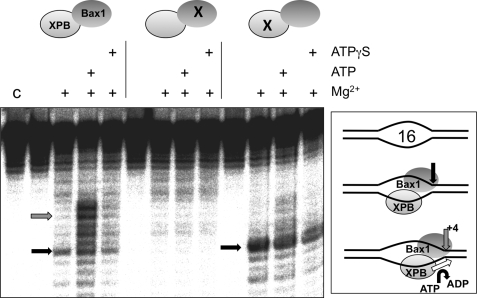
The XPB helicase in eukarya is thought to bind to DNA bubbles formed during transcription initiation or NER and to extend these bubbles by acting as a helicase or ATP-dependent conformational switch. We therefore tested the ability of the XPB-Bax1 complex to cleave a DNA duplex containing a 7-nucleotide centrally placed unpaired region (Bubble 7) (Fig. 1A). When ATP was present to support XPB activity, cleavage by Bax1 was observed 4–6 nucleotides 5′ of the ssDNA/dsDNA junction. This activity was lost when the predicted active site residue Asp301 of Bax1 was mutated to an alanine, confirming that the nuclease activity was specific for Bax1. DNA cleavage was also dependent on the activity of the XPB helicase, because no cleavage was observed either in the absence of ATP or when using a XPB variant with a mutation in the Walker A box (K96A). These data suggest that XPB and Bax1 function together as a helicase-nuclease partnership to unwind and cleave NER-type DNA substrates.
FIGURE 1.
XPB and Bax1 cooperate to cleave a model NER substrate. A, in the presence of ATP and Mg2+, the XPB-Bax1 complex cleaves a 7-nt DNA bubble substrate (Bubble 7) at three major sites located 4–6 bp 3′ of the ssDNA/dsDNA junction (black arrows). This activity is ablated when an active site residue of Bax1 is mutated (D301A; middle) and is also dependent on the activity of XPB as shown by mutation in the Walker A box of XPB (K96A; right). Control lane c, DNA alone; lane m, A + G sequence ladder. B, XPB-Bax1 cleaves the Bubble 7 substrate in the presence of ATP and magnesium, manganese, or cobalt cations. Lane C, control lane showing DNA alone. Quantification of the cleavage products yielded the following activities (relative to 100% for Mn2+): Mg2+, 24%; Ca2+, 1%; Mn2+, 100%; Co2+, 82%; Ni2+, 2%; Zn2+, 5%.
Reactions were carried out with a range of divalent metal ions to test the metal dependence of Bax1. Cleavage activity was detected in the presence of magnesium, manganese, and cobalt, with barely detectable activity in the presence of zinc and no activity in the presence of calcium or nickel (Fig. 1B). Overall, manganese yielded the highest nuclease activity. This spectrum of metal ion dependence is typical of the nuclease superfamily, consistent with the classification of Bax1 as a metal-dependent nuclease based on bioinformatic analysis (18). The three main cleavage sites were located 4–6 nucleotides into the duplex on the 3′-side of the bubble (black arrows). This suggests that XPB-Bax1 extends the unpaired region upon binding.
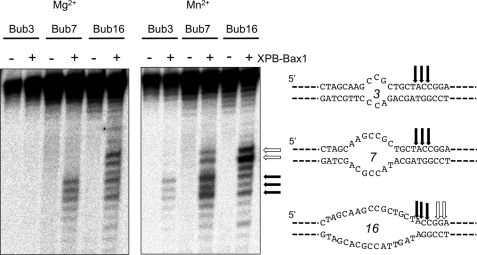
We next tested XPB-Bax1 against model DNA substrates with a range of bubble sizes (Fig. 2). No activity was observed against duplex or single-stranded DNA (data not shown). In the presence of magnesium and ATP, XPB-Bax1 cleaved bubbles of 7 and 16 nucleotides. The higher activity supported by manganese allowed detection of cleavage activity against a 3-nucleotide bubble substrate. All three bubble substrates had three cleavage sites in common (indicated by black arrows). This is consistent with a specific binding site size for XPB-Bax1 directed by the unpaired DNA of the bubble. For Bubble 16 and to a lesser extent Bubble 7, cleavage was observed further into the duplex region (white arrows), consistent with extension of the bubble by the XPB helicase.
FIGURE 2.
