Abstract
Innate immune defenses are essential for the control of virus infection and are triggered through host recognition of viral macromolecular motifs known as pathogen-associated molecular patterns (PAMPs) 1. Hepatitis C virus (HCV) is an RNA virus that replicates in the liver, and infects 200 million people 2. Infection is governed by hepatic immune defenses triggered by the cellular RIG-I helicase. RIG-I binds PAMP RNA and signals IRF-3 activation to induce the expression of α/β interferon (IFN) and antiviral/interferon-stimulated genes (ISGs) that limit infection 3–10. Here we identified the poly-uridine motif of the HCV genome 3’ nontranslated region (NTR) as the PAMP substrate of RIG-I, and show that this and similar homopoly-uridine motifs present in the genome of RNA viruses is the chief feature of RIG-I recognition and immune triggering 8. 5’ terminal triphosphate on the PAMP RNA was necessary but not sufficient for RIG-I binding, which was primarily dependent upon homopolymeric ribonucleotide composition, linear structure and length. The HCV PAMP RNA stimulated RIG-I-dependent signaling to induce a hepatic innate immune response in vivo, and triggered IFN and ISG expression to suppress HCV infection in vitro. These results provide a conceptual advance by identifying homopoly-uridine motfis present in the genome of HCV and other RNA viruses as the PAMP substrate of RIG-I, and define immunogenic features of the PAMP/RIG-I interaction that could be utilized as an immune adjuvant for vaccine and immunotherapy approaches.
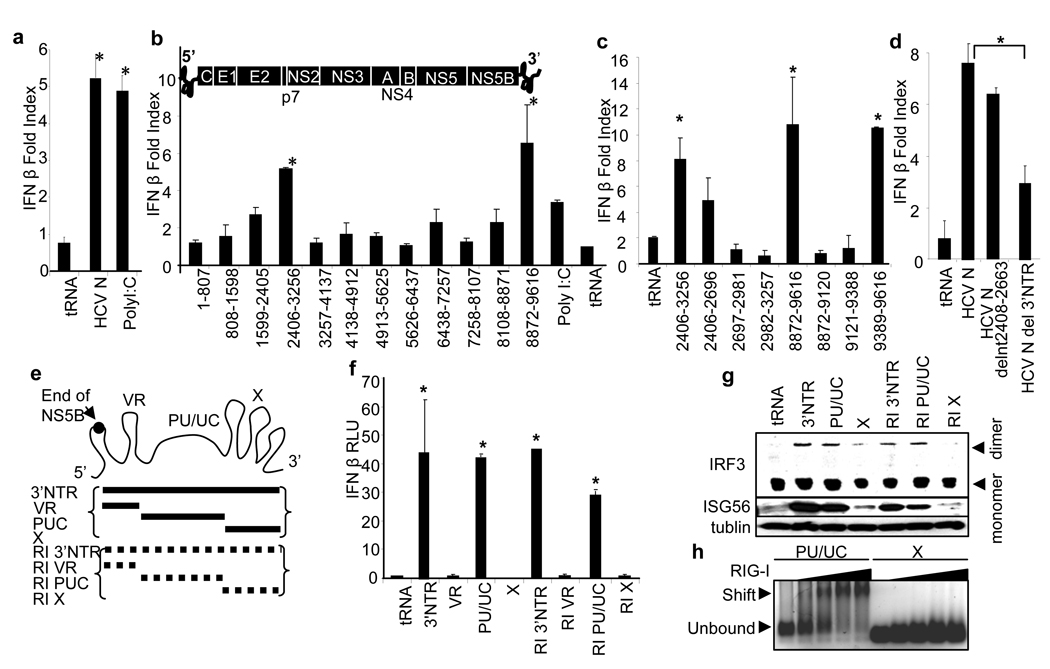
To determine the nature of the HCV PAMP RNA we conducted a functional screen to identify possible HCV PAMP RNA motifs. We assessed the ability of full length HCV genome RNA or contiguous HCV RNA segments to trigger the IFN-β promoter in transfected Huh7 cells. The full-length HCV genome RNA triggered innate immune signaling to induce the IFN-β promoter (Fig. 1a). Two regions of the HCV RNA, encoding nt 2406-3256 and nt 8872-9616, stimulated significant induction of the IFN-β promoter (Fig. 1b) with signaling activity respectively localized to nt 2406-2696 of the open reading frame and nt 9389-9619 encoding the 3’ NTR (Fig.1c). Deletion of nt 9389-9619 but not nt 2408-2663 from the HCV genome significantly attenuated signaling to the IFN-β promoter (Fig 1d). PAMP motifs are typically conserved among strains of a pathogen 1, and sequence comparison of multiple HCV genomes revealed global variability within nt 2406-3696 among virus strains but nt 9389-9616 encoded motifs of high conservation (Fig. S1) 11. Thus, the viral 3’ NTR might encode HCV PAMP motifs that trigger innate immune signaling in the host cell.
