Abstract
Extracellular hyperosmolarity, or osmotic stress, generally caused by differences in salt and macromolecule concentrations across the plasma membrane, occurs in lymphoid organs and at inflammatory sites. The response of immune cells to osmotic stress is regulated by nuclear factor of activated T cells 5 (NFAT5), a transcription factor that induces the expression of hyperosmolarity-responsive genes and stimulates cytokine production. We report that the guanine nucleotide exchange factor (GEF) Brx [also known as protein kinase A–anchoring protein 13 (AKAP13)] is essential for the expression of nfat5 in response to osmotic stress, thus transmitting the extracellular hyperosmolarity signal and enabling differentiation of splenic B cells and production of immunoglobulin. This process required the activity of p38 mitogen-activated protein kinase (MAPK) and NFAT5 and involved a physical interaction between Brx and c-Jun N-terminal kinase (JNK)–interacting protein 4 (JIP4), a scaffold molecule specific to activation of the p38 MAPK cascade. Our results indicate that Brx integrates the responses of immune cells to osmotic stress and inflammation by elevating intracellular osmolarity and stimulating the production of cytokines.
INTRODUCTION
Dehydration and hyperosmolarity, or osmotic stress, remain major challenges to land organisms because they evolved from the isotonic environment of the sea (1). Extracellular hyperosmolarity results in the extraction of water from cells and disturbs global cellular function by condensing or denaturing intracellular molecules and by altering subcellular architecture (1, 2). To counter this osmotic challenge, organisms have developed a conserved, yet incompletely understood, counter-regulatory mechanism that senses extracellular hyperosmolarity at the cell membrane and transduces this signal from the cytoplasm to the nucleus (1, 2). Osmotic stress stimulates the transcription of several genes that in turn cause intracellular accumulation of small organic osmolytes, such as sorbitol, myo-inositol, glycine, betaine, and taurine, which leads to increased intracellular osmotic pressure and thus the maintenance of isotonicity between the inside and outside of the cell (1, 2).
Saccharomyces cerevisiae has a signaling complex localized to the internal cytoplasmic membrane that uses osmotic sensors coupled with Rho-type small guanosine triphosphate (GTP)–binding proteins (G proteins) to activate the high osmolarity glycerol 1 (HOG1) protein, a yeast homolog of the mammalian p38 mitogen-activated protein kinase (MAPK) (3-6). Mammalian cells, such as those in the renal medulla that are continuously exposed to high concentrations of osmolytes, also use a multiprotein osmosensing complex that involves Rho-type small G proteins and p38 MAPK (1, 7-9). Activation of p38 MAPK in turn stimulates the expression and the transcriptional activity of a transcription factor, nuclear factor of activated T cells 5 [NFAT5, also known as tonicity enhancer binding protein (TonEBP)]. NFAT5 contains the Rel homology domain and shares a common Rel-like ancestor with rel, Dorsal, the nuclear factor κB (NF-κB) family proteins, and the other NFAT proteins (10-16). NFAT5 stimulates the transcription of hyperosmolarity-responsive genes, including those encoding aldose reductase (AR), the sodium-myo-inositol cotransporter (SMIT) and the sodium chloride–betaine cotransporter (BGT1), which are responsible for the production and uptake of small organic osmolytes (17).
In addition to activating a conserved adaptive mechanism to adjust to changes in osmotic imbalance, changes in extracellular osmolarity alter the function of the immune system. Infusion of hyperosmolar fluids into normal volunteers causes increased proliferation of T cells (18), whereas extracellular hyperosmolarity stimulates the production of many cytokines, including interleukin-1β (IL-1β), IL-6, IL-8, tumor necrosis factor–α (TNF-α), and lymphotoxin β (LT-β) (19-22). Further, some pathologic conditions are accompanied by extracellular (plasma) hyperosmolarity and immunologic deficits. These conditions include diabetes mellitus (310 to 330 mosmol/kg H20) (23), uremia (317.60 ± 6.29 mosmol/kg H20) (24), dehydration after exercise (305 ± 1 mosmol/kg H20) (25), heat stroke (296.5 ± 1.0 mosmol/kg H20) (26), and fatal burns (312 ± 22.1 mosmol/kg H20) (27). All of these conditions are associated with an altered immune response and cytokine secretion (23, 28-33). In addition, inflamed joints, intestine, and cornea are also sites of altered osmolarity (34-37). These findings indicate that an active response to osmotic stress is itself an essential component of the immune system.
Recent studies have confirmed that expression of nfat5 is highly induced in several tissues and cells upon their exposure to osmotic stress (12-14, 38) and that nfat5 is expressed in the thymus and the spleen (21, 38, 39). The tissue osmolarity of these organs is normally higher than that of serum (an increase of ~20 to 30 mosmol/kg H20) (38). Heterozygotic inactivation of the nfat5 allele in mice causes a marked reduction in the cellularity of the thymus and the spleen (38). These two observations indicate that expression of nfat5 is induced by physiologic hyperosmolarity and suggest that NFAT5 plays an essential role in normal lymphocyte proliferation in the thymus and spleen.
Rho-type small G proteins, specifically RhoA, Cdc42, and Rac1, act as second messengers of osmotic stress (3, 40). They also play important roles in reorganization of the cytoskeleton, embryonic development, and regulation of gene expression (40-43). These molecules exist in active GTP-bound and inactive guanosine diphosphate (GDP)–bound forms (41, 42) and activate downstream effector molecules through physical interactions (41). The guanine nucleotide exchange factors (GEFs) play essential roles through their activation of small G proteins in response to upstream stimuli and impart specificity to the response through their interactions with downstream effector molecules (44, 45). Many Rho-specific GEFs have been cloned (44, 45). We previously used the ligand-binding domain of the retinoic X receptor β as bait in an expression cloning strategy to identify a 1429-residue GEF called Brx [also known as protein kinase A–anchoring protein 13 (AKAP13) and AKAP-Lbc] (46). In addition to acting as a Rho family GEF, Brx also binds to nuclear hormone receptors through its C-terminal nuclear receptor–interacting domain (NRID) and enhances the transcriptional activity of estrogen receptor α (ERα) and ERβ and the glucocorticoid receptor (46-48). AKAP-Brx (Lbc), a larger splice variant of Brx with an additional 1389 amino acid residues, was subsequently reported (49). This protein has an N-terminal cyclic adenosine monophosphate (cAMP)–dependent protein kinase (PKA)–docking domain and a full Brx-type GEF domain and NRID in its C-terminal half (49). The presence of an NRID, or a PKA-docking domain, or both in Brx and AKAP-Brx suggests that splice variants of this GEF act as integrators or docking platforms of multiple signal transduction pathways, including those mediated by nuclear hormone receptors and PKA. These molecules orchestrate such independent signals through shared signaling cascades to effect distinct biologic actions.
In this report, we examined the role of Brx in the response of the immune system to osmotic stress, which is known to be mediated by small G proteins, and elucidated an intracellular signaling cascade that transmits the extracellular hyperosmolarity signal to the nucleus in immune cells. In mouse primary splenocytes and human Jurkat cells (a CD4+ T cell line), Brx activated specific small G proteins through its GEF domain, attracted an intermediate scaffold protein, c-Jun N-terminal kinase (JNK)–interacting protein 4 (JIP4), and stimulated the p38 MAPK cascade. Brx was required for inducing the expression of nfat5, the gene encoding the cytokine B cell–activating factor (BAFF), and downstream hyperosmolarity-responsive genes. These results reveal a role for Brx as an essential component in the immune response to osmotic stress in addition to its role in the adaptation of immune cells to protect themselves in hyperosmotic environments.
RESULTS
Mice haploinsufficient for brx have smaller spleens, fewer splenocytes, and altered follicular structure compared with wild-type mice
During our initial analysis of brx haploinsufficient (brx+/−) mice, we noticed that their spleens were smaller than those of wild-type (WT) mice. The spleens of both male and female brx+/− mice weighed ~20% less than those of age-, sex-, and weight-matched WT mice, consistent with our initial observations (Fig. 1A, left panel). Splenocytes obtained from brx+/−mice were also ~30% fewer in number than those of WT mice (Fig. 1A, right panel). As revealed by histological analysis, brx+/− mice had smaller splenic follicles than did WT mice, whereas there was no obvious alteration in the distribution of white or red pulp in their spleens (Fig. 1B). These results suggest that Brx plays a role in the development of splenic follicles and, thus, that brx+/− mice have defective immune function.
Fig. 1.
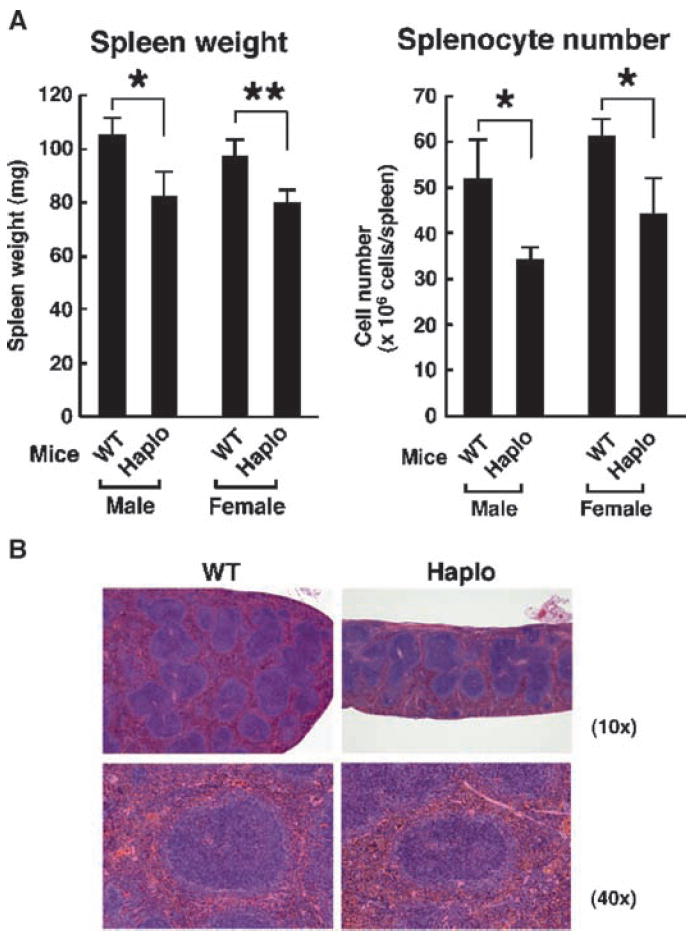
Characteristics of spleens from brx+/− mice. (A) The spleens from brx+/− (Haplo) mice weighed less and contained fewer splenocytes compared with those of WT mice. Data in the left panel represent the mean spleen weight, with error bars indicating the SEM (Haplo mice: male, n = 5; female, n = 7; WT mice: male, n = 5; female, n = 7), whereas those in the right panel represent the mean ± SEM of splenic mononuclear cells (Haplo mice: male, n = 5; female, n = 7; WT mice: male, n = 5; female, n = 7). *P < 0.05; **P < 0.01. (B) brx+/− mice exhibited an altered splenic follicular structure compared with that of WT mice. Results of hematoxylin and eosin (H&E) staining are shown and are representative of three experiments. The spleens of brx+/− mice had smaller follicles than did those of WT mice. Top and bottom panels show low- (10×) and high-magnification (40×) images, respectively.
Brx is required for the expression of nfat5 in the spleen
To better understand the molecular mechanisms underlying the defects observed in the spleens of brx+/− mice, we performed microarray analyses with total RNA purified from the spleens of brx+/− and WT mice. Among ~300 differentially expressed genes, nfat5 was the gene whose expression was most down-regulated in brx+/− compared with WT mice; expression of nfat5 in brx+/− mice was ~75% lower than that in WT mice (P = 0.047). In the initial microarray analyses, expression of messenger RNA (mRNA) of the taurine transporter (TauT), a well-known, osmotic stress–responsive, NFAT5 target gene, was less abundant in the spleens of brx+/− mice compared with that in WT mice (50). Because most osmotic stress–responsive genes were not represented on the array chip that we used, we measured the abundance of mRNAs of a panel of these genes in a reconstituted real-time, reverse transcription polymerase chain reaction (RT-PCR) assay. These experiments revealed that many of these genes, including those encoding AR, BGT1, SMIT, and TNF-α in addition to TauT, were less expressed in the spleens of brx+/− mice than in the spleens of WT mice (Table 1) (50, 51). Other genes that were expressed less in brx+/− mice included those encoding IL-17 receptor E, TNF-α receptor–associated factor 4 (TRAF4), the inositol 1,4,5-triphosphate receptor 1, the RAS p21 protein activator 2, various regulators of transcription and translation, and several other transcripts with unknown function.
Table 1.
Relative abundance of mRNAs of NFAT5-responsive genes in the spleens of brx+/− mice compared with those of WT mice.
| Gene | Relative mRNA abundance (mean ± SEM) |
|---|---|
| tnf-α | 0.51 ± 0.030** |
| ar | 0.70 ± 0.026** |
| taut | 0.57 ± 0.022* |
| smit | 0.39 ± 0.082* |
| bgt1 | 0.43 ± 0.054** |
| ata2 | 0.84 ± 0.200 |
| osp94 | 1.25 ± 0.182 |
| aqp2 | 0.75 ± 0.026 |
P < 0.05.