XPB-Bax1 cleaves a range of bubble substrates. XPB-Bax1 cleaves bubbles of 7 and 16 nt in the presence of magnesium and ATP and additionally a bubble of 3 nt in the presence of manganese and ATP. The cleavage sites are shown mapped for the three substrates. The black arrows show cleavage sites in common for all three bubbles. Cleavage further into the duplex 3′ of the bubble (white arrows) suggests further opening of the DNA by XBP.
The Helicase Activity of XPB Directs DNA Cleavage by Bax1
To confirm that DNA unwinding by XPB directs Bax1 cleavage, we looked in detail at XPB-Bax1 cleavage of the Bubble 16 substrate (Fig. 3). Unlike Bubble 7, this substrate was cleaved in the absence of XPB activity (either by omitting ATP, by substituting the non-hydrolyzable analogue ATPγS, or with the Walker A box mutant of XPB). Tellingly, under these conditions, cleavage was confined to a single site at the junction of ssDNA and dsDNA on the 3′-side of the bubble (black arrow). When ATP was included in the reaction, the main site of cleavage (representing 85% of the cleavage products) was observed to shift 4–6 nucleotides 3′ in the cleaved strand, as shown previously. These data suggest that the Bubble 16 substrate is large enough to allow binding and cleavage by XPB-Bax1 in the absence of XPB activity and suggest that Bax1 cleaves near the ssDNA/dsDNA junction. The known 3′ to 5′ directionality of XPB is consistent with binding on the bottom strand of the bubble, as shown in the schematic in Fig. 3 and discussed below.
FIGURE 3.
Bax1 cleaves at the junction between ssDNA and dsDNA, and bubbles are extended by XPB. With the Bubble 16 substrate, Bax1 cuts at the ssDNA/dsDNA junction when XPB helicase activity is inactivated due to lack of ATP or point mutation K96A (black arrows). When XPB is active, the point of cleavage moves 4–5 nt 3′ away from the edge of the bubble, consistent with DNA strand opening by XPB (gray arrows). No activity was observed when the Bax1 nuclease was inactivated by mutation (D301A). Lane C, control, DNA alone.
The Roles of XPB Domains in XPB-Bax1
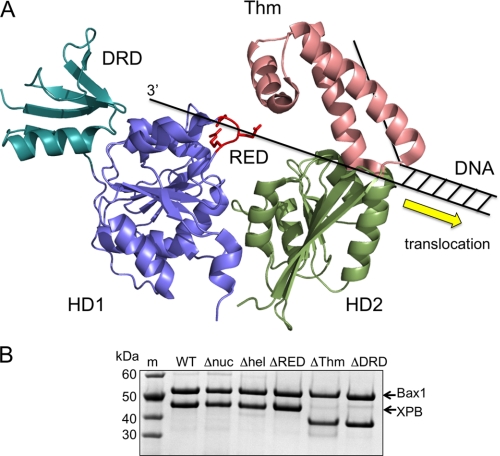
The structure of archaeal XPB revealed two helicase motor domains (HD1 and HD2), with an N-terminal DRD and a Thm domain arising from within HD2 (Fig. 4A). A conserved RED motif was also described as important for helicase activity. By analogy with other 3′ to 5′ helicases, strand separation by XPB is likely to occur close to helicase domain 2 (24). The most likely motif implicated in strand separation is therefore the Thm domain, with single-stranded DNA dragged across of the tops of the two motor domains, bringing it close to the position of the RED motif. Both the Thm domain and the RED motif have recently been implicated as important for binding of TFIIH to DNA damage sites (12).
FIGURE 4.
XPB domain structure and complex formation with Bax1. A, structural model of XPB from A. fulgidus, showing the two helicase motor domains (HD1 and HD2), the N-terminal DRD, and RED motif arising from HD1 and the Thm domain arising from HD2. XPB moves in a 3′ to 5′ direction on DNA and is predicted to disrupt the DNA duplex at HD2, possibly by the physical action of the Thm domain. B, a Coomassie Blue-stained SDS-polyacrylamide gel showing purified XPB-Bax1 complex after gel filtration. The wild type and all mutant variants of XPB and Bax1 co-purify in a 1:1 complex. m, molecular weight markers; W-T, wild type XPB-Bax1; Δnuc, Bax1 D301A mutant; Δhel, XPB K96A mutant; ΔRED, mutated RED motif in XPB; ΔThm, Thm domain in XPB deleted; ΔDRD, DRD of XPB deleted.
