Abstract
A screen for increased longevity in Caenorhabditis elegans has identified a transcription factor that programs cells for resistance to oxidative stress, DNA repair and cell cycle control. The mammalian orthologs of this factor are referred to as ‘Foxo’ for ‘Forkhead box’, with the second ‘o’ in the name denoting a subfamily of four members related by sequence. This family of factors is regulated by growth factors, oxidative stress or nutrient deprivation. Thus, it might readily control the inflammatory conflagration associated with infection-driven lymphocyte proliferation. Surprisingly, the first insights into Foxo-mediated immune regulation have instead revealed direct control of highly specialized genes of the adaptive immune system.
Foxo transcription factors regulate disparate transcriptional programs that affect cellular differentiation, survival, cell-cycle arrest, metabolism, stress resistance and tumor suppression. How do one, two or three widely expressed and highly similar transcription factors simultaneously regulate such diverse and fundamental functions of the cell? Before we begin to understand that, there is yet another level of complexity: Foxo factors also control specialized functions of differentiated tissues.
Cells of the immune system constitute the most dynamic populations found in metazoans. For example, after infection, a few hundred antigen-specific, quiescent T cells can proliferate more than 10,000-fold in a few days. As this population expansion proceeds, the rate of cell death progressively increases, eventually causing an equally dramatic die-off as the population finds a new equilibrium. All the while, the T cells in this rapidly fluctuating population differentiate into the various effector populations of the immune system, and surviving cells take on a memory phenotype. These are precisely the types of events that Foxo factors have been shown to regulate in many cell types, yet recent studies have shown that Foxo factors regulate homeostasis of the immune system in unpredictable ways. Although the Foxo signal-transduction pathways are conserved evolutionarily, they seem to have been co-opted in differentiated tissues for a variety of specialized functions.
This review first summarizes the many modes of regulation used by Foxo transcription factors and the present state of knowledge pertaining to cofactor associations. From this perspective, the presently known functions for Foxo factors in life and death associated with immune homeostasis are then covered.
Post-translational mechanisms of Foxo regulation
Three of the four known Foxo orthologs are widely expressed and similarly regulated and have overlapping targets of transcriptional regulation1. Foxo1 (FKHR), Foxo3 (FKHRL1) and Foxo4 (AFX or Mllt7) are all related to the Caenorhabditis elegans ortholog Daf16 and the Drosophila melanogaster ortholog dFoxo. A fourth factor identified by homology, Foxo6, is expressed only in the brain, and its cellular localization is alternatively regulated2. Two characteristics make Foxo1, Foxo3 and Foxo4 highly versatile in gene regulation. First, Foxo transcriptional activities are modified by phosphorylation, acetylation, methylation and O-linked glycosylation. These post-translational modifications alter Foxo intracellular localization, turnover, transactivation and transcriptional specificity and constitute a Foxo ‘code’3. Second, Foxo factors can associate with many different cofactor complexes to regulate context-dependent programs of gene expression4. Depending on their binding partners, they can act as direct or indirect transcriptional activators or repressors.
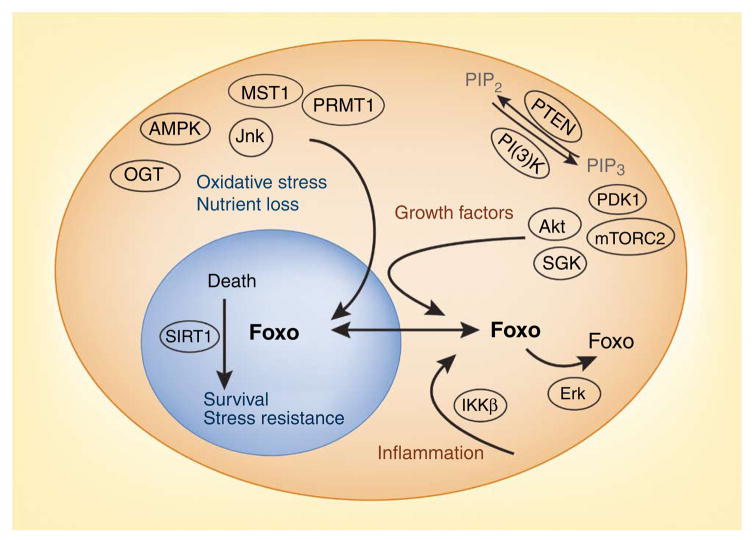
The simplest level of Foxo regulation, or at least the first to be described, depends on nuclear export controlled by the phosphatidyl- inositol-3-OH kinase (PI(3)K) pathway (Fig. 1). Activation of PI(3)K by the insulin receptor, among many others, results in the production of phosphatidylinositol-(3,4,5)-trisphosphate5, and this in turn causes localization of the kinase Akt to the plasma membrane6,7. Activation of Akt requires Thr308 phosphorylation induced by the phosphoinositide-dependent kinase PDK1 (refs. 8,9). Ser473 phosphorylation induced by mTORC2 (consisting of the kinase mTOR, Rictor, Sin1 and mLST8 complex) is required in vivo for Akt-mediated phosphorylation of a subset of targets, including the Foxo factors10,11. Foxo1, Foxo3 and Foxo4, like Daf16, have three conserved amino acids that are targets for phosphorylation by Akt or SGK (serum- and glucocorticoid-inducible kinase)12. The result of Foxo phosphorylation at these sites is nuclear exclusion. Phosphorylated Foxo factors have been shown to interact with the adaptor 14-3-3, which promotes relocalization to the cytoplasm, possibly through a conformational change that exposes the nuclear-export signal and masks the nuclear-localization signal13–15. In addition, there are two sites phosphorylated by caseine kinase 1 that may enhance interactions with nuclear-export molecules such as Crm1, Ran and exportin16. A second potentially important trigger of Foxo3 nuclear exclusion is the kinase IKKβ, which phosphorylates Foxo3 in humans and mice at Ser644 and is active in tumors that lack activated Akt17. Another signaling pathway able to induce Foxo inactivation involves a DNA-damage checkpoint induced by cyclin-dependent kinase 2–mediated phosphorylation of Foxo1; DNA damage–induced cell death is Foxo1 dependent18.