Figure 1. Identification of HCV PAMP RNA.
a–d, RNA-induced IFN-β promoter-luciferase activity in Huh7 cells, shown as mean fold-index induction (compared to non-treated cells; ±SD). Huh7 cells were transfected with 1 µg (0.4 pmol ) of HCV N (HCV 1b) genome RNA, 1 µg of poly inosine:cytosine (pIC) RNA (control) or with 1 µg ( 5–10 pmol) of of the indicated RNA species and harvested for dual luciferase assay 16 h later. HCV 1b refers to HCV genome RNA; tRNA, transfer RNA control; b–d, nt numbers encoded by HCV RNA constructs are shown. Bars are placed in their relative positions of each region within the HCV genome shown in b. The 5’ NTR, protein coding regions, and 3’ NTR are indicated. e, The HCV 3’ NTR motifs and respective RNA constructs. RI and broken lines denote replication intermediate. f, IFN-β promoter activation, shown here and in remaining figures as mean relative luciferase units (RLU; ±SD), triggered by 1 µg (20-150 pmole) of the indicated RNA species in transfected Huh7 cells. g, The abundance of IRF-3, ISG56 and tubulin (control) were measured by immunoblot. The upper panel shows the active IRF-3 dimer and inactive monomer forms separated by nondenaturing PAGE. h, RNA binding/gel-shift analysis of purified RIG-I with poly-U/UC or X region RNA (6 pmol) reacted with 0, 10, 20, 40, or 60 pmol of RIG-I protein. All RNAs contain 5’ppp. Asterisks indicate significant difference (P<0.01) as determined by Student’s T-test.
The HCV 3’ NTR is comprised of three regions: a variable region (VR) with potential secondary structure, a nonstructured poly-U/UC region containing polyuridine with interspersed ribocytidine, and the terminal X region containing three conserved stem-loop structures (Fig. 1e) 12. We evaluated the ability of RNA encoding the HCV 3’ NTR or each of its regions to trigger intracellular signaling. Since HCV RNA replicates through a negative-sense replication intermediate (RI) 2,13, we included analyses of signaling triggered by the RI counterparts of the 3’ NTR and its composite regions. RNA encoding the genomic or RI poly-U/UC region was sufficient to trigger signaling to the IFN-β promoter but neither the variable nor X region genomic and RI RNA induced promoter signaling (Fig. 1f). The HCV 3’ NTR and poly-U/UC but not the X region RNA motif similarly stimulated signaling when introduced into Hela cells (Fig. S3a). Moreover, in Huh7 cells genomic or RI 3’ NTR and poly-U/UC RNA each stimulated the formation of active IRF-3 dimers and expression of ISG56, an IRF-3 target gene 3, but X region RNA failed to trigger either (Fig 1g). The poly-U/UC but not X region RNA formed a stable complex with purified RIG-I (Fig. 1h). These results define the 100 nt poly-U/UC region of the HCV genome and RI RNA as the HCV PAMP motif and potential substrate of RIG-I signaling. We also found that the entire HCV 5’ NTR, which contains four major stem-loop structures comprising the viral internal ribosome entry site 14, was only a weak inducer of promoter signaling. However, prior treatment of cells with IFN-β to increase RIG-I levels 4 rendered responsiveness to signaling triggered by the HCV 5’ NTR or X region RNA (Fig. S3). Thus, dsRNA regions of the HCV RNA are not potent PAMPs but may confer signaling during the IFN response.