P < 0.01 (n = 4 mice of each type).
Because Brx activates members of the Rho family of small G proteins and because these proteins play an important but incompletely defined role in osmotic stress, we examined the abundance of NFAT5 and Brx mRNA and protein in splenocytes from brx+/− mice. The abundances of NFAT5 and Brx proteins were markedly reduced in brx+/−mice compared with that in WT mice (Fig. 2). As expected, the abundance of brx mRNA in brx+/− mice was ~50% less than that in WT mice. Consistent with the microarray assays, the abundance of nfat5 mRNA in splenocytes from brx+/− mice was ~25% of that of WT mice (Fig. 2B). Because the abundances of NFAT5 mRNA and protein in the brx+/− mice were greatly reduced, it appeared that both alleles of brx were required for the normal expression of nfat5 in splenocytes. In the renal medulla of brx+/− mice, the abundance of nfat5 mRNA was reduced compared with that in WT mice (Fig. 2C), suggesting that Brx is also required for optimal nfat5 expression in these cells, which are continuously exposed to hyperosmolar urine (1). In contrast to that of NFAT5, the abundances of mRNAs of NFAT1 to NFAT4 and of the NF-κB components RelA (p65) and p105 (p50) were not altered in the spleens of brx+/− mice (Table 2). Thus, Brx selectively regulates the expression of nfat5 among the transcription factors that harbor the Rel homology domain.
Fig. 2.
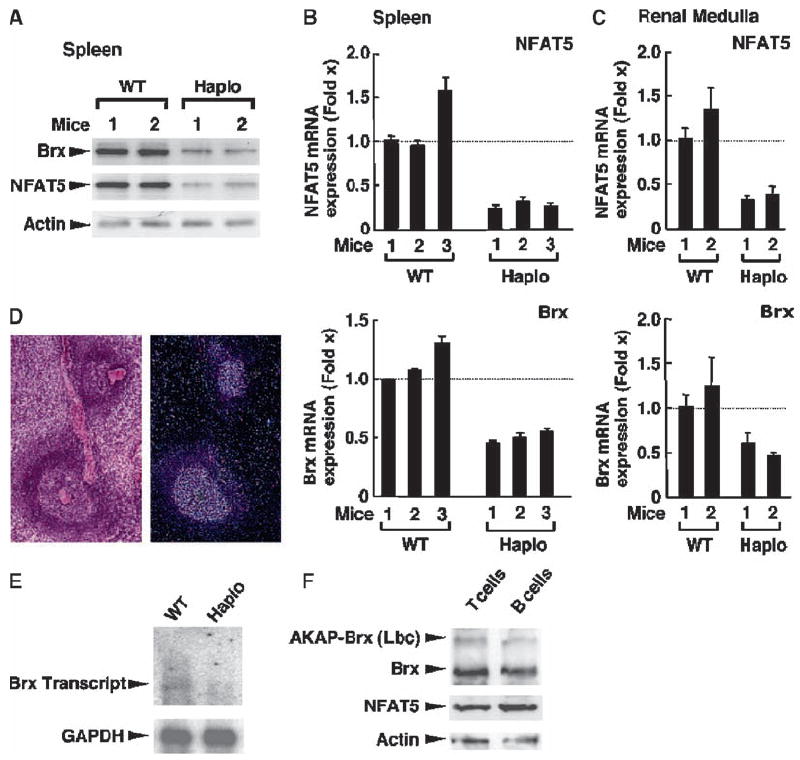
Splenocytes from brx+/− mice have a lower abundance of Brx and NFAT5 proteins and mRNAs compared with those of WT mice. (A) Western blotting analysis showed decreased abundance of Brx and NFAT5 proteins in splenocytes from brx+/− (Haplo) mice compared with those of WT mice. The abundance of actin was used as a loading control. Results shown are from two mice from each group. (B) Spleens from brx+/− mice expressed decreased amounts of brx and nfat5 mRNAs compared with those of WT mice. The abundances of brx and nfat5 mRNAs in the splenocytes of three WT and three brx+/− mice (each measured in triplicate) were evaluated with a SYBR Green–based real-time RT-PCR assay. Data shown represent the mean relative abundance of nfat5 (top) and brx (bottom) mRNAs shown as the fold difference compared with baseline (set by WT mouse 1). Error bars indicate SEM. (C) The renal medulla of brx+/− mice expressed less nfat5 mRNA compared with that of WT mice. The abundances of brx and nfat5 mRNAs in the renal medulla of two WT and two brx+/− mice were evaluated (each in triplicate) by real-time RT-PCR. Data shown represent the mean relative abundance of nfat5 mRNA (top) and brx mRNA (bottom) shown as the fold difference compared with baseline (set by WT mouse 1). Error bars indicate SEM. (D) Transcripts of brx are expressed in the white pulp of the spleen. Results of H&E staining of a section of spleen from a WT mouse (left) are representative of three experiments. In situ hybridization of another section from the same spleen shows the expression of brx transcripts (right). (E) The abundance of brx mRNA is reduced in the spleen of brx+/− mice compared with that in the spleen of WT mice. Purified mRNA from the spleens of WT and brx+/− mice were analyzed by Northern blotting for the expression of brx and gapdh mRNAs. (F) Brx is the major protein expressed from brx (akap13), whereas NFAT5 is equally abundant in T cells and B cells purified from the spleens of WT mice. Whole-cell homogenates from T cells and B cells obtained from WT splenocytes were resolved by 4% SDS-PAGE, blotted onto nitrocellulose membranes, and Brx, AKAP-Brx, NFAT5, and actin proteins were visualized by Western blotting analysis with anti-Brx, anti-NFAT5, or anti-actin antibodies, respectively. Experiments presented in (D) to (F) were repeated three times and representative images are shown.
Table 2.
Comparative expression of mRNAs of the NFATs and of NF-κB components in the spleens of brx+/− mice and WT mice.
| Gene | Relative mRNA expression (mean ± SEM) |
|---|---|
| nfat1 | 0.83 ± 0.957 |
| nfat2 | 0.99 ± 0.779 |
| nfat3 | 0.84 ± 0.105 |
| nfat14 | 1.11 ± 0.199 |
| nfat5 | 0.23 ± 0.066* |
| rela (p65) | 1.10 ± 0.146 |
| p105 (p50) | 1.08 ± 0.199 |
P < 0.01 (n = 4 mice of each type).
We next characterized the expression of brx in the spleen. In situ hybridization assays showed that brx transcripts were restricted to the white pulp of the WT spleen, where development, activation, and differentiation of B cells takes place (52, 53) (Fig. 2D). Northern blot analysis showed that a 6.7-kb brx transcript was present in the spleens of WT mice, whereas the larger transcript from akap13, which encodes AKAP-Brx, was almost undetectable (Fig. 2E). In comparison, WT mice expressed greater amounts of the 6.7-kb brx transcript. Brx and AKAP-Brx proteins were present in T cells and B cells from the spleens of WT mice, but Brx was the more abundant protein, consistent with the Northern analysis. The abundance of nfat5 mRNA was similar in T cells and B cells (Fig. 2F). Thus, among the Rho-type GEFs encoded by akap13, Brx, but not AKAP-Brx, was the most abundant protein found in the spleen.
brx+/− mice have altered splenocyte subpopulations, reduced production of immunoglobulin, and decreased amounts of circulating BAFF
Because the number of splenic cells in brx+/− mice was reduced compared with that in WT mice, we used flow cytometry to compare splenocytes from both types of mice (Fig. 3). The ratios of CD3+ T cells to CD19+ B cells (Fig. 3A) and CD4+ T cells to CD8+ T cells (Fig. 3B) were similar in brx+/− and WT mice. The B cell population, which contains cells expressing B220 or immunoglobulin M (IgM) as well as CD19, exhibited normal development in the spleens of brx+/− mice (Fig. 3, C and D); however, the proportion of mature, CD21+ transitional type 2 (T2) B cells was reduced compared with that in WT mice (Fig. 3E), whereas their precursors, T1 B cells, were reciprocally increased (Fig. 3F). These results suggest that the differentiation of cells of the B cell lineage was compromised in the spleens of brx+/− mice at the transition from T1 to T2 B cells, although overall development of B cells in the bone marrow appeared to be preserved.
Fig. 3.
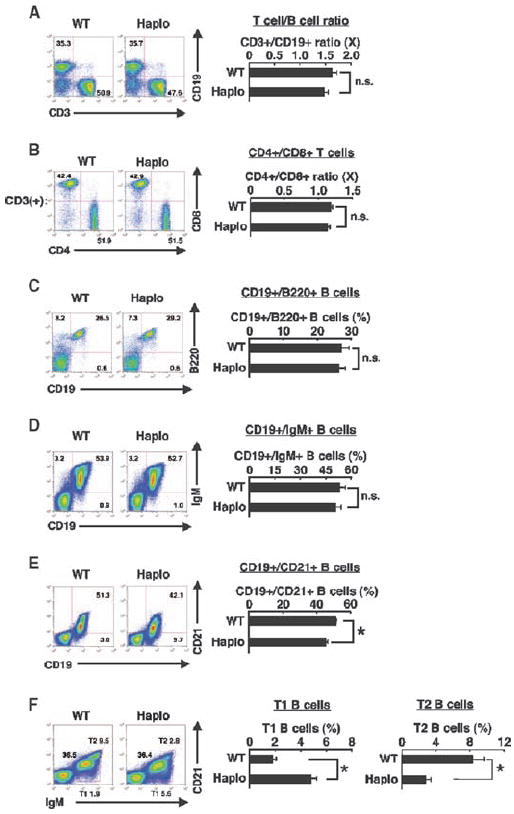
Flow cytometric analysis of subpopulations of splenocytes. Comparative analysis of splenocytes from WT and brx+/− (Haplo) mice based on their surface expression of (A) CD3 and CD19, (B) CD4 and CD8 (on CD3+ cells), (C) CD19 and B220, (D) CD19 and IgM, (E) CD19 and CD21, and (F) IgM and CD21. Representative flow cytometry experiments are shown in the left panels and pooled data from at least three independent experiments are shown in the right panels. Values shown are the means ± SEM of the ratios of the indicated cell populations when comparing WT to brx+/− mice. *P < 0.05; n.s., not significant (n = 3 to 5 mice).
These experiments showed that brx was highly expressed in the white pulp of the spleens of WT mice and that brx+/− mice exhibited altered follicular structure as well as defects in terminal B cell differentiation in the spleen. Because all these components of the spleen participate in the production of immunoglobulin (52, 53), we tested the effect of brx haplo-insufficiency on antigen-specific IgG1 production in response to immunization of brx+/− mice with ovalbumin (Fig. 4A). brx+/− mice produced reduced quantities of IgG1 compared with WT mice, suggesting that brx+/− mice had defective acquired humoral immunity. We suspected that BAFF might be involved because BAFF is necessary for immunoglobulin production from activated B cells and for the differentiation of B cells (especially from T1 to T2 B cells) (53-55). The abundance of baff mRNA in the spleen and serum concentrations of BAFF protein were substantially reduced in brx+/− mice compared with those of WT mice (Fig. 4B). Notably, the promoter region of murine baff (GenBank accession number EF100119) contains six putative NFAT5-response elements. Collectively, these results are consistent with the conclusion (see also below) that baff may be an NFAT5-responsive gene located downstream of the Brx-mediated signaling cascade, likely contributing to both the reduced production of immunoglobulin and the alteration of B cell differentiation in the spleen observed in brx+/− mice.
Fig. 4.
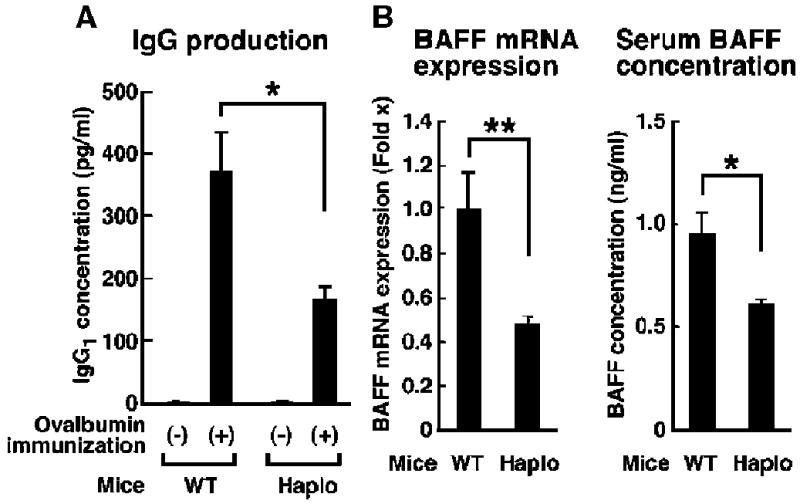
brx+/− mice exhibit impaired humoral immunity. (A) In response to immunization with ovalbumin, brx+/− mice produced lower quantities of ovalbumin-specific IgG1 than did WT mice. Results shown represent the mean serum concentrations ± SEM of ovalbumin-specific IgG1 in six mice from each group. *P < 0.05 (n = 6 mice). (B) brx+/− mice exhibited lower quantities of baff mRNA in the spleen (left) and a lower concentration of circulating BAFF (right) than did WT mice. Data shown indicate the mean fold difference ± SEM in the abundance of baff mRNA in the spleens of WT and brx+/− mice (left) and in the serum concentrations of BAFF (right) in four mice from each group. *P < 0.05; **P < 0.02.
