Abstract
The molecular mechanisms involved in epithelial ovarian cancer initiation and progression are just beginning to be elucidated. In particular, it has become evident that microRNAs (miRNAs), a class of molecules that post-trancriptionally regulates gene expression, play a major role in ovarian tumorigenesis. Several miRNA profiling studies have identified changes in miRNA patterns that take place during ovarian cancer development. While most deregulated miRNAs are down-regulated in cancer, and may therefore act as tumor suppressors, others are elevated and may represent novel oncogenes in this disease. A number of miRNAs identified as aberrantly expressed in ovarian carcinoma have been shown to have important functional roles in cancer development and may therefore represent targets for therapy. In addition, some of the miRNA patterns may have prognostic significance. The identification of functional targets represents a major hurdle in our understanding of miRNA function in ovarian carcinoma, but significant progress is being made. It is hoped that a better understanding of the miRNA expression and roles in ovarian cancer may provide new avenues for the detection, diagnosis, and therapy of this deadly disease.
Introduction
Ovarian cancer is the sixth most common gynecologic malignancy in women worldwide (over 230,000 new cases yearly), with a highly aggressive natural history and causing over 140,000 deaths every year (Garcia, et al. 2007). While the survival rates of women with early stage ovarian cancer are high, most cases are diagnosed late, when the likelihood of successful therapy is low (Ahmed, et al. 1996; Piver, et al. 1992; Wright, et al. 2009). Indeed, cytotoxic chemotherapy for ovarian carcinoma is often unsuccessful due to common resistance to the current chemotherapeutic regimens (Fung-Kee-Fung, et al. 2007). MicroRNAs (miRNAs), a recently discovered class of regulatory RNAs, are frequently deregulated in cancer and have been suggested to have important roles in cancer initiation and development. In ovarian carcinoma, various miRNAs have been found altered and some of these genes may represent ideal targets for detection, diagnosis, and/or therapy (Bartels & Tsongalis 2009). In addition, several research groups have observed altered expression of miRNAs in ovarian cancer and studied their roles in ovarian tumorigenesis. This review will focus on the recent advances in this exciting field.
MiRNAs
MiRNAs are small non-coding RNAs of 20–22 nucleotides, which were first discovered in C. elegans (Lee, et al. 1993), but have now been found to be present and highly conserved among a wide range of species (Wheeler, et al. 2009). Similar to other small regulatory RNAs, miRNAs are generally involved in post-transcriptional gene regulation. MiRNA genes are synthesized in the nucleus by DNA polymerase II as a long double stranded precursor called primary (pri)-miRNA, which is processed by two enzymes, Drosha and Pasha, into a precursor (pre)-miRNA, which is exported to the cytoplasm by exportin 5 (Bohnsack, et al. 2004; Cullen 2004; Zeng & Cullen 2004). Once the pre-miRNA reaches the cytoplasm, it is cleaved by Dicer into a ~22 nt long functional mature miRNA. The mature miRNA can then assemble into a ribonucleoprotein complex known as the RNA-induced silencing complexes (RISC) to participate in RNA interference (Pratt & MacRae 2009). While the exact composition of the RISC is not fully known and may be somewhat variable, the main components are: 1) members of the Argonaut family of proteins which bind directly to the small regulatory RNA and, 2) a small regulatory RNA (such as miRNA or siRNA) which directs the complex to its mRNA targets through direct base-pairing. Through the RISC, miRNAs can down-regulate their targets by inhibiting mRNA translation and/or promoting mRNA degradation. The mode of repression may depend, in part, on the level of complementarity of the miRNAs (with perfect or near-perfect complementarity favoring mRNA degradation). The mechanism of target regulation is complex, but recent work suggests a two-step process by which translation inhibition occurs first and is then followed by mRNA decay due to deadenylation of the mRNA (Fabian, et al. 2009). The extent of each of these processes may be variable, and this may account for the observation that target mRNAs are sometimes found to be inhibited exclusively at the mRNA stability level, exclusively at the translational level, or through a combination of both mechanisms.
The 5′ region of a miRNA, called the “seed region” (nucleotides 2–8 of the miRNA), is crucial for miRNA targeting and function, although other factors are also important (Bartel 2004; Grimson, et al. 2007). In mammals, miRNA target sites are often imperfect and located in the 3′UTR of the target genes (Gu, et al. 2009), although they can also be found in the 5′UTR or even in the coding region.. Because miRNAs do not require perfect complementarity for functional interactions with mRNA targets, a single miRNA can regulate multiple targets and conversely, multiple miRNAs are known to regulate individual mRNAs (Lewis, et al. 2003). In addition to down-regulating their target genes, miRNAs have also been reported to activate some targets (Vasudevan, et al. 2007).
While miRNA genes are typically expressed from their own promoters in intergenic regions, a significant portion of miRNA genes are located within other transcriptional units (Baskerville & Bartel 2005), usually in intronic regions, but sometimes in exonic regions as well. The exact mechanisms by which these intragenic miRNAs are processed still remain to be elucidated, but appear to be Drosha-independent. The expression of these miRNAs is coupled with their host genes and subject to the same regulatory controls (Baskerville & Bartel 2005). Intronic miRNAs can regulate genes involved in pathways of the host genes, adding to the complexity of these regulatory loops (Barik 2008).
miRNA targets
MiRNAs exert their functions through the regulation of specific sets of target mRNAs. Because of imperfect complementarity requirements and other factors that affect site accessibility, target prediction has been a particularly difficult task (John, et al. 2006). Several sequence-matching algorithms, such as Target Scan (Lewis et al. 2003), miRanda (John, et al. 2004), RNAHybrid (Rehmsmeier, et al. 2004), PicTar (Krek, et al. 2005), and DIANA Micro-T (Maragkakis, et al. 2009) have been developed and are an excellent starting point for identifying putative miRNA targets. The next generation target prediction and miRNA databases uses combinations of the programs above, as well as additional features such as sequence conservation and gene ontology (Nam, et al. 2008b; Roubelakis, et al. 2009). It is clear that experimental approaches are necessary in order to identify an accurate full complement of miRNA targets (Creighton, et al. 2008; John et al. 2006), and databases which include lists of experimentally-verified miRNA targets are being constructed (Sethupathy, et al. 2006). Similarly, a database that lists miRNAs that have been implicated in human disease is also available (Jiang, et al. 2009).
