Abstract
Cell growth is regulated by two antagonistic processes: TOR signaling and autophagy. These processes integrate signals including growth factors, amino acids, and energy status to ensure that cell growth is appropriate to environmental conditions. Autophagy responds indirectly to the cellular milieu as a downstream inhibitory target of TOR signaling and is also directly controlled by nutrient availability, cellular energy status, and cell stress. The control of cell growth by TOR signaling and autophagy are relevant to disease, as altered regulation of either pathway results in tumorigenesis. Here we give an overview of how TOR signaling and autophagy integrate nutritional status to regulate cell growth, how these pathways are coordinately regulated, and how dysfunction of this regulation might result in tumorigenesis.
Introduction
In order to reproduce efficiently, eukaryotic cells must grow in a manner appropriate to the extracellular milieu. For single-celled organisms, the decision to grow and divide is simple. In the presence of appropriate nutrients (glucose and amino acids), the cell will increase its size and mass and ultimately divide. Though the decision to grow and divide in multicellular organisms is complicated by the need for responsiveness to cell-to-cell signals and hormonal cues, much of the biological machinery required to respond to those cues remains highly conserved. One such conserved pathway is autophagy, which represents the major mechanism by which a cell catabolizes biological macromolecules (Levine and Klionsky, 2004). This can happen for small amounts of cytosol or specific proteins through direct fusion with the lysosome (i.e. microautophagy), the direct receptor-mediated import of cytosolic proteins containing specific recognition motif sequences into the lysosome (i.e. chaperone-mediated autophagy) or it can occur through the de novo formation of vesicles that engulf entire organelles and long-lived proteins (i.e. macroautophagy). The focus of this review will be the role of macroautophagy in the regulation of cellular growth in metazoans.
Although both increased cell size and cell proliferation contribute to cell growth, the rate of cell division is itself regulated by changes in cell size. Thus, the regulation of cell size is of paramount importance in regulating cellular growth. Increasing the mass of a cell requires the net synthesis of macromolcules including proteins, DNA, and RNA, which ultimately requires energy input. In contrast, the response to starvation or stress usually involves macromolecular remodeling, energy production, and the catabolism of existing macromolecules. The phosphatidylinositol 3-kinase (PI3K)/TOR pathway has emerged as the central conduit for integrating a variety of signaling pathways to promote cell growth and protein synthesis; autophagy is the primary pathway for catabolic activities (He and Klionsky, 2009; Hietakangas and Cohen, 2009). From a teleological perspective, an efficient system would couple the control of autophagy with the control of cell growth, and cues that promote one would inhibit the other, thus preventing inefficient use of energy and nutrients. Indeed, both autophagy and the PI3K signaling pathways have evolved to respond to a variety of cellular growth and nutrient signals including growth factors, the presence of amino acids, and the presence of glucose and energy. In addition, autophagy limits cellular growth and promotes survival during cellular stress. Given the tightly intertwined processes that are governed by TOR signaling and autophagy, previously postulated coordination and crosstalk between these pathways are now being confirmed.
Insulin/Growth factors
Anabolic hormones (e.g. insulin and growth factors) regulate cell growth by positively activating TOR signaling and inhibiting autophagy. Not surprisingly, many key signaling molecules are conserved between both pathways (e.g. AMPK, TSC2, and Vps34). Although TOR exists in two signaling complexes, TOR complex 1 (TORC1) and TOR complex 2 (TORC2), this discussion focuses on the rapamycin-sensitive TORC1 which constitutes the primary complex responsible for assessing the presence of nutrients and growth factor signals to control protein translation and cell growth (Loewith 2002; Hietakangas 2009). Insulin is a well-characterized growth factor that signals through the canonical PI3K-TOR pathway. In the presence of appropriate growth factors, receptor tyrosine kinases phosphorylate targets (e.g. IRS1) to stimulate Class 1 PI3Ks to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3) from phosphatidylinositol 4,5-bisphosphate (PIP2). The accumulation of PIP3 is inhibited by PTEN, phosphatase and tensin homolog deleted on chromosome ten, an important negative regulatory protein in the PI3K-TOR pathway. The availability of PIP3 at the membrane results in the activation of Akt which inhibits the tuberous sclerosis complex (TSC1/TSC2). This heterodimeric complex promotes the GTPase activity of Rheb, a Ras-related GTPase, which causes Rheb-GTP to become Rheb-GDP. Rheb-GTP activates TORC1 kinase activity while Rheb-GDP inhibits the complex. The net effect of growth factor stimulation is the activation of Rheb-GTP and subsequently TORC1. In addition to autophagy inhibition, some important downstream effects of the TORC1 pathway include the activation of ribosomal S6 protein kinase (S6K1), inhibition of elongation factor 4E binding protein (4E-BP1), and subsequent activation of the eukaryotic translation initiation factor 4E (eIF4E). These downstream effectors are critical for ribosome production, protein translation, and, ultimately, cell growth (Lum et al., 2005b; Wang and Proud, 2009).
