Abstract
Purpose
Currently, no validated blood-based assays accurately predict treatment response or outcome in melanoma patients. We hypothesized that methylation of tumor-related genes detected in serum DNA could predict disease outcome and therapeutic response in patients receiving concurrent biochemotherapy (BC) for metastatic melanoma.
Patients and Methods
American Joint Committee on Cancer stage IV melanoma patients (N = 50) had blood drawn before administration of BC. Patients (n = 47) were classified as BC responders or nonresponders. Responders (n = 23) demonstrated a complete or partial response following BC; nonresponders (n = 24) demonstrated progressive disease. Hypermethylation of Ras association domain family 1 (RASSF1A), retinoic acid receptor-β2 (RAR-β2), and O6-methylguanine DNA methyltransferase (MGMT) genes were assessed by methylation-specific polymerase chain reaction.
Results
Circulating methylated RASSF1A was significantly less frequent for responders (three of 23 patients; 13%) than nonresponders (10 of 24 patients; 42%; P = .028). Patients with RASSF1A, RAR-β2, or at least one serum methylated gene had significantly worse overall survival than patients with no methylated genes (log-rank, P = .013, .021, and .01, respectively). Methylated RASSF1A was the only factor that significantly correlated with overall survival and BC response (risk ratio, 2.38; 95% CI, 1.16 to 4.86; P = .018; odds ratio = 0.21; 95% CI, 0.05 to 0.90; P = .036).
Conclusion
Detection of circulating methylated DNA in serum can predict response to BC and disease outcome.
INTRODUCTION
Malignant cutaneous melanoma patients with American Joint Committee on Cancer (AJCC) stage IV disease have poor prognoses; the 5-year survival rate is less than 10%.1 For these patients, immunotherapy and chemotherapy are the most common treatment modalities but, thus far, significant clinical responses have been few. Biochemotherapy (BC), the use of chemotherapy in conjunction with immune modulators, has produced better response rates,2–9 but patient response is difficult to predict. Identifying molecular predictors of therapeutic response may permit physicians to treat those patients most likely to respond to therapy while sparing nonresponsive patients unnecessary treatment and its associated morbidity.
Hypermethylation of tumor-related gene promoter CpG islands plays a role in the development and progression of various cancers.10–20 The detection of hypermethylated genes in tumors has become an important approach in assessing candidate tumor-related gene inactivation.10–20 We first reported our screening of tumor-related gene hypermethylation in primary and metastatic melanoma tissues.21 Using a panel of candidate tumor suppressor genes, including RASSF1A, RAR-β2, and MGMT, at least one gene demonstrated promoter region CpG island hypermethylation in 75% of melanoma patients.21 Metastatic tumors demonstrated higher rates of tumor-related gene hypermethylation compared with primary tumors.21 Therefore, tumor-related gene methylation may be a useful molecular biomarker and could play a significant role in melanoma progression to systemic disease.
Circulating DNA in the serum of cancer patients may have clinical utility as a marker for disease surveillance.22–32 Fujiwara et al22 demonstrated a correlation between circulating DNA and disease progression in stages I to IV melanoma patients using 10 microsatellite markers on six chromosomes. Our group has shown that preoperative levels of allelic imbalance correlate with AJCC stage and predict response to surgical and adjuvant therapies.22 Fujimoto et al24 employed a panel of four microsatellite markers surrounding the 12q22–23 locus to detect loss of heterozygosity in the sera of stage IV patients. Previously we examined allelic imbalance in the blood of patients receiving BC for stage IV melanoma using nine microsatellite markers on seven chromosomes and showed an association between circulating DNA markers and response to adjuvant therapy.25 Although we have reported the presence of methylated tumor-related genes in serum in a pilot analysis of melanoma patients,21 no studies have assessed the prognostic or predictive significance of detecting methylated tumor-related DNA in the serum of melanoma patients receiving systemic therapy.
Identifying surrogate tumor-specific markers of response to systemic treatment in patients’ serum would be of tremendous clinical value. Currently, there are no validated blood-based assays that accurately predict treatment response or disease outcome in melanoma patients receiving systemic therapy. Not all stage IV patients are candidates for surgery, and tissue may be difficult to obtain using less invasive means. In these patients, it is critical to predict response to therapy using surrogate blood-based assays. There is a particular interest in identifying patients responsive to systemic treatment before the initiation of therapy. This way, patients not likely to respond to treatment may avoid nonbeneficial therapy and any untoward adverse effects. BC has achieved dramatic clinical responses in a significant number of patients,2,3 some of whom have survived for prolonged periods following treatment. Better selection of patients receiving BC treatment may boost the proportion of clinical responders, increase time to progression, and improve overall survival. We hypothesized that analysis of serum for circulating methylated tumor-related DNA could predict clinical response to BC in patients with AJCC stage IV melanoma.
