Abstract
Purpose
Circulating tumor cells (CTCs) in blood may be important in assessing tumor progression and treatment response. We hypothesized that quantitative real-time reverse transcriptase polymerase chain reaction using multimarker mRNA assays could detect CTCs and be used as a surrogate predictor of outcome in patients receiving neoadjuvant biochemotherapy (BC) for melanoma.
Patients and Methods
Blood specimens were collected at four sampling points from 63 patients enrolled on a prospective multicenter phase II trial of BC before and after surgical treatment of American Joint Committee on Cancer stage III melanoma. Each specimen was assessed by quantitative real-time reverse transcriptase polymerase chain reaction for expression of four melanoma-associated markers: melanoma antigen recognized by T cells 1; β1 → 4-N-acetylgalactosaminyltransferase; paired box homeotic gene transcription factor 3; and melanoma antigen gene-A3 family, and the changes of CTCs during treatment and prognostic effect of CTCs after overall treatment on recurrence and survival were investigated.
Results
At a median postoperative follow-up time of 30.4 months, 44 (70%) patients were clinically disease free. In relapse-free patients, the number of detected markers significantly decreased during preoperative BC (P = .036), during postoperative BC (P = .002), and during overall treatment (P < .0001). Marker detection after overall treatment was associated with significant decreases in relapse-free and overall survival (P < .0001). By multivariate analysis using a Cox proportional-hazards model, the number of markers detected after overall treatment was a significant independent prognostic factor for overall survival (risk ratio, 12.6; 95% CI, 3.16 to 50.5; P = .0003).
Conclusion
Serial monitoring of CTCs in blood may be useful for indicating systemic subclinical disease and predicting outcome of patients receiving neoadjuvant BC for metastatic melanoma.
INTRODUCTION
The metastasis of melanoma to regional lymph nodes and distant sites often portends a poor prognosis.1,2 These patients are candidates for adjuvant therapy because of their high risk of disease recurrence after complete surgical resection. Recent studies have suggested the benefit of biochemotherapy (BC) in advanced-stage melanoma.3–9 However, to date, no assays can accurately predict the survival of patients receiving neoadjuvant or adjuvant BC. Because previous studies have suggested that the presence of melanoma cells in blood is associated with metastasis and poor disease outcome,10–15 an assay to detect circulating tumor cells (CTCs) could be a predictive surrogate for subclinical disease. If so, its use would allow serial monitoring of patients during treatment and potential prediction of outcome.
The development of a quantitative real-time reverse transcriptase polymerase chain reaction (RT-PCR) assay has allowed rapid and reproducible quantitative analysis for detection of CTCs in blood.16 Investigators have reported the detection of CTCs in blood using both single and multimarker quantitative RT-PCR, but few studies have assessed quantitative RT-PCR as a predictive surrogate for treatment outcome.17–20 Heterogeneous expression of tumor genes and variable performance of the assays have posed major problems for detection of CTCs in blood. As we first reported, this heterogeneity of marker expression in blood and lymph nodes favors use of a multimarker RT-PCR assay instead of single-marker assays.13,14,21,22
We have developed a multimarker quantitative RT-PCR assay using four markers for primary and metastatic melanoma: melanoma antigen recognized by T cells-1 (MART-1), β1 → 4-N-acetylgalacto-saminyltransferase (GalNAc-T), paired box homeotic gene transcription factor 3 (PAX-3), and melanoma antigen gene-A3 family (MAGE-A3).23 MART-1 is a major melanocyte-differentiation antigen that is frequently expressed in melanoma cells and is not expressed in nonmelanoma malignancies.23,24 GalNAc-T has been investigated in melanoma and other tumors.25–27 PAX-3 is expressed in rhabdomyosarcoma, Ewing’s sarcoma, and melanoma.28,29 MAGE-A3 is commonly expressed in various tumors, but not in normal tissue except male germline cells and placenta.23,30 Use of quantitative RT-PCR analysis for these four markers can improve the identification of metastatic melanoma cells in paraffin-embedded sentinel lymph nodes and thus can upstage patients whose sentinel lymph nodes are negative by immunohistochemistry and hematoxylin and eosin staining.23
In this study we used our multimarker quantitative RT-PCR assay to detect CTCs in blood specimens from patients receiving BC before and after complete surgical resection of American Joint Committee on Cancer (AJCC) stage III melanoma. We hypothesized that multimarker mRNA detection could predict the presence of systemic subclinical disease and that changes in marker detection during and after treatment could be used as a surrogate predictor of treatment outcome.
PATIENTS AND METHODS
Patients
Patients for this quantitative RT-PCR study were selected from 92 patients with melanoma who were enrolled in a prospective multicenter trial of neoadjuvant BC. The 92 patients comprised 60 males and 32 females with a median age of 43 years (range, 17 to 76 years). All patients were pathologically diagnosed with AJCC stage III melanoma and treated with neoadjuvant BC and surgery between 1999 and 2002. A subset of patients from three centers, John Wayne Cancer Institute (Santa Monica, CA), University of Colorado Cancer Center (Aurora, CO), and Hubert H. Humphrey Cancer Center (Robbinsdale, MN), signed informed consent for the use of their blood specimens, and the quantitative RT-PCR study was approved and carried out in accordance with guidelines set forth by the individual institutional review boards.