Brx is required for osmotically induced expression of nfat5 in splenocytes and Jurkat cells
To further characterize defective splenocyte function, we exposed splenocytes to osmotic stress. Because hyperosmotic stimuli induce nfat5 expression in vivo and in vitro (1, 40), we evaluated the expression of nfat5 mRNA in splenocytes exposed to increasing concentrations of sodium chloride (NaCl), which induces hyperosmolarity (Fig. 5A). Incubation with 100 mM NaCl markedly induced nfat5 expression in splenocytes from WT mice, whereas this response to osmotic stress was blunted in cells obtained from brx+/− mice (Fig. 5A, left panel). The abundance of brx transcript in brx+/− mice was ~50% lower than that of WT mice and was not altered by incubation with hyperosmolar concentrations of NaCl (Fig. 5A, right panel). In the same samples, NaCl stimulated the expression of osmotic stress–responsive NFAT5 target genes, including those encoding AR, TauT, SMIT, and BGT1 in splenocytes from WT mice, whereas their expression was blunted in NaCl-treated splenocytes from brx+/− mice (Table 3). Although NaCl strongly induced the expression of nfat5, it did not affect the abundance of mRNAs of any of the other NFAT family members or of the NF-κB components, RelA (p65) and p105 (p50), in splenocytes from WT mice (Table 4). α-Amanitin, an inhibitor of RNA polymerase II (56), abolished the NaCl-induced increase in the expression of nfat5 mRNA in splenocytes from WT mice (Fig. 5B), indicating that osmotic stress activated transcription of the nfat5. Because the expression of baff mRNA in the spleen and the concentration of BAFF protein in the serum were reduced in brx+/− mice compared with those in WT mice, we examined the ability of splenocytes from these animals to secrete BAFF (Fig. 5C). Incubation of splenocytes from WT mice with hyperosmolar concentrations of NaCl resulted in increased concentrations of BAFF in the medium compared with that of untreated cells, but this increase was abrogated in splenocytes from brx+/− mice. NaCl also stimulated the expression of baff mRNA in splenocytes from WT mice, while its effect was significantly blunted in cells obtained from brx+/− cells (Fig. 5A, middle panel). These results support the conclusion that baff is an osmotic stress–responsive gene, which is possibly regulated by NFAT5 through Brx.
Fig. 5.
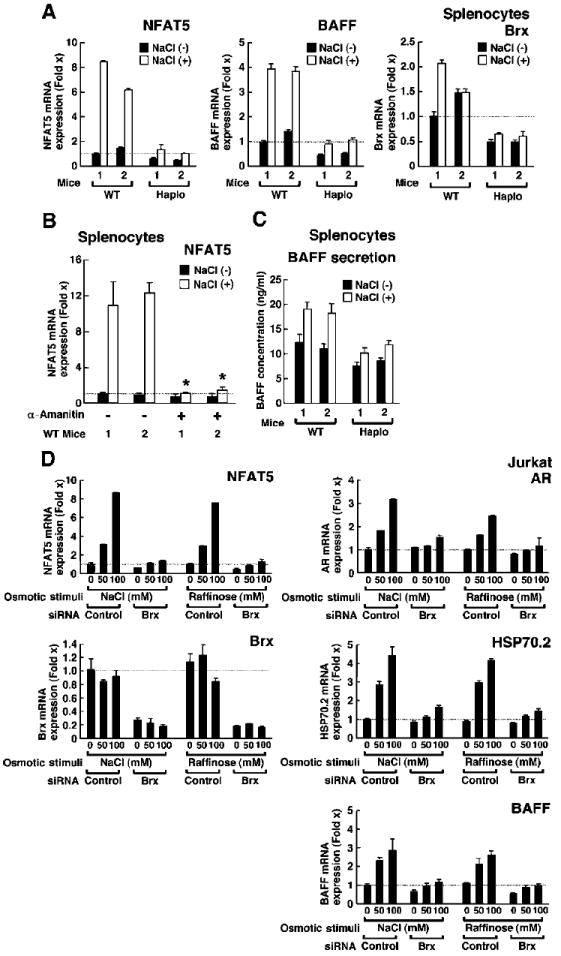
Brx is required for the increased expression of nfat5 and of endogenous NFAT5-responsive genes in response to osmotic stress. (A) Induction of the expression of nfat5 and baff mRNAs by osmotic stimulus was blunted in splenocytes from brx+/− (Haplo) mice compared with that in WT mice. Splenocytes from two WT and two brx+/− mice were incubated in the presence (open bars) or absence (filled bars) of 100 mM NaCl and the abundances of nfat5 mRNA (left), baff mRNA (middle), and brx mRNA (right) were determined by real-time RT-PCR analysis. Data shown represent the relative mean abundance ± SEM of nfat5, baff, and brx mRNAs shown as the fold induction over baseline (set by WT mouse 1 in the absence of NaCl). (B) Osmotic stimulus increases the abundance of nfat5 mRNA by stimulating its transcription in splenocytes. Triplicate samples of splenocytes from two WT mice were incubated with 100 mM NaCl in the absence or presence of 2 μM α-amanitin. Data shown represent the mean fold difference ± SEM in the abundance of nfat5 mRNA compared with baseline (set by splenocytes of WT mouse 1 cultured in the absence of NaCl and α-amanitin). *P < 0.01 (n = 3). (C) Secretion of BAFF is increased by an osmotic stimulus and is blunted in splenocytes from brx+/− mice compared with those from WT mice. Splenocytes obtained from two WT and two brx+/− mice were incubated in the presence (open bars) or absence (filled bars) of 100 mM NaCl and the concentration of BAFF secreted into the culture media was determined by ELISA (each mouse assayed in triplicate). Data shown represent the mean concentrations ± SEM of BAFF. (D) Brx is necessary for osmotic stimuli to induce the expression of nfat5 and of ar, hsp70-2, and baff mRNAs in Jurkat cells. Jurkat cells were transfected with control or Brx-specific siRNAs and incubated with the indicated concentrations of NaCl or raffinose. Data shown represent the mean fold difference ± SEM in the expression of nfat5, ar, hsp70-2, baff, and brx mRNAs compared with baseline (which was obtained from cells transfected with control siRNA and incubated in the absence of NaCl).
Table 3.
Induction of the expression of NFAT5-responsive genes in the splenocytes of WT and brx+/− mice by 100 mM NaCl.
| Gene | Fold induction of mRNA (mean ± SEM) | |
|---|---|---|
| WT splenocytes | brx+/− splenocytes | |
| ar | 6.55 ± 0.120 | 2.15 ± 0.151** |
| taut | 8.29 ± 0.784 | 1.96 ± 0.182** |
| smit | 3.84 ± 0.452 | 1.46 ± 0.223* |
| bgt1 | 3.41 ± 0.333 | 1.63 ± 0.142* |
P < 0.05.
P < 0.01 (n = 2 mice and measured in duplicate for each mouse).
Table 4.
NaCl-mediated expression of NFAT and NF-κB mRNAs in splenocytes of WT mice.
| Gene | Fold Induction in mRNA (mean ± SEM) |
|---|---|
| nfat1 | 1.06 ± 0.260 |
| nfat2 | 1.22 ± 0.344 |
| nfat3 | 0.89 ± 0.079 |
| nfat4 | 0.83 ± 0.141 |
| nfat5 | 7.56 ± 0.294* |
| rela (p65) | 1.43 ± 0.405 |
| p105 (p50) | 1.34 ± 0.324 |
P < 0.01 (measured in triplicate).
To determine whether the osmosensing signaling complex was specific to B cells, or whether it might also be present in T cells, we performed experiments with Jurkat cells by transfecting them with small interfering RNA (siRNA) specific for Brx and incubating them with increasing concentrations of NaCl or raffinose, a trisaccharide composed of galactose, fructose, and glucose (Fig. 5D). Brx-specific siRNA substantially reduced the abundance of brx mRNA both in the presence and in the absence of NaCl or raffinose (Fig. 5D, bottom left panel). These osmolytes stimulated nfat5 expression in Jurkat cells in the absence of Brx-specific siRNA, but this response to osmotic stress was nearly abolished by transfection of Brx-specific siRNA (Fig. 5D, top left panel). To test the possibility of an off-target effect by the Brx-specific siRNA, we examined two additional Brx-targeting siRNAs with different sequences and confirmed our previous results (fig. S1), indicating that knockdown of Brx by siRNA specifically resulted in the reduced expression of nfat5 mRNA.
Because NFAT5 stimulates the expression of the genes encoding heat shock protein 70-2 (HSP70-2) and AR by directly binding to their promoter regions (50, 51), we measured the expression of these genes in response to osmotic stress. We found that increasing concentrations of NaCl or raffinose strongly stimulated the expression of hsp70-2 and ar mRNAs, whereas induction of their expression was nearly abolished in the presence of Brx-specific siRNA (Fig. 5D, right top two panels). Although T cells are not a major source of BAFF, we also examined baff expression in Jurkat cells. Both NaCl and raffinose mildly stimulated the expression of baff mRNA compared with that in untreated cells, and Brx-specific siRNA markedly blocked this effect (Fig. 5D, right bottom panel). The expression of baff mRNA was also completely attenuated by siRNA-mediated knockdown of NFAT5 in Jurkat cells (Fig. 6A), indicating that BAFF is an osmotic stress–responsive gene downstream of NFAT5. We also found that NaCl stimulated the production of NFAT5 protein in a dose-dependent fashion, whereas transfection of Brx-specific siRNA efficiently suppressed the production of NFAT5 by hyperosmolar NaCl (Fig. 6B). Further, α-amanitin completely abolished the expression of nfat5 mRNA that was stimulated by hyperosmolar NaCl in Jurkat cells (Fig. 6C). Together, these results support the conclusion that Brx is essential and required for stimulation of nfat5 expression by osmotic stress and for NFAT5-mediated activation of endogenous genes in both primary splenocytes and Jurkat cells.
Fig. 6.
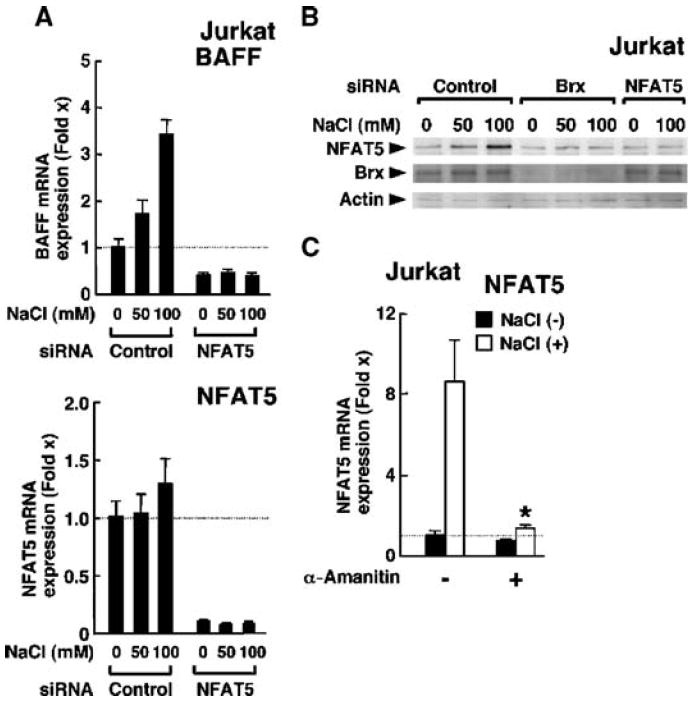
(A) Osmotic stimulus increases the expression of baff mRNA through the induction of nfat5 in Jurkat cells. Jurkat cells were transfected with control or NFAT5-specific siRNA and cultured in the indicated concentrations of NaCl. Data shown represent the mean fold difference ± SEM (n = 3) in the abundance of baff (top) and nfat5 (bottom) mRNAs compared with baseline (the value obtained from cells transfected with control siRNA and incubated in the absence of NaCl). (B) Brx is required for optimal production of NFAT5 protein in Jurkat cells in response to osmotic stimulus. Jurkat cells were transfected with control, Brx-specific siRNA, or NFAT5-specific siRNA and incubated with the indicated concentrations of NaCl. Cell homogenates were resolved by 4 to 20% SDS-PAGE and the abundances of NFAT5 (top gel), Brx (middle gel), and actin (bottom gel) were determined by Western blotting with their respective specific antibodies. Experiments were repeated three times and a representative image is shown. (C) Osmotic stimulus increases the expression of nfat5 in Jurkat cells. Jurkat cells were incubated with 100 mM NaCl in the absence or presence of 2 μM α-amanitin. Data shown represent the mean fold difference ± SEM in the abundance of nfat5 mRNA compared with baseline (the value obtained from Jurkat cells cultured in the absence of NaCl and α-amanitin). *P < 0.01 (the experiment was performed in triplicate).
