Abstract
The p53 tumor suppressor is a critical transcription factor for controlling cell growth and apoptosis during times of cellular stress. In this issue of Cancer Cell, Lain et al. have used a p53-responsive reporter gene as the readout for screening small-molecule activators of p53 that could potentially reduce tumor growth. Using this approach, tenovin-6 was identified as a potent SIRT1 and SIRT2 inhibitor that indirectly activated p53 at single-digit micromolar concentrations. The identification of a specific sirtuin inhibitor has broad implications in understanding sirtuin-p53 signaling and the development of novel chemotherapeutics.
The use of in vitro biochemical screening has been the traditional approach for drug discovery for a number of years. Structural activity relationship (SAR) studies and applied theories such as Lipinski’s “Rule of Five” are often used to guide medicinal chemistry to further develop these leads into “drug-like” compounds (Keller et al., 2006). Advancing compound screens directly to mammalian cell culture-based assays through the forward chemical genetics approach bypasses an initial biochemical screen and offers several advantages for development of hit compounds (Lokey, 2003). For example, the concentration of a hit compound identified using a mammalian cell-based screen is oftentimes the desired concentration for further in vivo analysis. In addition, use of a signaling pathway reporter system as a functional readout in combination with appropriate control assays can generally eliminate compounds that cause global cytotoxicity from potential hits. A major drawback of this type of approach resides in identifying the mechanism of action for a hit compound because several mechanisms can result in the desired cellular phenotype.
By using the well-characterized p53 pathway with defined transcriptional targets, Lain et al. have recently identified a small-molecule activator of p53 at single-digit micromolar concentrations and generated its derivatives (Lain et al., 2008). Furthermore, Lain et al. have defined the specific mechanism of action for these compounds and shown comparable potencies in both mammalian cell-based assays and highly aggressive melanoma xenograft tumors.
The p53 tumor suppressor is a well-characterized transcription factor that responds to a large variety of cellular stresses (Brooks and Gu, 2003). Using a p53-regulated reporter system, the authors identified a hit compound called tenovin-1 that reversibly increased p53 and p21CIP/WAF1 protein levels and decreased cellular growth in a panel of tumor cell lines tested. Additional observations suggested that the compound did not activate p53 indirectly through activation of a DNA damage response because it did not induce phosphorylation of p53 serine 15 or H2AX. The poor water solubility of tenovin-1 led to SAR studies and the development of a more water-soluble derivative, tenovin-6, that also had a slightly increased potency in p53 activation. Interestingly, although a functional p53 protein contributed to the growth inhibition of tumor cell lines both in vivo and in vitro by tenovins, it was not essential for the long-term killing effect. This suggested that tenovins targeted a factor or factors upstream of p53 that may affect other pathways of the cell growth circuitry.
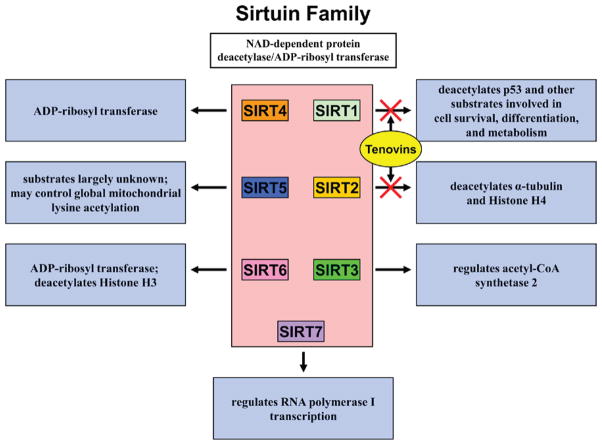
To identify the target of tenovins, the authors screened a collection of diploid S. cerevisiae strains containing heterozygous deletions that together covered over 94% of the protein-coding genes and identified SIR2 as a tenovin-6 target. SIR2 is a NAD+-dependent deacetylase gene and a yeast homolog of the mammalian sirtuin family that includes seven SIRT proteins (Michan and Sinclair, 2007). SIRT1 has been shown to be a direct NAD+-dependent deacetylase for a variety of proteins, including p53, that are involved in diverse cellular processes such as cellular metabolism, extended cellular life span, and differentiation (Figure 1) (Haigis and Guarente, 2006). SIRT2 is involved in the direct deacetylation of α-tubulin and histone H4. Tenovin-6 inhibited purified mammalian SIRT1 and SIRT2. In cell culture assays, tenovin-6 treatment resulted in an increased acetylation of p53 at lysine 382, a global increase in acetylation of histone H4 at lysine 16, and elevated α-tubulin acetylation. SIRT1 has previously been described as a potent negative regulator of p53 function, and the specific activation of p53 seen with Tenovin-6 underscores the importance of the SIRT1-p53 interaction (Luo et al., 2001; Vaziri et al., 2001). SIRT1 and SIRT2 are members of class I sirtuins, which includes SIRT3, but Tenovin-6 had limited inhibition of SIRT3. Overexpression of SIRT1 or SIRT2 weakened effects of tenovin-6 on p53 and α-tubulin acetylation, respectively, thus demonstrating the selectivity of this compound.