As indicated above, in addition to computational methods, several functional approaches are being used to identify actual targets of miRNAs. A straightforward approach consists of over-expressing or down-regulating specific miRNAs in cultured cells and examining the effects on mRNA and/or protein levels using gene expression profiling or proteomics approaches. This approach was used, for example, to identify mRNAs regulated by miR-1 and miR-124 (Lim, et al. 2005), and this was done by over-expressing those miRNAs and observing the resulting changes in gene expression using microarrays. While this type of experiments identifies regulation at the mRNA stability levels, translational regulation can be addressed by performing proteomics analyses (Grosshans & Filipowicz 2008). Stable isotope labeling by amino acids in cell culture (SILAC) followed by mass spectroscopy has been a particularly useful approach in the investigation of the translational effects of miRNAs. SILAC has been used to identify miR-1 targets in HeLa cells (Vinther, et al. 2006) and these targets exhibited significant overlap with targets identified using microarrays. Similarly, other studies confirmed that the levels of hundreds of proteins were affected following changes in the expression of specific miRNAs (Baek, et al. 2008; Selbach, et al. 2008). As a whole, these studies showed that miRNA expression often led to simultaneous mRNA degradation and protein translation inhibition of the targets, and that these effects were dependent on the presence of seed sequences in the 3′ UTR of the target mRNAs.
Finally, a number of biochemical methods have been designed to directly identify the interactions between the miRNAs and their corresponding targets. For example, labeled/tagged miRNAs have been used in pull-down experiments to identify mRNAs that these miRNAs can bind to (Hsu, et al. 2009; Orom & Lund 2007). In addition, it is well known that miRNAs are part of the RISC complex along with different members of the Argonaute (Ago) family, and this property has been used to enrich for targets of specific miRNAs. For example, following forced expression of miR-124a, immunoprecipitation of Ago2 led to the enrichment of known miR-124a targets (Karginov, et al. 2007). Interestingly, the mRNAs that were significantly enriched by immunoprecipitation included targets that were also down-regulated in total mRNA, and these targets were very likely to contain the seed site. Another approach consists of synthesizing cDNA clones from known mRNA templates using endogenous miRNAs as primers (Vatolin, et al. 2006). Sequencing of the synthesized cDNAs then allows the identification of the miRNAs as well as the binding sites on the mRNA.
miRNAs and cancer
miRNAs are conserved in distantly related organisms (Wheeler et al. 2009) suggesting important roles in vital cellular processes such as development, differentiation, cell cycle, apoptosis, metabolism, and proliferation (Flynt & Lai 2008). A possible link between miRNAs and cancer was first reported in chronic lymphocytic leukemia, where miR-15 and miR-16 were found to be deleted or down-regulated in the vast majority of tumors (Calin, et al. 2002). Since then, a large number of studies have found various miRNAs abnormally expressed in several human malignancies (Zhang, et al. 2007). Similar to their protein-coding counterparts, miRNAs involved in tumorigenesis can be classified as oncogenes or tumor suppressors, depending on their expression pattern and their function (Calin & Croce 2006a). Several mechanisms leading to abnormal expression of miRNAs in cancer have been reported, including chromosomal rearrangements (Calin & Croce 2007; Calin, et al. 2005; Tagawa & Seto 2005), genomic copy number change (Calin, et al. 2004; Giannakakis, et al. 2008; Zhang, et al. 2006), epigenetic modifications (Iorio, et al. 2007; Saito, et al. 2006), defects in miRNA biogenesis pathway (Kumar, et al. 2007), and regulation by transcriptional factors (He, et al. 2007).
Important insights into the mechanisms of miRNA function in cancer have been provided through the demonstration that miRNAs are involved in known oncogenic pathways. For example, the three human RAS oncogenes (H-, K-, and N-RAS) all contain let-7 sites in their 3′ UTR (Johnson, et al. 2005). Interestingly, the let-7 family of miRNAs, which are typically down-regulated in various tumors, have been shown to negatively regulate the RAS oncogenes in lung tumors, therefore acting as tumor suppressor genes (Johnson et al. 2005; Kumar, et al. 2008). Similarly, miR-15 and miR-16 have been shown to target the BCL2 oncogene, leading to its down-regulation and apoptosis in leukemic cells (Cimmino, et al. 2005). As an example of miRNAs acting as oncogenes, miR-221 and miR-222 can target and inhibit the expression of the p27KIP tumor suppressor (le Sage, et al. 2007). Indeed, high levels of these miRNAs were shown to maintain low p27 protein and elevated proliferation. Another oncogenic pathway, the p53 pathway, also includes miRNA components. In fact, the p53 tumor suppressor has been shown to transcriptionally induce miR-34 following genotoxic stress and this induction is important in mediating p53 function (Chang, et al. 2007; He et al. 2007; Raver-Shapira, et al. 2007; Tarasov, et al. 2007).