PATIENTS AND METHODS
Serum DNA Collection and Preparation
Fifty AJCC stage IV melanoma patients were treated with a concurrent BC regimen of dacarbazine (DTIC) or temozolomide, cisplatin, vinblastine, interferon α-2b, interleukin-2 (IL-2), and tamoxifen, as previously reported.2,3 Thirteen patients received maintenance treatment containing IL-2 and granulocyte-macrophage colony-stimulating factor (GM-CSF), if they had a partial response (PR) or stable disease (SD) response from the first concurrent BC (Table 1). Representative patients were selected and coded by the clinical coordinator and assessed in a blinded manner in laboratory and statistical analysis. The selection was based on response and nonresponse to BC, follow-up availability, completion of the BC trial, and specimen availability. The categorization of patients as responders or nonresponders was unrelated to their having received maintenance treatment with IL-2 or GM-CSF and was based entirely on their response to concurrent BC. Institutional review board approval from Saint John’s Health Center (Santa Monica, CA) and John Wayne Cancer Institute (Santa Monica, CA) were obtained before study initiation.
Table 1.
Clinical Demographics of BC Patients
| Characteristics | No. | % |
|---|---|---|
| Total patients | 50 | |
| Sex | ||
| Male | 38 | 76 |
| Female | 12 | 24 |
| Age, years | ||
| < 50 | 34 | 68 |
| ≥ 50 | 16 | 32 |
| ECOG status | ||
| 0 | 14 | 28 |
| 1 | 12 | 24 |
| 2 | 24 | 48 |
| BC response | ||
| Responder | ||
| CR | 13 | 26 |
| PR | 10 | 20 |
| Nonresponder | ||
| PD | 24 | 48 |
| LDH | ||
| ≤ 190 | 22 | |
| > 190 | 27 | |
| No. of metastatic sites | ||
| ≤ 2 | 28 | |
| > 2 | 21 | |
NOTE. Standard BC regimens (n = 25): dacarbazine (DTIC), cisplatin, vinblastine, interferon alfa-2b, IL-2, tamoxifen. Variation of BC protocol: (A) six patients without tamoxifen; (B) six patients with temozolomide substituted for DTIC; (C) 13 patients received subsequent maintenance of GM-CSF and IL-2.
Abbreviations: BC, biochemotherapy; ECOG, Eastern Cooperative Oncology Group; CR, complete remission; PR, partial response; PD, progressive disease; LDH, lactate dehydrogenase; IL-2, interleukin-2; GM-CSF, granulocyte-macrophage colony-stimulating factor.
Blood was drawn for serum before administration of BC (pre-BC serum). Patients were categorized as responders or non-responders on the basis of response criteria.2 Patients showing a complete response (CR; n = 13) or PR (n = 10) were included in the responder group (n = 23), whereas, patients demonstrating progressive disease were deemed nonresponders (n = 24). Patients exhibiting SD (n = 3) were neither considered responders nor nonresponders. One patient in the responder group was lost to follow-up and excluded from the survival analysis. Serum drawn from 40 healthy donors served as normal controls.
Serum DNA Extraction and Sodium Bisulfite Modification
Ten ml of blood was collected in serum separator tubes, centrifuged (3,000 rpm, 15 minutes), passed through a 13-mm serum filter (Fisher Scientific, Pittsburgh, PA), aliquoted, and cryopreserved at −30°C. DNA was extracted from serum as previously described.21 The DNA quantification assessment was performed using the PicoGreen quantification assay (PicoGreen; Molecular Probes, Eugene, OR).24
Extracted DNA was subjected to sodium bisulfite modification. Briefly, DNA from 500 μL of serum was supplemented with 1 μg salmon sperm DNA (Sigma, St Louis, MO) and denatured in 0.3 M NaOH for 3 minutes at 95°C. Overall, 550 μL of a 2.5 M sodium bisulfite/125 mmol/L hydroquinone solution was added. Samples were incubated under mineral oil in the dark for 3 hours at 60°C. Salts were removed using the Wizard DNA Clean-Up System (Wizard DNA Clean-Up System; Promega, Madison, WI) and samples were desulfonated in 0.3 M NaOH at 37°C for 15 minutes. Modified DNA was precipitated with ethanol using Pellet Paint NF (Pellet Paint NF; Novagen, Madison, WI) as a carrier and then resuspended in molecular grade H2O.
DNA Preparation From Paired Tumor Tissue
Paraffin-embedded tumor specimens from melanoma patients were obtained from the Division of Surgical Pathology at Saint John’s Health Center. Tumors represented distant nodal, subcutaneous, or visceral metastases in all cases. Of 50 melanoma patients with serum samples, 18 patients had paired paraffin-embedded metastatic tissues available for analysis.
Several 8-μm sections were cut from formalin-fixed, paraffin-embedded blocks as described previously.33 One section from each tumor block was deparaffinized, mounted on a glass slide, and stained with hematoxylin and eosin for microscopic analysis. Light microscopy was used to confirm the location of the tumor and to assess tissue homogeneity. Additional sections from the tumor block were mounted on glass slides and microdissected with the aid of light microscopy. Dissected tissues were digested with 50 μL of proteinase K containing lysis buffer at 50°C for 12 hours, followed by heat deactivation of proteinase K at 95°C for 10 minutes, as previously described. Extracted DNA was subjected to sodium bisulfite modification as described in the previous section.