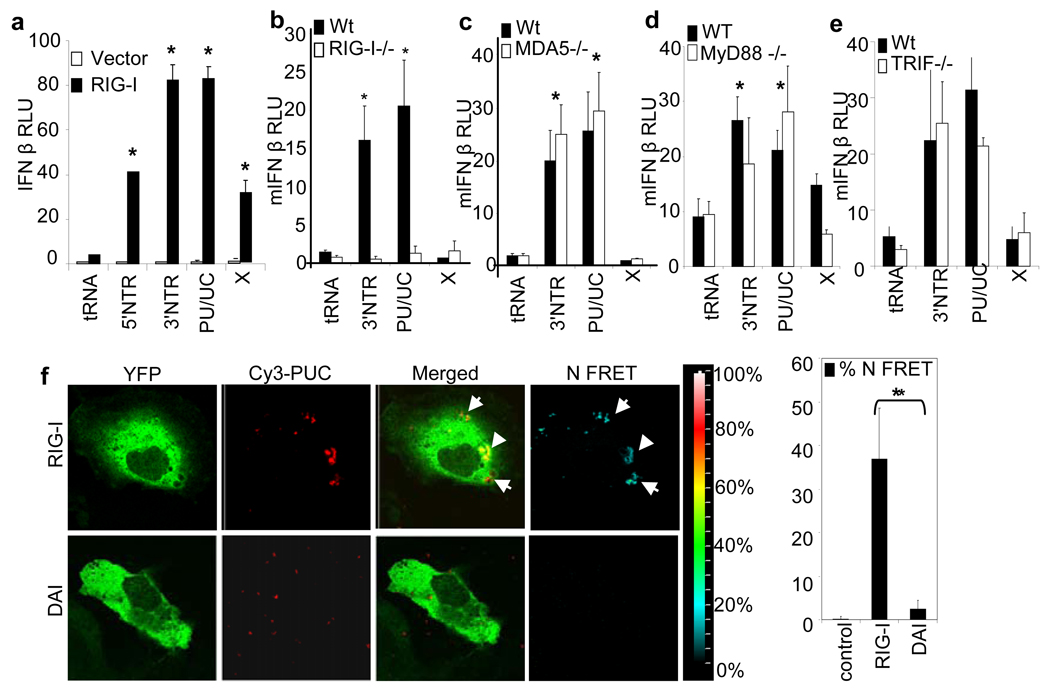
To determine the role of RIG-I or other PRR pathways in HCV PAMP signaling, we first examined IFN-β promoter induction in Huh7.5 cells encoding nonfunctional RIG-I 4. The cells were refractory to HCV RNA-induced signaling whereas their response was rescued and enhanced upon overexpression of wild type RIG-I (Fig. 2a). MDA5 is a PRR related to RIG-I that binds to dsRNA 15, whereas MyD88 and TRIF are essential adaptor proteins respectively used by Toll like receptor (TLR) 7/8 and TLR3, which are PRRs that recognizes poly-U RNA, single-stranded RNA or dsRNA 1,16. We examined PAMP signaling in mouse embryonic fibroblasts (MEFs) lacking RIG-I, MDA5, MyD88 or TRIF (Fig S4). When introduced into RIG-I−/− MEFs the HCV RNA failed to trigger promoter activation while 3’ NTR and poly-U/UC but not X region RNA stimulated signaling in wild type, MDA5−/−, MyD88−/− or TRIF−/− cells (Figs. 2b-e). In Huh7 cells, poly-U/UC RNA colocalized and mediated a specific interaction with RIG-I (Fig. 2f). Thus, RIG-I is the essential PRR that signals the innate immune response triggered by HCV poly-U/UC RNA independently of MDA5, MyD88 or TRIF-dependent PRR pathways.
Figure 2. RIG-I-specific HCV RNA PAMP recognition and signaling.
a–e, Induction of the IFN-β promoter in cells co-transfected with 1 µg (20-30pmol) of tRNA control or the indicated HCV RNA species. a, Huh7.5 cells, lacking functional RIG-I, were cotransfected with a plasmid encoding vector alone or RIG-I. b, Promoter signaling in wild-type (wt) and RIG-I−/−, c, MDA5−/−, d, MyD88−/− or e, TRIF−/− mouse embryo fibroblasts. Asterisk indicate a significant difference (P<0.01) from tRNA control. f, FRET analysis of Cy3-labeled poly-U/UC RNA (Cy3-PUC) interaction with YFP-RIG-I or YFP-DAI protein in co-transfected Huh7 cells. Panels show representative images of YFP, Cy3, merged fluorescence, and N FRET (corrected FRET). The color scale denotes N FRET levels. The bar graph at right shows the calculated values for RNA interaction with RIG-I or DAI. Control values are from the image area that has no colocalization signal. All RNAs contain 5’ppp.
