Abstract
Human and other mammalian thymidylate synthase (TS) enzymes have an N-terminal extension of about 27 amino acids which is not present in bacterial TSs. The extension, which is disordered in all reported crystal structures of TSs, has been considered to play a primary role in protein turnover but not in catalytic activity. In mammalian cells, the variant V3A has a half-life similar to that of wild type human TS (wt hTS) while V3T is much more stable; V3L, V3F and V3Y have half-lives approximately half of that for wt hTS. Catalytic turnover rates for most Val3 mutants are only slightly diminished, as expected. However, two mutants, V3L and V3F, have strongly compromised dUMP binding, with Km,app values increased by factors of 47 and 58, respectively. For V3L, this observation can be explained by stabilization of the inactive conformation of loop 181–197, which prevents substrate binding. In the crystal structure of V3L, electron density corresponding to a leucine residue is present in a position which stabilizes loop 181–197 in the inactive conformation. Since this density is not observed in other mutants and all other leucine residues are ordered in this structure, it is likely that this density represents Leu3. In the crystal structure of a binary complex V3F·FdUMP, the nucleotide is bound in an alternative mode to that proposed for the catalytic complex, indicating that the high Km,app value is caused not by stabilization of the inactive conformer but by substrate binding in a non-productive, inhibitory site. These observations show that the N-terminal extension affects the conformational state of the hTS catalytic region. Each of the mechanisms leading to the high Km,app values can be exploited to facilitate design of compounds acting as allosteric inhibitors of hTS.
Keywords: thymidylate synthase, protein turnover, conformational switching, allosteric inhibition, crystal structure
Thymidylate synthase (TS) is the essential enzyme that catalyzes the formation of dTMP from dUMP, using 5, 10-methylenetetrahydrofolate (mTHF) as a co substrate (1). The reaction is initiated by the active site cysteine, Cys195 in human TS (hTS), attacking the carbon-6 of dUMP. This leads to the formation of a covalent bond between TS and dUMP as well as activation of the carbon-5 of dUMP, generating a second covalent bond between dUMP and mTHF and ultimately a ternary catalytic complex. This reaction is the only de novo source of intracellular dTMP, although the thymidine salvage pathway may function as an alternative extracellular source of dTMP (2). Inhibition of hTS in rapidly dividing cells leads to nucleotide imbalance and ultimately results in apoptosis. For this reason, TS has been an important target in the chemotherapy of colon cancer and some other malignancies.
hTS is a homodimer of 313 amino acids, and in general, the TS amino acid sequences are very highly conserved. The major differences between mammalian TSs and those from bacterial sources is the presence of an N-terminal extension of approximately 25–29 amino acids and two insertions of 12 and 8 residues at positions 117 and 145, respectively (2). Unlike the sequence in the catalytic part of the molecule, the sequence of the N-terminal extension is poorly conserved. X-ray crystallography of the rat and human TS enzymes have shown that this region is intrinsically disordered (3,4,5,6). Although extensive sequence divergence has occurred during the evolution of the N-terminal regions of TS polypeptides in mammalian species, a disordered structure with a high proline content and high frequency of disorder-promoting residues compared to the rest of the TS molecule, has been conserved. Also a Pro residue at the penultimate site, is conserved in all species examined with the exception of mouse TS, (7).
The N-terminal extension has been shown to play an important role in determining the intracellular stability of hTS and to control its degradation. Biochemical and genetic evidence indicate that degradation of the hTS polypeptide is carried out by the 26S proteasome but does not require ubiquitinylation or the ubiquitinylation pathway (8). The N-terminal region, in particular, the disordered first 29 residues, directs the protein to the ubiquitin-independent degradation pathway (8, 9). Deletion of the first two to six residues results in very stable enzymes with half-lives greater than 48 hours. In addition, single amino acid substitutions at the penultimate site, Pro2, have a profound impact on the half-life of the enzyme (8, 9). Previous studies by Edman degradation experiments showed that the primary sequence of hTS begins with an unblocked Pro residue indicating that the protein undergoes posttranslational modification by Met excision (10). Analyses of hTS mutants with substitutions at Pro2 by MALDI-TOF showed that unstable mutants such as those with P2V and P2A substitutions undergo Met excision. On the other hand, stable mutants such as those wherein Pro2 has been replaced with the remaining amino acids, undergo either Nα-acetylation or both Met excision and Nα-acetylation resulting in a blocked N-terminus (7). These results suggest that a free, unblocked N-terminus is a requisite for efficient degradation of hTS by the proteasome. Modifications such as Nα-acetylation might stabilize the enzyme by hindering its ability to efficiently recognize and/or enter the proteasomal chamber (7).
The role of the N-terminus in directing protein degradation is underscored by its ability to destabilize E. coli TS (ecTS). This enzyme lacks the N-terminal extension and has a half-life of greater than 48 hours in mammalian cells. Fusion of the first 29 amino acids of hTS to the N-terminus of the ecTS reduced its half life to less than 4 hours (9).
Furthermore, fusion of the first 45 amino acids, which includes the disordered region and the adjacent alpha helix of hTS to the enhanced green fluorescent protein (eGFP), destabilized this structurally unrelated protein from a half-life greater than 48 hours to approximately 7 hours (7). Mutations in the N-terminus that affect the half-life of hTS exerted the same effects on the half-life of the N-terminal fusions with ecTS and eGFP, indicating that this region functions as a degron by promoting the degradation of an unrelated protein to which it is fused (7,9)
A unique feature of hTS is the existence of loop 181–197 in two conformations (3,4). One is similar to those found in TS enzymes from other sources and in complexes of hTS with substrate analogs (11); it is referred to as the “active conformation”. In the second “inactive” conformation, loop 181–197 is flipped 180° and the catalytic Cys195 is located outside the active site (4), making the enzyme inactive. Studies of conformational switching in solution indicated that the two conformations are represented to a similar extent and that binding of dUMP shifts the equilibrium towards the active conformation while binding of phosphate or sulfate ions shifts the equilibrium towards the inactive conformation (3). Crystallization of TS proteins is facilitated by the presence of phosphate or sulfate ions; therefore, all crystal structures of native wt hTS are in the inactive conformation. Two crystallization conditions that yielded good crystals of hTS are 1.8–2.0 M ammonium sulfate (AS) (12) and a relatively low ionic strength, 30 mM AS, with polyethylene glycol (PEG) as precipitant (13).