Figure 1.
Opposing influences on Foxo cellular localization and transcriptional activity. A major pathway of Foxo regulation consists of growth factor–mediated nuclear exclusion and inactivation (top right). Many tumors have been found to contain phosphorylated Akt or inflammation-driven phosphorylated IKKβ as a mechanism of inactivating Foxo tumor-suppressive activity. In addition, a major pathway of Ras-mediated transformation is the Erk-mediated phosphorylation of Foxo (bottom right), which results in its MDM2-mediated ubiquitination and degradation. Opposing these inactivating pathways is oxidative stress (top left), which results in either Jnk- or MST1-mediated Foxo phosphorylation. Nuclear Foxo transcriptional specificity can be altered by SIRT1-mediated deacetylation, which results in a prosurvival program of gene expression. OGT, O-linked β-N-acetylglucosamine transferase; PIP2, phosphatidylinositol-4, 5-bisphosphate; PIP3, phosphatidylinositol-3,4,5-trisphosphate.
Phosphorylation can also induce the proteosomal degradation of Foxo factors. Treatment of fibroblasts with epidermal growth factor or anisomycin increases phosphorylation of Foxo1, and this phosphorylation is diminished by the addition of inhibitors of the kinases p38 or Erk19 (Fig. 1). More recently, studies have shown that Erk-mediated phosphorylation of Foxo3 results in polyubiquitination of Foxo3 carried out by the E3 ligase MDM2, and this results in proteosomal degradation20. In fact, reestablishment of Foxo activity reverts cell growth control in tumors transformed by the GTPase Ras; the implication is that along with inactivation of the kinase GSK3β21, much of the oncogenic activity of the Ras-Erk pathway depends on inactivation of Foxo. In accord with that, the Foxo factors are important redundant tumor-suppressor molecules. Deletion of up to five alleles of the three genes results in remarkably modest neoplasia, whereas broad homozygous deletion of the three genes results in spontaneous hemangiomas and thymic lymphomas22.
Balancing the multiple avenues of Foxo inactivation described above are several opposing signaling pathways that stabilize nuclear Foxo localization. Protein arginine methyltransferases (PRMTs) catalyze methylation of the terminal nitrogens on the guanidinium side chains of arginine. There are 11 PRMT isoforms in all eukaryotes examined; they are involved in transcriptional regulation, DNA repair, protein interactions and signal transduction23. Studies have shown that oxidative stress causes induction of PRMT1, resulting in the methylation of two arginine residues present in Foxo1. This has the effect of directly inhibiting Akt-mediated phosphorylation and thus enhancing nuclear localization of Foxo1 and apoptosis24.
Oxidative stress or nutrient deprivation can also oppose nuclear export by alternative phosphorylation. Phosphorylation mediated by the kinase Jnk opposes Akt-mediated nuclear export of Foxo4, although the phosphorylation occurs in the transactivation domain25,26. In addition, oxidative stress in the form of hydrogen peroxide induces phosphorylation of Foxo3 mediated by the kinase MST1 on a site that is conserved in mammals and nematodes27 (Fig. 1). This phosphorylation disrupts interactions with 14-3-3 and promotes translocation to the nucleus. Whether Jnk and MST1 phosphorylate other Foxo isoforms is unknown at present. The result of Foxo nuclear localization is the induction of a program of gene expression whose purpose seems to be detoxification of reactive oxygen species28,29. This is clearly illustrated by the deletion of Foxo1, Foxo3 and Foxo4 in hematopoietic stem cells, which causes more production of reactive oxygen species and lower stem cell longevity30. In conditions of starvation, AMP-activated protein kinase phosphorylates Foxo factors at two or more sites, which results in the selective induction of genes important for metabolism and stress resistance31.
Foxo factors are further regulated by acetylation. Histone acetylation by calcium response element–binding–binding protein p300 or both may be necessary for Foxo transcriptional activity, yet acetylation of Foxo attenuates Foxo-DNA interactions32. The mammalian homologs of the yeast silent information regulator Sir2, including the longevity factor SIRT1, deacetylate Foxo factors and modify their transcriptional activity33. Binding of SIRT1 to Foxo3 results in the ‘preferential’ activation of genes involved in cell cycle arrest and resistance to oxidative stress but it causes less activation of proapoptotic genes34. The transcriptional specificity of Foxo genes can thus be regulated in different ways to promote life or death depending on the context—in this case, activation of SIRT1 (ref. 35; Fig. 1).
Another important post-translational modification is O-linked glycosylation in the form of modification of serine and threonine side chains by O-linked β-N-acetylglucosamine. All such glycosylations are catalyzed by O-linked β-N-acetylglucosamine transferase, whereas the substrate specificity of this transferase derives from its association with many adaptor proteins. It functions as a nutrient sensor, as its substrate, UDP-N-acetylglucosamine, rapidly responds to multiple metabolic pathways. Foxo1 is glycosylated as a result of hyperglycemia, with the result being more Foxo1 transcriptional activity36.