Detection of Gene Hypermethylation
Methylation status was assessed for each gene using two sets of fluorescent labeled primers designed to amplify methylated or unmethylated DNA sequences. Primer sequences are listed as methylated sense and antisense, followed by unmethylated sense and anti-sense sequences, with annealing temperatures and polymerase chain reaction (PCR) product size: RASSF1A methylated-specific forward, 5′-GTGTTAACGCGTTGCGTATC-3′ and reverse, 5′-AACCCCG-CGAACTAAAAACGA-3′ (60°C, 93 base pair [bp]); unmethylated-specific forward, 5′-TTTGGTTGGAGTGTGTTAATG-3′ and reverse, 5′-CAAACCCCACAAACTAAAAA CAA-3′(60°C, 150 bp), RAR-β2 methylated-specific forward, 5′-GAACGCGACCGATTCG AGT-3′ and reverse, 5′-GACCAATCCAACCGAAACG-3′ (59°C, 142bp);unmethylated-specific forward, 5′-GGATTGGGATGTTGA-GAATGT-3′ and reverse, 5′-CAACCAATCCAACCAAAACAA-3′ (59°C, 158 bp), MGMT methylated-specific forward, 5′-TTT-CGACGTTCGTAGGTTTTCGC-3′ and reverse, 5′-GCACTCTT CCGAAAACGAAACG –3′ (66°C, 81 bp), unmethylated-specific forward, 5′-TTTGT GTTTTGATGTTTGTAGGTTTTTGT-3′ and reverse, 5′-AACTCCACACTCTTCCA AAAACAAAAC-3′(66°C, 93 bp). Bisulfite-modified DNA was subjected to PCR amplification in a final reaction volume of 20 μL containing PCR buffer, 2.5 to 4.5 mmol/L MgCl2, dNTPs, 0.3 μmol/L primers, and 0.5 U of AmpliTaq Gold DNA polymerase (AmpliTaq Gold; Applied Biosystems, Foster, CA). PCR amplification was performed with an initial 10 minutes of incubation at 95°C, followed by 40 cycles of denaturation at 95°C for 30 seconds, annealing for 30 seconds, extension at 72°C for 30 seconds, and finally, 7 minutes hold at 72°C. Lymphocyte DNA obtained from healthy donors underwent sodium bisulfite modification and a universal unmethylated control synthesized by phi-29 DNA polymerase from normal DNA served as a positive unmethylated control.34 Unmodified lymphocyte DNA was used as a negative control for methylated and unmethylated reactions. SssI (New England Bio Labs, Beverly, MA) methylase-treated lymphocyte DNA was used as a positive methylated control. PCR products were visualized using capillary array electrophoresis (CEQ 8000XL; Beckman Coulter Inc, Fullerton, CA) in a 96-well microplate. Methylated and unmethylated products from each sample were assessed simultaneously by labeling forward primers with a choice of three Beckman Coulter WellRED dye-labeled phosphoramidites (Beckman Coulter WellRED; Genset Oligos, Boulder, CO). Forward methylated sequence-specific primers were labeled with D4pa dye, and forward unmethylated sequence specific primers were labeled with D2a dye. One methylated PCR product and one unmethylated PCR product were mixed with 40 μL loading buffer and a 0.5 μL dye-labeled size standard (Beckman Coulter Inc). Capillary array electrophoresis detects the different dyes and displays them in their respective colors. Each marker was optimized with methylated and unmethylated controls. Only those samples demonstrating a peak at the specific corresponding bp size marker for unmethylated DNA were considered unmethylated. Samples demonstrating a peak at the corresponding bp marker for methylated DNA were considered methylated.
Sequential Evaluation of Methylation
Among responders and nonresponders, methylation status during BC was assessed on day 5 of the first week, day 1 of the second week, and day 1 of the third week. The procedure of serum DNA extraction and detection for methylation was the same as described in the previous sections.
Statistical Analysis
The correlation between gene methylation status in serum, clinical factors, and BC response was assessed using the χ2 test. A logistic regression model was developed to correlate clinical factors and gene methylation status with response to BC. Survival length was determined from the first day of BC treatment to the date of death or last clinical follow-up. Survival curves were derived by the Kaplan-Meier method and the differences between curves were analyzed using the log-rank test. Cox’s proportional hazards regression model was used for univariate and multivariate analyses.13,35 Age, sex, Eastern Cooperative Oncology Group status, lactate dehydrogenase (LDH) level, number of metastatic sites, and gene methylation status were included in the model, and a stepwise method was used for variable selection. For comparison of circulating DNA levels, Wilcoxon rank sum test was used. A P value less than .05 (two-tailed) was considered significant.
RESULTS
After screening multiple tumor-related genes, we had reported frequent hypermethylation of RASSF1A, RAR-β2, and MGMT in metastatic melanoma tumor tissue.21 We chose to validate these three genes for circulating DNA in serum of melanoma patients because of their known frequency in advanced-stage melanomas.
Frequency of Circulating Methylated DNA in Serum
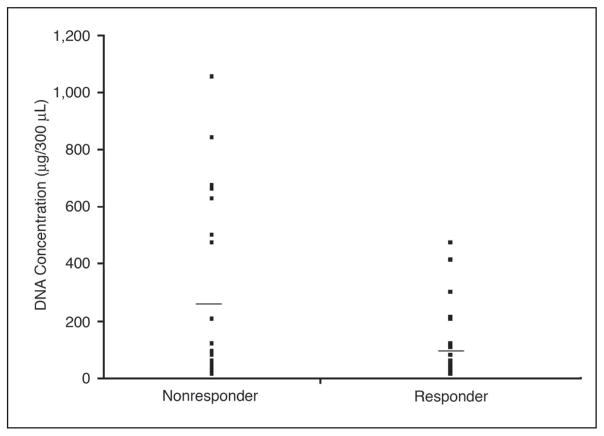
The average amount of DNA recovered from responders was a mean of 95.4 with an SE of 20.2 ng/300 μL, while the average DNA from nonresponders was a mean of 266.0 with an SE of 71.3 ng/300 μL. DNA from healthy volunteers averaged a mean of 78.9 with an SE of 15.4 ng/300 μL. The serum DNA concentrations in the nonresponder group were significantly higher (P = .03) than in the responder group (Fig 1).