There is evidence that miRNA expression patterns may be useful in cancer diagnosis and outcome prediction. Indeed, miRNA profiling of normal versus tumor tissues using various techniques has consistently shown a large number of deregulated miRNA genes, most of which are typically down-regulated (Calin & Croce 2006b). Interestingly, in a pioneering study that included 334 cancer samples from multiple cancers, the global expression patterns of miRNAs were found to be extremely accurate in distinguishing lineage and differentiation state (Lu, et al. 2005). Most of the changes were again found to be down-regulation of the miRNAs and a subsequent study showed that a general down-regulation of miRNAs in cells (achieved through a knockdown of miRNA processing enzymes) led to enhanced cellular transformation and tumorigenesis, suggesting a crucial roles for these genes in the process of transformation (Kumar et al. 2007).
MiRNA expression in ovarian carcinoma
A number of studies have used various gene expression profiling approaches to study miRNA expression in ovarian carcinoma. In order to clearly present these important data we will describe each study individually. The summary of the differentially expressed miRNAs for each study is included in Table 1.
Table 1.
miRNAs with altered expression in ovarian carcinoma
| miRNA | |||
|---|---|---|---|
| Up-regulated | down-regulated | Comparison | Reference |
| miR-221, miR-146b, miR-508 | let-7f, miR-106b, miR-134, miR-155, miR-21, miR-346, miR-422a, miR-424, miR-519a, miR-648, miR-662 | Ovarian carcinoma cell lines & tissues vs normal | (Dahiya et al. 2008) |
| miR-26b, miR-182, miR-103, miR-26a | miR-127, miR-134, miR-154*, miR-410, miR-377, miR-100, miR-432, miR-368, miR-154, miR-495, miR-376a, miR-323, miR-376b, miR-370, miR-299, let7d, miR-155, miR-140, miR-222, miR-337, miR-124a, miR-99a, miR-331, miR-104, miR-150, miR-184, miR-152, miR-145, miR-424, miR-224, miR-302c | EOC cell lines vs normal | (Zhang et al. 2008) |
| None | miR-509, miR-514, miR-513, miR-196, miR-376a, miR-184, miR-519d, miR-495, miR-424, miR-1, miR-368 miR-362 miR-22, miR-376b, miR-337, miR-133a, miR-508, miR-492, miR-137, miR-95, miR-448, miR-518, miR-491, miR-455, miR-365, miR-147, miR-488, miR-34a, miR--372, miR-202, miR-503, miR-520e, miR-410, miR-519e, miR-375, miR-346, miR-15a, miR-507, miR-450, miR-377, miR-34b, miR-518a, miR-432, miR-516 | Early-stage cancer vs late-stage cancer | (Zhang et al. 2008) |
| None | miR-514, miR-509, miR-508, miR-34c, miR-513, miR-368, miR-379, miR-154, miR-337, miR-507, miR-503, miR-376 | Low-grade cancer vs High-grade cancer | (Zhang et al. 2008) |
| miR-200b, miR-21, miR-200c, miR-141, miR-20a, miR-27a, miR-16, miR-93 | miR-145, miR-125b, miR-100, miR-99a, miR-26a, miR-10b, miR-143, miR-214, let-7b, miR-29a, miR-125a | Serous ovarian carcinoma vs normal ovarian tissues | (Nam et al. 2008a) |
| miR-182, miR-200c, miR-142-3p, miR-200b, miR-135b, miR-200a, miR-195, miR-126*, miR-26b, miR-10b, miR-126, miR-199b-5p, miR-107, miR-30b, miR-192, miR-335, miR-32, miR-20a, miR-30c, miR-143, miR-92a, miR-199b-3p, miR-99a, miR-26a, miR-18a, miR-16, miR-15a, miR-30e, miR-194, miR-29c, miR-30d, miR-106b, | miR-127-3p, miR-377*, miR-382, miR-493, miR-409-3p, miR-193a-5p, miR-210, miR-935, miR-100, miR-31, miR-22, miR-152, miR-379, miR-185, miR-221, miR-744, miR-21*, let-7a*, miR-574-5p, miR-31*, miR-130b, miR-149, miR-423-5p, miR-1308, miR-629, miR-320a | Stage III/IV epithelial ovarian carcinoma vs normal (only miRNAs differentially expressed between all OC subtypes vs normal are included in this table) | (Wyman et al. 2009) |
| miR-223, miR-206, let-7i, miR-30a3p, miR-368, miR-10b, miR-338, miR-195, miR-93, miR-23a, miR-185, miR-22, miR-339, miR-321, miR-29b, miR-186, miR-128a, miR-374, miR-193, miR-106b, miR-194, miR-370, miR-128b, miR-198, miR-224, miR-222, miR-29c, miR-21, miR-34c, miR-139, miR-197, miR-15a, miR-218, miR-106a, miR-340, miR-219, miR-155, miR-92, let-7g, miR-328, miR-149, miR-23b, miR-221, miR-150, miR-190, miR-107, miR-331, miR-181c, miR-133b | miR-326, miR-30d, miR-125b, miR-31, miR-99a, miR-100, miR-137, miR-9 | Primary Vs Recurrent serous papillary ovarian carcinomas | (Laios et al. 2008) |
| miR-200a, miR-200b, miR-200c, miR-141 | miR-140, miR-199a, miR-199b, miR-145, miR-143, miR-125a, miR-125b, miR-101, miR-212, miR-222 | Normal vs cancer | (Iorio et al. 2007) |
| miR-199a, miR-424, miR-302d, miR-320, miR-214, miR-200a, miR-29a | miR-493, miR-494, miR-125b, miR-100, let-7a, let-7b, let-7c | Normal vs primary tumors | (Yang et al. 2008a) |
Integrative analysis of miRNA changes in ovarian carcinoma (Zhang, et al. 2008)
An integrative genomic approach, which included miRNA microarray, array-based comparative genomic hybridization, cDNA microarray, and tissue array was used to evaluate miRNA changes in epithelial ovarian cancer. The authors found that both genomic losses and epigenetic alterations may be responsible for miRNA down-regulation. Out of 35 miRNAs deregulated between ovarian carcinoma and the normal controls (immortalized ovarian surface epithelial cells), 31 (88.6%) were down-regulated in cancer compared to non-cancer tissues. The down-regulated genes included miRNAs let-7d and miR-127, which had been previously implicated in cancer. Thirteen miRNAs were down-regulated in high-grade compared to low-grade tumors. Furthermore down-regulation was higher in late-stage cancers as compared to early stage, suggesting a tumor suppressor function for the down-regulated miRNAs. Among 44 miRNAs down-regulated in late stage tumors, three miRNAs, miR-15a, miR-34a, and miR-34b are believed to be tumor suppressors.