The central role of the PI3K-TOR pathway in regulating growth control is relevant to human disease as the loss of TORC inhibitors (e.g. PTEN, LKB1, TSC1/2) or constitutive activation of TORC (e.g. hyperactive PI3K or Ras signaling) results in both sporadic cancers and cancer predisposition syndromes (Samuels et al., 2004; Shaw and Cantley, 2006). For example, PTEN is mutated in Cowden’s disease, a disease characterized by mucocutaneous lesions (tricholemmomas, oral papillomas, and acral keratotic papules), and mutations in PTEN also predispose to a variety of other human cancers (Keniry and Parsons, 2008). The tuberous sclerosis complex 1 or 2 are mutated in the eponymous syndrome characterized by cutaneous fibromas and hamartomatous growth of a variety of tissues (Schwartz, 2007).
While growth factors are required for TORC activation, it is growth factor withdrawal that induces autophagy. Early evidence for a role for receptor tyrosine kinase signaling in autophagy inhibition came from genetic studies in C. elegans. The loss of C. elegans daf-2, an ortholog of the insulin-like receptor tyrosine kinase, induces autophagy to mediate constitutive dauer formation, lifespan extension, pathogen resistance and increased degradation of β-amyloid peptide (Florez-McClure et al., 2007; Hansen et al., 2008; Hars et al., 2007; Jia et al., 2009; Melendez et al., 2003). Similarly, in Drosophila, the deletion of its Insulin-Like Peptides (DILPs) induces autophagy and severe growth retardation (Zhang et al., 2009). Besides insulin signaling, the loss of other growth factor signaling also induces autophagy. For example, when bound to its receptor, interleukin-3 (IL-3) induces tyrosine phosphorylation and activates the MAPK cascade within hematopoietic cell lines, and IL-3 withdrawal from immortalized, apoptosis-deficient (Bax−/−Bak−/−) cells results in the induction of autophagy (Lum et al., 2005a). In addition, the loss of EGFR signaling or substrate detachment results in the induction of autophagy in breast epithelial cells (Fung et al., 2008).
The identification of mechanisms by which growth factor withdrawal induces autophagy provided one of the earliest links between autophagy regulation and TORC signaling; evidence from evolutionarily diverse model sytems has consistently demonstrated that the inhibition of autophagy by growth factors occurs through the activation of the Class I PI3K-TORC signaling pathway (Arico et al., 2001; Petiot et al., 2000; Rusten et al., 2004; Scott et al., 2004). Although TORC1 activation is necessary and sufficient for the repression of autophagy in the presence of growth factors, it has not been determined whether the de-repression of basal autophagy is adequate to explain growth-factor-withdrawal-induced autophagy, or whether additional signals (i.e. low energy status or limiting nutrient availability) that may act in parallel of TOR are required to activate autophagy more directly.
Amino acids
Perhaps even more evolutionarily conserved than their response to growth factors, TORC and autophagy are responsive to the presence of environmental nutrients. Even in the presence of adequate growth factor signals, TORC signaling requires the presence of intracellular amino acids, especially branched chain amino acids like leucine, for maximal TOR activation (Christie et al., 2002; Hara et al., 1998). The central role of amino acids in regulating cell growth was highlighted by the finding that L-glutamine positively regulates cell size by promoting the import of leucine and other essential amino acids through the SLC7A5/SLC3A2 bidirectional transporter (Nicklin et al., 2009). When intracellular amino acids are low, the binding of Rheb to mTORC1 is impaired in a manner that is independent of both TSC2 or even GTP binding by Rheb (Long et al., 2005; Nobukuni et al., 2005; Smith et al., 2005).