Fig 1.
DNA concentration in serum of responders and nonresponders to biochemotherapy (BC). Horizontal bar shows the mean DNA concentration (P = .03).
The serum DNA assay was established and optimized using normal healthy donor and melanoma patients’ serum, then applied to melanoma patients receiving BC. In the analysis of all 50 patients’ pre-BC sera, the frequencies of RASSF1A, RAR-β2, and MGMT methylation markers were 26% (13 of 50 patients), 20% (10 of 50 patients), and 10% (5 of 50 patients), respectively (Table 2). At least one marker was methylated in 17 of the 50 serum samples (34%), and at least two methylated markers were found in nine of the 50 serum samples (18%). Serum from healthy donors showed no detectable hypermethylation for all three genes.
Table 2.
Detection of Hypermethylated Genes in Serum of BC Patients
| Responders (n = 23) |
Nonresponders (n = 24) |
||||
|---|---|---|---|---|---|
| Gene | No. | % | No. | % | P |
| RAR-β2 | 2 | 9 | 8 | 33 | . 072 |
| RASSF1A | 3 | 13 | 10 | 42 | .028 |
| MGMT | 2 | 9 | 3 | 13 | .99 |
Abbreviation: BC, biochemotherapy.
Detection of Methylated DNA in Metastatic Tumors
Using the same methylation markers, we assessed available paraffin-embedded metastatic melanomas (n = 18; 10 lymph nodes, three lung, two liver, two subcutaneous, and one rectal) from the patients whose serum samples were assessed. Although all patients had distant melanoma metastases identifiable by routine radiographic and nuclear medicine imaging, only those undergoing elective surgery with palliative or curative intent were available for analysis. If methylated genes were detected in serum, they were also found to be methylated in the metastatic tumor. The frequencies of methylation in metastatic tumor specimens were: RASSF1A (nine of 18 patients; 50%), RAR-β2 (11 of 18 patients; 61%), and MGMT (two of 18 patients; 11%). Overall, 13 tumors (72%) demonstrated methylation of one or more genes and six of 18 tumors (33%) had two or more genes methylated.
Correlation to Disease Outcome
Tumor-related gene methylation in pre-BC serum was assessed to predict the patients most likely to respond to BC. The frequency of circulating RASSF1A methylated DNA for responders (three of 23 patients; 13%) was significantly lower (P = .028) than nonresponders (10 of 24 patients; 42%; Table 2). There was no significant difference (P = .72; P = .57) in the frequency of RAR-β2 and MGMT methylation in pre-BC serum between the responder (two of 23 patients; 9%; two of 23 patients; 9%) and the nonresponder group (eight of 24 patients; 33%; three of 24 patients; 13%). The frequency of at least one methylated DNA marker in responders (six of 23 patients; 26%) was lower (P = .16) than in nonresponders (11 of 24 patients; 46%). No other known prognostic factors were associated with BC response (Table 3).
Table 3.
Univariate Analysis of Response of BC Patients
| Responders |
||||
|---|---|---|---|---|
| Characteristic | No. | Total No. | % | P |
| Sex | .34 | |||
| Male | 19 | 36 | 53 | |
| Female | 4 | 11 | 36 | |
| Age, years | .17 | |||
| < 50 | 14 | 33 | 45 | |
| ≥ 50 | 9 | 14 | 64 | |
| ECOG status | .11 | |||
| 0–1 | 14 | 23 | 61 | |
| 2 | 9 | 24 | 38 | |
| LDH | .11 | |||
| ≤ 190 | 13 | 21 | 62 | |
| > 190 | 10 | 26 | 38 | |
| No. of metastatic sites | .29 | |||
| ≤ 2 | 15 | 27 | 56 | |
| > 2 | 8 | 20 | 40 | |
| RAR-β2 | .072 | |||
| U | 21 | 37 | 57 | |
| M | 2 | 10 | 20 | |
| RASSF1A | .028 | |||
| U | 20 | 34 | 59 | |
| M | 3 | 13 | 23 | |
| MGMT | .99 | |||
| U | 21 | 42 | 50 | |
| M | 2 | 5 | 40 | |
Abbreviations: BC, biochemotherapy; ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase; U, unmethylated; M, methylated.
The frequencies of corresponding paraffin-embedded tumor RASSF1A and RAR-β2 methylated DNA for responders (one of five patients; 20%; one of five patients; 20%, respectively) were lower (P = .30; P = .10, respectively) than for nonresponders (eight of 12 patients; 67%; nine of 12 patients; 75%, respectively). There was no significant difference (P = .99) in the frequency of MGMT methylation status in tumors between responders (zero of five patients; 0%) and nonresponders (two of 12 patients; 17%). The frequency of at least one methylated DNA marker for responders (one of five patients; 20%) was significantly lower (P = .01) than for nonresponders (11 of 12 patients; 92%).