The region containing miR-182 was amplified in 28.9% of EOC, implying an oncogene-type function for this miRNA, whereas, miR-15a, was deleted in 23.9% of EOC suggesting a tumor suppressive role. DNA copy number amplification and deletion were correlated with miR-182 and miR-15a expression, respectively, in both primary tumors and cell lines. However, DNA copy number changes could explain the expression of only 6 of the 33 abnormally expressed miRNAs. EOC cell lines that were treated with DNA demethylating and deacetylase HDAC inhibitors exhibited up-regulation of 16 miRNAs, which suggests epigenetic modification as another crucial factor determining the expression of miRNAs in EOC. In the tumors, loss of miR-377, miR-368, and miR-495 cluster, which is localized at Dlk1-Gtl2 domain, results in higher proliferation and shorter survival of the patients (Zhang et al. 2008)
Genome-wide analysis of miRNA copy number abnormalities in ovarian carcinoma (Zhang et al. 2006)
Using array-based comparative genomic hybridization, these authors found that genomic regions containing miRNA genes frequently exhibited copy number abnormalities. In particular, copy number losses of the region containing miR-218-1 and SLIT2 were observed in 15.5% of ovarian carcinomas. There was a positive correlation between miRNA copy number changes and the miRNA expression levels of 73.1% of the miRNA genes. In addition, experiments with demethylating agents suggested that up to 33% of miRNAs may be regulated by epigenetic mechanisms.
Hypoxia-responsive miRNAs in ovarian carcinoma (Giannakakis et al. 2008)
The screening of a panel of 157 miRNAs allowed for the identification of miR-210 as the most highly and consistently up-regulated miRNA following hypoxia induction. Interestingly, miR-210 is located at chromosome 11p15.5 within a frequent region of loss of heterozygosity in ovarian carcinoma. The gene copy number of miR-210 was reduced in 64% of ovarian carcinomas and these alterations were associated with decreased levels of miR-210, suggesting a tumor suppressive function for this gene.
MiRNA expression signature in ovarian carcinoma (Iorio et al. 2007)
In another study, 15 normal ovarian samples, 69 ovarian malignant tumors, and 5 ovarian carcinoma cell lines were studied by miRNA microarray analysis (Iorio et al. 2007). The unsupervised hierarchical clustering classified the samples into two distinct categories representing “normal” and “cancer samples/cell lines”. A total of 29 and 39 miRNAs were found to be aberrantly expressed by SAM and PAM analysis, respectively. MiR-200a and miR-141 were highly up-regulated whereas miR-199a, miR-140, miR-145, and miR-125b1 were most significantly down-regulated. Although some of the miRNAs were deregulated in all the subtypes, certain miRNAs could differentiate different subtypes (serous, endometrioid and clear cell) of ovarian carcinomas. For example, miR-200a and miR-200c were up-regulated in all three subtypes, miR-200b and miR-141 were up-regulated in serous as well as endometriod, miR-21, miR-203 and miR-205 were up-regulated only in endometrioid. Among the down-regulated miRNAs, miR-145 was down-regulated in serous and clear cell carcinomas, while miR-222 was down-regulated in both endometrioid and clear cell carcinomas. MiRNA expression profiles were not dependent on the grade or stage of the disease. The treatment of ovarian carcinoma cell lines with the demethylating agent 5-Aza-2′-Deoxycytidine, resulted in induction of miR-21, miR-203, miR-146b, miR-205, miR-30-5p, and miR-30c. Since miR-21, miR-203, and miR-205 were observed over-expressed in ovarian carcinoma, these experiments suggest that hypomethylation could be responsible for the observed expression patterns of these miRNAs in cancer. However, it was also interesting to observe that the most significantly up-regulated miRNAs belong to the same family and were localized in pairs: miR-200a and miR-200b at chromosome 1p36.33, and miR-200c and miR-141 at chromosome 12p13.31. This study is consistent with others showing that mechanisms of miRNA deregulation in ovarian carcinoma include copy number changes as well as epigenetic mechanisms (Zhang et al. 2006; Zhang et al. 2008).
Identification of differentially expressed miRNAs in ovarian carcinoma and identification of PTEN as a target of miR-214 (Yang, et al. 2008a)
Using miRNA microarrays, 36 miRNAs were found deregulated between normal ovarian cells and epithelial ovarian tumors, with miR-199a*, miR-214, miR-200a, and miR-100 being the most highly differentially expressed candidates. MiR-199a*, miR-214, and miR-200a were up-regulated in 53%, 56%, and 43% tumor tissues, respectively and their expression was associated with high-grade and late-stage tumors. MiR-100 was down-regulated in 76% of tumors. The authors selected miR-214 for further study and found PTEN as one of its potential targets, implicating this miRNA in the regulation of the AKT survival pathway.