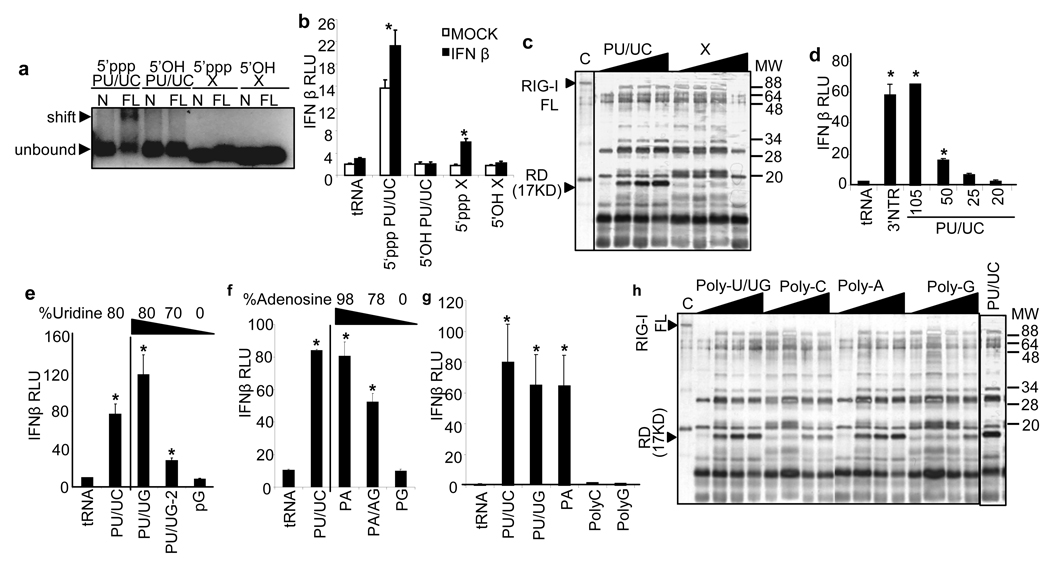
RIG-I binds to PAMP RNA containing 5’ppp through which the triphosphate end is proposed to anchor the RNA within charged residues of the RIG-I repressor domain (RD), causing a conformation change to displace the RD and release signaling autorepression 17–19. Gel-shift assays revealed that 5’ppp was required for poly-U/UC RNA binding by RIG-I but did not mediate stable RIG-I interaction with X region RNA (Fig. 3a). 5’ppp was required for IFN-β promoter signaling by poly-U/UC RNA, and supported low-level promoter induction triggered by X region RNA in IFN-treated cells (Fig. 3b). Since X region RNA failed to bind RIG-I but weakly triggered signaling, stable RIG-I/RNA interaction is most-likely required to release RIG-I autorepression. We therefore conducted limited trypsin digestion analysis of purified RIG-I alone or bound to poly-U/UC or X region RNA containing 5’ppp. This approach provides an assessment of RIG-I RD displacement in response to PAMP RNA binding wherein the displaced RD of signaling-active RIG-I presents as a protected 17 kDa fragment 17–19. As shown in Fig. 3c, RIG-I binding of poly-U/UC but not X region RNA rendered the protected 17 kDa RD fragment. These results indicate that 5’ppp is necessary but not sufficient for RNA binding by RIG-I wherein the HCV poly-U/UC RNA directs stable interaction with RIG-I in a 5’ppp-dependent manner that confers RIG-I signaling activation. We employed 5’ppp RNA in all further experiments.
Figure 3. Poly-uridine and poly-adenosine ribonucleotides are RIG-I ligands.