Vav1 is another GEF that plays an important role in T cell development and activation through T cell receptor (TCR)–mediated activation of extracellular signal–regulated kinase (ERK) and JNK MAPKs (57). For this reason, we examined whether Vav1 could mediate osmotic stress–induced increases in nfat5 mRNA expression in Jurkat cells (Fig. 7A). Transfection of Brx- or Vav1-specific siRNAs efficiently suppressed expression of their respective transcripts (Fig. 7A, right two panels). Consistent with our prior findings, Brx-specific siRNA greatly attenuated NaCl-induced expression of nfat5 mRNA, but Vav1-specific siRNA had no such effect (Fig. 7A, left panel). Thus, Brx, but not Vav1, mediated the response to osmotic stress to stimulate nfat5 expression in these cells. These results suggest that Brx and Vav1 function in distinct signaling pathways and that regions in Brx other than the GEF domain are likely required for lymphocytes to respond properly to osmotic stressors.
Fig. 7.
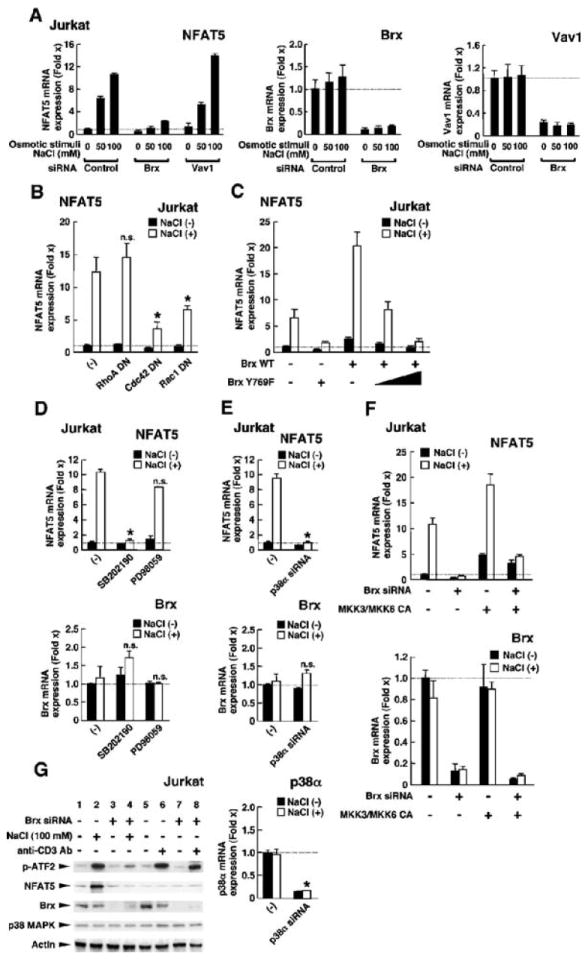
The GEF domain of Brx, as well as Cdc42, Rac1, and p38 MAPK, are all required for an osmotic stimulus to induce the expression of nfat5 mRNA. (A) Brx, but not Vav1, is necessary for the induction of nfat5 expression in Jurkat cells in response to osmotic stimuli. Jurkat cells were transfected with control, Brx-specific siRNA, or Vav1-specific siRNA and incubated with the indicated concentrations of NaCl. Data shown represent the mean fold difference ± SEM in the relative abundance of nfat5 (left), brx (middle), and vav1 (right) mRNAs compared with baseline (the value obtained from the cells transfected with control siRNA and incubated in the absence of NaCl). (B) DN mutants of Cdc42 and Rac1, but not RhoA, suppress induction of nfat5 expression in Jurkat cells in response to osmotic stimuli. Jurkat cells were transfected with plasmids expressing DN mutants of RhoA, Cdc42, or Rac1 (RhoAT19N, Cdc42T17N, and Rac1T17N, respectively) and incubated with 100 mM NaCl. Data shown represent the mean fold difference ± SEM in the relative abundance of nfat5 mRNA compared with baseline (the value obtained from cells transfected with control plasmid and incubated in the absence of NaCl). *P < 0.01; n.s., not significant (the experiment was peformed in triplicate). (C) Induction of the expression of nfat5 mRNA in Jurkat cells in response to an osmotic stimulus requires the GEF domain of Brx, whereas a GEF-defective mutant Brx acts in a DN manner. Jurkat cells were transfected with a plasmid encoding WT Brx or the GEF-inactive mutant Brx and incubated with 100 mM NaCl. Data shown represent the mean fold difference ± SEM in the relative abundance of nfat5 mRNA compared with baseline (the value obtained from cells transfected with control plasmid and incubated in the absence of NaCl). (D) p38 MAPK, but not ERK1/2, is necessary for an osmotic stimulus to induce the expression of nfat5 mRNA in Jurkat cells. Jurkat cells were incubated with 2 μM of the p38 MAPK inhibitor SB202190 or 10 μM of the MEK1 inhibitor PD98059 in the absence or presence of 100 mM NaCl. Data shown represent the mean fold difference ± SEM in the relative abundance of nfat5 (top) and brx (bottom) mRNAs compared with baseline. *P < 0.01; n.s.: not significant (the experiment was performed in triplicate). (E) siRNA against p38α abolishes osmotic stimulus–induced expression of nfat5 mRNA. Jurkat cells were transfected with control or p38α-specific siRNA and cultured in the absence or presence of 100 mM NaCl. Data shown represent the mean fold difference ± SEM in the relative abundance of nfat5 (top), brx (center), and p38α (bottom) mRNAs compared with baseline (cells transfected with control siRNA and cultured in the absence of NaCl). *P < 0.01; n.s., not significant (the experiment was performed in triplicate). (F) Expression of the CA forms of MKK3 and MKK6 rescues the attenuation of osmotic stimulus–induced nfat5 expression caused by siRNA-mediated knockdown of Brx. Jurkat cells were transfected with control or Brx-specific siRNA in the absence or presence of MKK3- or MKK6-expressing plasmids and were cultured with or without 100 mM NaCl. Data shown represent the mean fold difference ± SEM in the relative expression of nfat5 (top) and brx (bottom) mRNAs compared with baseline (cells transfected with control siRNA and cultured in the absence of NaCl). (G) Osmotic stimulus activates p38 MAPK through Brx in Jurkat cells. Jurkat cells were transfected with control or Brx-specific siRNA and cultured in the absence or presence of 100 mM NaCl or 10 μg/ml of anti-CD3. The kinase activity of p38 MAPK was evaluated by examining the abundance of phosphorylated ATF2 by Western blotting (top gel). The abundances of NFAT5, Brx, and p38 MAPK proteins were also examined (second to fourth gels) by Western blotting. Actin (bottom gel) was used as a loading control.
Several Rho-type small G proteins play roles in responses to osmotic stress, such as stimulating the expression of genes that encode osmolytes, remodeling the cytoskeleton, and producing various cytokines (40, 58-60). To identify the specific small G proteins that mediate the hyperosmolar NaCl–induced increase in nfat5 mRNA expression in Jurkat cells, we overexpressed dominant-negative (DN) forms of Rho family members whose activities have been well characterized (41) (Fig. 7B). Overexpression of the DN RhoA mutant RhoA Thr19→Asn did not affect the NaCl-induced increase in nfat5 mRNA expression, whereas expression of DN mutants of Cdc42 or Rac1 (Cdc42 Thr19→ Asn and Rac1 Thr19→Asn, respectively) strongly suppressed the effects of NaCl on nfat5 expression, with the Cdc42 Thr19→Asn mutant showing the most pronounced effect. Thus, Cdc42, and possibly Rac1, are required for the induction of nfat5 mRNA expression by osmotic stress in Jurkat cells.
To determine whether the GEF domain of Brx was required for osmotic stress–induced expression of nfat5 mRNA in Jurkat cells (Fig. 7C), we compared the effect of NaCl on nfat5 expression in cells expressing WT Brx with that in cells expressing the Brx mutant Tyr769→Phe, a previously described GEF-defective mutant of Brx (48, 49). Of note, the Tyr769→Phe Brx mutant behaved in a DN manner and suppressed the endogenous response to osmotic stress as well as that in cells expressing WT Brx. These findings indicate that the GEF domain of Brx is necessary for osmotic stress-induced increases in the expression of nfat5. Additionally, because the Tyr769→Phe mutant Brx exhibited DN activity, these findings suggest that Brx has a functional domain(s) distinct from its GEF domain that participates in mediating osmotic stress–induced stimulation of the expression of nfat5, possibly through communication with other molecules necessary for this response.
The p38 MAPK scaffold protein JIP4 binds to Brx and is necessary for osmotic stress–induced expression of nfat5
Several studies indicate that p38 MAPK plays a role in the response to osmotic stress (1, 8, 9). Subsequently, p38 MAPK was shown to activate NFAT5 (13, 61) and increase the expression of nfat5 mRNA in several cell types (12, 14). We suspected that osmosensing by Brx might be mediated through p38 MAPK because we previously reported that Brx enhances the transcriptional activity of ERβ through the activation of p38 MAPK (47). Therefore, we tested the effect of the p38 MAPK inhibitor SB202190 on hyperosmolar NaCl–stimulated expression of nfat5 mRNA in Jurkat cells (Fig. 7D). Incubation of Jurkat cells with SB202190 completely abolished NaCl-induced expression of nfat5 mRNA, whereas the MEK1 inhibitor PD98059, which inhibits the activation of ERK1/2 by MEK1, showed no effect. Transfection of Jurkat cells with a p38α-specific siRNA strongly suppressed the expression of p38α mRNA and also attenuated NaCl-induced expression of nfat5 mRNA (Fig. 7E). Transfection of Jurkat cells with the constitutively active (CA) forms of MAPK kinase 3 (MKK3) and MKK6, upstream activators of p38 MAPK (62, 63), partially rescued the Brx-specific siRNA-mediated reduction of NaCl-induced expression of nfat5 mRNA (Fig. 7F). Further, incubation with 100 mM NaCl activated the kinase activity of p38 MAPK and increased the abundance of NFAT5 protein in Jurkat cells, whereas transfection of cells with a Brx-specific siRNA completely abolished these changes (Fig. 7G). Treatment of Jurkat cells with an antibody to CD3, which stimulates the TCR leading to the activation of p38 MAPK (64), did not increase the expression of nfat5, although it fully activated p38 MAPK. Similarly, siRNA-mediated knockdown of Brx did not influence anti-CD3–mediated activation of p38 MAPK (Fig. 7G). These results indicate that p38 MAPK is required in Jurkat cells for the expression of nfat5 in response to osmotic stress. Further, Brx may direct diverse actions of p38 MAPK to particular component(s) of its intracellular signaling cascades that lead to the specific induction of the expression of nfat5.
The Brx-mediated selective induction of nfat5 expression through the activity of p38 MAPK and the ability of a Brx mutant to function in a DN manner (Fig. 7C) suggested that Brx might function as a scaffold to recruit and interact with other proteins in the p38 MAPK–mediated osmosensing complex. To identify candidate proteins in the complex, we performed GAL4-based yeast two-hybrid screens with the C-terminal portion of Brx as bait. Among ~70 putative positive clones, we found one clone that contained the coding sequence of JIP4. JIP4 functions as a scaffold for the activation of the p38 MAPK cascade, interacting with p38 MAPK, MKK3, and MKK6 (65, 66). In contrast to other JIP family members, JIP4 does not activate JNK (65). The JIP family proteins are essential for determining the signaling specificity of the MAPK family of proteins and govern the activation of pathway components by recruiting them to particular subcellular sites or to specific targets (66-68).
Given the important role of JIP4 in the p38 MAPK cascade, we mapped the portion of JIP4 that physically interacted with Brx in a reconstituted yeast two-hybrid assay (Fig. 8A). We constructed plasmids expressing different portions of JIP4 based on knowledge of its functional domains (65). Brx(1042–1429), which spans the C-terminal third of Brx, interacted with full-length JIP4 and the fragment JIP4(251–751), suggesting that Brx physically interacted with JIP4 at the domain flanked by amino acid residues 251 to 751. This portion of JIP4 contains multiple coiled-coil structures (65), a structural motif known to support protein-protein interactions between molecules (69). Because Brx interacted with JIP4 through a domain located C-terminally to its GEF domain, the GEF domain appears to be dispensable for this interaction.
Fig. 8.