The RED motif is not completely conserved across all archaeal XPB sequences (supplemental Fig. 1), and in fact the consensus sequence in archaea is ERXDG. Accordingly, to assess the importance of this motif, we made the double mutant E204A/R205A. The Thm domain was entirely deleted (mutant ΔThm) by removing amino acids Asp348–Val406 inclusive, joining amino acids 347–407 via a glycine residue. The N-terminal DRD (residues 1–53) was removed by the introduction of an NcoI site, including a new start codon at position 54 and subcloning to generate the ΔDRD mutant. The mutant XPB variants were co-expressed with Bax1 as for the wild type protein. Each mutant formed a stable complex with Bax1 (Fig. 4B).
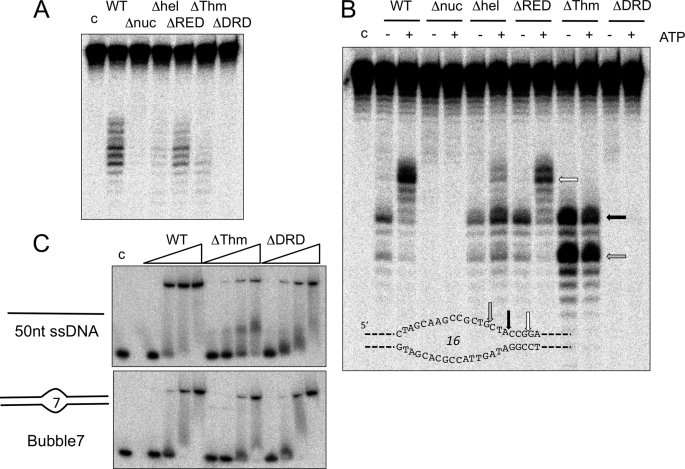
We next tested the ability of the mutant proteins to unwind and cleave substrates (Fig. 5). With Bubble 7, the ΔThm and ΔDRD mutants showed little or no Bax1 cleavage activity (<10% of wild type activity), similar to the situation where the helicase activity of XPB was disrupted, suggesting that these mutations interfere with the correct functioning of XPB in some way. The ΔRED mutant showed decreased but detectable substrate cleavage (40% of wild type activity), suggesting that this motif is involved in but not essential for the function of XPB in this context. More informative results were obtained for the Bubble 16 substrate (Fig. 5B). With this larger bubble, we showed previously that the helicase activity of XPB was not required for Bax1 cleavage near the ssDNA/dsDNA junction. The ΔRED mutant had activity comparable with the wild type protein (75%), with ATP-dependent cleavage sites introduced in the duplex region 3′ of the bubble, suggesting that the XPB helicase was at least partially active. In contrast, the ΔThm mutant did not support this invasion of the duplex region, suggesting that helicase activity was abrogated. Instead, there was strong cleavage within the ssDNA bubble (gray arrow) as well as at the ssDNA/dsDNA boundary. This suggests that the loss of the Thm domain has affected the ability of XPB to position Bax1 correctly at the DNA junction. Together, these observations are consistent with a role for the Thm domain at the DNA unwinding site, potentially acting as the “wedge” or “plowshare” that physically separates the duplex DNA. Finally, the ΔDRD mutant displayed very little cleavage activity (<2% of wild type activity) either in the presence or absence of ATP, suggesting a fundamental role in XPB-Bax1 function.
FIGURE 5.
Mutational analysis of XPB-Bax1 reveals subdomain function in binding and catalysis. A, activity of wild type (WT) and mutant versions of XPB-Bax1 on the Bubble 7 substrate in the presence of Mn2+ and ATP. B, activity of wild type and mutant versions of XPB-Bax1 on the Bubble 16 substrate in the presence of Mn2+, in the presence or absence of ATP. C, gel shift analysis of XPB-Bax1 binding to the Bubble 7 substrate and single-stranded DNA. The binding affinities of the wild type, ΔThm, and ΔDRD variants of XPB were compared by incubating 10 nm DNA with 50, 100, 250, and 500 nm XPB-Bax1. Lane c, DNA alone.