a, Gel shift analysis of complex formation between 25 pmol of purified N-RIG (control) or full-length RIG-I (FL) and 10 pmol of poly-U/UC (PU/UC) or X region RNA containing 5’ppp or 5’OH as indicated. Arrows denote position of unbound RNA and RNA/RIG-I complexes. b, Effect of 5’ppp on IFN-β promoter activity. Huh7 cells were either mock-treated or treated with IFN-β 8 h prior to transfection with 1 µg (30 pmol ) of RNA. c, Effect of poly-U/UC or X region RNA on RIG-I activation. The silver-stained gel image shows trypsin-digestion products of RIG-I that was pre-incubated with increasing amounts poly-U/UC or X region RNA. Arrows indicate positions of full length (FL) RIG-I and the 17 kDa trypsin-resistant RD of from RIG-I/RNA complexes. d, Effect of nt length of 1 µg (30-150 pmol) poly-U/UC 3’ truncation products on IFN-β promoter signaling in Huh7 cells. e–g, Effect of nt composition on IFN-β promoter signaling in Huh7 cells transfected with 1 µg (30 pmol ) of RNA. h, Effect of nt composition on RIG-I activation. The silver-stained gel image shows trypsin-digestion products of RIG-I that was pre-incubated with increasing amounts poly-U/UG, poly-C, poly-A, or poly-G RNA. Arrows indicate positions of full length RIG-I and the 17 kDa trypsin-resistant RD. We confirmed the 17 kDa fragment as the RIG-I RD by immunoblot analysis of the digestion products using an antiserum specific to the RIG-I carboxyl terminus (not shown), as previously described 17. Asterisks indicate significant difference (P<0.01) as determined by Student’s T-test.
The HCV polyU/UC region is a flexible motif among HCV strains and is essential for viral replication 13. In the HCV genotype 1b strain used in these experiments, the poly-U/UC region is comprised of 100 nt containing 78% uridine and 22% ribocytosine (Fig. S4). We evaluated the requirements of length and nt composition for RIG-I signaling by the poly-U/UC motif. A reduction of length through progressive 3’ truncation to 50 nt or less attenuated signaling in Huh7 cells (Fig. 3d), consistent with reduced RIG-I binding of short RNAs 20. Replacement of ribocytosine with riboguannine (poly-U/UG) had no impact on PAMP signaling but progressive replacement of uridine for riboguanine to below 80% uridine or 100% polyriboguanine (poly-G) reduced or abrogated PAMP signaling, respectively, in both Huh7 cells (Fig. 3e) and in Hela cells (Fig. S2b). Since the RI poly-U/UC RNA contains high polymeric riboadenosine composition, we also examined the impact of poly-A composition and RNA length on RIG-I signaling induced by 100 nt constructs (Fig. S4a). Synthetic poly-A RNA and poly-U/UC RNA equally induced signaling to the IFN-β promoter whereas reduced length to 50 nt or lower of the poly-A RNA significantly attenuated signaling activity (Fig S2c). Moreover, reduced riboadenosine content of the RI poly-U/UC RNA or alteration to poly-G comparatively attenuated or ablated signaling, respectively (Fig. 3f). In side-by-side analyses we found that poly-U/UC, poly-U/UG, and poly-A RNA, but neither poly-C nor poly-G RNA could trigger signaling to the IFN-β promoter (Fig. 3g). Whereas poly C and poly G RNA bound negligibly to RIG-I, poly-U/UC, poly-U/UG and poly-A RNA formed a stable complex with RIG-I (Fig. S4b) and and release of the RIG-I RD to the active conformation (Fig. 3h). These results demonstrate that 50-100 nt length polymeric uridine or riboadenosine motifs, including the PU/UC and RI PU/UC motif of HCV, constitute the PAMP motif that is efficiently recognized by RIG-I to trigger innate immune response.