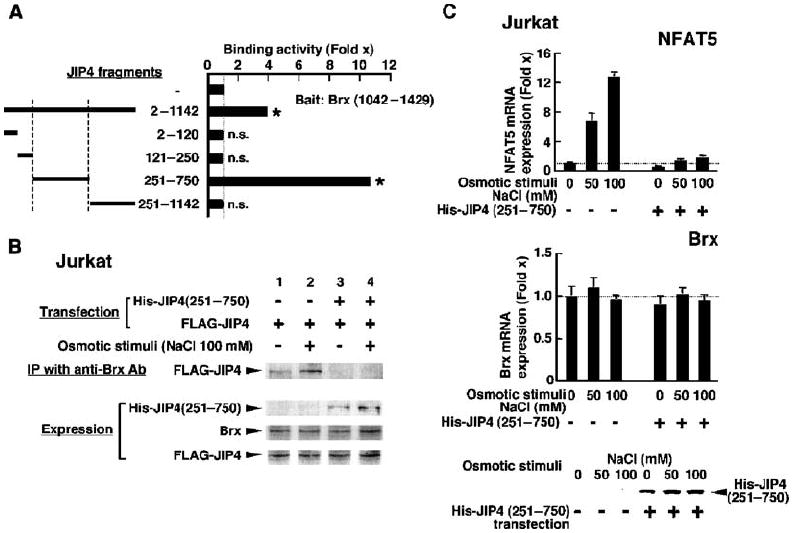
JIP4 is required for Brx to mediate osmotic stimulus–induced increases in the expression of nfat5 in Jurkat cells. (A) JIP4 interacts with Brx(1042–1429) through a region of JIP4 flanked by amino acid residues 251 to 750. Yeast two-hybrid assays were performed with plasmids expressing Brx(1042–1429) fused to the DBD of GAL4 and the indicated JIP4 fragment fused to the AD of GAL4. Data shown represent the mean fold increase in binding activity ± SEM obtained by dividing the corrected β-galactosidase obtained in the presence of the indicated JIP4-AD fragments by those obtained in its absence. *P < 0.01; n.s., not significant (the experiment was performed in triplicate). (B) Brx associates with JIP4 in Jurkat cells in an osmotic stimulus–dependent fashion and overexpression of JIP4(250–750) attenuates this association. Jurkat cells were transfected with a plasmid expressing FLAG-JIP4 with or without a plasmid expressing His-JIP4(251–750) and incubated with 100 mM NaCl. Coimmunoprecipitation was performed with anti-Brx, and Brx-associated FLAG-JIP4 was visualized with anti-FLAG. Exogenously or endogenously expressed His-JIP4(251–750), Brx, and FLAG-JIP4 were visualized by Western blotting of samples from 10% of the cell lysates used in the coimmunoprecipitation reaction (IP). (C) Overexpression of JIP4(251–750) attenuates osmotic stimulus–induced increases in the expression of nfat5 mRNA. Jurkat cells were transfected with a plasmid expressing His-JIP4(251–750) and incubated with the indicated concentrations of NaCl. Data shown represent the mean fold-difference ± SEM in the relative abundance of nfat5 (top) and brx (middle) mRNAs compared with baseline (from cells transfected with control siRNA and cultured in the absence of NaCl). His-JIP4(251–750) (bottom) was visualized in the whole-cell homogenates obtained in the same experiment by Western blotting with anti-His.
We next tested for an interaction between endogenous Brx and JIP4 proteins in Jurkat cells by coimmunoprecipitating JIP4 with an antibody to Brx (Fig. 8B). JIP4 was strongly coimmunoprecipitated with Brx in the presence of hyperosmolar NaCl, whereas overexpression of the JIP4(251–750) fragment, which harbors the Brx-binding site identified in the yeast two-hybrid assay, abolished the coimmunoprecipitation of JIP4 with Brx. Thus, JIP4 and Brx interacted with each other in an osmotic stimulus–dependent fashion through the mutual interaction domains identified in the yeast two-hybrid assay. Lastly, we examined the importance of JIP4 in the osmotic stimulus–induced increase in the expression of nfat5 mRNA in Jurkat cells (Fig. 8, C and D). Overexpression of the fragment JIP4(251–751), which inhibited the association between Brx and full-length JIP4 in the coimmunoprecipitation assay, strongly inhibited hyperosmolar NaCl–induced increases in the expression of nfat5 mRNA. These results indicate that recruitment of JIP4 is necessary for Brx to mediate osmotic stress–induced increases in the expression of nfat5 in these cells.
DISCUSSION
We found that brx+/− mice showed a marked reduction in the expression of nfat5 and osmotic stress–responsive genes in their spleens compared with those of WT mice. These mice showed mild alterations in follicular structure and contained fewer splenocytes than did WT mice, although their T cell/B cell, CD4+ T cell/CD8+ T cell, and B cell ratios were preserved. These features of brx+/− mice resemble those of nfat5+/− mice (38), suggesting that the phenotype observed in brx+/− mice is due in part to a reduction in nfat5 expression. These results indicate that Brx is required for basal nfat5 expression in the spleen. brx+/− mice showed reduced expression of nfat5 mRNA in the renal medulla, thus suggesting that Brx is necessary for the optimal expression of nfat5 in that tissue. Antigen-specific immunoglobulin production in response to ovalbumin injection was blunted in brx+/− mice compared with that in WT mice, possibly resulting from a reduction in the production of BAFF and the subsequent suppression of the differentiation of terminal B cells in the spleen. In reports by other investigators, T cells obtained from nfat5+/− mice, or from mice expressing a DN form of NFAT5, exhibited reduced proliferation under hyperosmotic culture conditions (38, 39) than did those of WT mice. Because the expression of nfat5 was markedly reduced in brx+/− mice, it is also possible that a dysfunction in T cells may have contributed to the reduced immunoglobulin production observed in these mice. Further, the defects in B cell differentiation that we have shown together with the reported dysfunction in T cells may cause the reduced numbers of splenocytes and the smaller spleens observed in brx+/− mice compared with those of WT mice.
At the cellular level, we found that Brx was necessary for osmotic stress–induced expression of nfat5 in mouse splenocytes and Jurkat cells. Splenocytes from brx+/− mice showed reduced expression of osmotic stress–responsive genes, and siRNA-mediated knockdown of Brx in Jurkat cells substantially blunted osmotic stress–induced expression of nfat5 mRNA and that of its downstream responsive genes ar, hsp70-2, and baff. Overexpression of DN forms of Cdc42 and Rac1, but not RhoA, strongly suppressed NaCl-induced expression of nfat5 mRNA, and the GEF domain of Brx was required for hyperosmolar NaCl to induce expression of nfat5. These results suggest that Brx efficiently activates these small G proteins through its GEF domain and thus stimulates the expression of nfat5 in response to osmotic stress (Fig. 9A).
Fig. 9.
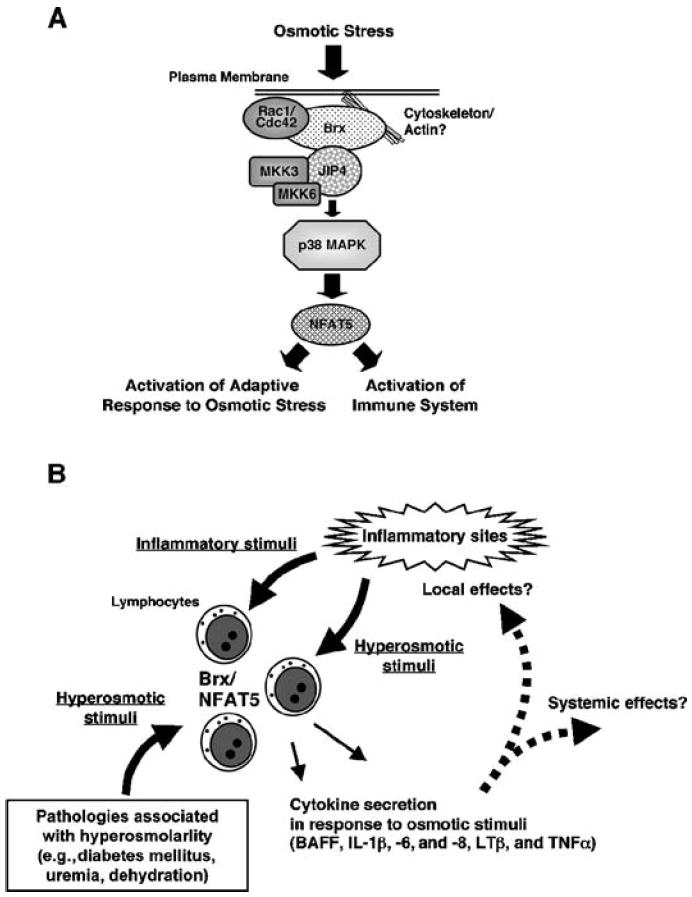
The Brx-mediated osmotic stress-activated signaling system and its implications for the immune response. (A) Intracellular signaling pathway of lymphocytes in response to osmotic stress. The sensing of and response to osmotic stress is largely mediated by Brx through the activation of Rho-type small G proteins and the subsequent stimulation of the p38 MAPK cascade and of NFAT5. NFAT5 participates in the elevation of intracellular osmolarity of immune cells and plays an essential role in the induction of various cytokines perhaps as a response to the hyperosmolar environment of inflammatory sites and several immune and other organs. (B) The Brx- and NFAT5-mediated response to osmotic stress in inflammatory sites and in pathologic conditions associated with extracellular hyperosmolarity. In addition to infectious agents, inflammatory cytokines, and other bioactive compounds released by inflamed tissues, inflammation activates lymphocytes by osmotic stress. The Brx- and NFAT5-mediated intracellular signaling system stimulates the secretion of cytokines to modulate local and systemic inflammatory reactions, while it induces hyperosmolarity-responsive genes to protect lymphocytes from the hyperosmolar environment of inflammatory sites. Pathologic conditions associated with extracellular hyperosmolarity may alter the various functions of lymphocytes through activation of Brx- and NFAT5-mediated signaling system.
We found that osmotic stress stimulated the expression of nfat5 by activating p38 MAPK, consistent with previous reports (12, 14). In agreement with this finding, Brx physically interacted with JIP4, a p38 MAPK–specific scaffold protein that serves as an activator of this kinase by communicating with its upstream kinases MKK3 and MKK6 (65). A JIP4 fragment consisting of amino acid residues 251 to 750 contained a domain involved in the physical interaction with Brx. Expression of this fragment abrogated the association between JIP4 and Brx and suppressed hyperosmolar NaCl–stimulated expression of nfat5. Thus, interaction with JIP4 was required for Brx to mediate osmotic stress–induced expression of nfat5. These results suggest that Brx is an essential protein in the immune system’s response to osmotic stress, activating small G proteins through its GEF domain and subsequently stimulating p38 MAPK by recruiting the scaffold protein JIP4. Further, because Brx specifically connected osmotic stress–stimulated p38 MAPK activity to expression of nfat5 but showed no effect on p38 MAPK activity induced by TCR-mediated activation of T cells, Brx may function as a molecular rectifier cooperating with JIP4, which directs the otherwise diverse actions of p38 MAPK into a specific cellular pathway for inducing the expression of nfat5 expression (70). Together, these studies reveal a role for Brx as a signal integrator in the osmotic stress–activating signaling pathway through small G proteins and p38 MAPK (Fig. 9A).
The signaling cascade(s) downstream of p38 MAPK that stimulate the transcription of the nfat5 is unknown. The ~1.5-kb promoter region of human nfat5 corresponding to the CpG island (GenBank accession number AF346509) contains two activating protein 1 (AP1)–binding sites and seven interferon-sensitive responsive elements (ISREs) upstream of the transcription initiation site, some of which are also conserved in the promoter region of murine nfat5 (GenBank accession number AF453571). p38 MAPK phosphorylates and activates c-Fos, a component of AP1, and the interferon regulatory factors (IRFs), which bind to AP1-binding sites and ISREs, respectively, and activate the transcription of their target genes (71, 72). Thus, it is possible that p38 MAPK phosphorylates these transcription factors to stimulate the expression of nfat5.
OSM, the osmosensing scaffold of MEKK3, is a small cytoplasmic protein that consists of 454 amino acid residues and mediates osmotic stress–induced activation of p38 MAPK in fibroblasts and kidney-derived epithelioid cells, such as the 293 and COS7 cell lines (8). OSM interacts with Rac and the kinases MEKK3 and MKK3, which are upstream of p38 MAPK, and has a domain similar to that of a number of adaptor and scaffold proteins, including JIPs, Disabled (Dab) proteins, and Numb proteins (8). Because the functions of OSM overlap to some extent with those of Brx, this scaffold protein might interact with Brx to mediate osmotic stress–induced activation of p38 MAPK and the subsequent induction of nfat5 expression, a possibility that remains to be tested.