The binding affinities of XPB-Bax1 variants for ssDNA and the Bubble 7 substrate were tested by electrophoretic gel mobility shift analysis (Fig. 5C). Previously, we demonstrated that S. solfataricus XPB bound relatively weakly to ssDNA, with an apparent dissociation constant of about 1 μm (16). By contrast, the XPB-Bax1 complex bound ssDNA an order of magnitude more tightly, with an apparent dissociation constant of about 100 nm (Fig. 5C). Slightly weaker binding (apparent KD of ∼250 nm) was observed for the ΔDRD and ΔThm mutants. The Bubble 7 substrate was bound with broadly comparable affinity (apparent KD values around 200 nm) by the wild type, ΔDRD, and ΔThm enzymes, whereas dsDNA was bound much more weakly by all three proteins (KD > 2 μm; data not shown). The enhanced binding affinity of XPB-Bax1 compared with XPB alone may be due either to ssDNA binding by Bax1 or to an alteration in the ssDNA binding properties of XPB when in complex with Bax1. These data rule out the possibility that the loss of activity observed for the ΔDRD mutation has resulted from a gross defect in DNA binding by the XPB-Bax1 complex. Potential roles for the XPB domains are examined in more detail under “Discussion.”
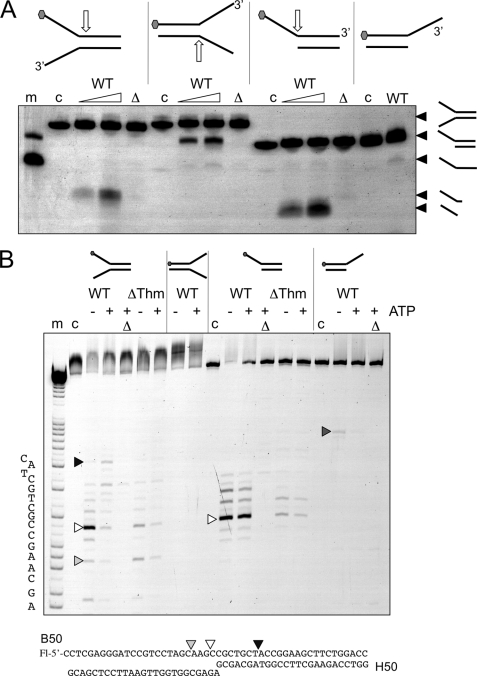
Finally, we compared the ability of the wild type and mutant XPB-Bax1 complexes to cleave model substrates, including splayed duplexes and single-stranded DNA overhangs (Fig. 6). Using native gel electrophoresis to analyze DNA products, we observed cleavage of splayed duplex substrates within the duplex region next to the 5′-arm by wild type XPB-Bax1 (Fig. 6A). This is consistent with the activity seen against bubble substrates. A model substrate with a 5′-ssDNA overhang was also cleaved, yielding smaller labeled products than for the equivalent splayed duplex, suggesting cleavage at the junction between the single- and double-stranded DNA rather than within the duplex region. A 3′-overhang substrate was not cleaved appreciably by XPB-Bax1. Importantly, no helicase activity was observed for any substrates, even when the nuclease activity was abrogated by the Δnuc mutation (Fig. 6A). This suggests strongly that XPB cannot destabilize a 25-bp duplex under these conditions, which would require unwinding of only about 12 bp of DNA, but rather catalyzes local unwinding near the junction, consistent with the data obtained from bubble structures.
FIGURE 6.
Cleavage of minimal substrates by XPB-Bax1. A, native gel electrophoresis showing cleavage of splayed duplex and overhang substrates. Oligonucleotide B50 with a 5′-fluorescein (gray hexagon) was annealed with a range of partner strands to generate minimal substrates. Approximate cleavage sites are indicated by white arrows. Cleavage products are indicated on the right of the gel. The wild type enzyme (WT) was incubated with each substrate for 5 or 20 min in the presence of ATP and MnCl2. The Δnuc mutant was incubated in the same buffer for 20 min. Control lanes are as follows. m, marker DNA showing migration of overhang and single-stranded B50 oligonucleotide; c, DNA substrate control; Δ, Δnuc mutant. B, the reaction products were also run on denaturing acrylamide TBE gels with (A + G) markers to map cleavage sites. All reactions were carried out in reaction buffer with 10 mm MnCl2 and ATP where indicated. m, A + G sequence markers; c, DNA alone; Δ, Δnuc mutant.