Thus, several pathways dominantly promote the nuclear, active form of Foxo1 instead of insulin- or growth factor–induced inactivation of Foxo1. This complex interplay of modifications seems to integrate the amount of growth factor receptor stimulation with the amount of oxidative stress and nutrient availability to determine the intracellular localization and transcriptional activity of Foxo factors (Fig. 1). Acetylation further alters Foxo transcriptional specificity. In addition to this intricate regulation, the specificity of Foxo-mediated transcriptional activity is determined by the association of Foxo factors with many different coactivator or corepressor complexes.
Transcriptional cofactors and targets
Foxo factors, like most transcription factors, mediate their activity as part of protein complexes able to influence sequence-specific chromatin interactions and transcriptional activity. In fact, Foxo1 is able to carry out chromatin opening as a ‘gateway’ transcription factor37. The association of Foxo factors with other transcription factors has been highlighted by experiments showing that Foxo3 regulates gene expression in three ways: sequence-specific gene activation; DNA binding–independent gene activation; and sequence-independent gene repression38. Gene array analyses of Foxo3 mutants unable to bind to canonical Foxo DNA-binding sequences have shown that regulation is maintained, at least in part, for certain classes of genes. This has been interpreted to show that Foxo factors bind to other DNA-binding factors and promote or suppress transcriptional activity. Such protein complexes may be mimicked by chromosomal translocations that result in transcripts that encode a transcriptional-activation domain from Foxo fused to a different DNA-binding domain. For example, one such translocation associated with rhab-domyosarcomas results in fusion of the genes encoding the transcription factors Pax3 and Foxo1, which produces transcripts containing the intact Pax3 DNA-binding domain and the Foxo1 transcriptional-activation domain39.
So far, at least 24 transcription factors have been shown to bind to Foxo factors4. These include nuclear hormone receptors, β-catenin, C/EBP, factors involved in the development of smooth and skeletal muscle, PPAR and the PPARγ coactivator PGC-1α, as well as Smad3 and Smad4 ‘downstream’ of transforming growth factor-β (TGF-β) signaling. The last is discussed in more detail below; a previously published review was also devoted to the topic of Foxo cofactor associations4. The target genes regulated by Foxo factors, whether by direct DNA binding or indirectly through other means, include a number of genes encoding molecules with demonstrated functions in the regulation of immunity (Table 1). Among the most prominent targets are genes encoding regulators of cell cycle, apoptosis, DNA repair, oxidative stress and, surprisingly, specialized functions of the immune system. A microarray analysis of genes affected by a Foxo1 deletion has shown that 187 genes are altered by twofold or more in both CD4+ T cells and CD8+ T cells, so there are many other potential targets40. Foxo factors also control the expression of genes involved in other physiological processes, such as metabolism41. Here I will focus on the possibility that one of the important ways Foxo factors function in the immune system is to coregulate many of the differentiation and survival pathways ascribed to TGF-β and its signaling network42.
Table 1.
Foxo targets
| Target | Ref. |
|---|---|
| Immune system specialized | |
| IL-7R (CD127) | 63,79 |
| KLF4 | 88 |
| KLF2 | 78 |
| S1P1 receptors EDG1 & EDG6 | 78 |
| RAG-1 & RAG-2 | 84 |
| Bcl-6 | 89 |
| Apoptosis | |
| Bim | 89 |
| Fas ligand | 90 |
| TRAIL | 91 |
| Oxidative stress & DNA repair | |
| Catalase | 92 |
| Mn-SOD | 28 |
| GADD45 | 93 |
| ATM | 94 |
| Cell cycle | |
| Cyclin G2 | 29 |
| Cyclin B | 95 |
| p15Ink4b | 96 |
| p19Ink4d | 96 |
| p21WAF1/Cip1 | 45 |
| p27KIP1 | 29 |
| Rb family member p130 | 97 |
| Plk | 95 |
Far left column indicates molecule encoded by target gene. S1P, sphingosine 1-phosphate; TRAIL, tumor necrosis factor receptor–related, apoptosis-inducing ligand; Mn-SOD, manganese-containing superoxide dismutase; Gadd45, growth arrest and DNA damage; ATM, ataxia telangiectasia mutated; Plk, polo-like kinase.
TGF-β cytokines are the most potent cytostatic factors known, and they have dramatic effects on immune regulation and T cell differentiation. After complex processing and secretion, mature TGF-β cytokines signal through the TGF-β type I receptor kinase (TGF-βRI) and TGF-β type II receptor kinase (TGF-βRII), which results in the phosphorylation of Smad2 and Smad3 (ref. 43). Phosphorylated nuclear Smad proteins associate with Smad4 to form a transcriptional activation complex44. Smad-mediated regulation of many transcriptional targets is mediated by association with Foxo factors; these targets include genes encoding the cell cycle inhibitors p15Ink4b and p21Cip1 (ref. 45). For example, the induction of p15Ink4b and the repression of c-Myc depend on Foxo, Smad4 and C/EBPβ46.
Mice with targeted mutation of the gene encoding TGF-β1 developed a florid autoimmune disease characterized by multiorgan inflammatory cell infiltration47,48. The mode of TGF-β action in the immune system has been worked out more clearly by the specific deletion of TGF-βRII in T cells. The phenotypic changes found in these mice include a rapidly progressive wasting disease, with death occurring by 5 weeks of age. Studies show that these mice have a defect in the survival of regulatory T cells (Treg cells), but this is not sufficient to explain the autoimmune infiltration. Instead, the absence of TGF-βRII signaling results in dysregulated, proliferating T cells regardless of the presence of wild-type Treg cells49. If Foxo factors are necessary for TGF-β signaling in the immune system, then mice with loss of Foxo function might be expected to develop a similar riotous cellular proliferation, ultimately leading to death.