Although the exact sensor(s) through which amino acids promote TORC1 activity remains ambiguous, recent studies have begun to reveal the mechanism of its amino acid-dependent signaling. The Rag-GTPases, a heterodimer of RagA/B-GTP with RagC/D-GDP, have been identified as direct binding partners of mTORC1 through Raptor and have been implicated in the amino acid sensitivity of TORC1 signaling (Kim et al., 2008; Sancak et al., 2008). This regulation is thought to occur not through the direct activation of mTOR, but rather through the subcellular relocalization of mTORC1 complexes to a vesicular compartment that also contains Rab7 (Sancak et al., 2008). It has been proposed that the Rag heterodimers promote the amino acid-dependent signaling of TORC1 by bringing the complex into the vicinity of farnesylated Rheb. However, the precise endomembrane localizations of both TORC1 and Rheb and the dynamics of their proposed localizations remain a matter of active investigation (Buerger et al., 2006; Drenan et al., 2004; Jiang and Vogt, 2008; Sancak et al., 2008).
An independent line of investigation has also hinted at the importance of lipid signaling and vesicle trafficking in amino acid-dependent TORC1 signaling. Vps34, a class III PI3 kinase which phosphorylates phosphatidylinositol (PI) to form phosphatidylinositol 3-phosphate (PI3P), has been identified as an amino acid-dependent activator of TORC (Byfield et al., 2005; Nobukuni et al., 2005). This role of Vps34 membrane signaling in the regulation of TORC1 activation is especially interesting given recent findings demonstrating a possible role for Beclin 1, an upstream regulator of autophagy and binding partner of Vps34 in diverse functions in vesicle sorting and membrane trafficking (Itakura et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). Curiously, Vps34 is not required for TORC activation in Drosophila, so it is unclear whether this regulation is evolutionarily conserved (Juhasz et al., 2008). A high research priority will therefore be to uncover the exact role of Vps34, Beclin 1, and other components of autophagosome/vesicle trafficking in crosstalk with TORC1 regulation and to further determine whether this is conserved in all metazoans.
While amino acids are required for TORC activation, the absence of amino acids induces autophagy through several mechanisms. Nutrient starvation activates autophagy indirectly through the inhibition of TORC1 signaling which acts to repress a complex containing Atg13, focal adhesion kinase-interacting protein 200 (FIP200), and unc-51 like kinases 1 and 2 (ULK1/2) in nutrient rich conditions; this regulation is likely important for the coordinated regulation of autophagy and TORC1 signaling (Hosokawa et al., 2009; Jung et al., 2009). However, signaling through the the PI3K-TOR pathway is only one of many ways in which autophagy is sensitive to amino acids.
Amino acid starvation has also been implicated in regulation of autophagy through the Raf kinase signaling cascade. In this cascade, in an amino acid-dependent manner, Raf-1 activates MEK1/2 (MAPK/ERK kinase), which activates ERK1/2 (extracellular signal regulated kinase 1 and ERK2 mitogen activated kinase). Erk kinases phosphorylate GAIP, a Gα interacting protein, whose phosphorylation promotes its GAP activity on the α subunit of the Gi3 protein, which ultimately promotes autophagy (Ogier-Denis et al., 2000; Pattingre et al., 2003; Shaw and Cantley, 2006). However, Raf is also downstream of Ras, which inhibits autophagy through the Class I PI3K-TOR signaling pathway (Furuta et al., 2004). It remains to be determined whether the contradictory regulation of autophagy by Ras through its Raf-1 and PI3K-TOR effector arms represents a biological checkpoint control or whether it is an artifact of particular cell lines.