By repeating the assays using denaturing gel electrophoresis, we were able to map the cleavage sites introduced by XPB-Bax1 precisely (Fig. 6B). First, for the splayed duplex with a labeled 5′-arm, in the absence of ATP the wild type protein cleaved predominantly close to the junction (white arrow). When ATP was present, the cleavage pattern changed, with new sites within the duplex region cleaved more predominantly (e.g. black arrow). The ΔThm mutant did not appear to position so precisely on the junction, evidenced by the stronger relative cleavage in the single-stranded arm (light gray arrow) and showed no ATP-dependent cleavage within the DNA duplex, consistent with a loss of helicase activity as shown previously for bubble substrates. On the 5′-overhang substrate, no ATP-dependent effect was observed, consistent with the model whereby XPB binds on the 3′-strand and moves 3′ to 5′ into the duplex. Very weak cutting of the 3′-flap substrate in the single-stranded region (dark gray arrow) may be consistent with the activity detected by the Kisker group (19) and could be due to Bax1 binding in the opposite orientation with respect to the junction.
DISCUSSION
Bax1, the Archaeal XPG?
The mechanism of NER in archaea has been the subject of much speculation since genome sequencing revealed the presence of eukaryal-type NER genes in the archaea (13, 25). Although the intervening years have seen good progress made in studying the structures and activities of individual NER proteins in archaea, little is known about how they function together to effect damage recognition and repair. We have shown that XBP and Bax1 form a stable complex (Fig. 7A) and function together to cleave model NER substrates on the 5′-side of DNA bubbles. In large bubble substrates, such as Bubble 16, Bax1 can function in the absence of XPB activity to cleave the DNA at the junction of ssDNA and dsDNA. For smaller bubbles, there is not enough ssDNA available to allow Bax1 to function unless the bubble size is increased by the action of XPB. XPB activity extends bubbles by at most 6–8 nucleotides, allowing Bax1 to engage the ssDNA and cleave it. Given the known 3′ to 5′ polarity of XPB, our data can only be explained by XPB binding on the bottom (undamaged) strand and extending the bubble on the “downstream” side (with respect to the lesion site), allowing Bax1 to introduce the cleavage site 3′ of the damage, as indicated in the schematic diagrams in Figs. 1 and 3. This is supported by the observation that minimal substrates with a 3′-strand for XPB loading are partially unwound before cleavage, whereas substrates lacking this strand are cleaved only at the junction point (Fig. 6). Fig. 6A also emphasizes the fact that XPB does not function as a canonical helicase because there was no evidence for unwinding of any of the DNA substrates tested by the XPB-Bax1 Δnuc variant. This is consistent with recent data suggesting that eukaryal XPB may be functioning as an ATP-dependent conformational switch rather than a canonical helicase (11).
FIGURE 7.
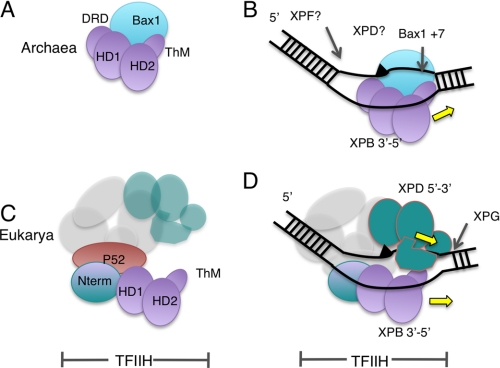
Model for XPB evolution and NER in archaea and eukarya. A, the structure of archaeal XPB revealed two helicase domains (HD1 and HD2), with an N-terminal DRD and a Thm domain arising from within HD2. There is a stable interaction with the nuclease Bax1 to form a helicase-nuclease machine. B, archaeal NER may involve duplex unwinding by XPB bound to the undamaged (bottom) strand allowing the Bax1 nuclease to cleave the damaged strand 3′ of the lesion. Further steps may be catalyzed by the archaeal XPD and XPF proteins. C, in eukarya, the interaction with Bax1 has been lost and replaced by p52, which anchors XPB to the TFIIH complex. The other TFIIH subunits required for NER are XPD, p34, p44, p62, and p8. D, a model for TFIIH binding during NER, showing XPB bound to the undamaged strand and XPD bound to the damaged strand, consistent with the archaeal scheme.