In the native hTS structure obtained at low AS concentration (inactive conformer), electron density consistent with a tert-butyl moiety was observed (13). This density is likely from a valine or a leucine from a disordered region and the corresponding residue interacts with a hydrophobic pocket which includes Trp182 and Leu187 from loop 181–197. The disordered regions in the structure correspond to the N-terminal extension, which contains Val3, and loop 108–129, which contains Leu118 and Leu121. The hydrophobic interactions between the tert-butyl and loop181–197 must stabilize the inactive conformer. A hTS variant, missing residues 7–29, crystallized with loop 181–197 in the active conformation at very similar crystallization conditions with 50 mM phosphate (14). Thus, the Δ7–29 deletion affects the conformation of loop 181–197. It appeared likely that this effect is mediated by Val3 and an expansion of the tert-butyl moiety into a strongly binding ligand should generate an allosteric inhibitor of hTS. We report here studies of hTS variants at position 3 which confirm the Val3 hypothesis.
MATERIALS AND METHODS
Bacterial Strains and Plasmids
The Escherichia coli strain TX61− (thyA−) and the pTS080 plasmid, expressing wt-hTS, were generously provided by Walter S. Dallas (Glaxo Wellcome, Research Triangle Park, NC) and have been described previously (15, 16). pTS080 contains resistance genes for ampicillin and tetracycline. TX61− is a thy− kanamycin-resistant mutant of BL21(DE3) and lacks detectable TS activity (15). For propagation, pTS080 was transformed into competent JM109 bacterial cells and grown in LB medium containing 50 µg/mL tetracycline. Plasmid DNA was isolated using the Qiagen Plasmid prep kit according to manufacturers’ instructions. Plasmid pJZ205 containing the hTS cDNA under the control of the SV40 promoter has been described previously (17) and was propagated in DH5-α bacterial strain.
Cells and Cell culture
The cell line RJK88.13, which is a TS-deficient derivative of V79 Chinese hamster lung cells, was obtained from Dr. Robert Nussbaum (University of Pennsylvania). These cells were maintained in Dulbecco’s modified Eagle’s medium (Cellgro) containing 4.5 g/L glucose and supplemented with 10% heat inactivated fetal bovine serum (Cellgro) and 10 µM thymidine. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere. To determine half-lives of TS mutants, cycloheximide (50 µg/mL Sigma Aldrich) was added to growing cells followed by harvest at the indicated time points.
Sited Directed Mutagenesis
The mutagenic primers were designed to be complementary to the sense strand of human TS cDNA. Mutagenic primers were synthesized and purified by Integrated DNA Technology (IDT, Coralville, IA). Site directed mutagenesis was performed using the Quick Change™ method (Stratagene, LaJolla,CA) with modifications as described previously (17). Mutations were confirmed by LiCor DNA sequence analysis (MUSC Biotechnology Resource Laboratory, Charleston, SC) of the entire TS gene. Plasmids containing the correct mutations were transformed into the TX61− bacterial strain and screened successively on LB plates containing 50 µg/mL tetracycline and 40 µg/mL kanamycin. Colonies were picked from the kanamycin plates, grown in 5mL LB containing 50 µg/mL of tetracycline, and used to make cell stocks for protein expression and purification.
For expression of Val3 mutants in RJK88.13 cells, plasmid pJZ205 was used as a template for mutagenesis as described above. Sequences of primers used to create V3A, V3T, V3L, V3Y, and V3F substitutions at codon 3 are available upon request. Following verification of mutations by sequencing, plasmids were stably transfected into TS-deficient RJK88.13 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, California) following manufacturer’s instructions. Stable transfectants were selected in media lacking thymidine and containing 5 µM dipyridamole, a nucleoside transport inhibitor, to block the thymidine salvage pathway. Stable transfectants were pooled together and grown in mass culture.
Cell-free Extracts
To prepare bacterial protein extracts, TX61 cells containing mutated pTSO80 plasmids were inoculated into 5 to 10 mL of LB containing 50 µg/mL tetracycline and grown overnight at 37°C. Cells in exponential growth were collected by centrifugation, resuspended in 300 µL of buffer A (50 mM Tris-HCl, pH 7.4, 1 mM EDTA and 14 mM β-ME) and sonicated using a Biosonik iV sonicator (Bronwill, Rochester, NY). The sonicated cells were centrifuged at 13,000 ×g at 4 °C for 10 minutes. The supernatant was separated from the cell debris.
To prepare extracts from stably transfected RJK88.13 cells, cells were split into 60 mm plates and cycloheximide (50 µg/mL) was added after 24 hours. Cells were harvested at the indicated time points and lysed by sonication (3 × 10 sec) in NET2 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.05% Nonidet P-40) containing 10 mM dithiothreitol (DTT), 2 mM β-mercaptoethanol, 5 mM phenylmethylsulfonyl fluoride (PMSF), 200 µg/mL aprotinin, 100 µg/mL pepstatin, and 50 µg/mL leupeptin. The crude lysates were centrifuged at 17,000 ×g for 1 h at 4°C and supernatant was separated from the cell debris. Total protein from all extracts was quantified using the Bradford BioRad assay reagent (18) with bovine serum albumin as standard.
Western Blotting
Total protein (15 – 50 µg per lane) was separated by electrophoresis in 12% SDS-PAGE gels and transferred to Immobilon-P PVDF membrane (Millipore, Bedford, MA) in buffer containing 25 mM Tris-base, 192 mM glycine, and 20% methanol with a Bio-Rad Trans-Blot SD apparatus (Bio-Rad, Hercules, CA). For immunodetection, standard procedures were employed. Cross-reacting protein was detected as described previously (19) by utilizing the murine anti-human TS monoclonal antibody, D3B31 (1:1000 dilution in TBS-T), as the primary antibody; goat anti-murine monoclonal antibody conjugated to horseradish peroxidase (1:2500 dilution in TBS-T) as the secondary antibody (Bio-Rad, Hercules, CA) and lumigen PS-3 acridan (ECL Plus, Amersham Pharmacia Biotech, Piscataway, NJ) as the horseradish peroxidase (HPR) substrate. The HPR reaction was visualized by chemiluminescence and autoradiography. Autoradiographs were digitized by scanning at 600 dpi using a Hewlett Packard C4100 Scanner. Protein levels were analyzed by densitometry using an Odyssey Infrared Imaging System (LI-COR, Lincoln, NE) or by ImageJ software (NIH, Bethesda, Maryland).
Analysis of Growth Rates
For qualitative analysis, TX61− cells transformed with pTS080 constructs, were streaked on agar-minimal medium plates in the presence or absence of 100 µg/mL thymidine overnight at 37°C. For quantitative analysis, transformants were incubated overnight at 37°C in minimal medium (20), supplemented with 100 µg/mL thymidine, 25 µg/mL kanamycin and 10 µg/mL tetracycline. The cultures were diluted 1:100 into minimal medium with or without 100 µg/mL thymidine and supplemented with 25 µg/mL kanamycin and 10 µg/mL tetracycline. The cultures were incubated at 37°C and optical densities were determined using a Klett-Summerson colorimeter with a green filter over a 9 hour period. Untransformed TX61− cells were used as a negative control.
Expression and Purification of Recombinant Human TS Enzymes
The recombinant proteins were purified as described previously (17).