Amino acid deprivation also activates autophagy by signaling through the eIF2α and ER stress/integrated stress response (Ron and Walter, 2007; Talloczy et al., 2002). In a pathway conserved from yeast to mammals, a limiting supply of amino acids results in uncharged tRNAs and the activation of Gcn2 (general control non-derepressible-2) and phosphorylation of eukaryotic translation initiation factor-2 on its α subunit (eIF2α) (Wek et al., 1995; Zhang et al., 2002). Phosphorylated eIF2α in turn inhibits eIF2B, a pentameric guanine nucleotide exchange factor, from recycling eIF2 to its active GTP-bound state. Phosphorylation of eIF2α also results in the upregulation of ATF4 (activating transcription factor 4), a transcription factor which in turn activates a transcriptional program to respond to diverse cellular stresses through the induction autophagy genes, amino acid transporters, anti-oxidant response proteins, and chaperones (Harding et al., 2000; Hinnebusch and Natarajan, 2002; Milani et al., 2009; Natarajan et al., 2001; Vattem and Wek, 2004). Given its broad role in response to cellular stress, it is not surprising that eIF2α is also a target for other stress kinases (e.g. PKR during viral infection, PERK during the unfolded protein response) and thus forms a common pathway for the activation of autophagy in response to amino acid starvation and other cellular insults (Talloczy et al., 2002).
Energy sensing/Glucose
In addition to amino acids, cells must also be supplied with glucose, fatty acids, or pyruvate to maintain a constant supply of ATP (Lum et al., 2005b). Both positive growth signaling and autophagy are closely linked to available energy in the cell. This is largely sensed through cellular levels of ATP. The primary energy-sensing pathway of the cell is the well-characterized AMPK pathway (Hardie, 2007). AMPK is an essential heterotrimeric kinase that is activated during times of energy depletion by increased ratios of AMP to ATP and inhibited by the presence of glycogen. When AMP is bound to the regulatory γ-subunit, LBK1/STK11 can phosphorylate and activate the catalytic α-subunit of AMPK. In contrast, the regulatory β-subunit can bind to glycogen to inhibit AMPK activity. The net effect of AMPK activation is the upregulation of the energy-producing pathways (e.g. Glut4 receptors, insulin sensitivity) and inhibition of energy storage pathways (e.g. glycogen synthesis, lipid synthesis). AMPK inhibits the TORC1 complex by phosphorylating TSC2, which then activates the GAP activity of TSC1/2 on Rheb-GTP favoring the formation of the Rheb-GDP, thereby inactivating TORC1 (Inoki et al., 2003). In a parallel inhibitory pathway, AMPK also phosphorylates and inhibits the TORC1-defining component, Raptor, leading to an interaction between Raptor and 14-3-3 that disrupts its binding to TOR (Gwinn et al., 2008). Mutations in LKB1/STK11 cause Peutz-Jeghers, which is characterized by pigmentary abnormalities and a predisposition to malignancy (Hardie, 2007; Inoki et al., 2003).
In a manner similar to growth factor withdrawal, the inhibition of TORC1 signaling by active AMPK can indirectly induce autophagy through a de-repression of the upstream regulators of the catabolic process. Evidence from yeast suggests that AMPK also has a more direct role in the induction of autophagy. Snf1, the yeast homolog of AMPK, promotes autophagy by acting on Atg1 and Atg13, yeast homologs of ULK and Atg13, respectively (Wang et al., 2001). However, a direct role for AMPK in autophagy activation in mammalian cells has not been demonstrated. In fact, early work in mammalian cells suggested that AMPK inhibits autophagy (Samari and Seglen, 1998). However, more recent work suggests that, similar to yeast, AMPK induces autophagy (Matsui et al., 2007; Meley et al.).
Cell stress
In addition to being responsive to cellular growth cues, autophagy is also critical for limiting cell growth and promoting cell survival in times of stress. Much of the cellular stress response has converged on signaling through the ER through the unfolded protein response. Mammalian cells possess three ER transmembrane receptors (IRE1α, ATF6, and PERK) that are responsible for transducing stress responses (Bernales et al., 2006; Ron and Walter, 2007). IRE1α and PERK are best characterized with respect to their ability to regulate autophagy.
IRE1α, inositol-requiring protein-1α, represents the most conserved core of the unfolded protein response. Upon activation by ER stress, IRE1α autophosphorylation induces a conformational change allowing it to bind to the adaptor protein tumour necrosis factor-α (TNF-α)-receptor-associated factor 2 (TRAF2) through its cytoplasmic domain (Urano et al., 2000). The recruitment of TRAF2 has been shown to be important in stimulating autophagy in response to the UPR via its activation of c-Jun NH(2)-terminal kinase (JNK) (Ogata et al., 2006). In turn, JNK-mediated phosphorylation of Bcl-2 and disruption of the Bcl-2/Beclin 1 complex, and JNK-mediated upregulation of Beclin 1 transcription likely contribute to the induction of autophagy (Li et al., 2009; Pattingre et al., 2009; Wei et al., 2008).