Archaeal Bax1 nuclease could be considered as the functional equivalent of the nuclease XPG (Fig. 7B). Initiation of this process from a DNA bubble as small as 3 nucleotides suggests that the XPB-Bax1 complex could initiate repair from very small regions of non-canonical duplex DNA (e.g. at a photoproduct site where base pairing is locally disturbed). The action of XPB in initiating the repair bubble may allow the XPD helicase to bind and extend the nascent bubble, a situation analogous to that suggested for eukaryal NER (12). Patch repair in the archaea may require the action of the 3′-flap nuclease XPF to complete the excision step (23, 26). One key question is how XPB is directed to bind to the undamaged strand, thus directing Bax1 to cleave the damaged strand. This may be determined by the protein(s) involved in the initial DNA damage detection step, analogous to the role of HR23B-XPC in eukarya. This step of NER in archaea is still not understood, although the SSB protein has been shown capable of unwinding damaged DNA in vitro (27). Reconstitution of the archaeal NER pathway in vitro is a key future goal.
The Kisker laboratory has recently reported that Bax1 and XPB from the euryarchaeote T. acidophilum form a 1:1 complex in vitro (19). Endonuclease activity ascribed to Bax1 was only observed in the absence of XPB and was specific for 3′-flaps with cleavage 4–6 nt from a dsDNA/ssDNA junction. In other words, the Bax1 activity was closer to XPF than to XPG. In contrast to our data, no endonuclease activity was detected in the presence of manganese, and bubble substrates were not cleaved (19). Sequence alignments demonstrate that Bax1 in the thermoplasmatales lacks many of the key residues conserved in other species, including the key nuclease active site motif. Given these differences and the lack of any activity for the T. acidophilum XPB-Bax1 complex, the relevance of these observations to the present study are hard to ascertain.
XPB Structure and the Role of the Accessory Domains
The A. fulgidus XPB crystal structure revealed an unusual conformation where helicase domain 2 was rotated through almost 180° with respect to helicase domain 1 compared with the “canonical” position that has been observed in all other Superfamily 1 and 2 helicase structures (17). It has been postulated that this conformational flexibility is important for the biological function of the XPB protein (12, 17). However, A. fulgidus XPB was crystallized in the absence of its cognate Bax1 partner, and it is therefore possible that the unusual structure observed by Fan et al. (17) was due to unusual conformational flexibility induced by the absence of the Bax1 subunit. It has been noted previously that XPB from S. solfataricus is heat-labile and prone to aggregation in the absence of its Bax1 partner (16). A definitive answer to this question will require further analysis of the conformational flexibility of XPB in the presence and absence of Bax1.
The XPB protein structure revealed two accessory domains named the Thm domain and the DRD (17). As we have already stated, by analogy with several other helicases, the position of the Thm domain above helicase domain 2 is consistent with a role in DNA duplex opening by XPB. Consistent with this, we note a loss of helicase activity and significant defects in helicase positioning at DNA junctions when the Thm domain is deleted. A similar domain (Domain 2) has been noted in the archaeal Hef helicase. Deletion of this domain resulted in the loss of helicase activity that was ascribed to an inability to target forked DNA structures in the deletion mutant (28).
The N-terminal DRD of A. fulgidus XPB is structurally related to the mismatch recognition domain of the MutS protein and has little or no intrinsic DNA binding affinity (17). Deletion of the DRD in S. solfataricus XPB does not disrupt the interaction with Bax1 or with DNA substrates but has a profound effect upon the activity of the complex. The role in damage detection suggested by Fan et al. (17) (and implicit in the name of the domain) is not relevant in our assays and thus cannot fully explain the function of the DRD. Together, these data suggest a subtle but crucial role for the DRD, perhaps in positioning XPB-Bax1 correctly on the DNA or supporting the nuclease activity of Bax1. The presence of an equivalent DRD in eukaryotic XPB was predicted based largely on the observation of sequence similarity of around 40% between the Archaeoglobus and human sequences in this region (17). However, the most highly conserved residues of the eukaryal XPB family in this region of the protein are not conserved in the archaeal XPBs (supplemental Fig. 2). Furthermore, secondary structure prediction by the JPRED server (29) yields an accurate match for the known structure of archaeal XPB but predicts a very different secondary structure for the equivalent region of eukaryotic XPBs (supplemental Fig. 2). Finally, eukaryal XPBs have an extra N-terminal domain that is important for interactions with eukarya-specific partner proteins. Therefore, on balance, we suggest that the DRD sequence is unique to archaeal XPB and plays a vital role in the functional interaction with the Bax1 protein. In eukaryal XPB, this domain, if it exists, may well have a different fold and function.