Treatment Program and Blood Procurement
All patients received two cycles of BC at 3-week intervals before surgery. The BC regimen comprised cisplatin 20 mg/m2 intravenously (IV) on days 1 to 4; dacarbazine 800 mg/m2 IV on day 1; vinblastine 1.6 mg/m2 IV on days 1 to 4; interleukin-2 (Chiron Corporation, Emeryville, CA) 9 MU/m2 IV over 24 hours on days 1 to 4; interferon alpha (Schering-Plough, Madison, NJ) 5 MU/m2 subcutaneously on days 1 to 5; and granulocyte colony-stimulating factor (Amgen Inc, Thousand Oaks, CA) 5 μg/kg subcutaneously on days 6 to 12. Patients then underwent therapeutic lymphadenectomy and began two cycles of BC within 42 days after surgery; the postoperative BC regimen was the same as the preoperative regimen. All patients were evaluated clinically and radiologically at specified time points during treatment and follow-up.
Peripheral blood was drawn immediately before preoperative BC (pre-BC; n = 63), before surgery (presurgery; n = 55), after surgery (postsurgery; n = 55), and after postoperative BC (post-BC; n = 58). The interval between each of the four sampling times was approximately 6 weeks, and all blood specimens were processed within 30 hours after drawing.
Standard Operation Procedure
Ten-milliliter blood samples were collected in sodium citrate– containing tubes, and the first several milliliters were discarded to eliminate skin-plug contamination as described previously.21,30 All blood specimens then were coded by a computer-generated number so that the quantitative RT-PCR study could be conducted in a blinded fashion. Total cells in blood were collected by using Purescript RBC lysis solution (Gentra, Minneapolis, MN) following the manufacturer instructions.
Tri-Reagent (Molecular Research Center, Cincinnati, OH) was used to isolate total cellular RNA from blood specimens as described previously.21,30 RNA was quantified and assessed for purity by ultraviolet spectrophotometry. Blood processing, RNA extraction, RT-PCR assay set-up, and post–RT-PCR product analysis were carried out in separate designated rooms to prevent cross contamination.
RT reactions were performed by using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) with oligo-dT primer.23,30 Multimarker quantitative RT-PCR assay was performed by using iCycler iQ RealTime Thermocycler Detection System (Bio-Rad Laboratories, Hercules, CA) as described previously.16,23 Primer and probe sequences were designed for the quantitative RT-PCR. Fluorescence resonance energy-transfer probe sequences were as follows: MART-1, 5′-FAM-TGCAGAACAGT-CACCACCACC-BHQ-1-3′ (FAM, carboxyfluorescein; BHQ, Black Hole Quencher; 2,5-di-tert-butyl-1,4-dihydroxybenzene); GalNAc-T, 5′-FAM-ATGAGGCTGCTTTCACTATCCGCA-BHQ-1-3′; PAX-3, 5′-FAM-CCAGACTGATTACGCGCTCTCCC-BHQ-1-3′; MAGE-A3, 5′-FAM-AGCTCCTGCCCACACTCCCGCCTGT-BHQ-1-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-FAM-CAGCAATGCCTCCTGCACCACCAA-BHQ-1-3′. We transferred 5 μL of cDNA from 250 ng of total RNA to a well of a 96-well PCR plate (Fisher Scientific, Pittsburgh, PA) in which 0.5 μmol/L of each primer, 0.3 μmol/L probe, 1 U of AmpliTaq Gold polymerase (Applied Biosystems, Branchburg, NJ), 200 μmol/L of each dNTP, 4.5 mmol/L MgCl2, and PCR buffer were applied to a final volume of 25 μL. Samples were amplified with a precycling hold at 95°C for 10 minutes, followed by 42 cycles of denaturation at 95°C for 1 minute, annealing at 55°C for 1 minute for GAPDH (59°C for MART-1, 62°C for GalNAc-T and PAX-3, and 58°C for MAGE-A3), and extension at 72°C for 1 minute.
The standard curve was generated by using a threshold cycle of nine serial dilutions of plasmid templates (10 to 108 copies). The threshold cycle of each sample was interpolated from the standard curve, and the number of mRNA copies was calculated by the iCycler iQ RealTime Detection System software (Bio-Rad Laboratories). Seventeen melanoma cell lines and peripheral blood leukocytes (PBLs) from 49 healthy donors were used to optimize the assay. Each quantitative RT-PCR assay was performed at least twice and included marker-positive (melanoma cell lines) and marker-negative (PBLs of healthy donors) controls and reagent controls (reagent alone without RNA or cDNA). The GAPDH gene was used as a control housekeeping gene. Any specimen with inadequate GAPDH mRNA was excluded from the study. The mean mRNA copy number was used for analysis.