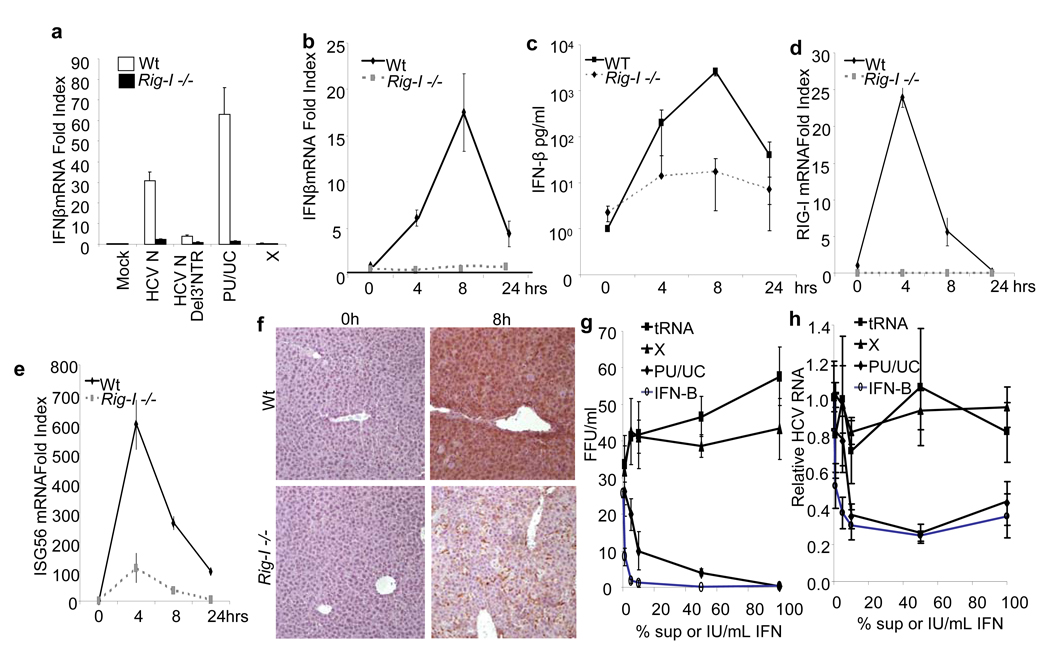
To determine if RIG-I recognizes the HCV poly-U/UC PAMP RNA to trigger hepatic innate immune defenses in vivo, we conducted RNA signaling analysis in wild type and RIG-I−/− mice. Intravenous administration of full-length HCV 1b genome stimulated hepatic IFN-β mRNA expression within 8 hrs in wild type mice but not in RIG-I−/− mice, and this occurred in a manner dependent on the viral 3’ NTR (Fig. 4a). Moreover, the poly-U/UC RNA but not the X region RNA motif was sufficient to trigger the hepatic IFN-β expression in wild type but not in RIG-I−/− mice. In time course studies we found that the poly-U/UC RNA motif induced a peak of hepatic IFN-β mRNA expression and IFN-β serum levels at 8 hrs post-injection in wild-type mice (Figs. 4b and 4c). This response associated with induced hepatic expression of RIG-I and ISG56 mRNA and tissue-wide expression of hepatic ISG54 (Figs. 4d–4f), similar to the hepatic response in HCV-infected patients 9,10. RIG-I−/− mice expressed only a low level of IFN-β and ISG56. The tissue-wide nature of hepatic ISG54 expression in wild-type mice suggests that paracrine signaling of IFN-β could play an important role in hepatic defenses against HCV. To test this idea we measured HCV production from Huh7 cells that were treated with IFN-β or supernatants collected from cultures transfected with HCV poly-U/UC RNA, X-region RNA, or tRNA (control). Poly-U/UC RNA triggered IFN-β expression in the transfected cells (data not shown), and only treatment with IFN-β or supernatant from the poly-U/UC-transfected cells induced a response that suppressed HCV infection (Figs. 4g and 4h). Thus, the poly-U/UC RNA is an HCV genome PAMP that is necessary and sufficient to trigger RIG-I signaling of the hepatic innate immune response. The actions of RIG-I signaling can induce an antiviral response directly (Fig. 1g) and through indirect, paracrine actions of IFN produced from HCV PAMP signaling (Figs. 4g and 4h).
Figure 4. HCV PAMP RNA triggers the hepatic innate immune response and anti-HCV defenses.
a–f, Wild-type or RIG-I−/− mice (n = 3) were hyrdrodynamically transfected intravenously with HCV RNA. a, mice received 100 μg of HCV 1b genome, HCV 1b genome lacking the 3’NTR (HCV 1b Δ 3’NTR), PU/UC RNA or X region RNA. Hepatic IFN-β mRNA expression was measured 8 hrs later. b–f, Wild-type or RIG-I −/− mice (n = 3) received 200 μg of poly-U/UC RNA or buffer control, and were sacrificed 4, 8 or 24 h later for comparative measurement of mRNA and protein expression. b, Liver-specific expression of IFN-β mRNA. c, Serum IFN-β protein levels. d, Liver-specific expression of RIG-I mRNA. e, Liver-specific expression of ISG56 mRNA. f, Immunohistochemcial stain of ISG56 protein expression in liver tissue sections. g and h, Paracrine antiviral effect of the innate immune response triggered by HCV PAMP RNA. g, Inhibition of HCV infection in pretreated cells. Triplicate cultures of Huh7.5 cells were treated with IFN-β or conditioned media collected from Huh7 cells transfected with the indicated RNA species for 12 h prior to HCV infection. The graph shows the number of infected cells (±SD) as determined by focus forming unit (FFU) assay at 48 h postinfection. h, Huh7.5 cells were infected with HCV for 48 h and then were treated with increasing concentrations of IFN-β or the indicated conditioned media for an additional 48 h. Intracellular HCV RNA levels relative to GAPDH were determined and are plotted as mean HCV RNA index (±SD) relative to infected, untreated cells.