Figure 1. Tenovins Inhibit SIRT1 and SIRT2 function.
The mammalian family of sirtuins consists of seven proteins with a variety of cellular functions. Tenovins specifically inhibit SIRT1 and SIRT2, two specific deacetylases for several downstream substrates including the tumor suppressor p53. Inhibition of SIRT1 and SIRT2 by tenovins at single-digit micromolar concentrations has a significant effect on the acetylation levels of their substrates.
The identification and characterization of a compound that activates p53 without activation of the DNA damage response pathway has strong implications for both the development of novel chemotherapeutics and further understanding of the p53 pathway. Approximately 25% of all human tumors retain a wild-type p53 gene and have most likely evolved other cellular pathways to circumvent p53 signaling (Lain and Lane, 2003). In these cases, use of small-molecule activators of p53 may attain sufficient levels of the protein for signaling cell growth arrest or apoptosis. Other compounds that activate p53 through direct interaction (RITA) or indirectly through Mdm2 inhibition (Nutlin-3A) have shown promising p53 activation and cell growth arrest properties in several different tumor cell lines tested. Compounds that specifically activate p53 through inhibition of upstream repressors, such as tenovins, may therefore be attractive chemotherapeutics as single agents or in combination with other specific p53 activators. In addition to their use in cancer research, the identification of inhibitors of SIRT1 and SIRT2 will be very useful for understanding their basic biological functions. Several inhibitors of sirtuin deacetylase activity have been described, including sirtinol, cambinol, and EX-527, though none at single-digit micromolar concentrations similar to Tenovins (Lain et al., 2008). The specificity and potency of tenovins on SIRT1 and SIRT2 and the resulting activation of p53 beg the further analysis and characterization of these compounds as therapeutics for cancers and other diseases. Specifically, direct SIRT2 inhibitors have been shown to be effective at reducing α-synuclein-mediated cytotoxicity in a cell-based Parkinson’s disease model (Outeiro et al., 2007). Further exploration of tenovins in the context of this model may be of particular interest for understanding the molecular link between sirtuins and neurodegeneration. Given the success and limited steps needed to develop a potent, nongenotoxic p53 activator, mammalian cell-based compound screens deserve consideration as high-throughput readouts of other cell signaling pathways as well. The work presented by Lain et al. also indicates the importance of SIRT1 in p53 activation and opens the door for further understanding of the biological functions of mammalian sirtuins.
References
- Brooks CL, Gu W. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Keller TH, Pichota A, Yin Z. Curr Opin Chem Biol. 2006;10:357–361. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Lain S, Lane D. Eur J Cancer. 2003;39:1053–1060. doi: 10.1016/s0959-8049(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, McCarthy A, Apple-yard V, Murray KE, Baker L, et al. Cancer Cell. 2008 doi: 10.1016/j.ccr.2008.03.004. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokey RS. Curr Opin Chem Biol. 2003;7:91–96. doi: 10.1016/s1367-5931(02)00002-9. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, et al. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Wein-berg RA. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]