As will become apparent, Foxo1 and Foxo3 have markedly distinct but possibly overlapping functions in the immune system. They affect not only homeostasis but also specialized functions in lymphocytes and myeloid cells that would not be predicted from the studies cited above. There is no evidence thus far to support the idea of a special function for Foxo4 in the immune system, and the lack of phenotypic changes associated with a Foxo4 genetic defect indicate that if one exists, it will be subtle or only manifest in the presence of other Foxo deletions.
Foxo3 in immune homeostasis
The tissue specificity of Foxo3 does not provide clues to its function, as it is consistently expressed in all cells of the body (http://biogps.gnf.org/#goto=genereport&id=2309). Nor can its function in immune homeostasis be inferred from observations of fibroblasts. Nonetheless, studies have shown that Foxo3 is able to mediate cell death in many cell types, including T cells and B cells. For example, expression of Foxo3, especially a mutant version confined to the nucleus, causes apoptosis in the tumor suppressor PTEN–deficient T lymphoma Jurkat29,50. One study has shown death to be dependent on expression of the ligand for the cell surface receptor Fas50, whereas another has shown G1 arrest to result from p27Kip1 expression29. A second mechanism affecting T cell apoptosis is the induction of the proapoptotic molecule Bim. The culture-adapted CTLL-2 cell line requires interleukin 2 (IL-2) to exclude Foxo3 from the nucleus and prevent Bim-induced cell death; a similar result occurs in Ba/F3 B cells grown in IL-3 (refs. 51,52). Subsequent studies of PTEN-deficient primary T cells have shown that IL-2 withdrawal induces the expression of Bim and Puma, two death-effector molecules, in a Foxo3-dependent manner53. In addition, memory T cells have higher expression of phosphorylated Foxo3 and consequently lower expression of Bim54. In addition, ‘elite controller’ human immunodeficiency virus–positive people (that is, people who despite being infected do not progress to AIDS) have persistent memory T cells as a direct consequence of Foxo3 inactivation55. From these and other studies56–58, a conclusion would be that Foxo3 mediates the death of T cells and B cells, possibly in response to the limiting growth factor availability during the contraction phase of an immune response59.
Consistent with that idea, the first characterization of Foxo3- deficient mice revealed autoimmunity characterized by spontaneous lymphoproliferation, hyperactivated T cells, constitutive activity of the transcription factor NF-κB, and lymphocyte organ infiltration60. These mice were created by retroviral gene-trap techniques and were made using embryonic stem cells from mice of strain 129 (Foxo3Trap mice). A difficulty with this tidy story is that it has not held up to further experimentation. Mice generated separately from another retroviral gene- trap project (Foxo3Kca mice), along with a third strain generated by targeted recombination (Foxo3− mice), have also been analyzed61,62. The original histological analysis of 36 tissues from Foxo3Kca mice found no changes relative to wild-type mice, with the exception of abnormal ovary development61. B cell development and function in these mice are no different from those of wild-type littermates63, although a subsequent analysis of Foxo3− mice has shown some decrease in the number of in pre-B cells and circulating B cells64. Confounding the initial expectations, T cells from Foxo3Kca and Foxo3− mice are not different from those of their wild-type littermates in the rate of proliferation or survival after mitogen stimulation. In addition, there is no evidence of constitutive NF-κB activation or lymphocyte organ infiltration65. In fact, the mice are remarkably normal. Despite the work showing involvement of Foxo3 in lymphocyte death, and the phenotype of Foxo3Trap mice, the preponderance of evidence suggests that loss of Foxo3 itself does not result in lymphoproliferative disease, and hence it may not have a unique function in the natural survival of lymphocytes. A speculation is that a genetic polymorphism found in 129 mice acts together with the loss of Foxo3 to dysregulate T cell homeostasis and cause autoimmune disease66–68. Still, deletion of Foxo3 has been shown to have other important effects on immune regulation.
Infection of Foxo3-deficient mice with lymphocytic choriomeningitis virus results in an increase of two- to threefold in the total number of splenic virus-specific T cells, whereas adoptive-transfer and bone marrow–reconstitution experiments have shown that this increase is not T cell intrinsic but is of hematopoietic origin. Dendritic cells and granulocytes are more abundant in Foxo3-deficient mice, and Foxo3-deficient dendritic cells overproduce IL-6, which results in enhanced survival of T cells in vivo and in tissue culture. Experiments have shown that signaling through CD80 and CD86 (B7) causes nuclear localization of Foxo3 in dendritic cells from nonobese diabetic mice, which results in the production of manganese-containing superoxide dismutase69. Consistent with that finding, B7 signaling mediated by ligation with a fusion of the immunomodulatory receptor CTLA-4 and immunoglobulin causes nuclear localization of Foxo3 and inhibition of Toll-like receptor 7–mediated production of IL-6 and tumor necrosis factor. In the absence of Foxo3, CTLA-4–immunoglobulin has little, if any, effect. The conclusion is that one of the functions of CTLA-4 is to resolve inflammatory responses by signaling through B7 and overriding the nuclear exclusion of Foxo factors mediated by IKKβ and/or Akt17. The origin of this signal override is unknown, but a conjecture is that IKKα and the noncanonical NF-κB pathway could be involved70,71. These experiments show that the only nonredundant function for Foxo3 in a primary immune response is to suppress inflammatory cytokine production by dendritic cells. As discussed below, Foxo3 may also have overlapping functions with Foxo1.