Enzyme Kinetics
Enzyme activity was measured spectrophotometrically by monitoring the absorbance change accompanying the conversion of mTHF to DHF (ε340 = 6.4 mM−1 cm−1) using a Shimadzu UV 1601 spectrophotometer (Shimadzu Corporation, Japan) Measurements were conducted at 37°C in buffer A. One unit of enzyme activity is defined as the amount of enzyme required to synthesize 1 µmole of thymidylate per minute. For determination of kinetic constants for dUMP, initial velocities were measured by utilizing 40 nM of purified protein, 0.15 mM of mTHF, and dUMP concentrations ranging from 25 µM to values between 100 and 400 µM. The reaction mixture, dUMP and buffer A, was preincubated for 3 min at 37°C prior to addition of enzyme, immediately followed by addition of the mixture to a prewarmed cuvette containing mTHF. Data presented are averaged from three independent determinations. Kinetic constants were determined by fitting the data by nonlinear least squares analysis to the hyperbolic form of the Michaelis-Menten rate equation using the program KaleidaGraph WIN (v.3.6x Synergy Software, Reading, PA).
Crystallization
High-salt crystals of four Val 3 mutants were grown using the hanging drop method in high ammonium sulfate (AS) conditions as described previously (3). Briefly, 100 mM Tris-base, pH 8.5, 20 mM BME (2-Mercaptoethanol), and 32–40% ammonium sulfate (AS) was used.
For co-crystallization experiments, the proteins were incubated with 2 mM dUMP or FdUMP overnight and then separated from excess ligand by sephadex G-50–80 gel filtration (Sigma). The final protein concentrations were 8.0 mg/mL, 4.5 mg/mL, and 6.5 mg/mL, for V3L, V3F, and V3Y respectively. V3L low-salt crystals were grown in 100 mM Tris-base, pH 8.5, 20 mM BME, 30 mM AS, and 16% polyethylene glycol (PEG) 4000. V3F low-salt crystals were grown in 100 mM Tris-base, pH 8.0, 20 mM BME, 30 mM AS, and 24% PEG 4000. V3Y low-salt crystals were grown in 100 mM Tris-base, pH 8.0, 20 mM BME, 30 mM AS and 24% PEG 4000. Crystals were transferred to artificial mother liquor additionally containing 20% ethylene glycol and flash frozen in liquid nitrogen vapors.
X-ray Diffraction Data Collection and Structure Determination
Data were collected on ID22 or BM22 beamlines at Argonne National Laboratory and processed with HKL2000 software (21). Data collection and processing statistics are listed in Table 1. The high-AS structures of V3T, V3F, and V3L were solved using molecular replacement with the CNS software (22) using the hTS structure (1YPV) as the search model. The crystal structure of co-crystallized V3F was solved with the AMoRe software (23) from CCP4 suite of programs (24) using the R163K structure (PDB code 2RD8), which represents the active conformer (25), as the starting model. The crystal structures of V3L and V3Y at low-salt conditions were solved by molecular replacement to compensate for slightly different unit cell dimensions using the V3F structure as the starting model. Structure rebuilding and subsequent refinements were performed using the Turbo-Frodo software (26) and CNS, respectively. The final refinements of the high-AS structures were carried out using Refmac5 (27), and the final refinements of low-salt structures were performed using CNS. Superpositions were calculated using the LSQKAB software (28) from the CCP4 suite of programs. Figures 3, 5, and 6 were prepared using Turbo-Frodo. Figures 4, 7, 8a, and 9 were made using MOLSCRIPT (29) and Raster3D (30). R-factors of the final models are presented in Table 1 along with data collection and other refinement statistics.
Table 1.
Crystallographic Data and Refinement Statistics for Val3 mutants Structures
| Co-crystallized V3F |
Co-crystallized V3L |
Co-crystallized V3Y |
||
|---|---|---|---|---|
| X-Ray Source | APS SER-CAT 22ID | APS SER-CAT 22ID | APS SER-CAT 22ID | |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 | |
| Space group | C2 | P31 2 1 | C2 | |
| Unit cell dimensions: | a (Å) | 157.43 | 96.349 | 157.56 |
| b (Å) | 94.64 | 96.349 | 94.86 | |
| c (Å) | 131.99 | 83.56 | 130.86 | |
| Resolution Range (Å) (highest shell) | 44.0-2.75 (2.85-2.75) | 50.00-3.0 (3.11-3.0) | 50.00-2.75 (2.85-2.75) | |
| Mosaicity (degree) | 0.86 | 0.48 | 1.5 | |
| Completeness (%),(highest shell) | 78.5(26.4) | 75.5(22.6) | 88.2(67.0) | |
| Total linear R-merge | 0.085 | 0.195 | 0.086 | |
| Reflections Observed | 33960 | 6685 | 36191 | |
| I over average sigma(I) | 15.5 | 5.3 | 13.5 | |
| Redundancy, (highest shell) | 2.9 (1.2) | 2.5 (1.1) | 1.6 (1.4) | |
| Rmsd bonds (Å) | 0.009 | 0.012 | 0.053 | |
| Rmsd angles (°) | 1.5 | 2.3 | 4.4 | |
| Ramachandran Statistics | ||||
| Residues in most favored regions | 753 (78.1%) | 161(72.2%) | 704(73.2%) | |
| Residues in additionally allowed regions | 210 (21.8%) | 59(26.5%) | 257(26.7%) | |
| Residues in generaously allowed regions | 1 (0.1%) | 3(1.3%) | 1(0.1%) | |
| Residues in disallowed regions | 0 | 0 | 0 | |
| Rc | 0.232 (CNS) | 0.252 (CNS) | 0.237 (CNS) | |
| Rfreed | 0.258 (CNS) | 0.298 (CNS) | 0.254 (CNS) | |
| V3L (high salt) | V3F (high salt) | V3T(high salt) | V3Y(high salt) | ||
|---|---|---|---|---|---|
| X-Ray Source | APS SER-CAT 22ID | APS SER-CAT 22ID | APS SER-CAT 22ID | APS SER-CAT 22BM | |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 | 1.00000 | |
| Space group | P31 2 1 | P31 2 1 | P31 2 1 | P31 2 1 | |