In a parallel pathway to JNK activation, IRE1α–TRAF2 complexes also recruit IκB kinase (IKK) (Hu et al., 2006). Active IKK induces autophagy in a nuclear factor-κB (NF-κB) transcription-independent manner (Criollo et al., 2009). Strikingly, the absence of IKK results in impaired induction of autophagy suggesting that IKK-mediated induction of autophagy is not redundant with other ER stress signaling pathways. Although IKK activation appears to inhibit TOR signaling and induce AMPK and JNK, it is still unclear precisely how IKK activation promotes autophagy.
In contrast to IRE1α, the induction of autophagy by PERK appears to be transcription-dependent. In response to unfolded proteins, PERK can phosphorylate eIF2α which then activates the transcriptional upregulation of autophagy as noted previously (Kouroku et al., 2007). As already noted, other kinases capable of sensing cell stress, including GCN2 and interferon-induced double-stranded RNA-dependent protein kinase R (PKR), can also activate eIF2α in response to conditions like amino acid deprivation and viral infection (Kouroku et al., 2007; Talloczy et al., 2002). Finally, as the major site of intracellular calcium storage, ER stress also results in the release of intracellular calcium stores. This release may result in the activation of autophagy through the CaMKKβ-dependent activation of AMPK (Hoyer-Hansen et al., 2007).
Because the ER has the ability to integrate diverse cell stress signals including starvation, hypoxia, misfolded proteins, and infection, it is teleologically well-placed as a sensor to limit cell growth during times of stress. A prominent role for ER stress in limiting cell growth is consistent with a current model in which the ER is thought to be the source of membranes for autophagosomal structures (Axe et al., 2008). Although it is known that Bcl-2 inhibits autophagy through a direct interaction with the BH3 domain of Beclin 1, the precise mechanistic details of this inhibition are unclear (Pattingre et al., 2005; Sinha et al., 2008). Given that ER-localized Bcl-2 is the form that inhibits Beclin 1 autophagy function, and the recent finding that preautophagosomal structures originate from the ER membrane, it is possible that Bcl-2 sequesters Beclin 1 in ER membrane subdomains away from preautophagosomal structures in the ER (Axe et al., 2008). According to this model, in response to a variety of cell stressors (e.g. nutrient starvation, unfolded proteins, viral proteins, etc.), the phosphorylation of Bcl-2 and its subsequent dissociation from the Beclin 1/Vps34 complex may contribute to the initiation of autophagosomal membrane formation.
Crosstalk between autophagy and TOR signaling
Direct interactions between the TORC1 and autophagy pathways help to coordinate the respective anabolic and catabolic processes. As noted above, part of the coordinate regulation occurs through TORC1-dependent phosphorylation and inactivation of a complex containing Atg13, FIP200, and ULK1/2 (Hosokawa et al., 2009; Jung et al., 2009). When TOR signaling is suppressed (e.g. growth factor withdrawal, starvation, or pharmalogical treatment), ULK becomes activated and phosphorylates and activates Atg13 and FIP200 to induce autophagy. As the ULK-Atg13-FIP200 complex functions upstream in autophagy, it is likely that the direct inhibitory phosphorylation of this complex by TORC1 represents an important signaling step. The precise mechanism through which the ULK-Atg13-FIP200 complex induces autophagy in metazoans remains to be determined.
Although the regulation of autophagy is clearly downstream of TOR signaling, there is increasing evidence that autophagy may also regulate TOR signaling. Positive regulators of autophagy inhibit activation of the TOR pathway (e.g. ULK1, Atg13) as assessed by the phosphorylation of downstream targets of TOR like S6K1 (Jung et al., 2009; Lee et al., 2007). The finding that Vps34 may be important for the amino acid sensitive signaling through TORC suggests additional coordination in the regulation of autophagy and cellular growth (Byfield et al., 2005; Nobukuni et al., 2005). At first glance, it would appear that the cell growth-promoting properties of Vps34 would seem to conflict with its role in initiating autophagic vesicle nucleation to regulate autophagy. However, this apparent discrepancy may represent the function of distinct complexes of Beclin 1/Vps34 with different functions. For example, Vps34 forms a complex with Beclin 1 and Atg14 (also known as Atg14L, Barkor) to positively regulate autophagosome formation and maturation (Itakura et al., 2008; Matsunaga et al., 2009). Beclin 1 also forms Atg14-independent distinct complexes with UVRAG and Rubicon with possible roles in the regulation of autophagosome/endosome maturation; however, the precise functions of these complexes is unclear (Liang et al., 2006; Liang et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009).