Foxo1
Foxo1 is also widely expressed, but its expression is highly variable in different tissues. In humans and mice, Foxo1 is most highly expressed in T cells, B cells and ovaries (http://biogps.gnf.org/#goto=genereport&id=2308), but until very recently there have been few studies on Foxo1 in the immune system. The first study showed that Foxo1 is expressed in thymocyte subsets, but expression of an inducible dominant negative Foxo1 mutant did not have an effect on thymocyte development72. In mature T cells, an active form of Akt has been shown to inactivate Foxo1, and this correlates with enhanced cell cycle progression73. One natural pathway for activation of PI(3)K and Akt is mediated by IL-7, which causes higher expression of the antiapoptotic molecule Bcl-2 and lower expression of p27Kip1, as well as hyperphosphorylation of the transcription factor Rb74. Another pathway is through ligation of the T cell antigen receptor, which mediates sustained PI(3)K activation and localization to the immunological synapse75,76. Illustrating the potential for Foxo1 and Foxo3 redundancy, another study has demonstrated that enforced expression of either Foxo1 or Foxo3 in B cells results in partial cell cycle arrest in G1 and more apoptosis77.
None of the studies discussed above anticipated the specialized function for Foxo1 in T cells and B cells. Studies of PTEN-deficient Jurkat cells have shown that the introduction of a constitutively nuclear mutant of Foxo1 causes the induction of L-selectin, two sphingosine 1-phosphate receptors, and KLF2, a transcription factor that controls lymphocyte homing. Chromatin immunoprecipitation has further demonstrated that Foxo1 binds to the promoter of the gene encoding KLF2 (ref. 78). Those results have been confirmed and extended in mice with T cell–specific deletion of Foxo1. Such T cells have lower expression of KLF2 and the homing molecules L-selectin and CCR7 (ref. 79). Expression of the egress receptor S1P1 is also diminished in these T cells (Y. Kerdiles, unpublished data). In accord with those results, acute knockout of Foxo1 results in highly defective T cell homing to the lymph nodes79. Further analysis of Foxo1-deficient T cells has revealed a survival defect and lower expression of Bcl-2; these defects are caused by a complete loss of expression of the IL-7 receptor α-chain (IL-7Rα; CD127)40,79. Consistent with those results, mice with T cell–specific Foxo1 deletion have few naive T cells but excess cells with an activated or memory phenotype. In addition, chromatin-immunoprecipitation experiments have shown that Foxo1 binds to an enhancer 3.5 kilobases upstream of the Il7r transcription start site79.
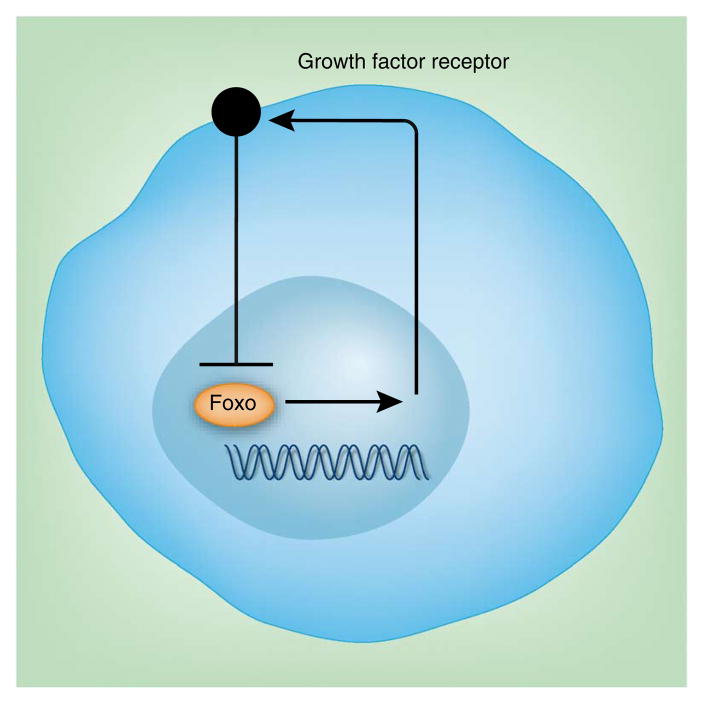
An important point is that Foxo1 is essential in the previously described IL-7-regulated negative-feedback control of IL-7Rα expression79,80. The addition of IL-7 to T cells decreases the occupancy of Foxo1 on the Il7r enhancer and, as described above, Foxo1 is essential for IL-7Rα expression. This is analogous to the insulin-regulated negative-feedback control of the insulin receptor81 and, as discussed below, it may be a common unifying theme of Foxo transcriptional control (Fig. 2).
Figure 2.
A typical negative feedback loop controlled by Foxo-regulated transcription. Genes encoding growth factor receptors controlled by Foxo will be subject to downregulation in conditions of abundant growth factors. The net effect is a self-limited growth factor–mediated signaling. Examples include the insulin receptor and the IL-7 receptor.
The PI(3)K pathway controls T cell homing through the regulation of KLF2 (ref. 82), and this would seem to be consistent with the function of PI(3)K in the inactivation of Foxo1. As KLF2 is apparently a direct target of Foxo1, the simple model is that PI(3)K signaling inactivates Foxo1, which in turn causes a decrease in KLF2 transcription. The difficulty is that KLF2 downregulation has also been shown to be sensitive to rapamycin, a specific inhibitor of mTORC1, whereas Foxo1 inactivation is dependent on mTORC2 and is thus thought to be insensitive to the effects of rapamycin83. This apparent contradiction has not yet been resolved.