| Unit cell dimensions: | a (Å) | 95.546 | 96.022 | 95.616 | 95.70 |
| b (Å) | 95.546 | 96.022 | 95.616 | 95.70 | |
| c (Å) | 80.932 | 80.723 | 81.616 | 81.94 | |
| Resolution Range (Å) (highest shell) | 50.0-1.56 (1.62-1.56) | 50.0-1.75 (1.81-1.75) | 50-2.050 (2.12-2.05) | 37.0-1.86 (1.93-1.86) | |
| Mosaicity (degree) | 0.26 | 0.83 | 0.29 | 0.19 | |
| Completeness (%),(highest shell) | 86.8 (54.4) | 86.0 (45.4) | 95.2 (76.5) | 91.6 (57.5) | |
| Total linear R-merge | 0.062 | 0.084 | 0.063 | 0.061 | |
| Reflections Observed | 53034 | 37576 | 26097 | 33719 | |
| I over average sigma(I) | 16.1 | 11.6 | 17.7 | 35.5 | |
| Redundancy, (highest shell) | 11.2 (3.2) | 4.9 (2.0) | 5.4 (5.1) | 5.1 (2.1) | |
| Rmsd bonds (Å) | 0.017 | 0.027 | 0.006 | 0.023 | |
| Rmsd angles (°) | 1.6 | 2.3 | 1.2 | 2.0 | |
| Ramachandran Statistics | |||||
| Residues in most favored regions | 210 (89.0%) | 204 (89.1%) | 109 (87.6%) | 189 (88.7%) | |
| Residues in additionally allowed regions | 24 (10.2%) | 24 (10.5%) | 26 (12.0%) | 22 (10.3%) | |
| Residues in generaously allowed regions | 2 (0.8%) | 1 (0.4%) | 1 (0.5%) | 2 (0.9%) | |
| Residues in disallowed regions | 0 | 0 | 0 | 0 | |
| R-value (%) | 0.209 (refmac) | 0.217 (refmac) | 0.201 (refmac) | 0.230 (refmac) | |
| Rfree-value (%) | 0.233 (refmac) | 0.253 (refmac) | 0.235 (refmac) | 0.268 (refmac) | |
RESULTS
Growth Analysis of Bacterial Transformants
TX61− cells lack TS protein due to insertional disruption of the thyA gene (15). In order to grow, the cells must be provided with exogenous thymidine or transformed with a plasmid construct expressing a functional TS. All hTS mutants constructed provided sufficient TS activity to support the growth of TX61− cells as indicated by the formation of colonies in media lacking thymidine. In contrast, untransformed TX61− cells failed to grow in the thymidine deficient medium. The growth rates of TX61− transformed with wild-type and mutant TSs in minimal media lacking thymidine compared to untransformed TX61− cells (in the presence of thymidine) are shown in supplementary Figure 1. The growth rates of all mutants were similar except for V3A which exhibited an extended lag phase and a slight decrease in growth rate. Cell-free extracts from all transformants were analyzed by Western blotting (supplementary Figure 2). The data appear to indicate that the decreased growth rate of V3A is not due to its significantly lower expression compared to the other mutants.
Intracellular Protein Stability
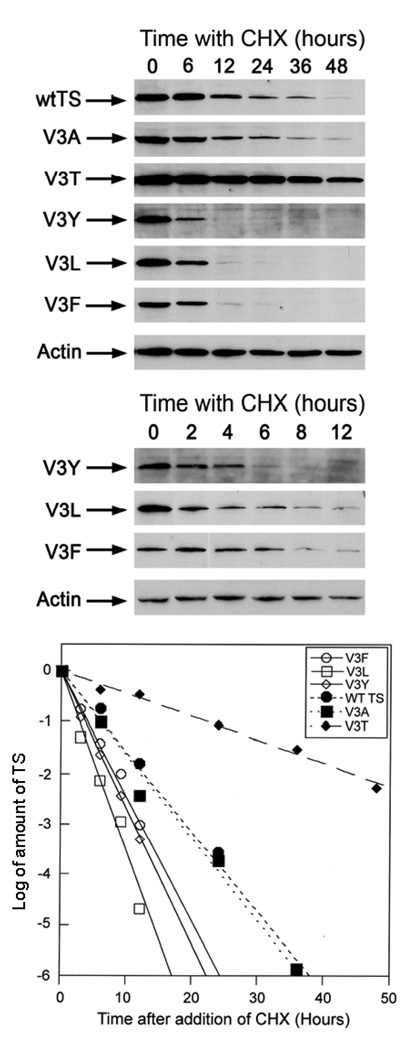
To determine the effects of mutations at Val3 on intracellular protein stability, TS-deficient Chinese hamster lung cells (RJK88.13) were stably transfected with plasmid pJZ205 expressing wt hTS or the various mutant forms of hTS. All mutants were capable of supporting the growth of these cells in the absence of exogenous thymidine and in the presence of the nucleoside transport inhibitor, dipyridamole, providing the basis for selection of stably transfected cells. The cultured cells were treated with cycloheximide to stop protein synthesis, and steady state levels of wt and mutant hTS proteins were determined by immunoblotting at the indicated time points after cycloheximide addition. The results presented in Figure 1 show that, with the exception of V3A, Val3 substitutions have large effects on the half-lives of the TS proteins. The wt-TS and V3A, had half-lives of approximately 6 hours while the V3T mutant had a half-life of approximately 24 hours. In contrast, the V3Y, V3L, V3F were less stable and had half-lives of 2.5, 2.5 and 3.5 hours, respectively (Fig. 1). Sequence based analysis using the program Terminator (31) very strongly predicts that all Val3 mutants, like wt-hTS, have a free Pro2 at the N-terminus.
Figure 1.
Intracellular stability of wild type and mutants with substitutions in Val3 of hTS. Stably transfected cells were treated with cycloheximide at the indicated time points and TS levels were analyzed by immunoblotting using an anti-human TS monoclonal antibody (D3B31). Blots were stripped and re-probed with actin as an internal control to normalize quantitation using ImageJ software. Half-lives were determined from the shown log2 plots of hTS levels (normalized against the actin content in each lane) determined by densitometry. Top panel: Western blot analysis of wt and Val3 mutant hTS protein levels after treatment with cycoheximide (CHX) for 0, 6,12,24,36, and 48 hours; Middle panel: Western blot analyses of V3Y, V3L, and V3F mutants after treatment with CHX for 0,2,4,6,8 and12 hours; Bottom panel: Log2 plot of the relative amount of protein in the band for hTS, (each unit corresponds to one half-life) for wt and Val3 mutants after treatment with CHX at the indicated time points.
Steady-State Kinetic Properties
Kinetic parameters of mutants were analyzed under conditions in which the concentration of mTHF is fixed at 5-fold the Km value for wt-hTS. The resulting km,app for dUMP and V[dUMP] are shown in Table 2. In general, the values of Km,app are higher and V[dUMP] are lower for the mutant enzymes. Most mutants are only modestly impaired; however, V3F and V3L have Km,app values that are higher by factors of 58 and 47, respectively, relative to wt-hTS. The variation in V[dUMP] values is much lower and all mutants are at least half as active as the wild type enzyme.
Table 2.