MiRNA expression profiles in serous ovarian carcinoma (Nam, et al. 2008a)
In this study, 23 miRNAs were found aberrantly expressed in at least 60% of ovarian cancer samples. Among 11 up-regulated miRNAs, miR-21 topped the list showing expression in 85% samples, whereas among 12 down-regulated miR-125b was most significantly and exhibited down-regulation in 95% tumor samples. 50% of miRNAs reported in this study were also found in the Iorio study (Iorio et al. 2007). The most significantly altered miRNAs in both studies were: miR-200a/b/c, miR-141, miR-21, miR-145, miR-99a, let-7, and miR-125b.
Identification of miRNAs that are differentially expressed in ovarian carcinoma: importance of the let-7 family (Dahiya, et al. 2008)
Our laboratory investigated miRNA expression profiles in 34 ovarian carcinoma tissues as well as 10 ovarian carcinoma cell lines. A total of 25 up-regulated and 31 down-regulated miRNAs were identified. We also found 5 up-regulated and 23 down-regulated miRNAs in ovarian carcinoma cell lines compared with non-neoplastic cells. Fourteen miRNAs were deregulated in both tissues and cell lines (Table 1). MiR-221 was the most highly elevated miRNA in both tissues and cell lines (9-fold and 7-fold respectively), while miR-21 was significantly decreased in both sample types (3-fold and 9-fold, respectively). Among the different let-7 family members, let-7e and let-7f showed more than 2-fold deregulation in at least 60% of the tumor samples. The other let-7 family members (let-7g, let-7d, let-7c, let-7a-e, let-7i, let-7a, and let-7b were not down-regulated as consistently, but each one of them was found decreased 2-fold or more in at least 20% of the tumors. Overall, 94% of the tumors had at least one let-7 family member down-regulated at least 2-fold. Cell lines exhibited down-regulation of the let-7 family members as well. This study suggested an important role for let-7 family members in ovarian carcinoma.
Comprehensive analysis of miRNA repertoire in ovarian carcinoma using massively parallel sequencing technologies (Wyman, et al. 2009)
Using novel massively parallel sequencing technology (454 sequencing) to sequence small RNA cDNA libraries derived from epithelial ovarian cancers and normal samples, this group was able to generate comprehensive miRNA digital profiles for these tissues. In their data, the investigators identified 498 previously annotated miRNAs, six novel ones and 39 candidate miRNAs. They found 124 miRNAs that were differentially expressed in cancers of various subtypes compared to the normal ovarian cells. A subset of 37 miRNAs was over-expressed in all epithelial ovarian cancer subtypes and 21 were under-expressed (Table 1). Among those, several were validated by RT-PCR. The validated over-expressed miRNAs included several members of the miR-200 family, while the down-regulated genes included miR-100, miR-210, miR-22, and miR-222.
Roles of miRNAs in ovarian carcinoma chemotherapy
Cisplatin is the most efficacious chemotherapeutic agent against ovarian carcinoma with initial response rates varying between 40% and 80% (Ozols & Young 1984). Platinum-based combination therapy, especially carboplatin/paclitaxel, offers a modest but significant improvement over cisplatin alone and this regimen is now standard for women with advanced epithelial ovarian cancer (McGuire, et al. 1996). Unfortunately, many women with tumors that initially respond to chemotherapy eventually relapse with drug-resistant disease (Ozols & Young 1984). Overall, fewer than 25% of the women diagnosed with advanced ovarian carcinoma will show progression-free survival after 4 years, in spite of treatment (McGuire et al. 1996). In this context, a better understanding of drug resistance may lead to the development of novel approaches for the treatment of ovarian and other cancers. A number of investigators have now reported possible roles for miRNA in the establishment of drug resistance in ovarian cancer
In a recent report where primary and recurrent cases of ovarian cancers were compared, 60 miRNAs were found deregulated more than 2-fold between primary and recurrent disease (Laios, et al. 2008). In contrast to several studies reporting a majority of miRNAs to be down-regulated in cancer tissues, a marked up-regulation of miRNAs in recurrent compared to primary tumors was observed (52 miRNAs showed over-expression and 8 under-expression of >2 fold in recurrent versus primary tumors). MiR-223 (up) and miR-9 (down) were the most highly deregulated genes in the recurrent versus primary samples (Laios et al. 2008).
When cisplatin resistance was specifically examined for miRNA expression, and tumors from responders were compared to those of non-responders, 34 statistically significant changes were found (24 miRNAs were higher in the non-response group and 10 were higher in the complete response group) (Yang, et al. 2008b). Let-7i was the most down-regulated miRNA in the chemotherapy-resistant patients. In addition, functional analyses confirmed that reduced let-7i expression increased the resistance of ovarian and breast cancer cells to cisplatin. In another study, elevation of miR-214 was found to be responsible for development of resistance against cisplatin (Yang et al. 2008a). MiR-214 also has anti-apoptotic functions, as blocking miR-214 expression made A2780 cells more sensitive to cisplatin-induced apoptosis (Yang et al. 2008a). Another expression profiling study in a panel of cisplatin-, paclitaxel-, and cyclosporin A-resistant ovarian carcinoma cells revealed expression of let-7e, miR-30c, miR-125b, miR-130a, and miR-335 in all the resistant cell lines (Sorrentino, et al. 2008). While analyzing their downstream targets, the investigators found a direct relationship between down-regulation of miR-130a with up-regulation of M-CSF, a gene already known to be up-regulated in ovarian carcinoma.