Extracellular hyperosmolarity reduces cellular volume by causing the outward movement of intracellular water from cells; thus, a substantial number of reports indicate that osmotic stress activates the cellular adaptive response by directly influencing the cytoskeleton, which, in this case, functions as a sensor of the cell volume (40, 73-75). Rho-type small G proteins and their GEFs, on the other hand, are central players in the formation and remodeling of cytoskeletal organization (40). Cytokine- or TCR-mediated stimulation of lymphocytes dramatically alters cytoskeletal structure through the activation of GEFs, and such modification of the cytoskeleton is critical for their biological functions (57-59, 76). We previously found that the abundance of AKAP13 protein is substantially increased by mechanical stress in leiomyoma cells and is associated with actin fibers—pivotal components of the cytoskeleton (77). We also used yeast two-hybrid screens to show that the C-terminal portion of Brx interacts with many actin-associated molecules, such as filamin A and B, catenin α, and vimentin, in addition to JIP4. Because mechanical stress dramatically alters the cytoskeleton similarly to that by osmotic stress (78, 79), it is possible that Brx is activated by osmotic stress through modulation of the cytoskeleton (Fig. 9A). Alternatively, in response to osmotic stress, Brx might interact with as yet unidentified eukaryotic osmosensor molecules that are orthologous to the yeast cell-surface osmoreceptors Sln1 and Sho1 (3). Indeed, the yeast Rho-type GEFs Rom1p and Rom2p interact with the hyperosmolarity sensor Mid2 and activate Rho1 to stimulate the MAPK pathway, which leads to alteration of cell osmolarity and remodeling of the cytoskeleton (3, 80).
The sensing of and response to osmotic stress is essential for the maintenance of proper immune function (21, 38, 39). Brx-mediated induction of the expression of nfat5 on osmotic stress participates in the elevation of intracellular osmolarity and plays an essential role in the production of various cytokines as an adaptive response to the hyperosmolar environment of inflammatory sites and several immune and other organs (Fig. 9B). Because extracellular hyperosmolarity is associated with several pathologic conditions, such as diabetes mellitus and uremia, pharmacologic manipulation of the Brx-NFAT5 signaling system provides a potential means for treating various immunological complications associated with these diseases.
MATERIALS AND METHODS
Plasmids
pBK/RSV-FLAG–based plasmids expressing either WT Brx or the Tyr769→Phe mutant of Brx were described previously (48). pGBKT7-Brx(1042–1429), expressing Brx(1042–1429) fused to the DNA binding domain of GAL4, was constructed by subcloning the complementary DNA (cDNA) of the indicated Brx fragment into pGBKT7 (Clontech, Palo Alto, CA). pCDNA3-RhoAT19N and pCDNA3-Rac1T17N, which express DN forms of human RhoA and Rac1, respectively, were produced by PCR-assisted mutagenesis reactions with pCDNA3-RhoA and pCDNA3-Rac1 as templates (University of Missouri-Rolla cDNA Resource Center, Rolla, MO). pCDNA3-Cdc42N17N, which expresses a DN form of human Cdc42, was described previously (48). pCDNA3-FLAG-JIP4 was donated by R. J. Davis (Howard Hughes Medical Institute and University of Massachusetts Medical School, Worcester, MA) (65). pCDNA3.1His/A-JIP4(251–750) was created by subcloning the corresponding portion of JIP4 cDNA into pCDNA3.1His/A (Invitrogen, Carlsbad, CA). pGAD424-JIP4(2–1142), pGAD424-JIP4 (2–120), pGAD424-JIP4(121–250), pGAD424-JIP4(251–750), and pGAD424-JIP4(751–1142), which express the indicated fragments of JIP4 fused to the activation domain (AD) of GAL4, were produced by inserting the corresponding JIP4 cDNAs into pGAD424 (Clontech). pRc/RSV-FLAG-MKK3(glu) and pCDNA3-FLAG-MKK6(glu) were purchased from Addgene, Inc. (Cambridge, MA). These plasmids express the CA forms of human MKK3 or MKK6, respectively. MKK3(glu) contains Glu in place of Ser189 and Thr193, whereas MKK6(glu) contains Glu in place of Ser207 and Thr211 (62, 63). The parent plasmids pCDNA3 and pRc/RSV (Invitrogen) were used as negative controls in transfections.
Generation of brx+/− mice
Targeted disruption of brx was performed using standard homologous recombination by insertion of a Neor cassette resulting in elimination of the exon encoding amino acid residues 752 to 1044 of Brx (46, 48). These residues are required for GEF activity and are conserved throughout Rho GEF family members, as previously described (81). Details of the methods used for the targeted disruption of brx (akap13) and the detailed phenotyping of brx−/− mice will be presented elsewhere. Briefly, murine akap13 cDNA clones were isolated from a splenic cDNA library (Stratagene, La Jolla, CA) and used to identify a genomic fragment containing the GEF region of mAKAP13 from a bacterial artificial chromosome library (GenomeSystems Inc., St. Louis, MO). The targeting vector RMV1-pPNT was constructed by insertion of a 1.6-kb Not I–Xho I fragment into the 5′ arm and a 5.0-kb Eco RI–Eco RI fragment into the 3′ arm of murine akap13, thus replacing a critical exon in the region encoding the Dbl homology (DH) domain and 5.39 kb of flanking genomic sequence with the PGK-Neor cassette. The targeting vector was sequenced, linearized with Hind III, and electroporated into 129 embryonic stem (ES) cells. The targeted event on the 5′ end of the gene was confirmed by Southern blot hybridization of Bam HI–digested DNA and hybridization with a 32P-labeled 5′ probe. Targeting of the 3′ region was confirmed with overlapping primer pairs. Mice homozygous for the null brx allele did not express brx-related mRNAs or proteins, including any fragment forms, and died in utero (82). Mice haploinsufficient for the brx allele (brx+/−) appeared grossly normal. Genotypes of adult, embryonic, and neonatal mice were confirmed by analysis of Southern blots and by PCR. Three-week- to 2-month-old female mice were used in all experiments.
Preparation of splenocytes
Spleens were dissected from brx+/− and WT mice, crushed, and passed through nylon cell strainers (Becton Dickinson, Franklin Lakes, NJ). The obtained extracts were treated with ACK Lysing Buffer (Quality Biological, Inc., Gaithersburg, MD) to remove contaminating red blood cells and washed with RPMI 1640 medium three times; the remaining splenocytes were used for further analyses, as described previously (48, 83). T cells and B cells were further purified from splenocytes with the mouse pan T cell isolation kit and the mouse B cell isolation kit (Miltenyi Biotec Inc., Auburn, CA), respectively.
Microarray analyses
Total RNAs were purified from brx+/− and WT mice with the RNeasy Midi kit (QIAGEN Inc., Valencia, CA) and labeled with the 3DNA Array 9000 Expression Array Detection kit (Genisphere Inc., Hatfield, PA), which involves the hybridization of the reverse-transcribed cDNA and a fluorescent 3DNA reagent to the microarray. The fluorescent 3DNA reagent hybridizes to the cDNA through a “capture sequence” that is complementary to the 5′ end of the RT primer. The mouse 23K cDNA microarrays were made as described previously (84, 85). Briefly, the cDNA clone library of the National Institute of Aging (NIA) mouse 23K gene sets (15K set and 7.4K set) was originally provided by M. Ko (NIA, Baltimore, MD). The plasmids were extracted from bacterial clones of the library. Each cDNA fragment of the NIA 23K mouse cDNA clones was amplified by PCR and printed by the GeneMachine OmniGrid 100 Microarrayer (Genomic Solutions, Ann Arbor, MI) on polylysine-coated glass slides, with 96 highly expressed genes as indicators for the beginning and end of the printing. After washing of the unbound 3DNA reagent according to a routine protocol, the array chips were scanned to obtain hybridized images with the ScanArray Express microarray scanner (Perkin Elmer Life Sciences, Inc., Boston, MA) (86). To gain more statistical power, all the comparisons were converted to the log 2–transformed ratio of brx+/− mice versus WT mice after LOWESS normalization, and one-sample t test was applied to the six gene expression values (from a total of six hybridizations, one forward and one reverse hybridization for each of the three pairs of WT and brx+/− mice) of each gene. Final candidate genes were obtained by setting the P value cutoff at 0.05. The microarray data discussed in this publication have been deposited in the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE10117.
In situ hybridization and Northern blotting
In situ hybridization and Northern blotting to detect brx transcripts in the spleen were performed as previously described (87). The 1.7-kb fragment of brx cDNA (nucleotides 3126 to 4859), which encodes the C-terminal portion of Brx and a subsequent untranslated region (46), was used in these assays. The gapdh probe was described previously (87). For Northern blots, mRNA was further purified from total RNA with the micro ploy(A) purist kit (Applied Biosystems/Ambion, Austin, TX).
Flow cytometric analysis of splenocytes
Antibodies against CD3 (145-2C11), CD19 (1D3), CD8 (53-6.7), CD4 (GK1.5), B220 (RA3-6B2), IgM (R6-60.2), and CD21 (7G6) were purchased from BD Biosciences (Franklin Lakes, NJ). Cells stained with the appropriate antibodies were analyzed on a FACS Calibur flow cytometer (BD Biosciences). Fluorescent events were collected and analyzed with Flow Jo software (Tree Star Inc., Ashland, OR), as previously described (83). Splenocytes obtained from six WT mice and six brx +/−mice were tested, and representative profiles and the means ± SEM of the specific cell populations from at least three independent experiments are shown in Fig. 3.
Immunization of mice with ovalbumin and quantification of specific IgG and BAFF
The following animal study was approved by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Animal Care and Use Committee (proposal number ASP06-002). Six WT and six brx+/− mice were immunized subcutaneously with 20 μg of ovalbumin (Sigma-Aldrich, St. Louis, MO) together with 5 μg of lipopolysaccharide (Sigma-Aldrich) and 150 μl of incomplete Freund’s adjuvant (Sigma-Aldrich). Three weeks after the immunization, sera from the mice were obtained and the amount of ovalbumin-specific IgG1 was measured with a specific enzyme-linked immunosorbent assay (ELISA) assay with a rat antibody to mouse IgG1 and a goat antibody to rat IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnologies, Inc., Santa Cruz, CA). Monoclonal antibody to chicken egg albumin (mouse IgG1: Clone OVA-14, Sigma-Aldrich) was used as a standard. Serum concentrations of BAFF were determined with the mouse BAFF/BLyS/TNFSF13B Quantikine ELISA kit (R & D Systems Inc., Minneapolis, MN).
Stimulation of splenocytes and Jurkat cells with osmotic agents
Splenocytes (1 × 105 cells/ml) obtained from WT or brx+/− mice or Jurkat cells (1 × 105 cells/ml) were incubated in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) in the presence or absence of the indicated concentrations of NaCl or raffinose for 8 hours. Cells were harvested by centrifugation at 500g for 5 min and total RNA was purified with the RNeasy Mini Kit (Qiagen). For Western blots and coimmunoprecipitation assays, cells were lysed in buffer containing 50 mM tris-HCl (pH 7.4), 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, and one Complete Tablet (Roche Diagnostics Corp., Indianapolis, IN) per 50 ml, and whole-cell homogenates were obtained by centrifugation of the lysates at 10,000g for 15 min (88). Jurkat cells were transfected with siRNAs specific for Brx, NFAT5, p38 MAPK α isoforms, or Vav1 or with plasmids expressing Brx or other small G proteins with the Nucleofector System (Amaxa GmbH, Cologne, Germany) with solution V and program A-17 (Amaxa GmbH) (89). We achieved nearly 80% transfection efficiency with this condition. The first Brx-specific siRNA used was described previously (48), and the latter three siRNAs were purchased from Santa Cruz Biotechnologies, Inc. Twenty-four hours after transfection, the cells were exposed to the indicated concentrations of NaCl or raffinose. After an additional 8 hours, cells were lysed for Western blots or coimmunoprecipitation assays, or total RNA was purified as described above for real-time RT-PCR analysis. In some experiments, Jurkat cells were incubated with the p38 MAPK inhibitor SB202190 (2 μM) or the MEK1 inhibitor PD98059 (10 μM) (Sigma-Aldrich) in the absence or presence of 100 mM NaCl. After 8 hours of incubation, total RNA was harvested for real-time RT-PCR analysis.
Real-time RT-PCR analysis
Total RNA was reverse-transcribed into cDNA and the abundances of mRNAs for the genes encoding AQP2, AR, ATA2, BAFF, BGT1, Brx, HSP70-2, p38α, NFAT1 to NFAT5, NF-κB RelA (p65), NF-κB p105 (p50), Osp94, SMIT, TauT, TNF-α, and Vav1 were determined by real-time RT-PCR assays performed in triplicate or quadruplicate with the SYBR Green PCR Master Mix (Applied Biosystems) in the 7500 real-time PCR System (Applied Biosystems) as previously described (90). The primer pairs used are shown in table S1. The obtained Ct (threshold cycle) values for each of the targets were normalized by those of the acidic ribosomal phosphoprotein P0 (RPLP0) and their relative abundance was presented as fold induction over baseline.
Western blotting and coimmunoprecipitation assays
Proteins from whole-cell homogenates of splenocytes or Jurkat cells transfected or treated with the indicated agents were resolved by 4 to 20% SDS–polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. The presence of Brx, FLAG-JIP4, His-JIP4(251–750), NFAT5, and actin on these membranes was detected with antibodies to Brx, FLAG M2, His, NFAT5, and actin (Santa Cruz Biotechnology, Inc. and Sigma-Aldrich), respectively (48). Coimmunoprecipitation assays were performed with whole-cell homogenates obtained from Jurkat cells, which had been transfected with the indicated plasmids. Briefly, proteins associated with Brx were coimmunoprecipitated with anti-Brx and the protein-antibody complexes were collected with Protein Agarose A/G PLUS (Santa Cruz Biotechnology, Inc.). Associated proteins were resolved by 4 to 20% SDS-PAGE and blotted onto nitrocellulose membranes, and Brx-associated FLAG-JIP4 was detected with anti-FLAG. To evaluate exogenously or endogenously expressed His-JIP4(251–750), FLAG-JIP4, or Brx, 10% of the volume of total cell lysates used in the coimmunoprecipitation reaction was resolved by SDS-PAGE and these molecules were visualized on Western blots as described above.