Steady-state kinetic constants for wild-type hTS and hTS mutants at position 3 [dUMP] was varied from 25 µM up to 400 µM.
| hTS | V (s−1) |
Km,app (µM) |
V/Km (s−1 µM−1) |
|---|---|---|---|
| wild-type | 3.04±0.02 | 2.3±0.2 | 1.3 |
| V3A | 0.96±0.05 | 22±5 | 4.3×10−2 |
| V3F | 1.6±0.3 | 134±6 | 1.2×10−2 |
| V3L | 2.6±0.2 | 109±2 | 2.4×10−2 |
| V3T | 1.8±0.1 | 11±3 | 0.17 |
| V3Y | 2.6±0.1 | 4.4±0.2 | 0.60 |
Crystal Structures
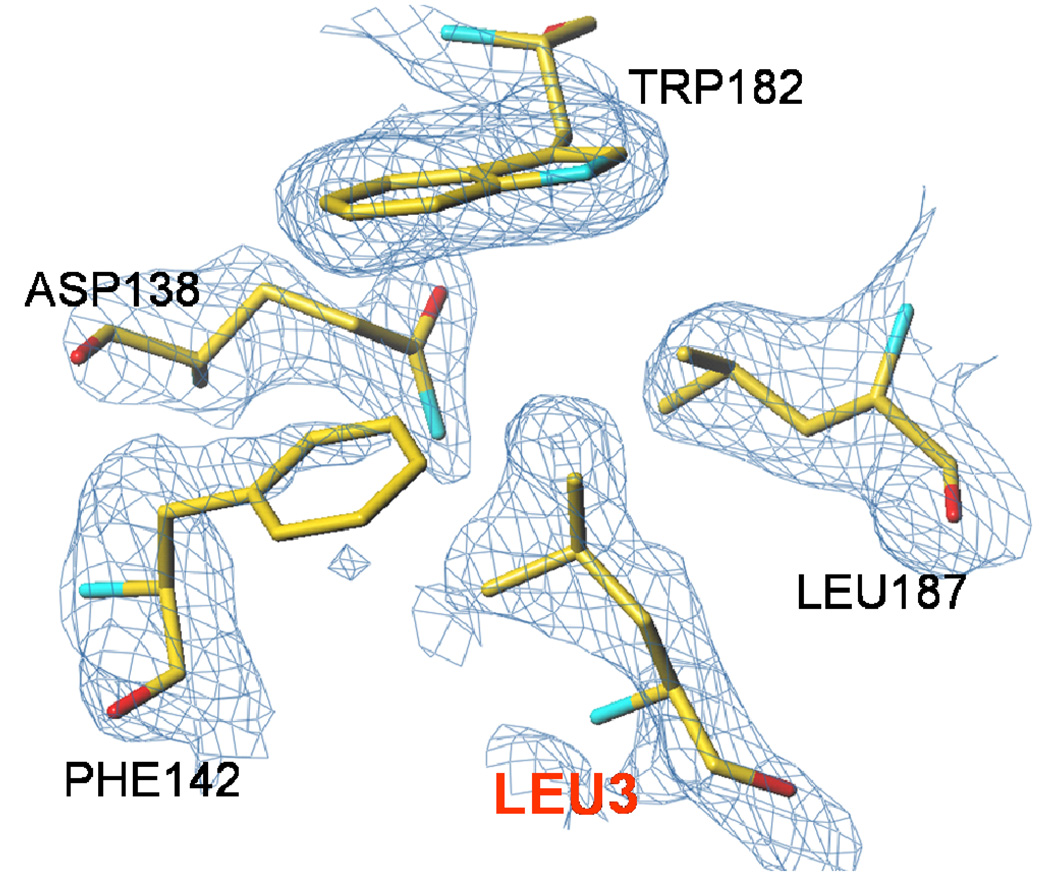
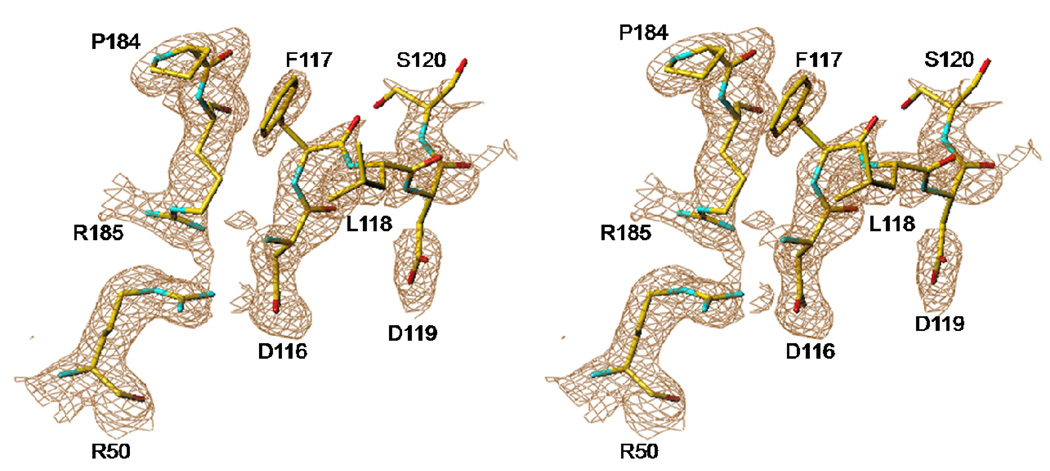
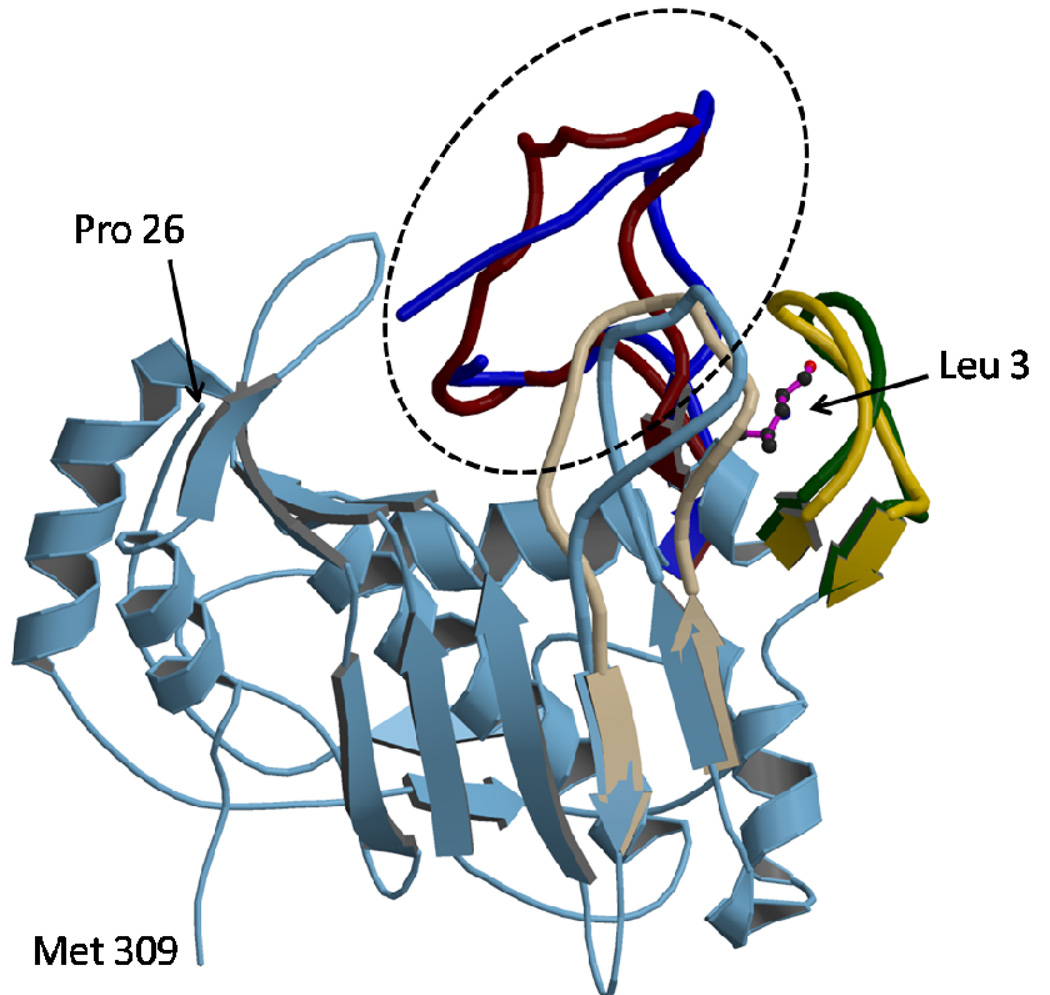
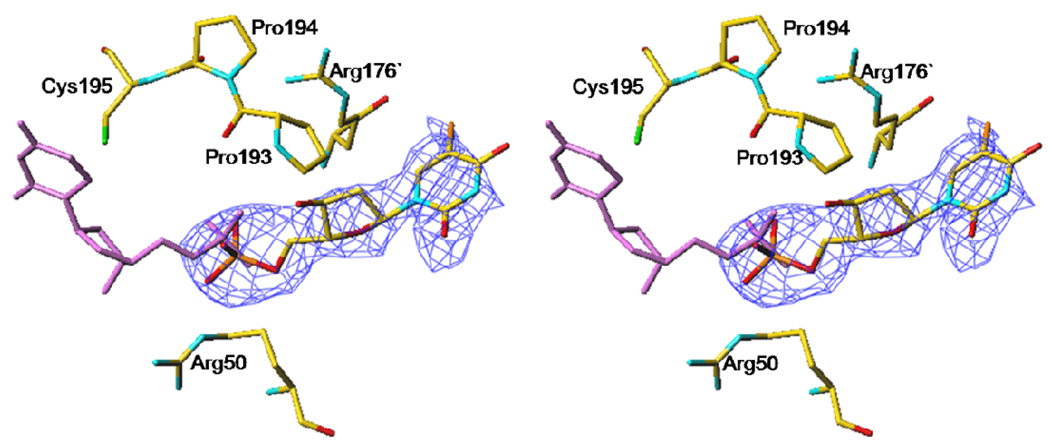
All crystals obtained at high AS concentrations (1.3–1.6 M) are of excellent scattering power; the V3L diffraction to 1.6 Å resolution is the best ever reported for hTS, likely through additional stabilization of the inactive conformation (vide infra). Loops 181–197 are in the inactive conformation, as expected, since this conformation is stabilized by sulfate ions. The high-AS structure of V3L showed well-defined electron density at the site at which a tert-butyl was observed previously (13). This density, shown in Figure 2, corresponds to a leucine rather than a valine. The distance between Pro26, the first residue observed at the N-terminus, and the putative Leu3 is 43 Å, which is approximately half of the length of 23 residues in an extended conformation. No evidence for a bound moiety of any type in this site was observed in the other structures reported here. The electron density for a fragment of loop 100–130 is shown in Figure 3. The loop, which is disordered in other structures containing loop 181–197 in the inactive conformation, was mostly observed (only residues from 104 to 108 are missing). This clearly eliminates Leu118 or Leu121 as possible ligands in the hydrophobic pocket at Trp182 and Leu187. Interestingly, the conformation of this loop in the high-AS structure of V3L is somewhat different from that observed in the active conformation (shown in Figure 4). In the high-AS structures of V3F, this new conformation of the loop was also observed.