MiRNA targets and pathways in ovarian carcinoma
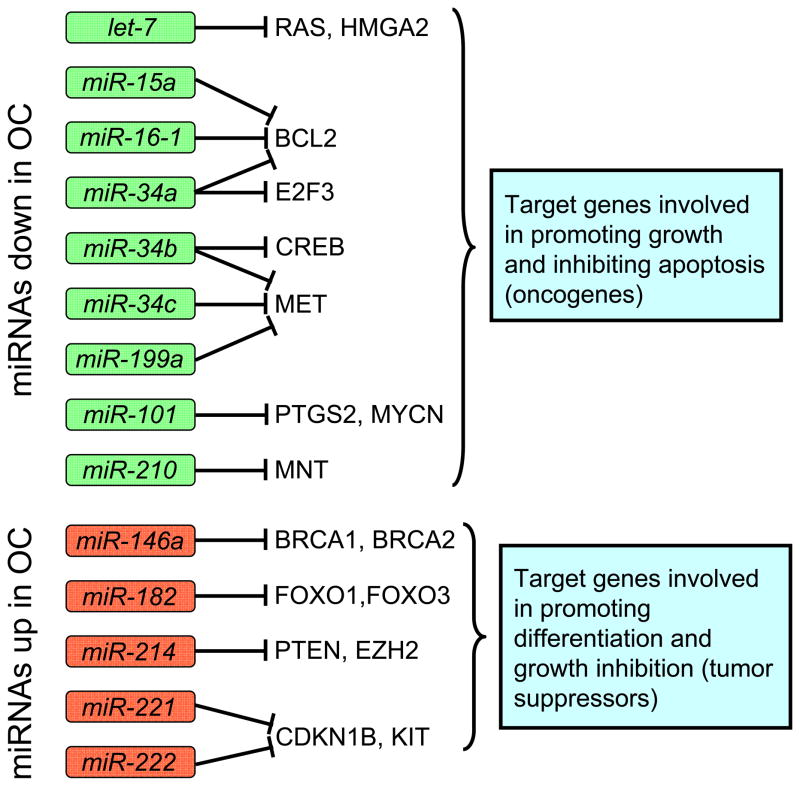
There have been relatively few miRNA targets that have been specifically shown to be relevant to ovarian tumorigenesis, but work in other systems allow us to make educated guesses concerning the pathways and targets of some of the miRNAs found aberrantly expressed in ovarian carcinoma (Figure 1). For example, the RAS oncogenes are well-known targets of the let-7 cluster (Johnson et al. 2005), which has been reported to be down-regulated in ovarian carcinoma by several groups (Table 1). Other let-7 targets, such as HMGA2, CDK6, and cMYC, are likely relevant to tumorigenesis (Peter 2009). Similarly, the tumor suppressor gene WT1, which encodes a transcription factor, is a putative target of miR-212.
Figure 1.
Schematic representation of selected known targets for miRNAs that are frequently altered in ovarian carcinoma. The miRNAs frequently down-regulated in ovarian carcinoma (shown in green boxes) typically target genes that have growth promoting functions, while miRNAs that are up-regulated (red boxes) target genes that have negative effects on cell growth. These miRNAs may represent targets for therapeutic interventions.
Unlike the previous examples, which were elucidated in other systems, miR-214 was shown to target PTEN in ovarian carcinoma, explaining its ability to regulate survival and drug resistance (Yang et al. 2008a). The p53 pathway is another pathway with important miRNA components that was studied in ovarian carcinoma. MiR-34b and miR-34c, two known transcriptional targets of p53, have been shown to be down-regulated in ovarian carcinoma (Zhang et al. 2008). Interestingly, their over-expression can suppress proliferation and colony formation in soft agar in neoplastic epithelial ovarian cells, providing functional relevance of this pathway in ovarian tumorigenesis (Corney, et al. 2007). MiR-146 was found elevated in epithelial ovarian cancer (Volinia, et al. 2006) and may target BRCA1 and BRCA2 (Shen, et al. 2008), two well-documented tumor suppressors in breast and ovarian cancers.
In a recent study, putative targets for a number of miRNA genes were identified using a microarray approach in ovarian carcinoma cells (Dahiya et al. 2008). Intriguingly, the experimental targets varied depending on the cell line used for the experiment, suggesting a significant influence of the molecular background on miRNA target selection. It will therefore be important to methodically study each miRNAs in several ovarian models in order to fully understand their roles in ovarian carcinoma. These findings also imply that targets identified in other cell models may not necessarily be relevant to ovarian cancer.
MiRNAs in detection and diagnosis of ovarian carcinoma
A number of studies have shown that miRNAs may be useful in predicting ovarian carcinoma outcome. For example, it was shown that patients with low let-7a-3 methylation had worse overall survival than those with high methylation (Lu, et al. 2007). In another study, the miR-200b-429 cluster, which harbors miR-200a, miR-200b, and miR-429, could predict poor survival when the miR-200 genes were expressed at low levels (Hu, et al. 2009). The targets of miR-200 miRNAs have known roles in cancer development. A promising study reported that the HMGA2/let-7 ratio was able to categorize ovarian carcinoma patients into 2 groups with significantly different prognosis (Shell, et al. 2007). The group with lower HMGA2/let-7 ratio exhibited increased 5-year progression free survival (~40%) compared to the group with a higher HMGA2/let-7 ratio (<10%). Another study showed that miR-214, miR-199*, and miR-200a were associated with high-grade and late-stage tumors (Yang et al. 2008a). Tumors with higher expression of miR-200a had a median overall survival of 27.5 months compared with 61 months for those with no significant expression (Nam et al. 2008a). Interestingly, polymorphisms in miRNAs may affect their function and be of prognostic value. For example, the precursor mir-146a, which targets BRCA1 and BRCA2, exhibits a G to C polymorphism in ovarian and breast cancer (Shen et al. 2008). This polymorphism results in a change from G:U pair to a C:U mismatch in the miRNA stem region. Ovarian cancer patients showing G to C polymorphism were typically diagnosed younger than the patients having common mir-146a allele (Shen et al. 2008). Additionally, with the presence of the variant allele, there was increased production of mature mir-146a, which may be responsible for early onset of the disease.