In a logistic regression multivariable analysis, the presence of serum methylated RASSF1A was the only factor that significantly correlated with response to BC (odds ratio = 0.21; 95% CI, 0.05 to 0.90; P = .036).
Patients with serum methylation of only RAR-β2, methylation of only RASSF1A, or methylation of at least one marker had significantly worse overall survival compared with patients with no methylated genes (log-rank test; P = .021, .013, and .010, respectively; Figs 2A to 2C). Responders had significantly better overall survival compared with nonresponders (log-rank test; P < .0001).
Fig 2.
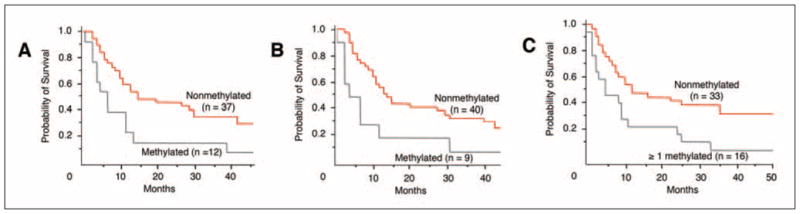
(A) Kaplan-Meier survival curves of biochemotherapy (BC) patients: Correlation of pre-BC serum RASSF1A methylation status with overall survival (log-rank test, P = .013). Methylated: Patients with serum methylation of RASSF1A. Nonmethylated: Patients with no serum methylation of RASSF1A. (B) Correlation of pre-BC serum RAR-β2 methylation status with overall survival (log-rank test, P = .02). Methylated: Patients with serum methylation of RAR-β2. Nonmethylated: Patients with no serum methylation of RAR-β2. (C) Correlation of pre-BC serum methylation of at least one marker with overall survival (log-rank test, P = .01). ≥ 1 methylated: Patients with serum methylation of at least one marker. Nonmethylated: Patients with no serum methylation of genes.
Using a Cox’s proportional hazards regression model, serum methylation of only RAR-β2, methylation of only RASSF1A, methylation of at least one marker, or LDH more than 190 significantly correlated with survival (RAR-β2 methylation, 95% CI, 3 to 7; P = .021; RASSF1A methylation, 95% CI, 4 to 12; P = .013; methylation of at least one marker, 95% CI, 4 to 14; P = .01; LDH > 190, 95% CI, 5 to 13; P = .015) in univariate analyses (Table 4). Other prognostic factors were not significant. In a multivariate analysis, RASSF1A methylation of serum DNA was the only independent factor predicting overall survival (risk ratio, 2.38; 95% CI, 1.16 to 4.86; P = .018).
Table 4.
Univariate Analysis of Survival in BC Patients
| Deaths |
Median Survival |
||||
|---|---|---|---|---|---|
| Characteristic | No. | Total No. | Months | 95% CI | Log-Rank Test (P) |
| Sex | .70 | ||||
| Male | 28 | 37 | 13 | 7 to 30 | |
| Female | 9 | 12 | 19 | 10 to 39 | |
| Age, years | .10 | ||||
| < 50 | 28 | 34 | 11 | 7 to 20 | |
| ≥ 50 | 9 | 15 | 30 | 9 to NA | |
| ECOG status | .12 | ||||
| 0–1 | 18 | 25 | 27 | 11 to 42 | |
| 2 | 19 | 24 | 8 | 4 to 14 | |
| LDH | .015 | ||||
| ≤ 190 | 13 | 21 | 29 | 13 to NA | |
| > 190 | 24 | 28 | 9.5 | 5 to 13 | |
| No. of metastatic sites | .28 | ||||
| ≤ 2 | 20 | 28 | 14.5 | 10 to 39 | |
| > 2 | 17 | 21 | 10 | 5 to 20 | |
| RAR-β2 | .021 | ||||
| U | 29 | 40 | 14.5 | 11 to 30 | |
| M | 8 | 9 | 4 | 3 to 7 | |
| RASSF1A | .013 | ||||
| U | 26 | 37 | 15 | 10 to 42 | |
| M | 11 | 12 | 6 | 4 to 12 | |
| MGMT | .064 | ||||
| U | 32 | 44 | 13.5 | 9 to 30 | |
| M | 5 | 5 | 7 | 3 to 29 | |
| No. of methylated genes | .01 | ||||
| 0 | 22 | 33 | 15 | 10 to 42 | |
| ≥ 1 | 15 | 16 | 7 | 4 to 14 | |
Abbreviations: BC, biochemotherapy; NA, not applicable; ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase; U, unmethylated; M, methylated.
Sequential Blood Evaluation of Methylation
To test if the assay would be more sensitive when used at multiple time points during the administration of BC, the assay was performed using available serum samples obtained during the first week of the one to three cycles of BC. At least one marker was subsequently found to be hypermethylated in six patients among 13 nonresponders whose pre-BC serum circulating DNA was initially unmethylated. This raised the methylation frequency from 46% (11 of 24 patients) to 71% (17 of 24 patients). Contrary to the results obtained in nonresponders, four responders with available serum drawn at identical time periods as nonresponders revealed no methylated DNA.