Another interesting interface between PI3K-TOR signaling and autophagy resides in the regulation of PI and its derivatives by different classes of PI3Ks. Class I and Class III PI3Ks share structural similarities and both exist as a heterodimers of a catalytic (p110 and Vps34) and a regulatory (p85 and Vps15) subunit, respectively. Both PI3Ks are crucial upstream regulatory kinases in their respective signaling cascades. In PI3K-TOR signaling, the conversion of PIP2 to PIP3 is essential for the downstream activation of the PIP3-dependent kinases, PDK and Akt. Similarly, in autophagy, the conversion of PI to PI3P by the Beclin 1/Vps34 complex appears to be one of the earlier, if not the earliest, signaling event in the budding of the omegasome from the endoplasmic reticulum and the eventual formation of the autophagosome (Axe et al., 2008). In another interesting parallel, both signaling pathways possess lipid phosphatases (PTEN and Jumpy), which inhibit the activation of their respective pathways (Vergne et al., 2009). Although the role of PIs in PI3K-TOR signaling and in autophagy has been studied, little is known about how the two pathways may interface at the level of lipid signaling. For example, it will be a critical to determine whether the activity of Beclin 1/Vps34 at autophagosomes in generating PI3P ultimately affects the generation of PIP3 by Class I PI3Ks. Given the known crosstalk between Vps34 and TORC signaling, the possible role of PI derivatives in this crosstalk demands further attention.
Distinct from its activity in generating PI3P, Beclin 1/Vps34 may function in other roles to affect TORC signaling. For example, given the recent the recent finding that TORC1 relocalizes to Rab7-containing vesicles upon activation, it is possible that autophagy and cell growth could be coordinated through the action of different Beclin 1/Vps34 complexes on different subsets of the endomembrane system. In amino acid and growth factor replete conditions, the Beclin 1/Vps34 complexes would hypothetically act on endosomes/lysozomes to regulate TOR signaling complexes by ensuring the appropriate maturation and recycling of TORC1-, Rheb-GTP-, Rag-GTPase-containing vesicles. In contrast, upon depletion of amino acids, the Beclin 1/Vps34 complexes might localize to specific sites in the endoplasmic reticulum (ER) membranes where they would promote the generation of PI3P to initiate the formation of the autophagic isolation membrane. Clearly, more experiments are necessary to understand the role of the Beclin 1/Vps34 complex in regulation of endomembrane trafficking and how it may affect TORC signaling.
Emerging functions of autophagy in cell cycle control
As cell division contributes to cell growth, cell cycle checkpoints are a critical regulatory point in the growth control of eukaryotes. The decision to enter S phase from G1 is promoted by cyclin-dependent kinase (CDK)/cyclin complexes (e.g. cyclin D/Cdk4 and cyclin E/Cdk2) which are inhibited by CDK inhibitors (CDKIs) including p16 and p27. The levels of both CDK/cyclins and CDKIs are regulated through ubiquitination (Sherr and Roberts, 1999). TORC1 signaling has been shown to contribute to cell cycle regulation through affecting the levels of both cyclins and CDKIs (Wang and Proud, 2009).
Less is known about the relationship between the cell cycle and autophagy. Experiments using pharmacologically synchronized cells have demonstrated that autophagy is most active in the G1 and S phases of the cell cycle, while it is inhibited in mitosis (Eskelinen et al., 2002; Tasdemir et al., 2007). Specific regulators of the cell cycle have also been shown to affect autophagy. For example, p14ARF, an alternative reading frame product of the p16 locus and inhibitor of G1 cell cycle progression, binds to Bcl-XL, a Bcl-2 anti-apoptotic family member; this binding disrupts Beclin 1/ Bcl-XL binding and induces autophagy (Pimkina et al., 2009). Also, the overexpression of the CDKI, p27, is sufficient to induce autophagy (Liang et al., 2007). Interestingly, both low energy status (via AMPK) and amino acid starvation have both been shown to stabilize p27 (Leung-Pineda et al., 2004; Liang et al., 2007) suggesting a possible role for p27 in the physiological regulation of autophagy. However, our understanding of the impact of the cell cycle on autophagy is in its infancy, and the mechanism through which cell cycle regulators, like p14ARF and p27, inhibit autophagy remain an important area of continued research.