Mice with T cell–specific Foxo1 deficiency also show aspects of autoimmunity. Radiation chimeras generated with bone marrow from a donor with T cell–specific Foxo1 deletion develop spontaneous colitis associated with a lack of Foxp3+ Treg cells. This pathology is ‘rescued’ in chimeras reconstituted with a mixture of wild-type and Foxo1-deficient bone marrow and correlates with the presence of Treg cells contributed by the wild-type bone marrow40. Why radiation chimeras develop colitis when mice with T cell–specific Foxo1 deficiency themselves do not was not determined; however, the loss of Treg cells in Foxo1-deficient bone marrow–radiation chimeras might be explained if Treg cell development requires TGF-β signaling and thus Foxo1 (ref. 42). As for autoimmunity and complementation in mixed–bone marrow chimeras, this has been confirmed separately (Y. Kerdiles, unpublished observations). In addition, mice with T cell–specific loss of Foxo1 show an increase in germinal center B cells and memory B cells that is consistent with the observed increases in antibodies to DNA (Y. Kerdiles, unpublished observations).
Contemporaneous studies have shown that Foxo1 has equally specialized functions in B cells. It directly regulates expression of the recombination-activating genes Rag1 and Rag2 throughout early B cell development, and transcription of these genes is repressed by Akt activation84. Further studies of mice lacking Foxo1 at various stages of B cell development have shown that early deletion (mediated by Cre recombinase expressed under the control of Cd79a) causes a partial block at the transition from pre-pro–B cell to early pro–B cell and that very few B cells progress beyond the cycling pre–B cell stage. This has been found to be due to a lack of IL-7Rα expression necessary for the survival and proliferation of early B cells and accessibility of the variable heavy-chain region to RAG-mediated rearrangements63. Later deletion (mediated by Cre recombinase expressed under the control of Cd19) causes a block at the pre–B cell stage due to diminished expression of Rag1 and Rag2. This results in an excess of small resting pre–B cells negative for immunoglobulin M and immunoglobulin D in peripheral lymphoid organs. Late deletion of Foxo1 (mediated by Cre recombinase expressed under the control of Cr2) results in a deficiency of lymph node B cells due to homing defects resulting from loss of L-selectin expression. Interestingly, activated B cells also lack activation-induced cytidine deaminase and are thus unable to undergo class-switch recombination63. The implication here is that the PI(3)K pathway must be attenuated and Foxo1 must be expressed for B cells to transition through multiple checkpoints all the way to class-switch recombination. Either B cell antigen receptor or IL-7Rα signaling occurring during B cell development can modulate expression of Rag1 and Rag2, possibly as a means of coordinating gene rearrangements with developmental stage or productive rearrangements of B cell antigen receptor heavy- and light-chain genes. Likewise, class-switch recombination would not take place as long as B cells experience activation of the PI(3)K pathway, as would be expected to occur in the presence of strong CD19-CD21 costimulation85.
Foxo redundant functions
A question is whether the role of Foxo1 in T and B cells is limited to control of cell type–specific, highly specialized functions or whether Foxo1 also regulates cell cycle control, apoptosis and response to oxidative stress. In experiments designed to address the requirement for Foxo1 in T cell quiescence and differentiation, Rag1−/− mice expressing the OT-II T cell antigen receptor transgene were bred to mice with T cell–specific deletion of Foxo1. In this case, T cells numbers were diminshed to 10% of wild-type and all had a naive phenotype40,79. Thus, loss of Foxo1 by itself is not sufficient to allow uncontrolled T cell population expansion in the absence of signals from the T cell antigen receptor; perhaps other Foxo factors are involved in maintaining T cell quiescence. However, in other experiments, Foxo1, Foxo3 and Foxo4 were deleted in hematopoietic progenitors of 4-week-old mice with Cre recombinase expressed under the control of Mx1. After a further 4–5 weeks, there was moderate expansion of some myeloid and lymphoid subsets but no uncontrolled growth30. In accord with those results, mice deficient in Foxo-targeted cell cycle inhibitors have various phenotypes, although none show uncontrolled cellular proliferation86. These results are therefore consistent with the idea that the mild autoimmune or lymphoproliferative disease caused by the deletion of Foxo1 in T cells is not cell intrinsic but is due to the loss of specific immunological control mechanisms, including but not limited to induced Treg cells. A guess is that the mechanism underlying this deficiency is a defect in TGF-β signaling.
A question is how the dynamics of T cell or B cell population expansion might be altered after acute deletion of Foxo1, Foxo3 and Foxo4 in mature lymphocytes. One possibility is that deletion of Foxo1 in T cells and Foxo3 in dendritic cells could act in synergy to produce spontaneous autoreactivity or at least altered kinetics of lymphocyte population expansion. Alternatively, whereas genetic ablation of Foxo3 or Foxo4 may not result in a notable phenotype in T cells, they may nonetheless serve a function in quiescence that can only be revealed by the loss of Foxo1. Clearly there is a need to further analyze lymphocyte dynamics in the absence of multiple Foxo genes.