Statistical Analysis
This study was designed to investigate the changes on CTCs during treatment course and prognostic effect of circulating tumor burden after overall treatment on disease relapse and survival. The primary outcomes were the number of markers detected after overall treatment, relapse of the disease, and survival. The mRNA copies and the detection rate of individual markers at each time point were considered as secondary end points.
The detection of individual markers at each time point was tabulated. The Wilcoxon signed-rank test was used to compare the number of markers during treatment. The Mann-Whitney U test was used to assess the difference of markers between relapse and relapse-free patients. Relapse-free survival (RFS) after lymphadenectomy and overall survival (OS) from the start of BC (pre-BC) were used for outcome measurement. A Cox proportional-hazards model was developed to examine the association of markers detected with RFS and OS and used for multivariate analysis. Known clinical and pathologic risk factors such as age, sex, primary tumor site, Breslow tumor thickness, ulceration, AJCC primary tumor (T) stage, regional lymph node (N) stage, stage III grouping (IIIA, IIIB, and IIIC), previous treatment status, detection of individual markers, and number of multimarkers after overall treatment were included in the model. A stepwise method was used for prognostic variable selection. The log-rank test was used to compare RFS and OS among patients with individual marker detection and patients with zero, one, two or more detectable markers after overall treatment. Survival curves were generated by using the Kaplan-Meier method.
For the secondary outcomes, McNemar’s test was used to compare the detection of individual markers between any two time points, and the Wilcoxon signed-rank test was used to examine the change of mRNA copies during treatment course. The analysis was performed by using SAS statistical software (SAS Institute, Cary, NC), and all tests were two sided with a significance level of ≤ .05.
RESULTS
Eligible Patients for Quantitative RT-PCR Study
Our quantitative RT-PCR study included 63 of the 92 patients enrolled on the clinical trial; the remaining 29 patients were excluded because of lack of blood procurement (21 patients), rapid disease progression (four patients), or severe toxicity during neoadjuvant BC (four patients). A multimarker quantitative RT-PCR assay was performed on 231 blood samples collected from the 63 patients (41 males and 22 females; median age, 42 years [range, 17 to 76 years]; median postoperative follow-up, 30.4 months).
Standard Curves and Specificity of Multimarker Quantitative RT-PCR Assay
The standard curves showed the expected linear increase of signal with the logarithm of the copy number (data not shown). PCR efficiency assessed from the slopes of the curves was between 90% and 100%. The correlation coefficient for all standard curves in the study was ≥ 0.99. MART-1, GalNAc-T, PAX-3, and MAGE-A3 mRNA were not detected in blood specimens from the 49 healthy donors under the optimized conditions, but were frequently detected in melanoma cell lines.16 Individual markers were detected in one to five melanoma cells diluted in 107 PBLs of healthy donors; the coefficient of variation was 1.8% to 22% for triplicate results (intra-assay variation) and 14% to 34% between assays for individual markers.16
Change in Multimarker mRNA Detection During Treatment
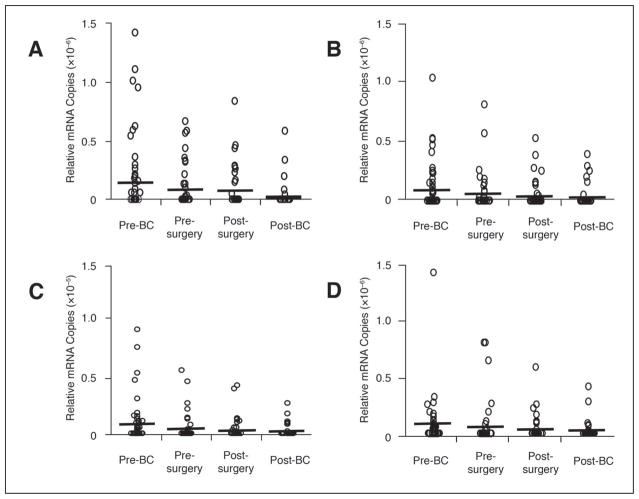
GAPDH expression was detected in all blood specimens; the absolute copy number per 250 ng of total RNA ranged from 9.21 × 104 to 1.83 × 108 (median, 1.75 × 107). The range of relative mRNA copies (absolute mRNA copies of each marker/absolute mRNA copies of GAPDH) was 0 to 8.4 × 10−6 overall and 10−8 to 10−6 for marker-positive specimens (Fig 1). During treatment, relative mRNA copies of all markers gradually decreased to levels significantly below pretreatment levels (MART-1, P = .0006; GalNAc-T, P = .005; PAX-3, P = .002; MAGE-A3, P = .013). Changes in relative mRNA copy level during each sampling interval were not significant except for a significant decrease in the PAX-3 mRNA copy level during preoperative BC (P = .032).
Fig 1.
Copy levels of individual mRNA markers in blood specimens obtained before, during, and after treatment. The relative mRNA copy number of each marker after treatment was significantly lower than its copy number before treatment: melanoma antigen recognized by T cells 1 (MART-1), P = .0006; β1 → 4-N-acetylgalactosaminyltransferase (GalNAc-T), P = .005; paired box homeotic gene transcription factor 3 (PAX-3), P = .002; melanoma antigen gene-A3 family (MAGE-A3), P = .013. Bars indicate mean copy numbers.