Implications for Eukaryal NER
Eukaryal XPB is involved in both NER and transcription initiation. The observation that archaeal XPB is coupled functionally with a DNA endonuclease makes it more likely that the ancestral role of XPB was in excision repair. In the eukaryal lineage, XPB evolved a close interaction with the p52 subunit of TFIIH, which is required for XPB helicase activity (10) (Fig. 7C). The ancestral form of TFIIH may thus have been co-opted by the RNA polymerase II transcriptional apparatus in the eukaryal lineage. In this scenario, the archaeal Bax1 nuclease was replaced with XPG, which interacts with TFIIH during NER (30, 31).
The placement of archaeal XPB on the undamaged strand, moving 3′ to 5′ and thus opening up the damaged strand on the 3′-side of the lesion for cleavage by Bax1, also has implications for eukaryal NER (Fig. 7D). The repair complexes formed by the succession of NER proteins binding around the DNA damage site in eukarya are understood incompletely. It is clear that HR23B and XPC are the first proteins to recognize the damaged DNA in global NER and that these proteins help recruit the TFIIH complex to the damage site (32). TFIIH opens up the DNA further, displaces HR23B-XPC, and helps recruit the XPA and RPA proteins to the repair site (33). XPD has been proposed to bind to the damaged strand and play a role in confirming the presence of DNA damage, perhaps by stalling at the lesion (reviewed in Ref. 34), but direct evidence in support of this hypothesis is rather limited, and archaeal XPD has been shown recently to be capable of translocating past DNA lesions (35).
There are two fundamental options for XPB binding to DNA during NER. First, XPB could bind on the 5′-side of the damaged strand and thus move 3′ to 5′ away from the lesion, defining the boundary for the 5′ cut by XPF-ERCC1. Second, XPB could bind the undamaged strand, in which case its 3′ to 5′ polarity would move it in tandem with XPD on the opposite strand toward the 3′-side of the repair bubble. The data for eukaryal NER are inconclusive (e.g. photocross-linking of XPB to a cisplatin lesion revealed extensive contacts with sites both 5′ and 3′ of the DNA lesion) (33).
In conclusion, the archaeal XPB-Bax1 complex is an interesting new example of a helicase-nuclease DNA-processing machine. Our data suggest an ancestral role for XPB in extending NER substrates by strictly limited translocation along the undamaged strand, generating a span of ssDNA on the damaged strand for attack by a structure-specific nuclease. This model is consistent with recent studies showing that the activity of eukaryal XPB is essential for the recruitment of TFIIH to damage sites. Thus, the detailed characterization of the archaeal enzymes can extend our understanding of eukaryal NER, where the additional complexity of the system has presented a barrier to detailed analysis.
Supplementary Material
Acknowledgments
We thank Paul Talbot for technical support and the University of St. Andrews Mass Spectrometry Unit for expert service.
This work was supported by Cancer Research UK Grant C22223.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- NER
- nucleotide excision repair
- ssDNA
- single-stranded DNA
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- dsDNA
- double-stranded DNA
- Thm
- thumb
- DRD
- damage recognition domain
- nt
- nucleotide(s).