Figure 2.
Stabilization of the inactive conformation in V3L. The 2FoFc electron density map is contoured at 1 σ level. There is a likely leucine residue, labeled Leu3, interacting with loop 181–197 in the inactive conformation.
Figure 3.
Stereo view of a fragment of the new conformation of loop 116–120 observed in V3L high salt structure.
Figure 4.
Comparison of loops in hTS-V3L, which is in an inactive conformation, with loops from hTS in the active conformation (from the inhibitory complex hTS·dUMP·raltitraxed, PDB 1HVY). Loop 100–130 is in dark blue (V3L) and brown (complex) and is highlighted by the oval. Loop 144–154 is in dark green in V3L and orange in the complex. Loop 181–197, which contains the catalytic Cys195, is shown in the active conformation (from the complex) in gray and in the inactive conformation (from V3L) in light blue which is also used for the constant part of the molecule.
Perhaps due to the high resolution of the data, a previously unidentified piece of density close to the catalytic site was also observed. It likely represents a partially occupied peptide chain for which the sequence could not be identified; consequently, a polyalanine model was fitted to the density (shown in supplementary Figure 3). No non-protein polymers were present in the crystallization media. This polyalanine fragment can be seen in all the inactive structures of the Val3 mutants obtained at high-salt conditions. The disordered C-terminus, residues 310–313, and loop 104–108 are too short to generate this density, leaving only the N-terminus, residues 1–25, as a possible source. Alternatively, the density may arise from a non-covalently bound proteolytic fragment. The conformations of loops and the new fragments from all structures are summarized in Table 3.
Table 3.