Common characteristics of Foxo regulation
What are the common characteristics of genes regulated by Foxo? Although the PI(3)K pathway causes the induction of a wide variety of genes, it programs an equally profound loss of gene expression87, and I surmise that in many examples of receptor-mediated signaling that cause a PI(3)K-dependent loss of gene expression, Foxo factors may have been evolutionarily recruited to provide the necessary transcriptional regulation (Fig. 2). In mammals, genes as diverse as those encoding RAG-1, RAG-2, activation-induced cytidine deaminase, IL-7Rα and KLF2 are limited in expression by Foxo1, probably in conjunction with varying transcriptional coregulators and with context-specific post-translational modifications. In contrast, with tumor regression as an extreme example of the loss of cell cycle control, there seems to be a great deal of redundancy in Foxo function22. By this logic, one proposal is that specialized tasks may be controlled by individual factors, whereas genes involved in basic cellular functions such as cell cycle progression, oxidative stress reduction or survival may be controlled redundantly by two or three Foxo factors. In this way, specialized functions can be controlled independently from generalized functions, and only when all Foxo molecules are inactivated will there occur a loss of cell cycle inhibitors or reactive oxygen intermediate detoxifying enzymes. The implication is that individual Foxo factors may be inactivated in different ways in different contexts. Of course, this idea would apply only to vertebrates with multiple Foxo genes, because in nematodes and flies, only a single Foxo ortholog is present. In any event, the studies so far show clearly that Foxo molecules serve important functions in immune homeostasis, and future work will determine how these functions integrate with other physiological processes that alter Foxo activity, such as diet- induced changes in insulin, insulin-like growth factor or other PI(3)K-activating cytokines.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Burgering BM. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258–2262. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs FM, et al. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278:35959–35967. doi: 10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- 3.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 4.van der Vos KE, Coffer PJ. FOXO-binding partners: it takes two to tango. Oncogene. 2008;27:2289–2299. doi: 10.1038/onc.2008.22. [DOI] [PubMed] [Google Scholar]
- 5.Sun XJ, et al. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 6.Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 7.Franke TF, et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 8.Alessi DR, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 9.Stokoe D, et al. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 10.Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 11.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Burgering BM, Kops GJ. Cell cycle and death control: long live Forkheads. Trends Biochem Sci. 2002;27:352–360. doi: 10.1016/s0968-0004(02)02113-8. [DOI] [PubMed] [Google Scholar]
- 13.Rena G, Prescott AR, Guo S, Cohen P, Unterman TG. Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targetting. Biochem J. 2001;354:605–612. doi: 10.1042/0264-6021:3540605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brownawell AM, Kops GJ, Macara IG, Burgering BM. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol Cell Biol. 2001;21:3534–3546. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunet A, et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156:817–828. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rena G, et al. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 2002;21:2263–2271. doi: 10.1093/emboj/21.9.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu MC, et al. IκB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 18.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–297. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 19.Asada S, et al. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell Signal. 2007;19:519–527. doi: 10.1016/j.cellsig.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Yang JY, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Q, et al. Erk associates with and primes GSK-3β for its inactivation resulting in upregulation of β-catenin. Mol Cell. 2005;19:159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamagata K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Essers MA, et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 27.Lehtinen MK, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 28.Kops GJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 29.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 30.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Greer EL, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 32.Fukuoka M, et al. Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int J Mol Med. 2003;12:503–508. [PubMed] [Google Scholar]
- 33.Yang Y, Hou H, Haller EM, Nicosia SV, Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005;24:1021–1032. doi: 10.1038/sj.emboj.7600570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 35.Haigis MC, Guarente LP. Mammalian sirtuins–emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 36.Housley MP, et al. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283:16283–16292. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatta M, Cirillo LA. Chromatin opening and stable perturbation of core histone: DNA contacts by FoxO1. J Biol Chem. 2007;282:35583–35593. doi: 10.1074/jbc.M704735200. [DOI] [PubMed] [Google Scholar]
- 38.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 39.Galili N, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 40.Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, et al. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 42.Li MO, Flavell RA. TGF-β: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massague J, Gomis RR. The logic of TGFβ signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, Massague J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 45.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 46.Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 47.Shull MM, et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulkarni AB, et al. Transforming growth factor β1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci USA. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-β controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 50.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 51.Stahl M, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 52.Dijkers PF, et al. FKHR-L1 can act as a critical effector of cell death induced by cytokine withdrawal: protein kinase B-enhanced cell survival through maintenance of mitochondrial integrity. J Cell Biol. 2002;156:531–542. doi: 10.1083/jcb.200108084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You H, et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riou C, et al. Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. J Exp Med. 2007;204:79–91. doi: 10.1084/jem.20061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Grevenynghe J, et al. Transcription factor FOXO3a controls the persistence of memory CD4+ T cells during HIV infection. Nat Med. 2008;14:266–274. doi: 10.1038/nm1728. [DOI] [PubMed] [Google Scholar]