REFERENCES
- 1.Evans E., Moggs J. G., Hwang J. R., Egly J. M., Wood R. D. (1997) EMBO J. 16, 6559–6573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tirode F., Busso D., Coin F., Egly J. M. (1999) Mol. Cell 3, 87–95 [DOI] [PubMed] [Google Scholar]
- 3.Lehmann A. R. (2003) Biochimie 85, 1101–1111 [DOI] [PubMed] [Google Scholar]
- 4.Andressoo J. O., Weeda G., de Wit J., Mitchell J. R., Beems R. B., van Steeg H., van der Horst G. T., Hoeijmakers J. H. (2009) Mol. Cell. Biol. 29, 1276–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guzder S. N., Sung P., Bailly V., Prakash L., Prakash S. (1994) Nature 369, 578–581 [DOI] [PubMed] [Google Scholar]
- 6.Coin F., Marinoni J. C., Rodolfo C., Fribourg S., Pedrini A. M., Egly J. M. (1998) Nat. Genet. 20, 184–188 [DOI] [PubMed] [Google Scholar]
- 7.Naegeli H., Modrich P., Friedberg E. C. (1993) J. Biol. Chem. 268, 10386–10392 [PubMed] [Google Scholar]
- 8.Rudolf J., Makrantoni V., Ingledew W. J., Stark M. J., White M. F. (2006) Mol. Cell 23, 801–808 [DOI] [PubMed] [Google Scholar]
- 9.Fan L., Fuss J. O., Cheng Q. J., Arvai A. S., Hammel M., Roberts V. A., Cooper P. K., Tainer J. A. (2008) Cell 133, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jawhari A., Lainé J. P., Dubaele S., Lamour V., Poterszman A., Coin F., Moras D., Egly J. M. (2002) J. Biol. Chem. 277, 31761–31767 [DOI] [PubMed] [Google Scholar]
- 11.Coin F., Oksenych V., Egly J. M. (2007) Mol. Cell 26, 245–256 [DOI] [PubMed] [Google Scholar]
- 12.Oksenych V., de Jesus B. B., Zhovmer A., Egly J. M., Coin F. (2009) EMBO J. 28, 2971–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White M. F. (2007) in Archaea: Evolution, Physiology, and Molecular Biology (Garrett R. A., Klenk H. P. eds) pp. 171–184, Blackwell Publishing Ltd., Oxford [Google Scholar]
- 14.Liu H., Rudolf J., Johnson K. A., McMahon S. A., Oke M., Carter L., McRobbie A. M., Brown S. E., Naismith J. H., White M. F. (2008) Cell 133, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolski S. C., Kuper J., Hänzelmann P., Truglio J. J., Croteau D. L., Van Houten B., Kisker C. (2008) PLoS Biol. 6, e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards J. D., Cubeddu L., Roberts J., Liu H., White M. F. (2008) J. Mol. Biol. 376, 634–644 [DOI] [PubMed] [Google Scholar]
- 17.Fan L., Arvai A. S., Cooper P. K., Iwai S., Hanaoka F., Tainer J. A. (2006) Mol. Cell 22, 27–37 [DOI] [PubMed] [Google Scholar]
- 18.Kinch L. N., Ginalski K., Rychlewski L., Grishin N. V. (2005) Nucleic Acids Res. 33, 3598–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth H. M., Tessmer I., Van Houten B., Kisker C. (2009) J. Biol. Chem. 284, 32272–32278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wurtzel O., Sapra R., Chen F., Zhu Y., Simmons B. A., Sorek R. (2010) Genome Res 20, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H., Naismith J. H. (2009) Protein Expr. Purif. 63, 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kvaratskhelia M., White M. F. (2000) J. Mol. Biol. 295, 193–202 [DOI] [PubMed] [Google Scholar]
- 23.Roberts J. A., White M. F. (2005) Nucleic Acids Res. 33, 6662–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singleton M. R., Dillingham M. S., Wigley D. B. (2007) Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 25.Grogan D. W. (2000) Trends Microbiol. 8, 180–185 [DOI] [PubMed] [Google Scholar]
- 26.Roberts J. A., White M. F. (2005) J. Biol. Chem. 280, 5924–5928 [DOI] [PubMed] [Google Scholar]
- 27.Cubeddu L., White M. F. (2005) J. Mol. Biol. 353, 507–516 [DOI] [PubMed] [Google Scholar]
- 28.Nishino T., Komori K., Tsuchiya D., Ishino Y., Morikawa K. (2005) Structure 13, 143–153 [DOI] [PubMed] [Google Scholar]
- 29.Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Araújo S. J., Nigg E. A., Wood R. D. (2001) Mol. Cell. Biol. 21, 2281–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winkler G. S., Sugasawa K., Eker A. P., de Laat W. L., Hoeijmakers J. H. (2001) Biochemistry 40, 160–165 [DOI] [PubMed] [Google Scholar]
- 32.Min J. H., Pavletich N. P. (2007) Nature 449, 570–575 [DOI] [PubMed] [Google Scholar]
- 33.Tapias A., Auriol J., Forget D., Enzlin J. H., Schärer O. D., Coin F., Coulombe B., Egly J. M. (2004) J. Biol. Chem. 279, 19074–19083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schärer O. D. (2007) Mol. Cell 28, 184–186 [DOI] [PubMed] [Google Scholar]
- 35.Rudolf J., Rouillon C., Schwarz-Linek U., White M. F. (2010) Nucleic Acids Res. 38, 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.