Patients (n = 21) with more than one bleed available within a week were assessed for consistency in DNA processing of serum and PCR analysis. Three markers (RASSF1A, RAR-β2, and MGMT) were assessed and identical results were obtained 75 of 81 times (93%).
DISCUSSION
In this study, RASSF1A was the most frequently hypermethylated gene, supporting our prior report of high rates of RASSF1A methylation in free circulating DNA from patients with metastatic melanoma.21 Among the three markers assayed, RASSF1A was the most useful for predicting BC response and overall survival. RASSF1A may play a role in terminating the cell cycle by inhibiting cyclin D accumulation.36 Recent reports indicate that RASSF1A regulates microtubule and genomic stability.37 Resistance to cisplatin and tamoxifen, both of which are components of the BC regimen used in our study, has been associated with RASSF1A hypermethylation in other malignancies.38,39 RASSF1A serum hypermethylation has demonstrated prognostic significance, but not predictive utility, before the initiation of adjuvant treatment in breast cancer patients.39 Thus, the association between RASSF1A hypermethylation and poor clinical response to BC may, in part, be mediated by melanoma resistance to the tamoxifen and cisplatin components of the regimen. Our findings support the role of RASSF1A as a tumor-related gene and its use as a surrogate marker for BC response.
The second most frequently hypermethylated gene in the current study was RAR-β2. RAR-β2 is a member of the nuclear retinoid receptor of genes, a family of retinoic acid receptors that are infrequently expressed in carcinomas.40,41 RAR-β2 is involved in the regulation of cellular growth inhibition and apoptosis by unknown mechanisms.40,41 We demonstrated a significant correlation between the hypermethylation of RAR-β2 in serum DNA and survival. Overall response approached a significant value. Expression of RAR-β2 in tumor cells susceptible to RAR-β2 mediated apoptosis may be an important determinant of BC response in melanoma patients.
MGMT is a DNA repair gene that protects mammalian cells from spontaneous G:C to A:T translations.42 Previously, MGMT was found to be the third most frequently methylated gene among multiple tumor-related genes screened.21 In the present study, there was no significant correlation between MGMT methylation and response to BC. A previous report has indicated MGMT methylation predicted increased tumor responsiveness to alkylating agents in glioma.43 In our BC regimen, which includes the alkylating agent DTIC, prognosis may be determined by responsiveness to this agent and loss of MGMT expression by epigenetic inactivation. Consistent with our previous study, the frequency of methylation among the three markers studied in serum and tumor tissue was lowest for MGMT. Our findings verify prior reports that serum MGMT methylation occurs less frequently than for RASSF1A and RAR-β2 in melanoma. Such a low serum detection rate precludes the use of MGMT hypermethylation as a surrogate marker of disease outcome or therapeutic response.
Metastatic melanoma is characterized by tumor heterogeneity.44 Using multiple methylation markers compensates for cell-to-cell variation in individual marker expression and increases the overall assay sensitivity. In this study, however, the detection of RASSF1A gene hypermethylation was the best predictor of BC response and overall survival in a multivariate analysis. It is possible that the incorporation of new candidate methylation markers and tumor-related genes in a multimarker methylation panel with RASSF1A could further improve the assay’s efficacy. The search for such markers is currently being investigated in our laboratory.
Sequential examination of initially unmethylated non-responder sera detected methylated tumor-related genes in six of 13 specimens sampled during the early phase of BC treatment, while no methylation was detected among initially unmethylated responders. The release of intact tumor-related DNA occurs in actively growing tumors. Destruction of tumor cells by immuno- or chemotherapy is primarily through apoptosis-related events, resulting in the release of small characteristic fragments of DNA. As a result, the tumor-related DNA released in patients responding to immuno- or chemotherapy is cleared rapidly and not readily detected. Tumors of nonresponder patients are growing during BC, and intact, larger DNA is released into the blood stream. DNA can also be released directly from cells as a result of physical trauma or cell necrosis. Further studies are underway to verify the clinical utility of genetic analysis in serial serum specimens from patients with melanoma and the mechanism of DNA release during treatment.
Responders showed a significantly lower circulating DNA level than nonresponders. Because these DNA levels were measured before treatment, they were not influenced by BC administration and are consistent with prior studies.28 The use of circulating methylated tumor-related genes as an adjunct to serum DNA markers to predict BC response requires further assessment. The approach to measuring small amounts of free circulating DNA varies among institutions, making comparisons difficult.26 Verifying the significance of circulating DNA levels between responders and nonresponders will likely require a large-scale multi-center study.
This study demonstrates the utility of detecting circulating methylated tumor-related genes in serum as a potentially predictive marker of response to BC and overall survival. To our knowledge, this is the first study to assess the association between circulating methylated tumor-related genes and disease outcome during treatment in cancer patients. Circulating methylated tumor-related genes may be used as a predictive marker of melanoma progression before the initiation of adjuvant therapy. This study requires confirmation with other forms of therapy to determine its universal efficacy in predicting treatment response. This is ongoing in a prospective clinical trial in melanoma.
Acknowledgments
We thank Gwen Berry for her expert editorial assistance, Lori Wilson, MD, for logistical support, and the clinical and laboratory staff of the Angeles Clinic and Research Institute for their participation in this study.
Supported by National Institutes of Health, National Cancer Institute Project II Grants No. P0 CA029605 and CA012582, and R33-CA100314.