Similarly, individual marker-detection rates dropped significantly during overall treatment (MART-1, P = .001; GalNAc-T, P = .005; PAX-3, P = .001; MAGE-A3, P = .004), reflecting a gradual, nonsignificant decrease during each sampling interval (Table 1). The number of markers detected in each specimen also decreased significantly during overall treatment (P < .0001), reflecting significant decreases during preoperative and postoperative BC (P = .046 and .008, respectively), but not during surgery. Before treatment, blood specimens from 47 (75%) of 63 patients expressed at least one marker, and specimens from 22 (35%) patients expressed more than one marker. After overall treatment, specimens from 41 (65%) patients had no markers, and only five (8%) specimens expressed more than one marker.
Table 1.
Detection of Markers in Blood Sampled at Specific Intervals During Treatment
| Pre-BC (n = 63) |
Presurgery (n = 55) |
Postsurgery (n = 55) |
Post-BC (n = 58) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | P* | |
| Marker | |||||||||
| MART-1 | 23 | 37 | 15 | 27 | 10 | 18 | 5 | 9 | .001 |
| GalNAc-T | 18 | 29 | 12 | 22 | 10 | 18 | 6 | 10 | .005 |
| PAX-3 | 23 | 37 | 11 | 20 | 11 | 20 | 7 | 12 | .001 |
| MAGE-A3 | 22 | 35 | 10 | 18 | 11 | 20 | 7 | 12 | .004 |
| No. of markers detected | |||||||||
| 0 | 16 | 25 | 25 | 46 | 22 | 40 | 41 | 70 | < .0001 |
| 1 | 25 | 39 | 16 | 29 | 26 | 47 | 12 | 20 | |
| 2 | 6 | 10 | 10 | 18 | 5 | 9 | 3 | 6 | |
| 3 | 15 | 24 | 4 | 7 | 2 | 4 | 1 | 2 | |
| 4 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 2 | |
Abbreviation: BC, biochemotherapy.
The P value was calculated by using McNemar’s test for each marker and the Wilcoxon signed-rank test for number of markers.
Multimarker mRNAs As a Predictor of Disease Relapse and Survival
At a median postoperative follow-up of 30.4 months (range, 4.6 to 52.4 months), 19 (30%) of 63 patients had experienced a relapse, and 44 (70%) were clinically disease free. After treatment, the marker-detection rate after treatment clearly distinguished patients who had a relapse and those who were relapse-free (61% and 15%, respectively). The number of markers was significantly lower in relapse-free patients (P = .001), reflecting a significant overall decrease (P < .0001) and significant decreases during preoperative and postoperative BC (P = .036 and P < .0001, respectively) (Table 2). In patients with relapse, there was no significant difference between any two sampling points; 18 (95%) of 19 patients expressed at least one marker during treatment, and those who expressed at least two markers after treatment developed relapse within 7 months (Table 3).
Table 2.
Multimarker Detection Correlated With Disease Outcome
| Pre-BC |
Presurgery |
Postsurgery |
Post-BC |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of Markers Detected | No. | % | No. | % | No. | % | No. | % | P* |
| No relapse (n = 44) | |||||||||
| 0 | 12 | 27 | 17 | 44 | 17 | 42 | 34 | 85 | < .0001 |
| 1 | 15 | 34 | 12 | 31 | 19 | 48 | 5 | 13 | |
| 2 | 5 | 12 | 9 | 23 | 3 | 8 | 1 | 2 | |
| 3 | 12 | 27 | 1 | 2 | 1 | 2 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Relapse (n = 19) | |||||||||
| 0 | 4 | 21 | 8 | 50 | 5 | 33 | 7 | 39 | .8 |
| 1 | 10 | 53 | 4 | 25 | 7 | 47 | 7 | 39 | |
| 2 | 1 | 5 | 1 | 6 | 2 | 13 | 2 | 10 | |
| 3 | 3 | 16 | 3 | 19 | 1 | 7 | 1 | 6 | |
| 4 | 1 | 5 | 0 | 0 | 0 | 0 | 1 | 6 | |
Abbreviation: BC, biochemotherapy.
The P value was calculated by using the Wilcoxon signed-rank test.
Table 3.