The summary of loop conformations and observed ligands in crystal structures
| High salt unliganded structures | Low salt co-crystallization structures |
||||||
|---|---|---|---|---|---|---|---|
| Crystals | V3Lhs | V3Fhs | V3Yhs | V3Ths | V3L+ dUMP |
V3F+ FdUMP |
V3Y+ FdUMP |
| Ligand present |
Leu3 | no | no | no | no | FdUMP | no |
| Loop180–198 | inactive | inactive | inactive | inactive | inactive | active | active |
| Loop109–130 | New conformation |
New conformation |
disordered | disordered | disordered | Active conformation |
Active conformation |
| Polyalanine | yes | yes | yes | yes | no | no | no |
| Space group | P3121 | P3121 | P3121 | P3121 | P3121 | C2 | C2 |
| Resolution | 1.56 | 1.75 | 1.86 | 2.05 | 3.0 | 2.75 | 2.75 |
The crystals obtained at low-AS conditions were of significantly lower scattering power than those obtained at high-AS concentrations; consequently, only their major features are discussed. In the V3L structure, the inactive conformation was observed, and there was no bound nucleotide, even though the enzyme was incubated with dUMP prior to crystallization. In contrast, V3F co-crystallized with FdUMP in low-salt conditions, yielding the enzyme in the active conformation. One subunit from the dimer binds FdUMP and the other binds a sulfate ion, reflecting the reported negative cooperativity of the enzyme. The FdUMP binding mode is, however, different from that previously observed for nucleotides. While the phosphate group binds in the standard site, the uridylate moiety is pointed in an opposite direction (shown in Figure 5). V3Y also co-crystallized with FdUMP in low-AS conditions, yielding the enzyme in the active conformation. However, the density for FdUMP is not clear, indicative of disorder; consequently, only a sulfate ion was modeled in each subunit. This likely reflects a low propensity of V3Y for nucleotide binding in the unique mode observed for V3F, as indicated by the disparate Km,app values.
Figure 5.
Stereo view of FdUMP binding site in V3F complex. Omit electron density is contoured at 1 σ level. In purple is the superposition, based on Cα positions, of dUMP bound in the inhibitory complex hTS·dUMP·raltitrexed (10).
DISCUSSION
The prevailing opinion about the N-terminal extension is that it has non-enzymatic functions. This hypothesis is supported by the poor sequence conservation of this region, including the most drastic difference, a complete lack of the extension in prokaryotes. The results reported here do not challenge this view. Rather, they indicate that such extensions can easily interact with the catalytic site of the enzyme and affect its enzymatic properties. Such interactions need not be strong and specific to be significant due to a high local concentration resulting from its covalent attachment. Therefore, the extensions are apparently under evolutionary pressure to achieve their physiological function without affecting other parts of the enzyme.
For all mutants studied here, the values of V[dUMP]t are only slightly reduced, indicating that after substrate binding occurs, the catalytic machinery is essentially unaffected by the Val3 substitutions. The Km,app values in some mutants are, however, quite different, especially for a variant residue that is not directly involved in substrate binding. In the inhibitory complex hTS•dUMP•raltitrexed, which should be a good model for the catalytic complex, dUMP is sequestered from the solvent, and it is highly unlikely that the N-terminus, which is disordered, interacts directly with the substrate (11). For the two mutants that stand out in terms of significantly increased Km,app values, the mechanisms by which the dUMP binding is affected appear to be entirely different. For V3L, the equilibrium between the active and inactive conformations of loop 181–197 and the conformation of loop 100–130 appear to be affected. It is surprising that the effects of a very conservative change, V3L, are so large. They likely are related to enhanced binding of the Leu3 side chain in the hydrophobic pocket formed by loop 181–197. If so, small molecules binding in this pocket should function as allosteric inhibitors stabilizing the inactive conformer. Potential advantages of such inhibitors versus active site directed inhibitors have been discussed (3, 32).
Like V3L, V3F has strongly compromised dUMP binding. In the V3F high-salt structure, no density is observed in the hydrophobic pocket at loop 181–197, indicating that the mechanism through which Km,app is increased is different. The structure of V3F·FdUMP complex suggests that the alternative mode of nucleotide binding, which uses a part of the active site and is thus inhibitory, is responsible for this effect. Again this complex may be used as a starting point for the structure-based development of a new class of inhibitors. Despite these differences, V3L and V3F have a common structural feature: the conformation of 109–130 module. This region is ordered and its conformation is different than that observed in the active conformation of wt-hTS (Figure 4). In the inactive conformer of native wt-hTS and in V3Y and V3T, this module is disordered. It is likely that the disorder is due to a mixture of conformations observed in the active conformation and that observed in V3L and V3F.
The N-terminus of hTS has been shown to be an important structural determinant that controls protein turnover in mammalian cells. Deletions of six, or as few as one amino acid, result in stabilization of the enzyme with up to five-fold increases in half-life (8,9). In addition, insertions at the N-terminus and certain substitutions at the penultimate proline residue of TS result in enzyme stabilization (9) due to posttranslational modifications by Nα-acetylation (7) that may block entry into the proteasomal core for degradation. The altered turnover rates for Val3 mutants confirm the role of N-terminus and show that a residue in position 3 is almost as important as in position 2. The observed variability among the mutants demonstrates the difficulty in pinpointing structure-function relationships. The relatively conservative substitution in V3T slowed down intracellular degradation by a factor of four. Three other substitutions studied here, including V3Y, which also introduces a hydroxyl function, had opposite effects and one substitution had no effect. Among 21 mammalian TS molecules that have been sequenced, only 4 have Val at position 3. Ala3 is the most common, being found in 14 species; 2 have Thr3 and one has His3 (7). Thus, none of mutants for which we observed lower stability has been reported in mammalian cells.
TS enzymes from E. coli and the majority of other bacteria have similar catalytic efficiency as mammalian enzymes despite lacking the N-terminal extension. Moreover, the extension is only poorly conserved in mammals. These observations suggested that the extension evolved to serve in roles that are not related to catalysis. Indeed, mutational analyses of the hTS N-terminus indicate that it plays a critical role in determining intracellular stability by targeting the protein for degradation by ubiquitin-independent proteasomal degradation pathway (7, 8, 9). This function is accomplished by maintaining an intrinsic disorder in the N-terminus rather than by sequence conservation (7). The studies of Val3 mutants indicate that, in addition to this role, the N-terminus may also affect the catalytic machinery of hTS, likely by altering conformational equilibria in this highly flexible protein and suggesting that variability in the N-terminus may affect a range of molecular properties. The studies of Val3 mutants reveal new conformational states of hTS and show correlations between eukaryotic inserts and the N-terminus. Since these regions of the molecule are only poorly conserved, the ongoing efforts to generate inhibitors of TS that are species-specific (33) gain additional credibility.
Supplementary Material
Abbreviations used
- TS
thymidylate synthase
- hTS
human TS
- FdUMP
5-fluoro-2’-deoxyuridine monophosphate
- dUMP
2’-deoxyuridine 5’-monophosphate
- dTMP
2’-deoxythymidine 5’-monophosphate
- CH2H4folate or mTHF
5,10-methylenetetrahydrofolate
- EDTA
ethylenediaminetetraacetic acid
- BME
β-mercaptoethanol
- PDPA
1,3-propanediphosphonic acid
- PDB
Protein Data Bank
- PEG
polyethylene glycol
Footnotes
This work was supported by NIH Grant CA 76560.