Our results provide new insights into the features of RNA PAMP specificity of RIG-I wherein nt composition consisting of viral genome RNA poly-U and respective RI RNA poly-A of greater than 50 nt length is a major determinant that confers RIG-I binding and signaling. 5’ppp was necessary but not sufficient for stable binding of HCV PAMP RNA by RIG-I. In terms of the HCV genome, well-defined internal RNA interactions of the 5’ and 3’ ends 21,22 could provide 5’ppp and PAMP motif proximity for stable RIG-I binding. The poly-U/UC motif is an essential determinant of HCV replication fitness 21. Thus, while the virus must maintain this motif for its viability, the host takes advantage of this requirement and targets the poly-U/UC region as a discriminator of PAMP RNA through RIG-I interaction. Poly-U and/or poly-A motifs are present in localized regions in the genome of RNA viruses known to trigger RIG-I signaling (Table S1) 5,8,19. We found that 5’ppp genomic poly-U/A rich RNA motifs encoded in the rabies virus leader, Ebola virus 3’ region, or the measles virus leader sequence each triggered signaling to the IFN-β promoter in Huh7 cells but GC-rich RNA motifs from each viral genome did not trigger a significant response (Figs. S5a and S5b). We also found that pppT3-63, Tri-GFP and EGFP #2 T7 RNAs, previously described as RIG-I substrates and comprised of 50% or fewer A/U nts 5,18,23 , could induce only weak signaling to the IFN-β promoter compared to the poly-U/UC RNA . Thus, A/U composition and poly-U motifs are major determinants of viral PAMP RNA recognition by RIG-I. Cellular RNAs also contain poly-U and poly-A motifs but mRNAs are typically capped and are bound by poly-A binding proteins 24 while ribosomal RNA are “masked” as ribonucleoprotein complexes 25. These features and the context of 5’ppp with viral poly-U and poly-A motifs serve to identify self from nonself RNA by governing RIG-I recognition, wherein, non-self recognition of the HCV PAMP RNA triggers a hepatic innate immune response. These observations provide a possible explanation of why the 25% or more of all HCV-exposed people clear acute infection 2 and why HCV needs to evade innate immunity through the viral NS3/4A protease targeting of the RIG-I pathway 26,27. RIG-I substrates such as the poly-U/UC RNA or structurally-similar compounds could provide therapeutic application as immune adjuvants similar to TLR agonists 28, and offer innate immune stimulatory properties that may improve IFN-based therapy for HCV through paracrine immune actions that limit infection 2.
Methods Summary
RNA
RNA was synthesized from plasmid DNA, PCR products or annealed DNA oligonucleotides encoding the T7 promoter using the T7 Megascript kit or MEGAshortscript kit (Ambion). Full-length HCV RNA and subgenomic constructs were respectively produced from plasmid DNA or T7 promoter-linked PCR products generated from cloned HCV N (A gift from S. Lemon) or Con1 genome RNA (genotype 1b) 29. Chemically synthesized 5’OH RNAs were purchased from Fidelitysystems. RNA transfection was performed using the Transmessenger kit (QIAGEN). 1 µg of RNA was transfected into 1x105 cells. This mass of RNA is equivalent to the following number of moles: Full-length HCV 1b genome, HCV 1b Δ2408-2663 and HCV 1b Δ3’NTR: 0.4 pmol; HCV 1b subgenomic RNA constructs: 5 or10 pmol, HCV genotype 1b (Con1) 5’NTR, 3’NTR (20 pmol), and all other RNA constructs: 30 pmol. RNA concentrations in the transfection mix were 5-10 µg/ml. RNA expression was determined by RT-quantitative PCR assay. RNA delivery efficiency was assessed as described in Fig S6.