Marker Detection and Duration of Relapse-Free Survival in Patients With Relapse
| Patient | Pre-BC (No. of markers detected) | Pre-Surgery (No. of markers detected) | Post-Surgery (No. of markers detected) | Post-BC (No. of markers detected) | Relapse-Free Survival (months) |
|---|---|---|---|---|---|
| 1 | 0 | NA | NA | 3 | 2.80 |
| 2 | 1 | 1 | 0 | 0 | 20.50 |
| 3 | 1 | 0 | 1 | 1 | 31.30 |
| 4 | 1 | 1 | 1 | 0 | 44.10 |
| 5 | 0 | 1 | 0 | NA | 28.23 |
| 6 | 1 | 3 | 1 | 1 | 12.56 |
| 7 | 2 | 0 | 1 | 0 | 23.90 |
| 8 | 3 | 0 | 0 | 1 | 14.73 |
| 9 | 0 | 3 | 1 | 1 | 6.93 |
| 10 | 3 | 0 | 0 | 1 | 9.60 |
| 11 | 1 | 1 | 1 | 1 | 31.67 |
| 12 | 1 | 0 | 3 | 4 | 6.57 |
| 13 | 4 | 0 | 2 | 0 | 14.37 |
| 14 | 1 | 2 | NA | 0 | 28.70 |
| 15 | 1 | 0 | 2 | 1 | 22.83 |
| 16 | 1 | 0 | 1 | 2 | 5.83 |
| 17 | 0 | NA | NA | 0 | 13.03 |
| 18 | 1 | 3 | NA | 2 | 3.87 |
| 19 | 3 | NA | 0 | 0 | 8.90 |
Abbreviations: BC, biochemotherapy; NA, not applicable.
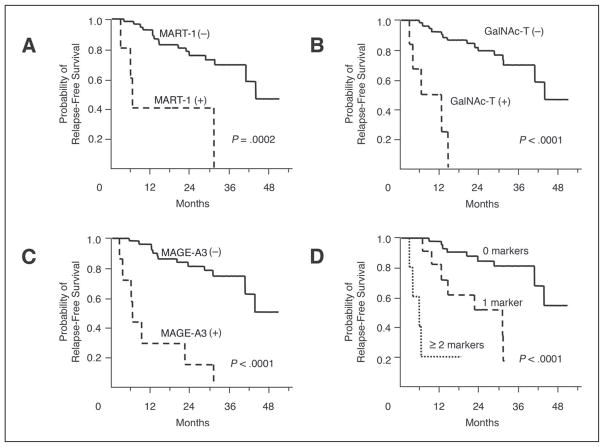
Before treatment, marker detection had no correlation with sex, age, primary site, Breslow tumor thickness, ulceration, pT stage, pN stage, stage III grouping (IIIA, IIIB, and IIIC), and previous treatment status. After treatment, RFS significantly decreased when blood specimens were positive for MART-1, GalNAc-T, and/or MAGE-A3 (P = .0003, P < .0001, and P < .0001, respectively; Fig 2). The size of the decrease was directly correlated with the number of positive markers (P < .0001) (Fig 2). For patients with no positive markers (n = 41), seven patients had experienced a relapse, and the estimated RFS rates were 97.6% ± 2.4% (estimate± SE) for 12 months and 89.8% ± 4.9% for 24 months. For patients with one positive marker (n = 12), seven had experienced a relapse, and the estimated RFS rates were 81.8% ± 11.6% for 12 months and 62.3% ± 15.0% for 24 months. Of patients with two or more positive markers (n = 5), four had experienced a relapse, and the estimated RFS rates was 20.0% ± 17.9% for 12 months. Cox proportional-hazards model analysis selected the detection of MART-1 (risk ratio, 10.2; 95% CI, 1.91 to 54.1; P = .007), GalNAc-T (risk ratio, 6.56; 95% CI, 1.43 to 30.0; P = .015), and MAGE-A3 (risk ratio, 33.6; 95% CI, 7.82 to 144.6; P < .0001) after overall treatment as the significant prognostic factors for RFS. No other factors were selected in the Cox proportional-hazards model except age (> 50 v ≤ 50 years; risk ratio, 8.25; 95% CI, 2.01 to 33.8; P = .003). Marker detection in blood specimens obtained before or during treatment was not correlated with RFS.
Fig 2.
Kaplan-Meier curves of relapse-free survival based on marker detection after treatment.
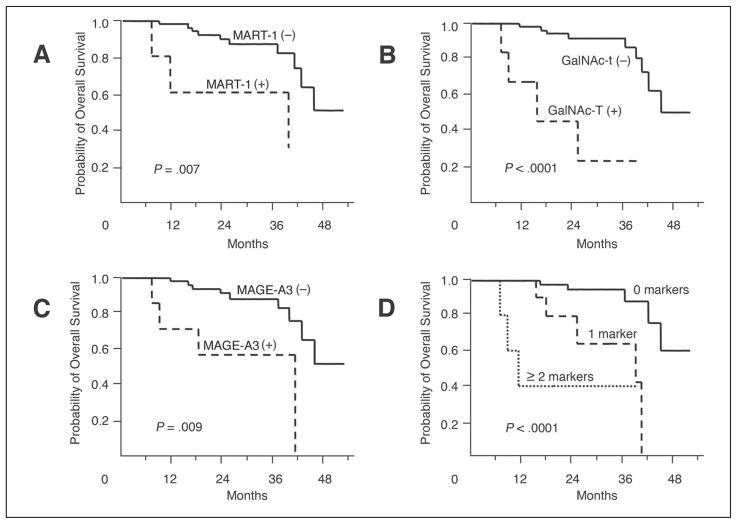
After treatment, OS decreased significantly if blood specimens were positive for MART-1, GalNAc-T, and/or MAGE-A3 (P = .007, P < .0001, and P = .009, respectively; Fig 3). Again, the level of decrease was directly correlated with the number of positive markers (P < .0001; Fig 3). For patients with no positive markers, no patients died within 12 months, and four patients died during the follow-up period; the estimated OS was 94.5% ± 3.8% (estimate ± SE) for 24 months. For patients with one positive marker, no patients died within 12 months, and five died; the estimated OS was 80.0% ± 12.7% for 24 months. For patients with two or more positive markers, three patients died within 12 months, and the estimated 1-year survival rate was 40.0% ± 21.9%. Using a Cox proportional-hazards regression model, the number of positive markers after treatment (risk ratio, 12.6; 95% CI, 3.16 to 50.5; P = .0003) was selected as a significant independent prognostic factor for OS. No other factors were selected in the Cox proportional model except age (> 50 v ≤ 50 years; risk ratio, 8.19; 95% CI, 2.30 to 28.5; P = .001).