Crystal coordinates have been deposited in the Protein Data Bank as entries 3EAW, 3EBU, 3GH0, 3GH2, 3EF9, 3EJL and 3GG5 for high-salt V3Y, high-salt V3T, high-salt V3L, high salt V3F, low salt V3L, V3F•FdUMP complex and low salt V3Y, respectively. Data were collected at the SER-CAT 22-ID and 22-BM beamlines at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
SUPPLEMENTARY INFORMATION AVAILABLE
Growth curves for the mutants, images of SDS-PAGE gels, activity measurements as a function of dUMP concentrations, and stereo view of the polyalanine fragment from the V3F structure are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Humphreys GK, Greenberg DM. Studies on the conversion of deoxyuridylic acid to thymidylic acid by a soluble extract from rat thymus. Arch. Biochem. Biophys. 1958;78:275–287. doi: 10.1016/0003-9861(58)90351-5. [DOI] [PubMed] [Google Scholar]
- 2.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 3.Phan J, Steadman DJ, Koli S, Ding WC, Minor W, Dunlap RB, Berger SH, Lebioda L. Structure of human thymidylate synthase suggests advantages of chemotherapy with noncompetitive inhibitors. J. Biol. Chem. 2001;276:14170–14177. doi: 10.1074/jbc.M009493200. [DOI] [PubMed] [Google Scholar]
- 4.Schiffer CA, Clifton IJ, Davisson VJ, Santi DV, Stroud RM. Crystal structure of thymidylate synthase: a structural mechanism for guiding substrates into the active site. Biochemistry. 1995;34:16279–16287. doi: 10.1021/bi00050a007. [DOI] [PubMed] [Google Scholar]
- 5.Sotelo-Mundo RR, Ciesla J, Dzik JM, Rode w, Maley F, Maley GF, Hardy LW, Montfort WR. Crystal structures of rat thymidylate synthase inhibited by Tomudex, a potent anticancer drug. Biochemistry. 1999;38:1087–1094. doi: 10.1021/bi981881d. [DOI] [PubMed] [Google Scholar]
- 6.Phan J, Koli S, Minor W, Dunlap RB, Berger SH, Lebioda L. Human Thymidylate synthase is in the closed conformation when complexed with dUMP and Raltitrexed, an antifolate drug. Biochemisty. 2001;40:1897–1902. doi: 10.1021/bi002413i. [DOI] [PubMed] [Google Scholar]
- 7.Peña MMO, Melo SP, Xing Y, White K, Barbour K, Berger FGB. The Intrinsically disordered N-terminal domain of Thymidylate synthase targets the enzyme to the Ubiquitin-independent proteasomal degradation pathway. J. Biol. Chem. 2009;284:31597–31607. doi: 10.1074/jbc.M109.038455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pena MMO, Xing YY, Koli S, Berger FG. Role of N-terminal residues in the ubiquitin-independent degradation of human thymidylate synthase. Biochem. J. 2006;394:355–363. doi: 10.1042/BJ20051479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsthoefel AM, Pena MMO, Xing YY, Rafique Z, Berger FG. Structural determinants for the intracellular degradation of human thymidylate synthase. Biochemistry. 2006;43:1972–1979. doi: 10.1021/bi035894p. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu K, Ayusawa D, Takeishi K, Seno T. Purification and NH2-terminal amino acid sequence of human thymidylate synthase in an overproducing transformant of mouse FM3A cells. J. Biochem. 1985;97:845–850. doi: 10.1093/oxfordjournals.jbchem.a135125. [DOI] [PubMed] [Google Scholar]
- 11.Phan J, Koli S, Minor W, Dunlap RB, Berger SH, Lebioda L. Human thymidylate synthase is in the closed conformation when complexed with dUMP and raltitrexed, an antifolate drug. Biochemistry. 2001;40:1897–1902. doi: 10.1021/bi002413i. [DOI] [PubMed] [Google Scholar]
- 12.Schiffer CA, Davisson VJ, Santi DV, Stroud RM. Crystallization of human thymidylate synthase. J. Mol. Biol. 1991;219:161–163. doi: 10.1016/0022-2836(91)90558-n. [DOI] [PubMed] [Google Scholar]
- 13.Lovelace LL, Minor W, Lebioda L. Structure of human thymidylate synthase under low-salt conditions. Acta Crystallogr. 2005;D61:622–627. doi: 10.1107/S0907444905005895. [DOI] [PubMed] [Google Scholar]
- 14.Almog R, Waddling CA, Maley F, Maley GF, Van Roey P. Crystal structure of a deletion mutant of human thymidylate synthase Δ(7–29.) and its ternary complex with Tomudex and dUMP. Protein Sci. 2001;10:988–996. doi: 10.1110/ps.47601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dev IK, Yates BB, Leong J, Dallas WS. Functional role of cysteine-146 in Escherichia coli thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 1988;85:1472–1476. doi: 10.1073/pnas.85.5.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dev IK, Dallas WS, Ferone R, Hanlon M, McKee DD, Yates BB. Mode of binding of folate analogs to thymidylate synthase - evidence for a 2 asymmetric but interactive substrate-binding sites. J. Biol. Chem. 1994;269:1873–1882. [PubMed] [Google Scholar]
- 17.Lovelace LL, Johnson SR, Gibson LM, Bell BJ, Berger SH, Lebioda L. Variants of human thymidylate synthase with loop 181–197 stabilized in the inactive conformation. Protein Sci. 2009;18:1628–1636. doi: 10.1002/pro.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Steadman DJ, Zhao P-S, Spencer HT, Dunlap RB, Berger SH. A structural role for glutamine 214 in human thymidylate synthase. Biochemistry. 1998;37:7089–7095. doi: 10.1021/bi9725428. [DOI] [PubMed] [Google Scholar]
- 20.Williams AW, Dunlap RB, Berger SH. A hydroxyl group at residue 216 is essential for catalysis by human thymidylate synthase. Biochemistry. 1998;37:7096–7102. doi: 10.1021/bi972562+. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;D50:157–163. [Google Scholar]
- 24.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Gibson LM, Lovelace LL, Lebioda L. The R163K mutant of human thymidylate synthase is stabilized in an active conformation: asymmetry and reactivity of Cys195. Biochemistry. 2008;47:4636–3643. doi: 10.1021/bi7019386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roussel A, Cambillau C. “Turbo Frodo” Silicon Graphics Geometry Partners Directory. Mountain View, CA: Silicon Graphics; 1991. p. 86. [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. 1976;A32:922–923. [Google Scholar]
- 29.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 30.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 31.Meinnel T, Peynot P, Giglione C. Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochemie. 2005;87:701–712. doi: 10.1016/j.biochi.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Berger SH, Berger FG, Lebioda L. Effects of ligand binding and conformational switching on intracellular stability of human thymidylate synthase. Biochim. Biophys. Acta. 2004;1696:15–22. doi: 10.1016/j.bbapap.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Ferrari S, Losasso V, Costi MP. Sequence-Based Identification of Specific Drug Target Regions in the Thymidylate Synthase Enzyme Family. ChemMedChem. 2008;3:392–401. doi: 10.1002/cmdc.200700215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.