In addition to ties to the canonical cell cycle, there is also evidence for a direct role for autophagy in executing cellular senescence. Cellular senescence is a form of irreversible cell cycle arrest; it can be induced by exogenous DNA damage, telomere depletion, or oncogene (e.g. Ras or MEK) overexpression. This pathway is proposed to contribute to tumor suppression by inhibiting the proliferation of otherwise damaged cells. Oncogene expression induces the transcriptional upregulation of many mediators of autophagy, and the inhibition of autophagy delays the onset of senescence (Young et al., 2009).
Although there is no direct evidence that the genetic disruption or pharmacologic inhibition of autophagy has strong effects on cell cycle regulation, experimental mouse models indicate that deficiency of some autophagy proteins such as Beclin 1 and Ambra1 results in increased cell proliferation (Fimia et al., 2007; Qu et al., 2003). Given the emerging important role for autophagy in the degradation of ubiquitinated targets through specific receptor molecules (e.g. p62/SQSTM1), it is possible that autophagy plays a role in the degradation of cell signaling molecules subject to ubiquitination (Kirkin et al., 2009). The loss of autophagy might result in the abnormal persistence of these cell cycle regulators (e.g. CDKIs) and result in aberrant cell cycle entry. While there is growing evidence for a role of autophagy in the control of cell proliferation, more work is required to define the mechanism(s) by which this occurs.
Autophagy and tumor suppression
One clinically important sequelae of dysfunction of cellular growth control is tumorigenesis. Although the role of hyperactivation of the PI3K/TOR pathway in promoting tumorigenesis is well-established, the contribution of autophagy to tumor suppression is only now being established (Shaw and Cantley, 2006). Activators of TOR signaling (e.g. Class I PI3K, Akt, and Ras) function as oncogenes while inhibitors (e.g. TSC1/2, PTEN, LKB1/AMPK) function as tumor suppressors. In contrast, activators of autophagy (e.g. LKB1/AMPK, p27, DAPk, PTEN, TSC1/2) function as tumor suppressors while inhibitors of autophagy (e.g. Bcl-2, Akt, activated Class I PI3K) function as oncogenes. Of note, the relationship of autophagy to the established tumor suppressor p53 is less clear. Stress-induced (e.g. DNA damage) activation of p53 induces autophagy, while basal levels of p53 appear to inhibit autophagy (Feng et al., 2005; Levine and Abrams, 2008; Tasdemir et al., 2008). This apparent discrepancy may relate to the diverse, context-specific functions of p53 in multiple signaling pathways that influence autophagy or to differing nuclear and cytoplasmic functions of p53 (Levine and Abrams, 2008). Interestingly, about 1/3 of colon cancer-associated mutated forms of p53 accumulate in the cytoplasm, and inhibit autophagy, suggesting a mechanism by which mutation of p53 in human cancer may impair autophagy (Morselli et al., 2008). Thus, despite some conflicting data regarding p53-dependent control of autophagy, taken together, the preponderance of evidence suggests that autophagy may be a critical downstream effector of multiple signaling pathways relevant to tumorigenesis.
Several studies in mammalian systems have confirmed the importance of autophagy execution genes in tumor suppression. Beclin 1 and UVRAG (a potential activator of Beclin 1-dependent autophagy) inhibit tumor cell growth in vitro and tumor xenograft formation in vivo (Liang et al., 1999 Nature; Koneri K et al. 2007; Liang et al. 2006). Mouse models demonstrate that the monoallelic loss of beclin 1 or Ambra1 or bi-allelic loss of Bif-1 results in increased spontaneous tumorigenesis (Qu et al., 2003; Takahashi et al., 2007) (personal communication, F. Cecconi) and Atg4c−/− null mice demonstrate increased chemically-induced fibrosarcomas. (Marino et al., 2007). Furthermore, monoallelic deletions of UVRAG are common in human colon cancer (Ionov et al., 2004) and monoallelic deletions of beclin 1 are common in human breast, ovarian, and prostate cancer (Aita et al., 1999; Ionov et al., 2004). In addition, decreased Beclin 1 expression has been linked to advanced tumor grade and/or poor survival prognosis in several types of human cancer, including gastric carcinoma, hepatocellular carcinoma, colon cancer, ovarian cancer, and brain tumors (Ahn et al., 2007; Miracco et al., 2007; Shen et al., 2008; Won et al., 2010). Thus, there is emerging data suggesting that loss of autophagy execution gene function may contribute to tumorigenesis.