Fig 3.
Kaplan-Meier curves of overall survival based on marker detection after treatment.
DISCUSSION
Recent studies have shown the importance of CTCs in blood; however, most have focused on correlation with staging and tumor burden.10,31 We previously demonstrated the significance of CTCs for prediction of disease outcome in patients with AJCC stage III melanoma receiving melanoma vaccine.13,14 Because CTCs may indicate systemic subclinical disease, their quantitative real-time detection can represent a surrogate marker for predicting outcome to adjuvant therapy. In this study, we demonstrated that a multimarker quantitative RT-PCR assay could detect CTCs in the blood of patients with AJCC stage III melanoma who were receiving BC before and after surgery, and changes in multimarker detection were correlated with disease progression and OS.
The four mRNA markers selected for this study are found frequently in blood specimens from patients with melanoma, but not in blood from healthy individuals.16 Because metastatic melanoma tumors are heterogenous in melanoma-associated marker expression,21,23 a combination of markers can compensate for variations in individual marker expression; thus, we expect detection of tumor cells to be greatly increased and false-negative results reduced. Detection rates were higher with the multimarker quantitative RT-PCR assay than with any individual marker assay. These findings indicate that a single-marker assay in blood has limited clinical utility.21,32,33
The number of positive markers in each blood specimen significantly decreased across all three sampling intervals and during preoperative and postoperative BC. The nonsignificant decrease after surgery probably reflects residual tumor burden. CTCs were detected in more than half of the patients after surgery, consistent with the high incidence of disease relapse in patients with AJCC stage III melanoma. These findings also indicate the need for postoperative monitoring to detect systemic subclinical disease in patients who have undergone complete resection of stage III melanoma.34
The change in marker-detection rates was clearly different between relapse-free patients and patients who had a relapse. Preoperative and postoperative BC significantly reduced the number of markers only in relapse-free patients. Marker detection after treatment also correlated with OS. These results suggest that serial monitoring of CTCs might be used to predict disease outcome. The fact that surgery did not significantly affect marker-detection rates in relapse-free patients or in patients who had a relapse suggests that subclinical systemic disease had already been established before operative intervention. The Cox proportional model did not select the histopathologic factor (pT and/or pN stage) as a prognostic factor, and these findings might indicate that assessment of primary and regional lymph nodes according to TNM staging criteria could not accurately predict the disease relapse and OS in patients receiving neoadjuvant treatment because of modification of tumor burden by chemotherapeutic and/or immunotherapeutic drugs.
From a clinical standpoint, the development of a tool to identify high-risk patients and to monitor the response to adjuvant therapy in patients who are clinically disease free would represent significant progress in the management of patients with melanoma. Although most studies of CTCs in melanoma have used specimens obtained at only one time point, serial assessment can detect CTC changes during different phases of treatment, which makes CTC assessment a promising method to detect real-time subclinical tumor spreading. Although we did not measure clinical response to treatment, our assay system may have the potential to identify which component of treatment is most effective and which needs to be improved. As treatment regimens become multimodal and multiphasic, there will be an urgent need for clinically relevant surrogate markers that can be used to monitor response and predict outcome.
Most studies for assessment of predictive markers in patients treated with neoadjuvant therapy have used tumor tissues. However, static assessment of primary and meta-static tumor specimens after neoadjuvant therapy does not indicate whether tumor cells are being shed or whether treatment is reducing metastasis. In contrast, dynamic assessment of serially obtained blood specimens allows molecular evaluation of tumor-cell shedding during treatment and is highly important to evaluate efficacy in controlling systemic disease. This study supports the use of molecular markers as surrogates for disease progression and suggests that our assay system could minimize sample variation from different sites and thereby increase the feasibility of multicenter studies.
Acknowledgments
Supported in part by the National Institutes of Health, National Cancer Institute P01 Grants CA 29605 Project II and CA 12528 Project II; the Martin H. Weil Fund; and research grant 05-977 from the Chiron Corporation (Emeryville, CA).