RNA signaling analysis
IFN-β promoter-luciferase analyses of transfected cells were conducted as described 17. Protein expression and the abundance of IRF-3 dimer and monomeric forms in transfected cells were determined by immunoblot analysis of extracts 17.
RIG-I purification and RNA binding analysis
Full-length RIG or RIG-I aa 1-228 (N-RIG) were expressed in E. coli and purified by column chromatography. RIG-I/RNA complexes were assessed by gel-shift assay of purified RIG-I reacted with RNA, separated by agarose gel electrophoresis, and visualized by Sybr Green staining. RIG-I activation/conformation shift was analyzed using the limited trypsin digestion method as described 17. FRET analysis of Cy3-poly-U/UC RNA interaction with YFP-RIG-I or YFP-DAI protein constructs was conducted using N-FRET on a Ziess confocal microscope 30. Serum IFN-β levels were measured by ELISA (PBL, Inc.)
Mice
Wt and RIG-I-/- mice 15 were from Dr. S. Akira. RNA was transfected in mice using the lipid based in vivo RNA transfection reagent (Altogen).
Statistical analysis
Differences between groups were analyzed for statistical significance by the Student's t-test.
Supplementary Material
Acknowledgements
We thank Dr. T. Fujita helpful discussion of our data, and Dr. S. Horner for helpful discussion and manuscript review. We thank Drs. T. Fujita, S. Lemon, G. Sen, C. Rice, T. Taniguchi, A. Miyawaki, S. Akira for reagents, R. Hirai for technical consultations and S. Thomas and G. Martin for technical assistance. Supported by funds from the State of Washington, and National Institutes of Health grants AI060389, AI40035, the Burroughs-Wellcome Fund, and a gift from Mr. and Mrs. R. Batcheldor.
Reference List
- 1.Saito T, Gale M. Principles of intracellular viral recognition. Current Opinion in Immunology. 2007;19:17–23. doi: 10.1016/j.coi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Lauer GM, Walker BD. Medical progress: Hepatitis C virus infection. New England Journal of Medicine. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 3.Gale M, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- 4.Stumper R, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. Journal of Virology. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornung V, et al. 5 '-triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5 '-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature Immunology. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 8.Loo YM, et al. Distinct RIG-I and MDA5 signaling regulation by RNA viruses in innate immunity. Journal of Virology. 2007;27:697. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau DT, et al. Interferon regulatory factor-3 activation, hepatic interferon-stimulated gene expression, and immune cell infiltration in hepatitis C virus patients. Hepatology. 2008 doi: 10.1002/hep.22076. [DOI] [PubMed] [Google Scholar]
- 10.Smith MW, et al. Gene expression patterns that correlate with hepatitis C and early progression to fibrosis in liver transplant recipients. Gastroenterology. 2006;130:179–187. doi: 10.1053/j.gastro.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Simmonds P. Genetic diversity and evolution of hepatitis C virus - 15 years on. Journal of General Virology. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 12.Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3 ' nontranslated region are essential for virus replication in vivo. Journal of Virology. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi MK, Lemon SM. 3 ' Nontranslated RNA signals required for replication of hepatitis C virus RNA. Journal of Virology. 2003;77:3557–3568. doi: 10.1128/JVI.77.6.3557-3568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda M, et al. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 15.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 16.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 17.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahasi K, et al. Nonself RNA-Sensing Mechanism of RIG-I Helicase and Activation of Antiviral Immune Responses. Molecular Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 19.Cui S, et al. The C-Terminal Regulatory Domain Is the RNA 5'-Triphosphate Sensor of RIG-I. Molecular Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 20.Marques JT, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nature Biotechnology. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 21.You S, Rice CM. 3' RNA elements in hepatitis C virus replication: kissing partners and long poly(U) J Virol. 2008;82:184–195. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito T, Lai MMC. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3 '-untranslated sequence. Virology. 1999;254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 23.Kim DH, et al. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nature Biotechnology. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 24.Afonina E, Stauber R, Pavlakis GN. The human Poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. Journal of Biological Chemistry. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 25.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 angstrom resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 26.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 27.Loo YM, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tse K, Horner AA. Update on toll-like receptor-directed therapies for human disease. Annals of the Rheumatic Diseases. 2007;66:77–80. doi: 10.1136/ard.2007.078998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beard MR, et al. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–324. doi: 10.1002/hep.510300137. [DOI] [PubMed] [Google Scholar]
- 30.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–U14. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.