Assay of p38 MAPK activity
The kinase activity of p38 MAPK was measured with the p38 MAP Kinase Assay Kit (Cell Signaling, Danvers, MA). Briefly, Jurkat cells were transfected with control or Brx-specific siRNA and incubated in the absence or presence of 10 μg/ml of anti-CD3 (clone OKT3, eBioscience, Inc., San Diego, CA). Cells were lysed and phosphorylated p38 MAPK was immunoprecipitated with the antibody to phosphop38 MAPK (Thr180/Tyr184). Precipitated proteins were incubated with activating transcription factor 2 (ATF2), and phosphorylation of ATF2 at Thr71 was evaluated on Western blots with an antibody that reacts specifically with ATF2 phosphorylated at Thr71. Aliquots of the cells were also lysed and used for evaluating the abundance of actin, Brx, p38 MAPK, and NFAT5 in Western blots with their specific antibodies.
Yeast two-hybrid screening and assay
The GAL4-based yeast two-hybrid screening assay was performed with pGBKT7-Brx(1042–1429) as the bait plasmid in the human HeLa cell cDNA library (Clontech) according to the manufacturer’s instructions (89). For reconstituted yeast two-hybrid assays, the yeast strain Y190 (Clontech) was transformed with the pGBKT7-Brx(1042–1429) and pGAD424-JIP4 plasmids as indicated. Transformed yeast cells were further cultured in appropriate selection media and β-galactosidase activity was measured in the cell suspension, as previously described (89). The β-galactosidase activity was normalized for an optical density value at 600 nm. The fold induction in enzyme activity was calculated by the ratio of adjusted β-galactosidase values of cells transformed in the presence versus the absence of pGAD424-based plasmids expressing the indicated JIP4 fragments.
Statistical analysis
Statistical analysis was carried out by analysis of variance followed by Student’s t test with Bonferroni correction for multiple comparisons or unpaired Student’s t test with the two-tailed P value.
Supplementary Material
Footnotes
www.sciencesignaling.org/cgi/content/full/2/57/ra5/DC1
Fig. S1. Brx-specific siRNAs suppress the expression of brx mRNA and attenuate osmotic stimulus–induced expression of nfat5 mRNA in Jurkat cells.
Table S1. Primer pairs used in real-time RT-PCR assays.
REFERENCES AND NOTES
- 1.Burg MB, Kwon ED, Kultz D. Regulation of gene expression by hypertonicity. Annu Rev Physiol. 1997;59:437–455. doi: 10.1146/annurev.physiol.59.1.437. [DOI] [PubMed] [Google Scholar]
- 2.Häussinger D. The role of cellular hydration in the regulation of cell function. Biochem J. 1996;313:697–710. doi: 10.1042/bj3130697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatebayashi K, Yamamoto K, Tanaka K, Tomida T, Maruoka T, Kasukawa E, Saito H. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 2006;25:3033–3044. doi: 10.1038/sj.emboj.7601192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raitt DC, Posas F, Saito H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000;19:4623–4631. doi: 10.1093/emboj/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 7.Lunn JA, Rozengurt E. Hyperosmotic stress induces rapid focal adhesion kinase phosphorylation at tyrosines 397 and 577. Role of Src family kinases and Rho family GTPases. J Biol Chem. 2004;279:45266–45278. doi: 10.1074/jbc.M314132200. [DOI] [PubMed] [Google Scholar]
- 8.Uhlik MT, Abell AN, Johnson NL, Sun W, Cuevas BD, Lobel-Rice KE, Horne EA, Dell’Acqua ML, Johnson GL. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat Cell Biol. 2003;5:1104–1110. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- 9.Sheikh-Hamad D, Di Mari J, Suki WN, Safirstein R, Watts BA, III, Rouse D. p38 kinase activity is essential for osmotic induction of mRNAs for HSP70 and transporter for organic solute betaine in Madin-Darby canine kidney cells. J Biol Chem. 1998;273:1832–1837. doi: 10.1074/jbc.273.3.1832. [DOI] [PubMed] [Google Scholar]
- 10.Aramburu J, Drews-Elger K, Estrada-Gelonch A, Minguillon J, Morancho B, Santiago V, López-Rodríguez C. Regulation of the hypertonic stress response and other cellular functions by the Rel-like transcription factor NFAT5. Biochem Pharmacol. 2006;72:1597–1604. doi: 10.1016/j.bcp.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Macián F, López-Rodríguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 12.Tsai TT, Guttapalli A, Agrawal A, Albert TJ, Shapiro IM, Risbud MV. MEK/ERK signaling controls osmoregulation of nucleus pulposus cells of the intervertebral disc by transactivation of TonEBP/OREBP. J Bone Miner Res. 2007;22:965–974. doi: 10.1359/jbmr.070322. [DOI] [PubMed] [Google Scholar]
- 13.Morancho B, Minguillón J, Molkentin JD, López-Rodríguez C, Aramburu J. Analysis of the transcriptional activity of endogenous NFAT5 in primary cells using transgenic NFAT-luciferase reporter mice. BMC Mol Biol. 2008;9:13. doi: 10.1186/1471-2199-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, Kim M, Im YS, Choi W, Byeon SH, Lee HK. NFAT5 induction and its role in hyperosmolar stressed human limbal epithelial cells. Invest Ophthalmol Vis Sci. 2008;49:1827–1835. doi: 10.1167/iovs.07-1142. [DOI] [PubMed] [Google Scholar]
- 15.Graef IA, Gastier JM, Francke U, Crabtree GR. Evolutionary relationships among Rel domains indicate functional diversification by recombination. Proc Natl Acad Sci U S A. 2001;98:5740–5745. doi: 10.1073/pnas.101602398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keyser P, Borge-Renberg K, Hultmark D. The Drosophila NFAT homolog is involved in salt stress tolerance. Insect Biochem Mol Biol. 2007;37:356–362. doi: 10.1016/j.ibmb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Tonicity-responsive enhancer binding protein, a Rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci U S A. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coimbra R, Junger WG, Liu FC, Loomis WH, Hoyt DB. Hypertonic/hyperoncotic fluids reverse prostaglandin E2 (PGE2)–induced T-cell suppression. SHOCK. 1995;4:45–49. doi: 10.1097/00024382-199507000-00007. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Yanaka A, Tauchi M, Suzuki H, Shibahara T, Matsui H, Nakahara A, Tanaka N. Hyperosmotic stress enhances interleukin-1β expression in Helicobacter pylori–infected murine gastric epithelial cells in vitro. J Gastroenterol Hepatol. 2006;21:759–766. doi: 10.1111/j.1440-1746.2006.04293.x. [DOI] [PubMed] [Google Scholar]
- 20.Hubert A, Cauliez B, Chedeville A, Husson A, Lavoinne A. Osmotic stress, a pro-inflammatory signal in Caco-2 cells. Biochimie. 2004;86:533–541. doi: 10.1016/j.biochi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 21.López-Rodríguez C, Aramburu J, Jin L, Rakeman AS, Michino M, Rao A. Bridging the NFAT and NF-κB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity. 2001;15:47–58. doi: 10.1016/s1074-7613(01)00165-0. [DOI] [PubMed] [Google Scholar]
- 22.Esensten JH, Tsytsykova AV, Lopez-Rodriguez C, Ligeiro FA, Rao A, Goldfeld AE. NFAT5 binds to the TNF promoter distinctly from NFATp, c, 3 and 4, and activates TNF transcription during hypertonic stress alone. Nucleic Acids Res. 2005;33:3845–3854. doi: 10.1093/nar/gki701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman WH, Helman SW, Passmore G. Acute activation of peripheral lymphocytes during treatment of diabetic ketoacidosis. J Diabetes Complications. 2001;15:144–149. doi: 10.1016/s1056-8727(00)00142-2. [DOI] [PubMed] [Google Scholar]
- 24.Wu SG, Jeng FR, Wei SY, Su CZ, Chung TC, Chang WJ, Chang HW. Red blood cell osmotic fragility in chronically hemodialyzed patients. Nephron. 1998;78:28–32. doi: 10.1159/000044878. [DOI] [PubMed] [Google Scholar]
- 25.Mitono H, Endoh H, Okazaki K, Ichinose T, Masuki S, Takamata A, Nose H. Acute hypoosmolality attenuates the suppression of cutaneous vasodilation with increased exercise intensity. J Appl Physiol. 2005;99:902–908. doi: 10.1152/japplphysiol.00156.2005. [DOI] [PubMed] [Google Scholar]
- 26.Ito T, Itoh T, Hayano T, Yamauchi K, Takamata A. Plasma hyperosmolality augments peripheral vascular response to baroreceptor unloading during heat stress. Am J Physiol Regul Integr Comp Physiol. 2005;289:R432–R440. doi: 10.1152/ajpregu.00027.2004. [DOI] [PubMed] [Google Scholar]
- 27.Holtfreter B, Bandt C, Kuhn SO, Grunwald U, Lehmann C, Schutt C, Grundling M. Serum osmolality and outcome in intensive care unit patients. Acta Anaesthesiol Scand. 2006;50:970–977. doi: 10.1111/j.1399-6576.2006.01096.x. [DOI] [PubMed] [Google Scholar]
- 28.Wong CK, Szeto CC, Chan MH, Leung CB, Li PK, Lam CW. Elevation of pro-inflammatory cytokines, C-reactive protein and cardiac troponin T in chronic renal failure patients on dialysis. Immunol Invest. 2007;36:47–57. doi: 10.1080/08820130600745505. [DOI] [PubMed] [Google Scholar]
- 29.Yano A, Nakao K, Sarai A, Akagi S, Kihara T, Morimoto H, Nakamura A, Hiramatsu M, Nagake Y, Makino H. Elevated serum interleukin-18 levels might reflect the high risk of hospitalization in patients on peritoneal dialysis. Nephrology (Carlton) 2005;10:576–582. doi: 10.1111/j.1440-1797.2005.00497.x. [DOI] [PubMed] [Google Scholar]
- 30.Nakanishi I, Moutabarrik A, Okada N, Kitamura E, Hayashi A, Syouji T, Namiki M, Ishibashi M, Zaid D, Tsubakihara Y. Interleukin-8 in chronic renal failure and dialysis patients. Nephrol Dial Transplant. 1994;9:1435–1442. [PubMed] [Google Scholar]
- 31.Descamps-Latscha B, Herbelin A, Nguyen AT, Roux-Lombard P, Zingraff J, Moynot A, Verger C, Dahmane D, de Groote D, Jungers P, Dayer J-M. Balance between IL-1β, TNF-α, and their specific inhibitors in chronic renal failure and maintenance dialysis. Relationships with activation markers of T cells, B cells, and monocytes. J Immunol. 1995;154:882–892. [PubMed] [Google Scholar]
- 32.Ostrowski K, Schjerling P, Pedersen BK. Physical activity and plasma interleukin-6 in humans—effect of intensity of exercise. Eur J Appl Physiol. 2000;83:512–515. doi: 10.1007/s004210000312. [DOI] [PubMed] [Google Scholar]
- 33.Bird MD, Kovacs EJ. Organ-specific inflammation following acute ethanol and burn injury. J Leukoc Biol. 2008;84:607–613. doi: 10.1189/jlb.1107766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shanfield S, Campbell P, Baumgarten M, Bloebaum R, Sarmiento A. Synovial fluid osmolality in osteoarthritis and rheumatoid arthritis. Clin Orthop Relat Res. 1988:289–295. [PubMed] [Google Scholar]
- 35.Németh ZH, Deitch EA, Szabó C, Haskó G. Hyperosmotic stress induces nuclear factor-κB activation and interleukin-8 production in human intestinal epithelial cells. Am J Pathol. 2002;161:987–996. doi: 10.1016/s0002-9440(10)64259-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vernia P, Gnaedinger A, Hauck W, Breuer RI. Organic anions and the diarrhea of inflammatory bowel disease. Dig Dis Sci. 1988;33:1353–1358. doi: 10.1007/BF01536987. [DOI] [PubMed] [Google Scholar]
- 37.Foulks GN. The correlation between the tear film lipid layer and dry eye disease. Surv Ophthalmol. 2007;52:369–374. doi: 10.1016/j.survophthal.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Go WY, Liu X, Roti MA, Liu F, Ho SN. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc Natl Acad Sci U S A. 2004;101:10673–10678. doi: 10.1073/pnas.0403139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trama J, Go WY, Ho SN. The osmoprotective function of the NFAT5 transcription factor in T cell development and activation. J Immunol. 2002;169:5477–5488. doi: 10.4049/jimmunol.169.10.5477. [DOI] [PubMed] [Google Scholar]