Footnotes
Authors’ Disclosures of Potential Conflicts of Interest
Although all authors completed the disclosure declaration, the following authors or their immediate family members indicated a financial interest. No conflict exists for drugs or devices used in a study if they are not being evaluated as part of the investigation. For a detailed description of the disclosure categories, or for more information about ASCO’s conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
| Authors | Employment | Leadership | Consultant | Stock | Honoraria | Research Funds | Testimony | Other |
|---|---|---|---|---|---|---|---|---|
| Steven J. O’Day | Chiron (A); Berlex (A) | Chiron (C); Berlex (C); Shering (C) | ||||||
| Rene Gonzalez | Chiron (A); Shering (A) | Amgen (A) | Chiron (A); Amgen (A); Shering (A) | Chiron (B); Amgen (B); Shering (B) | ||||
| Dollar Amount Codes (A) 3 $10,000 (B) $10,000–99,999 (C) ≥ $100,000 (N/R) Not Required | ||||||||
References
- 1.Balch CM, Buzaid AC, Soong SJ, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–3648. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 2.Greene FL, Page DL, Fleming ID, et al., editors. AJCC Cancer Staging Manual. 6. New York, NY: Springer-Verlag; 2002. [Google Scholar]
- 3.O’Day SJ, Gammon G, Boasberg PD, et al. Advantages of concurrent biochemotherapy modified by decrescendo interleukin-2, granulocyte colony-stimulating factor, and tamoxifen for patients with metastatic melanoma. J Clin Oncol. 1999;17:2752–2761. doi: 10.1200/JCO.1999.17.9.2752. [DOI] [PubMed] [Google Scholar]
- 4.Richards JM, Gale D, Mehta N, et al. Combination of chemotherapy with interleukin-2 and interferon alfa for the treatment of meta-static melanoma. J Clin Oncol. 1999;17:651–657. doi: 10.1200/JCO.1999.17.2.651. [DOI] [PubMed] [Google Scholar]
- 5.Gibbs P, Iannucci A, Becker M, et al. A phase II study of biochemotherapy for the treatment of metastatic malignant melanoma. Melanoma Res. 2000;10:171–179. [PubMed] [Google Scholar]
- 6.Atkins MB, Gollob JA, Sosman JA, et al. A phase II pilot trial of concurrent biochemotherapy with cisplatin, vinblastine, temozolomide, interleukin 2, and IFN-alpha 2B in patients with metastatic melanoma. Clin Cancer Res. 2002;8:3075–3081. [PubMed] [Google Scholar]
- 7.Khayat D, Bernard-Marty C, Meric JB, et al. Biochemotherapy for advanced melanoma: Maybe it is real. J Clin Oncol. 2002;20:2411–2414. doi: 10.1200/JCO.2002.20.10.2411. [DOI] [PubMed] [Google Scholar]
- 8.Gibbs P, Anderson C, Pearlman N, et al. A phase II study of neoadjuvant biochemotherapy for stage III melanoma. Cancer. 2002;94:470–476. doi: 10.1002/cncr.10186. [DOI] [PubMed] [Google Scholar]
- 9.Eton O, Legha SS, Bedikian AY, et al. Sequential biochemotherapy versus chemotherapy for metastatic melanoma: Results from a phase III randomized trial. J Clin Oncol. 2002;20:2045–2052. doi: 10.1200/JCO.2002.07.044. [DOI] [PubMed] [Google Scholar]
- 10.Pantel K, Cote RJ, Fodstad O. Detection and clinical importance of micrometastatic disease. J Natl Cancer Inst. 1999;91:1113–1124. doi: 10.1093/jnci/91.13.1113. [DOI] [PubMed] [Google Scholar]
- 11.Palmieri G, Strazzullo M, Ascierto PA, et al. Polymerase chain reaction-based detection of circulating melanoma cells as an effective marker of tumor progression. Melanoma Cooperative Group. J Clin Oncol. 1999;17:304–311. doi: 10.1200/JCO.1999.17.1.304. [DOI] [PubMed] [Google Scholar]
- 12.Mellado B, Gutierrez L, Castel T, et al. Prognostic significance of the detection of circulating malignant cells by reverse transcriptase-polymerase chain reaction in long-term clinically disease-free melanoma patients. Clin Cancer Res. 1999;5:1843–1848. [PubMed] [Google Scholar]
- 13.Hoon DS, Bostick P, Kuo C, et al. Molecular markers in blood as surrogate prognostic indicators of melanoma recurrence. Cancer Res. 2000;60:2253–2257. [PubMed] [Google Scholar]
- 14.Wascher RA, Morton DL, Kuo C, et al. Molecular tumor markers in the blood: Early prediction of disease outcome in melanoma patients treated with a melanoma vaccine. J Clin Oncol. 2003;21:2558–2563. doi: 10.1200/JCO.2003.06.110. [DOI] [PubMed] [Google Scholar]
- 15.Taback B, Morton DL, O’Day SJ, et al. The clinical utility of multimarker RT-PCR in the detection of occult metastasis in patients with melanoma. Recent Results Cancer Res. 2001;158:78–92. doi: 10.1007/978-3-642-59537-0_8. [DOI] [PubMed] [Google Scholar]