- 56.Pandiyan P, et al. CD152 (CTLA-4) determines the unequal resistance of Th1 and Th2 cells against activation-induced cell death by a mechanism requiring PI3 kinase function. J Exp Med. 2004;199:831–842. doi: 10.1084/jem.20031058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asselin-Labat ML, et al. FoxO3 mediates antagonistic effects of glucocorticoids and interleukin-2 on glucocorticoid-induced leucine zipper expression. Mol Endocrinol. 2005;19:1752–1764. doi: 10.1210/me.2004-0206. [DOI] [PubMed] [Google Scholar]
- 58.Hur EM, et al. Osteopontin-induced relapse and progression of autoimmune brain disease through enhanced survival of activated T cells. Nat Immunol. 2007;8:74–83. doi: 10.1038/ni1415. [DOI] [PubMed] [Google Scholar]
- 59.Marrack P, Kappler J. Control of T cell viability. Annu Rev Immunol. 2004;22:765–787. doi: 10.1146/annurev.immunol.22.012703.104554. [DOI] [PubMed] [Google Scholar]
- 60.Lin L, Hron JD, Peng SL. Regulation of NF-κB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 61.Hosaka T, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci USA. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- 63.Dengler HS, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hinman RM, et al. Foxo3−/− mice demonstrate reduced numbers of pre-B and recirculating B cells but normal splenic B cell sub-population distribution. Int Immunol. 2009;21:831–842. doi: 10.1093/intimm/dxp049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dejean AS, et al. Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat Immunol. 2009;10 :504–513. doi: 10.1038/ni.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Botto M, et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19:56–59. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 67.Bickerstaff MC, et al. Serum amyloid P component controls chromatin degradation and prevents antinuclear autoimmunity. Nat Med. 1999;5:694–697. doi: 10.1038/9544. [DOI] [PubMed] [Google Scholar]
- 68.Santiago-Raber ML, et al. Role of cyclin kinase inhibitor p21 in systemic autoimmunity. J Immunol. 2001;167:4067–4074. doi: 10.4049/jimmunol.167.7.4067. [DOI] [PubMed] [Google Scholar]
- 69.Fallarino F, et al. CTLA-4-Ig activates forkhead transcription factors and protects dendritic cells from oxidative stress in nonobese diabetic mice. J Exp Med. 2004;200:1051–1062. doi: 10.1084/jem.20040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 71.Puccetti P, Grohmann U. IDO and regulatory T cells: a role for reverse signalling and non-canonical NF-κB activation. Nat Rev Immunol. 2007;7:817–823. doi: 10.1038/nri2163. [DOI] [PubMed] [Google Scholar]
- 72.Leenders H, Whiffield S, Benoist C, Mathis D. Role of the forkhead transcription family member, FKHR, in thymocyte differentiation. Eur J Immunol. 2000;30:2980–2990. doi: 10.1002/1521-4141(200010)30:10<2980::AID-IMMU2980>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 73.Patra AK, Na SY, Bommhardt U. Active protein kinase B regulates TCR responsiveness by modulating cytoplasmic-nuclear localization of NFAT and NF-κB proteins. J Immunol. 2004;172:4812–4820. doi: 10.4049/jimmunol.172.8.4812. [DOI] [PubMed] [Google Scholar]
- 74.Barata JT, et al. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J Exp Med. 2004;200:659–669. doi: 10.1084/jem.20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Costello PS, Gallagher M, Cantrell DA. Sustained and dynamic inositol lipid metabolism inside and outside the immunological synapse. Nat Immunol. 2002;3:1082–1089. doi: 10.1038/ni848. [DOI] [PubMed] [Google Scholar]
- 76.Fabre S, et al. Stable activation of phosphatidylinositol 3-kinase in the T cell immunological synapse stimulates Akt signaling to FoxO1 nuclear exclusion and cell growth control. J Immunol. 2005;174:4161–4171. doi: 10.4049/jimmunol.174.7.4161. [DOI] [PubMed] [Google Scholar]
- 77.Yusuf I, Zhu X, Kharas MG, Chen J, Fruman DA. Optimal B-cell proliferation requires phosphoinositide 3-kinase-dependent inactivation of FOXO transcription factors. Blood. 2004;104:784–787. doi: 10.1182/blood-2003-09-3071. [DOI] [PubMed] [Google Scholar]
- 78.Fabre S, et al. FOXO1 regulates L-selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181:2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 79.Kerdiles YM, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park JH, et al. Suppression of IL7Rα transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity. 2004;21:289–302. doi: 10.1016/j.immuni.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 81.Puig O, Tjian R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005;19:2435–2446. doi: 10.1101/gad.1340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sinclair LV, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 84.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buhl AM, Pleiman CM, Rickert RC, Cambier JC. Qualitative regulation of B cell antigen receptor signaling by CD19: selective requirement for PI3-kinase activation, inositol-1,4,5-trisphosphate production and Ca2+ mobilization. J Exp Med. 1997;186:1897–1910. doi: 10.1084/jem.186.11.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ciemerych MA, Sicinski P. Cell cycle in mouse development. Oncogene. 2005;24:2877–2898. doi: 10.1038/sj.onc.1208608. [DOI] [PubMed] [Google Scholar]
- 87.Li G, et al. PTEN deletion leads to up-regulation of a secreted growth factor pleiotrophin. J Biol Chem. 2006;281:10663–10668. doi: 10.1074/jbc.M512509200. [DOI] [PubMed] [Google Scholar]
- 88.Yusuf I, et al. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008;20:671–681. doi: 10.1093/intimm/dxn024. [DOI] [PubMed] [Google Scholar]
- 89.Tang TT, et al. The forkhead transcription factor AFX activates apoptosis by induction of the BCL-6 transcriptional repressor. J Biol Chem. 2002;277:14255–14265. doi: 10.1074/jbc.M110901200. [DOI] [PubMed] [Google Scholar]
- 90.Jonsson H, Allen P, Peng SL. Inflammatory arthritis requires Foxo3a to prevent Fas ligand-induced neutrophil apoptosis. Nat Med. 2005;11:666–671. doi: 10.1038/nm1248. [DOI] [PubMed] [Google Scholar]
- 91.Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J Biol Chem. 2002;277:47928–47937. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- 92.Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295:2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- 93.Tran H, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 94.Yalcin S, et al. Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J Biol Chem. 2008;283:25692–25705. doi: 10.1074/jbc.M800517200. [DOI] [PubMed] [Google Scholar]
- 95.Alvarez B, Martinez-A C, Burgering BM, Carrera AC. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature. 2001;413:744–747. doi: 10.1038/35099574. [DOI] [PubMed] [Google Scholar]
- 96.Katayama K, Nakamura A, Sugimoto Y, Tsuruo T, Fujita N. FOXO transcription factor-dependent p15INK4b and p19INK4d expression. Oncogene. 2008;27:1677–1686. doi: 10.1038/sj.onc.1210813. [DOI] [PubMed] [Google Scholar]
- 97.Kops GJ, et al. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol Cell Biol. 2002;22:2025–2036. doi: 10.1128/MCB.22.7.2025-2036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]