Interestingly, miRNAs can also be detected in serum samples (Feng, et al. 2008; Lawrie, et al. 2008; Lodes, et al. 2009), suggesting that they may represent useful detection biomarkers. By RT-PCR analysis, miR-21, miR-92, miR-93, miR-126, and miR-29a were found up-regulated, while miR-155, miR-127, and miR-99b were found down-regulated in serum collected from ovarian carcinoma patients (Resnick, et al. 2009). Up-regulation of miR-21, miR-92, and miR-93 in the serum of 3 cancer patients with normal CA-125 level suggests that miRNAs may be complementary to current detection approaches. Interestingly, circulating tumor-derived exosomes (small lipid vesicles) have been found to contain miRNAs that could potentially be used as detection and/or diagnostic markers (Taylor & Gercel-Taylor 2008). The use of exosomes-derived miRNAs in diagnosis may circumvent the need for biopsy material.
Therapeutic potential of miRNAs
Because of their properties, miRNAs have been suggested as possible therapeutic tools. The approaches suggested include manipulating the expression of tumor suppressor or oncogenic miRNAs (Mishra & Merlino 2009), using specific miRNA to down-regulate oncogenic genes (Cimmino et al. 2005), and using miRNA to confer tissue specificity to transgene expression in gene therapy (Brown & Naldini 2009). When considering altering miRNA expression for therapeutic purposes, the regulation of multiple genes by a single miRNA suggests the possible targeting of pathways involved in the development or progression of the disease. Normal levels of tumor suppressor miRNAs that are down-regulated in cancer could potentially be restored by over-expression (Mishra & Merlino 2009). MiRNAs can be introduced into cells by infecting cells with retrovirus or lentivirus expressing the miRNA of interest. This method has the advantage of making the system inducible or cell-specific, thereby reducing toxicity. For miRNAs that are over-expressed in cancer, miRNA inhibitors can be used to modulate their levels. For example, sequence-specific antisense oligonucleotides against an individual miRNA can be transfected into mammalian cells, preventing the miRNA from binding its targets. (Horwich & Zamore 2008; Hutvagner, et al. 2004; Meister, et al. 2004). Another approach consists of high expression of artificial target genes containing multiple tandem binding sites for the miRNA of interest (Ebert, et al. 2007). This competitive inhibition approach, known as “miRNA sponge”, results in the release of miRNA-mediated repression of endogenous targets (Ebert et al. 2007). While the specific targets useful for ovarian cancer therapy remain to be determined, miR-31 inhibition using a “sponge” strategy was recently shown to inhibit breast cancer metastasis in vivo (Valastyan, et al. 2009). The determination of useful miRNA targets for therapy will likely be a major focus of future research in ovarian cancer.
In addition, miRNAs against known oncogenes could potentially be used to repress the expression of these genes in tumors. For example, miR-15 and miR-16 over-expression was shown to induce apoptosis in leukemic cells through the ability of these miRNAs to target BCL2 (Cimmino et al. 2005). Similarly, Mir-34a was shown to target E2F3 and induce apoptosis in neuroblastoma cells (Welch, et al. 2007). In ovarian carcinoma, let-7 is commonly down-regulated and was shown to target HMGA2 (Shell et al. 2007), a gene that may be responsible for de-differentiation during ovarian carcinoma progression (Park, et al. 2007; Shell et al. 2007). Re-introduction of let-7 has therefore been suggested as a possible approach for the therapy of ovarian carcinoma (Park et al. 2007).
Finally, miRNA may be useful in ensuring tight tissue-specific control of transgene expression in gene therapy. Indeed, by incorporating carefully-chosen specific miRNA target sites into a therapeutic mRNA, it may be possible to inhibit this mRNA in tissues where its expression is not wanted, thereby minimizing its toxicity and side effects (Brown & Naldini 2009). In cancer, expression of a toxic gene such as thymidine kinase, or re-expression of a tumor suppressor gene, could potentially be made more specific by including, in these transgenes, sites for miRNAs that are expressed in normal tissues, but not in cancer cells. Clearly, a detailed knowledge of miRNA expression in normal and neoplastic tissues will be crucial for the success of these approaches.
Perspectives
While a large amount of information has been gain on the roles and possible therapeutic use of miRNAs in ovarian carcinoma, much remains to be done. In particular, more thorough miRNA expression profiling will be necessary to clarify expression in ovarian carcinoma of various grades, stages, or drug resistance status. The next step, the identification of relevant targets, will likely be a tedious task, complicated by the fact that miRNAs can have several functional targets and that these targets may be dependent on several factors, including the expression of other miRNAs. Once relevant miRNAs and functional targets are identified, the investigation of possible clinical use for these molecules will represent the next frontier, and may, ultimately, lead to novel strategies for ovarian cancer detection and therapy.