Footnotes
Presented at the 41st Annual Meeting of the American Society of Clinical Oncology, Orlando, Florida, May 13–17, 2005 (T.M., ASCO Merit Award).
Authors’ Disclosures of Potential Conflicts of Interest
Although all authors completed the disclosure declaration, the following author or immediate family members indicated a financial interest. No conflict exists for drugs or devices used in a study if they are not being evaluated as part of the investigation. For a detailed description of the disclosure categories, or for more information about ASCO’s conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
| Authors | Employment | Leadership | Consultant | Stock | Honoraria | Research Funds | Testimony | Other |
|---|---|---|---|---|---|---|---|---|
| Steven J. O’Day | Chiron (C); Berlex (C); Schering-Plough (C) | |||||||
| Dollar Amount Codes (A) < $10,000 (B) $10,000–99,999 (C) ≥ $100,000 (N/R) Not Required | ||||||||
References
- 1.Greene FL, Fleming ID, Fritz AG, et al. AJCC Cancer Staging Manual. 6. New York, NY: Springer-Verlag; 2002. pp. 229–254. [Google Scholar]
- 2.O’Day SJ, Boasberg PD, Piro L, et al. Maintenance biotherapy for metastatic melanoma with interleukin-2 and granulocyte macrophage-colony stimulating factor improves survival for patients responding to induction concurrent biochemotherapy. Clin Cancer Res. 2002;8:2775–2781. [PubMed] [Google Scholar]
- 3.O’Day SJ, Gammon G, Boasberg PD, et al. Advantages of concurrent biochemotherapy modified by decrescendo interleukin-2, granulocyte colony-stimulating factor, and tamoxifen for patients with metastatic melanoma. J Clin Oncol. 1999;17:2752–2761. doi: 10.1200/JCO.1999.17.9.2752. [DOI] [PubMed] [Google Scholar]
- 4.Gollob JA, Veenstra KG, Parker RA, et al. Phase I trial of concurrent twice-weekly recombinant human interleukin-12 plus low-dose IL-2 in patients with melanoma or renal cell carcinoma. J Clin Oncol. 2003;21:2564–2573. doi: 10.1200/JCO.2003.12.119. [DOI] [PubMed] [Google Scholar]
- 5.O’Day SJ, Atkins MB, Weber J. A phase II multi-center trial of maintenance biotherapy (MBT) after induction concurrent biochemotherapy (BCT) for patients (Pts) with metastatic melanoma. J Clin Oncol. 23(suppl; abstr 7503):710s, 205. doi: 10.1200/JCO.2008.20.3075. [DOI] [PubMed] [Google Scholar]
- 6.Dutcher J. Current status of interleukin-2 therapy for metastatic renal cell carcinoma and metastatic melanoma. Oncology (Williston Park) 2002;16(suppl 13):4–10. [PubMed] [Google Scholar]
- 7.Khayat D, Bernard-Marty C, Meric JB, et al. Biochemotherapy for advanced melanoma: Maybe it is real. J Clin Oncol. 2002;20:2411–2414. doi: 10.1200/JCO.2002.20.10.2411. [DOI] [PubMed] [Google Scholar]
- 8.Hakansson A, Gustafsson B, Krysander L, et al. Biochemotherapy of metastatic malignant melanoma: Predictive value of tumour-infiltrating lymphocytes. Br J Cancer. 2001;85:1871–1877. doi: 10.1054/bjoc.2001.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asemissen AM, Scheibenbogen C, Letsch A, et al. Addition of histamine to interleukin 2 treatment augments type 1 T-cell responses in patients with melanoma in vivo: Immunologic results from a randomized clinical trial of interleukin 2 with or without histamine (MP 104) Clin Cancer Res. 2005;11:290–297. [PubMed] [Google Scholar]
- 10.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: Epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 12.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 13.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 14.Jeronimo C, Henrique R, Hoque MO, et al. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8478. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- 15.Hoque MO, Begum S, Topaloglu O, et al. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64:5511–5517. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- 16.Jeronimo C, Henrique R, Hoque MO, et al. Quantitative RARbeta2 hypermethylation: A promising prostate cancer marker. Clin Cancer Res. 2004;10:4010–4014. doi: 10.1158/1078-0432.CCR-03-0643. [DOI] [PubMed] [Google Scholar]
- 17.Xing M, Cohen Y, Mambo E, et al. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res. 2004;64:1664–1668. doi: 10.1158/0008-5472.can-03-3242. [DOI] [PubMed] [Google Scholar]
- 18.Umetani N, Mori T, Koyanagi K, et al. Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene. 2005;24:4721–4727. doi: 10.1038/sj.onc.1208538. [DOI] [PubMed] [Google Scholar]
- 19.Shinozaki M, Hoon DS, Giuliano AE, et al. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin Cancer Res. 2005;11:2156–2162. doi: 10.1158/1078-0432.CCR-04-1810. [DOI] [PubMed] [Google Scholar]
- 20.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 21.Hoon DS, Spugnardi M, Kuo C, et al. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–4022. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara Y, Chi DD, Wang H, et al. Plasma DNA microsatellites as tumor-specific markers and indicators of tumor progression in melanoma patients. Cancer Res. 1999;59:1567–1571. [PubMed] [Google Scholar]