- 16.Koyanagi K, Kuo C, Nakagawa T, et al. Multimarker quantitative real-time PCR detection of circulating melanoma cells in peripheral blood: Relation to disease stage in melanoma patients. Clin Chem. 2005;51:981–988. doi: 10.1373/clinchem.2004.045096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stathopoulou A, Gizi A, Perraki M, et al. Real-time quantification of CK-19 mRNA-positive cells in peripheral blood of breast cancer patients using the lightcycler system. Clin Cancer Res. 2003;9:5145–5151. [PubMed] [Google Scholar]
- 18.Keilholz U, Goldin-Lang P, Bechrakis NE, et al. Quantitative detection of circulating tumor cells in cutaneous and ocular melanoma and quality assessment by real-time reverse transcriptase-polymerase chain reaction. Clin Cancer Res. 2004;10:1605–1612. doi: 10.1158/1078-0432.ccr-0610-3. [DOI] [PubMed] [Google Scholar]
- 19.Schuster R, Max N, Mann B, et al. Quantitative real-time RT-PCR for detection of disseminated tumor cells in peripheral blood of patients with colorectal cancer using different mRNA markers. Int J Cancer. 2004;108:219–227. doi: 10.1002/ijc.11547. [DOI] [PubMed] [Google Scholar]
- 20.Howe JG, Crouch J, Cooper D, et al. Real-time quantitative reverse transcription-PCR for cyclin D1 mRNA in blood, marrow, and tissue specimens for diagnosis of mantle cell lymphoma. Clin Chem. 2004;50:80–87. doi: 10.1373/clinchem.2003.024695. [DOI] [PubMed] [Google Scholar]
- 21.Hoon DS, Wang Y, Dale PS, et al. Detection of occult melanoma cells in blood with a multiple-marker polymerase chain reaction assay. J Clin Oncol. 1995;13:2109–2116. doi: 10.1200/JCO.1995.13.8.2109. [DOI] [PubMed] [Google Scholar]
- 22.Sarantou T, Chi DD, Garrison DA, et al. Melanoma-associated antigens as messenger RNA detection markers for melanoma. Cancer Res. 1997;57:1371–1376. [PubMed] [Google Scholar]
- 23.Takeuchi H, Morton DL, Kuo C, et al. Prognostic significance of molecular upstaging of paraffin-embedded sentinel lymph nodes in melanoma patients. J Clin Oncol. 2004;22:2671–2680. doi: 10.1200/JCO.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawakami Y, Eliyahu S, Delgado CH, et al. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchida T, Saxton RE, Morton DL, et al. Gangliosides of human melanoma. J Natl Cancer Inst. 1987;78:45–54. doi: 10.1093/jnci/78.1.45. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchida T, Saxton RE, Irie RF. Gangliosides of human melanoma: GM2 and tumorigenicity. J Natl Cancer Inst. 1987;78:55–60. doi: 10.1093/jnci/78.1.55. [DOI] [PubMed] [Google Scholar]
- 27.Kuo CT, Bostick PJ, Irie RF, et al. Assessment of messenger RNA of beta 1 → 4-N-acetylgalactosaminyl-transferase as a molecular marker for metastatic melanoma. Clin Cancer Res. 1998;4:411–418. [PubMed] [Google Scholar]
- 28.Walther C, Guenet JL, Simon D, et al. Pax: A murine multigene family of paired box-containing genes. Genomics. 1991;11:424–434. doi: 10.1016/0888-7543(91)90151-4. [DOI] [PubMed] [Google Scholar]
- 29.Scholl FA, Kamarashev J, Murmann OV, et al. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001;61:823–826. [PubMed] [Google Scholar]
- 30.Miyashiro I, Kuo C, Huynh K, et al. Molecular strategy for detecting metastatic cancers with use of multiple tumor-specific MAGE-A genes. Clin Chem. 2001;47:505–512. [PubMed] [Google Scholar]
- 31.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 32.Jung FA, Buzaid AC, Ross MI, et al. Evaluation of tyrosinase mRNA as a tumor marker in the blood of melanoma patients. J Clin Oncol. 1997;15:2826–2831. doi: 10.1200/JCO.1997.15.8.2826. [DOI] [PubMed] [Google Scholar]
- 33.Glaser R, Rass K, Seiter S, et al. Detection of circulating melanoma cells by specific amplification of tyrosinase complementary DNA is not a reliable tumor marker in melanoma patients: A clinical two-center study. J Clin Oncol. 1997;15:2818–2825. doi: 10.1200/JCO.1997.15.8.2818. [DOI] [PubMed] [Google Scholar]
- 34.Voit C, Kron M, Rademaker J, et al. Molecular staging in stage II and III melanoma patients and its effect on long-term survival. J Clin Oncol. 2005;23:1218–1227. doi: 10.1200/JCO.2005.04.098. [DOI] [PubMed] [Google Scholar]