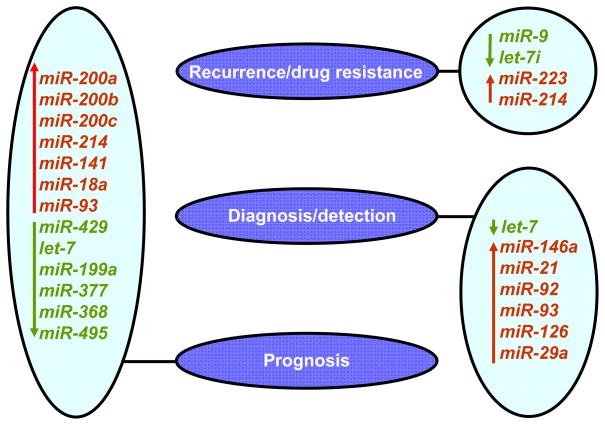
Figure 2.
MiRNAs with potential clinical use in ovarian carcinoma. List of miRNAs with potential roles in recurrence/drug resistance (Laios et al. 2008; Yang et al. 2008a; Yang, et al. 2008b), diagnosis/detection (Shell et al. 2007; Shen et al. 2008), prognosis (Nam et al. 2008a; Yang et al. 2008a; Zhang et al. 2008) of ovarian cancer. MiRNAs indicated in green are typically down-regulated in ovarian carcinoma, while miRNAs indicated in red are up-regulated.
Acknowledgments
We thank Drs Ashani Weeraratna and Myriam Gorospe, as well as the members of our laboratory for helpful comments on the manuscript. This work was supported entirely by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
References
- Ahmed FY, Wiltshaw E, Ahern RP, Nicol B, Shepherd J, Blake P, Fisher C, Gore ME. Natural history and prognosis of untreated stage I epithelial ovarian carcinoma. J Clin Onc. 1996;14:2968–2975. doi: 10.1200/JCO.1996.14.11.2968. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–5241. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. Rna. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006a;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006b:6857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J Clin Invest. 2007;117:2059–2066. doi: 10.1172/JCI32577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. Rna. 2008;14:2290–2296. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004:16861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih Ie M, Zhang Y, Wood W, 3rd, Becker KG, Morin PJ. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Li G, Gentil-Perret A, Tostain J, Genin C. Elevated serum-circulating RNA in patients with conventional renal cell cancer. Anticancer Res. 2008;28:321–326. [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung-Kee-Fung M, Oliver T, Elit L, Oza A, Hirte HW, Bryson P. Optimal chemotherapy treatment for women with recurrent ovarian cancer. Curr Oncol. 2007;14:195–208. doi: 10.3747/co.2007.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Jemal A, Ward EM, Center MM, Hao Y, Siegel R, Thun MJ. Global Cancer Facts, Figures 2007. Atlanta, GA: American Cancer Society; 2007. [Google Scholar]
- Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O’Brien-Jenkins A, Katsaros D, Weber BL, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Filipowicz W. Proteomics joins the search for microRNA targets. Cell. 2008;134:560–562. doi: 10.1016/j.cell.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Zamore PD. Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat Protoc. 2008;3:1537–1549. doi: 10.1038/nprot.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu RJ, Yang HJ, Tsai HJ. Labeled microRNA pull-down assay system: an experimental approach for high-throughput identification of microRNA-target mRNAs. Nucleic Acids Res. 2009;37:e77. doi: 10.1093/nar/gkp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Sander C, Marks DS. Prediction of human microRNA targets. Methods Mol Biol. 2006;342:101–113. doi: 10.1385/1-59745-123-1:101. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Laios A, O’Toole S, Flavin R, Martin C, Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. Embo J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS ONE. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med. 1996;334:1–6. doi: 10.1056/NEJM199601043340101. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. Rna. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ, Merlino G. MicroRNA reexpression as differentiation therapy in cancer. J Clin Invest. 2009;119:2119–2123. doi: 10.1172/JCI40107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008a;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008b;36:D159–164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods. 2007;43:162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Ozols RF, Young RC. Chemotherapy of ovarian cancer. Semin Oncol. 1984;11:251–263. [PubMed] [Google Scholar]
- Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piver MS, Malfetano J, Baker TR, Hempling RE. Five-year survival for stage IC or stage I grade 3 epithelial ovarian cancer treated with cisplatin-based chemotherapy. Gynecol Oncol. 1992;46:357–360. doi: 10.1016/0090-8258(92)90232-8. [DOI] [PubMed] [Google Scholar]
- Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- Roubelakis MG, Zotos P, Papachristoudis G, Michalopoulos I, Pappa KI, Anagnou NP, Kossida S. Human microRNA target analysis and gene ontology clustering by GOmir, a novel stand-alone application. BMC Bioinformatics. 2009;10(Suppl 6):S20. doi: 10.1186/1471-2105-10-S6-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–1966. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111:478–486. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Vasudevan S, Tong Y, Steitz JA. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science. 2007 doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vatolin S, Navaratne K, Weil RJ. A novel method to detect functional microRNA targets. J Mol Biol. 2006;358:983–996. doi: 10.1016/j.jmb.2006.02.063. [DOI] [PubMed] [Google Scholar]
- Vinther J, Hedegaard MM, Gardner PP, Andersen JS, Arctander P. Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucleic Acids Res. 2006;34:e107. doi: 10.1093/nar/gkl590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- Wright JD, Shah M, Mathew L, Burke WM, Culhane J, Goldman N, Schiff PB, Herzog TJ. Fertility preservation in young women with epithelial ovarian cancer. Cancer. 2009 doi: 10.1002/cncr.24461. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O’Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS, Tewari M. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008a;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008b;68:10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171:728–738. doi: 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]