- 23.Taback B, Hoon DS. Circulating nucleic acids and proteomics of plasma/serum: Clinical utility. Ann N Y Acad Sci. 2004;1022:1–8. doi: 10.1196/annals.1318.002. [DOI] [PubMed] [Google Scholar]
- 24.Fujimoto A, O’Day SJ, Taback B, et al. Allelic imbalance on 12q22–23 in serum circulating DNA of melanoma patients predicts disease outcome. Cancer Res. 2004;64:4085–4088. doi: 10.1158/0008-5472.CAN-04-0957. [DOI] [PubMed] [Google Scholar]
- 25.Taback B, O’Day SJ, Boasberg PD, et al. Circulating DNA microsatellites: Molecular determinants of response to biochemotherapy in patients with metastatic melanoma. J Natl Cancer Inst. 2004;96:152–156. doi: 10.1093/jnci/djh011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gautschi O, Bigosch C, Huegli B, et al. Circulating deoxyribonucleic acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J Clin Oncol. 2004;22:4157–4164. doi: 10.1200/JCO.2004.11.123. [DOI] [PubMed] [Google Scholar]
- 27.Dulaimi E, Hillinck J, Ibanez de Caceres I, et al. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin Cancer Res. 2004;10:6189–6193. doi: 10.1158/1078-0432.CCR-04-0597. [DOI] [PubMed] [Google Scholar]
- 28.Holdenrieder S, Stieber P, von Pawel J, et al. Circulating nucleosomes predict the response to chemotherapy in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2004;10:5981–5987. doi: 10.1158/1078-0432.CCR-04-0625. [DOI] [PubMed] [Google Scholar]
- 29.Nawroz-Danish H, Eisenberger CF, Yoo GH, et al. Microsatellite analysis of serum DNA in patients with head and neck cancer. Int J Cancer. 2004;111:96–100. doi: 10.1002/ijc.20240. [DOI] [PubMed] [Google Scholar]
- 30.Capone RB, Pai SI, Koch WM, et al. Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin Cancer Res. 2000;6:4171–4175. [PubMed] [Google Scholar]
- 31.Takeuchi H, Fujimoto A, Hoon DS. Detection of mitochondrial DNA alterations in plasma of malignant melanoma patients. Ann N Y Acad Sci. 2004;1022:50–54. doi: 10.1196/annals.1318.009. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama T, Taback B, Nguyen DH, et al. Clinical significance of circulating DNA microsatellite markers in plasma of melanoma patients. Ann N Y Acad Sci. 2000;906:87–98. doi: 10.1111/j.1749-6632.2000.tb06596.x. [DOI] [PubMed] [Google Scholar]
- 33.Umetani N, Fujimoto A, Takeuchi H, et al. Allelic imbalance of APAF-1 locus at 12q23 is related to progression of colorectal carcinoma. Oncogene. 2004;23:8292–8300. doi: 10.1038/sj.onc.1208022. [DOI] [PubMed] [Google Scholar]
- 34.Umetani N, de Maat MF, Mori T, et al. Synthesis of universal unmethylated control DNA by nested whole genome amplification with phi29 DNA polymerase. Biochem Biophys Res Commun. 2005;329:219–223. doi: 10.1016/j.bbrc.2005.01.088. [DOI] [PubMed] [Google Scholar]
- 35.Hoon DS, Bostick P, Kuo C, et al. Molecular markers in blood as surrogate prognostic indicators of melanoma recurrence. Cancer Res. 2000;60:2253–2257. [PubMed] [Google Scholar]
- 36.Shivakumar L, Minna J, Sakamaki T, et al. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309–4318. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vos MD, Martinez A, Elam C, et al. A role for the RASSF1A tumor suppressor in the regulation of tubulin polymerization and genomic stability. Cancer Res. 2004;64:4244–4250. doi: 10.1158/0008-5472.CAN-04-0339. [DOI] [PubMed] [Google Scholar]
- 38.Koul S, McKiernan JM, Narayan G, et al. Role of promoter hypermethylation in cisplatin treatment response of male germ cell tumors. Mol Cancer. 2004;3:16. doi: 10.1186/1476-4598-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiegl H, Millinger S, Mueller-Holzner E, et al. Circulating tumor-specific DNA: A marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Res. 2005;65:1141–1145. doi: 10.1158/0008-5472.CAN-04-2438. [DOI] [PubMed] [Google Scholar]
- 40.Fackler MJ, McVeigh M, Evron E, et al. DNA methylation of RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int J Cancer. 2003;107:970–975. doi: 10.1002/ijc.11508. [DOI] [PubMed] [Google Scholar]
- 41.Youssef EM, Lotan D, Issa JP, et al. Hypermethylation of the retinoic acid receptor-beta(2) gene in head and neck carcinogenesis. Clin Cancer Res. 2004;10:1733–1742. doi: 10.1158/1078-0432.ccr-0989-3. [DOI] [PubMed] [Google Scholar]
- 42.Christmann M, Pick M, Lage H, et al. Acquired resistance of melanoma cells to the antineoplastic agent fotemustine is caused by reactivation of the DNA repair gene MGMT. Int J Cancer. 2001;92:123–129. [PubMed] [Google Scholar]
- 43.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 44.Haqq C, Nosrati M, Sudilovsky D, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]