- 40.Di Ciano-Oliveira C, Thirone AC, Szaszi K, Kapus A. Osmotic stress and the cytoskeleton: The R(h)ole of Rho GTPases. Acta Physiol (Oxf) 2006;187:257–272. doi: 10.1111/j.1748-1716.2006.01535.x. [DOI] [PubMed] [Google Scholar]
- 41.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 42.Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- 43.Aznar S, Lacal JC. Rho signals to cell growth and apoptosis. Cancer Lett. 2001;165:1–10. doi: 10.1016/s0304-3835(01)00412-8. [DOI] [PubMed] [Google Scholar]
- 44.Cherfils J, Chardin P. GEFs: Structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: Turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 46.Rubino D, Driggers P, Arbit D, Kemp L, Miller B, Coso O, Pagliai K, Gray K, Gutkind S, Segars J. Characterization of Brx, a novel Dbl family member that modulates estrogen receptor action. Oncogene. 1998;16:2513–2526. doi: 10.1038/sj.onc.1201783. [DOI] [PubMed] [Google Scholar]
- 47.Driggers PH, Segars JH, Rubino DM. The proto-oncoprotein Brx activates estrogen receptor β by a p38 mitogen-activated protein kinase pathway. J Biol Chem. 2001;276:46792–46797. doi: 10.1074/jbc.M106927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kino T, Souvatzoglou E, Charmandari E, Ichijo T, Driggers P, Mayers C, Alatsatianos A, Manoli I, Westphal H, Chrousos GP, Segars JH. Rho family guanine nucleotide exchange factor Brx couples extracellular signals to the glucocorticoid signaling system. J Biol Chem. 2006;281:9118–9126. doi: 10.1074/jbc.M509339200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diviani D, Soderling J, Scott JD. AKAP-Lbc anchors protein kinase A and nucleates Gα12-selective Rho-mediated stress fiber formation. J Biol Chem. 2001;276:44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- 50.Ho SN. Intracellular water homeostasis and the mammalian cellular osmotic stress response. J Cell Physiol. 2006;206:9–15. doi: 10.1002/jcp.20445. [DOI] [PubMed] [Google Scholar]
- 51.Woo SK, Lee SD, Na KY, Park WK, Kwon HM. TonEBP/NFAT5 stimulates transcription of HSP70 in response to hypertonicity. Mol Cell Biol. 2002;22:5753–5760. doi: 10.1128/MCB.22.16.5753-5760.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MacLennan IC, Toellner KM, Cunningham AF, Serre K, Sze DM, Zuniga E, Cook MC, Vinuesa CG. Extrafollicular antibody responses. Immunol Rev. 2003;194:8–18. doi: 10.1034/j.1600-065x.2003.00058.x. [DOI] [PubMed] [Google Scholar]
- 54.Su TT, Guo B, Wei B, Braun J, Rawlings DJ. Signaling in transitional type 2 B cells is critical for peripheral B-cell development. Immunol Rev. 2004;197:161–178. doi: 10.1111/j.0105-2896.2004.0102.x. [DOI] [PubMed] [Google Scholar]
- 55.Do RK, Chen-Kiang S. Mechanism of BLyS action in B cell immunity. Cytokine Growth Factor Rev. 2002;13:19–25. doi: 10.1016/s1359-6101(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 56.Khobta A, Ferri F, Lotito L, Montecucco A, Rossi R, Capranico G. Early effects of topoisomerase I inhibition on RNA polymerase II along transcribed genes in human cells. J Mol Biol. 2006;357:127–138. doi: 10.1016/j.jmb.2005.12.069. [DOI] [PubMed] [Google Scholar]
- 57.Tybulewicz VL. Vav-family proteins in T-cell signalling. Curr Opin Immunol. 2005;17:267–274. doi: 10.1016/j.coi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Ridley AJ, Allen WE, Peppelenbosch M, Jones GE. Rho family proteins and cell migration. Biochem Soc Symp. 1999;65:111–123. [PubMed] [Google Scholar]
- 59.Billadeau DD, Nolz JC, Gomez TS. Regulation of T-cell activation by the cytoskeleton. Nat Rev Immunol. 2007;7:131–143. doi: 10.1038/nri2021. [DOI] [PubMed] [Google Scholar]
- 60.Zhao D, Pothoulakis C. Rho GTPases as therapeutic targets for the treatment of inflammatory diseases. Expert Opin Ther Targets. 2003;7:583–592. doi: 10.1517/14728222.7.5.583. [DOI] [PubMed] [Google Scholar]
- 61.Ko BC, Lam AK, Kapus A, Fan L, Chung SK, Chung SS. Fyn and p38 signaling are both required for maximal hypertonic activation of the osmotic response element–binding protein/tonicity-responsive enhancer-binding protein (OREBP/TonEBP) J Biol Chem. 2002;277:46085–46092. doi: 10.1074/jbc.M208138200. [DOI] [PubMed] [Google Scholar]
- 62.Dérijard B, Raingeaud J, Barrett T, Wu IH, Han J, Ulevitch RJ, Davis RJ. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 63.Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hehner SP, Hofmann TG, Dienz O, Droge W, Schmitz ML. Tyrosine-phosphorylated Vav1 as a point of integration for T-cell receptor- and CD28-mediated activation of JNK, p38, and interleukin-2 transcription. J Biol Chem. 2000;275:18160–18171. doi: 10.1074/jbc.275.24.18160. [DOI] [PubMed] [Google Scholar]
- 65.Kelkar N, Standen CL, Davis RJ. Role of the JIP4 scaffold protein in the regulation of mitogen-activated protein kinase signaling pathways. Mol Cell Biol. 2005;25:2733–2743. doi: 10.1128/MCB.25.7.2733-2743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitmarsh AJ. The JIP family of MAPK scaffold proteins. Biochem Soc Trans. 2006;34:828–832. doi: 10.1042/BST0340828. [DOI] [PubMed] [Google Scholar]
- 67.Yasuda J, Whitmarsh AJ, Cavanagh J, Sharma M, Davis RJ. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol Cell Biol. 1999;19:7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda J, Davis RJ. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 69.Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 70.Shi Y, Gaestel M. In the cellular garden of forking paths: How p38 MAPKs signal for downstream assistance. Biol Chem. 2002;383:1519–1536. doi: 10.1515/BC.2002.173. [DOI] [PubMed] [Google Scholar]
- 71.Tanos T, Marinissen MJ, Leskow FC, Hochbaum D, Martinetto H, Gutkind JS, Coso OA. Phosphorylation of c-Fos by members of the p38 MAPK family. Role in the AP-1 response to UV light. J Biol Chem. 2005;280:18842–18852. doi: 10.1074/jbc.M500620200. [DOI] [PubMed] [Google Scholar]
- 72.Li Y, Sassano A, Majchrzak B, Deb DK, Levy DE, Gaestel M, Nebreda AR, Fish EN, Platanias LC. Role of p38α Map kinase in Type I interferon signaling. J Biol Chem. 2004;279:970–979. doi: 10.1074/jbc.M309927200. [DOI] [PubMed] [Google Scholar]
- 73.Pedersen SF, Hoffmann EK, Mills JW. The cytoskeleton and cell volume regulation. Comp Biochem Physiol A Mol Integr Physiol. 2001;130:385–399. doi: 10.1016/s1095-6433(01)00429-9. [DOI] [PubMed] [Google Scholar]
- 74.Henson JH. Relationships between the actin cytoskeleton and cell volume regulation. Microsc Res Tech. 1999;47:155–162. doi: 10.1002/(SICI)1097-0029(19991015)47:2<155::AID-JEMT7>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 75.Hoffmann EK, Dunham PB. Membrane mechanisms and intracellular signalling in cell volume regulation. Int Rev Cytol. 1995;161:173–262. doi: 10.1016/s0074-7696(08)62498-5. [DOI] [PubMed] [Google Scholar]
- 76.Jones GE, Allen WE, Ridley AJ. The Rho GTPases in macrophage motility and chemotaxis. Cell Adhes Commun. 1998;6:237–245. doi: 10.3109/15419069809004479. [DOI] [PubMed] [Google Scholar]
- 77.Rogers R, Norian J, Malik M, Christman G, Abu-Asab M, Chen F, Korecki C, Iatridis J, Catherino WH, Tuan RS, Dhillon N, Leppert P, Segars JH. Mechanical homeostasis is altered in uterine leiomyoma. Am J Obstet Gynecol. 2008;198:474.e1–474.e11. doi: 10.1016/j.ajog.2007.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li C, Xu Q. Mechanical stress–initiated signal transduction in vascular smooth muscle cells in vitro and in vivo. Cell Signal. 2007;19:881–891. doi: 10.1016/j.cellsig.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 79.Wang JH, Thampatty BP, Lin JS, Im HJ. Mechanoregulation of gene expression in fibroblasts. Gene. 2007;391:1–15. doi: 10.1016/j.gene.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manning BD, Padmanabha R, Snyder M. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1829–1844. doi: 10.1091/mbc.8.10.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 82.McDaniel K, Mayers C, Venere M, Driggers P, Westphal H, Gorivodsky M, Segars J. Brx, a cytoplasmic protein capable of augmenting estrogen action, is essential for murine development. J Soc Gynecol Investig. 2004;11:199A. [Google Scholar]
- 83.Takatori H, Nakajima H, Kagami S, Hirose K, Suto A, Suzuki K, Kubo M, Yoshimura A, Saito Y, Iwamoto I. Stat5a inhibits IL-12–induced Th1 cell differentiation through the induction of suppressor of cytokine signaling 3 expression. J Immunol. 2005;174:4105–4112. doi: 10.4049/jimmunol.174.7.4105. [DOI] [PubMed] [Google Scholar]
- 84.Su YA, Trent JM. Isolation of tumor suppressor genes in melanoma by cDNA microarray. In: Nickoloff BJ, editor. Melanoma Techniques and Protocols: Molecular Diagnosis, Treatment, and Monitoring. Vol. 61. Humana Press; Totowa, NJ: 2001. pp. 15–29. [DOI] [PubMed] [Google Scholar]
- 85.Bai X, Wu J, Zhang Q, Alesci S, Manoli I, Blackman MR, Chrousos GP, Goldstein AL, Rennert OM, Su YA. Third-generation human mitochondria–focused cDNA microarray and its bioinformatic tools for analysis of gene expression. Biotechniques. 2007;42:365–375. doi: 10.2144/000112388. [DOI] [PubMed] [Google Scholar]
- 86.Manoli I, Le H, Alesci S, McFann KK, Su YA, Kino T, Chrousos GP, Blackman MR. Monoamine oxidase-A is a major target gene for glucocorticoids in human skeletal muscle cells. FASEB J. 2005;19:1359–1361. doi: 10.1096/fj.04-3660fje. [DOI] [PubMed] [Google Scholar]
- 87.Ramayya MS, Zhou J, Kino T, Segars JH, Bondy CA, Chrousos GP. Steroidogenic factor 1 messenger ribonucleic acid expression in steroidogenic and non-steroidogenic human tissues: Northern blot and in situ hybridization studies. J Clin Endocrinol Metab. 1997;82:1799–1806. doi: 10.1210/jcem.82.6.3967. [DOI] [PubMed] [Google Scholar]
- 88.Kino T, Ichijo T, Amin ND, Kesavapany S, Wang Y, Kim N, Rao S, Player A, Zheng YL, Garabedian MJ, Kawasaki E, Pant HC, Chrousos GP. Cyclin-dependent kinase 5 differentially regulates the transcriptional activity of the glucocorticoid receptor through phosphorylation: Clinical implications for the nervous system response to glucocorticoids and stress. Mol Endocrinol. 2007;21:1552–1568. doi: 10.1210/me.2006-0345. [DOI] [PubMed] [Google Scholar]
- 89.Kino T, Tiulpakov A, Ichijo T, Chheng L, Kozasa T, Chrousos GP. G protein β interacts with the glucocorticoid receptor and suppresses its transcriptional activity in the nucleus. J Cell Biol. 2005;169:885–896. doi: 10.1083/jcb.200409150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ichijo T, Voutetakis A, Cotrim AP, Bhattachryya N, Fujii M, Chrousos GP, Kino T. The Smad6-histone deacetylase 3 complex silences the transcriptional activity of the glucocorticoid receptor: Potential clinical implications. J Biol Chem. 2005;280:42067–42077. doi: 10.1074/jbc.M509338200. [DOI] [PubMed] [Google Scholar]
- 91.This study was funded by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Development, the National Center for Complementary and Alternative Medicine, the National Institute of Arthritis and Mus-culoskeletal and Skin Diseases, and the National Cancer Institute. Y. A. Su was supported by the NIH research contract NIH-NIDDK-06-925 from the NICHD Intramural Research Programs (263MK611483) and the Catherine Birch McCormick Genomics Center (813154). We thank R. J. Davis for providing the plasmids, J. Zhou and C. A. Bondy for performing in situ hybridization, and H. Lu, N. Nader, S. Shrivastav, T. Shibata, C. Mayers, and K. Zachman for their superb technical assistance.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