While the tumor suppressor function of autophagy is undisputed, the mechanistic details of how autophagy functions in tumor suppression are still unclear. The role of autophagy in tumor suppression can be explained, in part, by its ability to prevent chromosome instability. The loss of autophagy genes (e.g. monoallelic loss of beclin 1, or biallelic loss of Atg5) promotes DNA damage, gene amplification, and aneuploidy in cell culture and tumor xenograft models (Mathew et al. 2007; Karantza-Wadsoworth etal., 2007). It is postulated that autophagy protects cells from genomic instability by promoting the degradation of p62 and damaged organelles (e.g. mitochondria), and by decreasing the accumulation of damaging ROS (Karantza-Wadsworth et al., 2007; Mathew et al., 2009; Mathew et al., 2007). It remains to be determined whether autophagy also regulates cellular proteins that directly function in DNA damage repair. Regardless of the mechanism by which autophagy prevents genomic instability, this function of autophagy likely represents a critical part of its ability to prevent tumorigenesis.
However, in addition to preventing genomic instability, there is evidence that autophagy may function in additional parallel pathways to prevent tumorigenesis. Specifically, increased proliferation of mammary epithelial cells and splenic lymphocytes has been noted in beclin 1 heterozygous-deficient mice and increased neural cell proliferation has been noted in Ambra1-deficient embryonic mice (Fimia et al., 2007; Qu et al., 2003). The mechanism by which autophagy inhibits cell proliferation is not clear. However, several known functions of autophagy could theoretically contribute. First, autophagy has been proposed to play a role in executing cellular senescence (Young et al., 2009), so the loss of autophagy might impair the induction of this tumor suppressor mechanism in response to inappropriate mitogenic signals or DNA damage. Second, a postulated role for autophagy in limiting cell cycle progression could also be a tumor suppressor mechanism. Third, as noted above, autophagy or specific autophagic proteins may function in limiting TOR signaling, so the partial loss of autophagy could result in inappropriate signaling through the TOR pathway, which is known to occur in tumorigenesis. Finally, the loss of autophagy may also promote tumorigenesis by providing selective pressures for cells that inappropriately amplify cellular growth signals. In a setting of nutrient deprivation, individual cells of a multicellular organism usually survive through the preferred method of autophagy and the catabolism of endogenous macromolecules for the generation of energy and nutrients. Alternatively, individual cells could mutate and survive by developing a competitive advantage in nutrient uptake to the detriment of its adjacent sibling cells. Mutations that improve nutrient uptake have been shown to be tumorigenic (e.g. BRAF or KRAS mutations) (Yun et al., 2009). In this speculative model, autophagy-deficient cells would be under constant selective pressure for oncogenic mutations to overcome their nutrient limitation.
Conclusion
Given its central role in integrating environmental signals like the presence of sufficient nutrients, energy, and growth factors, the PI3K/TOR signaling pathway is often viewed as the principal mediator of cellular growth control. More recently, autophagy has emerged as a crucial player in the negative regulation of cellular growth. Although autophagy is regulated as a downstream target of TORC signaling, autophagy can also regulate cell growth in response to distinct stimuli like amino acid depletion, energy deprivation, and ER stress, independently of PI3K/TOR signaling. Thus, TORC signaling and autophagy actually represent parallel, but contrasting pathways that function together in a coordinated manner to maintain homeostasis and growth control. The loss of such control by either dysfunction of TOR signaling and/or autophagy likely underlies the pathogenesis of most human cancers. However, the precise mechanisms by which autophagy acts in cell growth control and tumor suppression require further discovery and “digestion.”
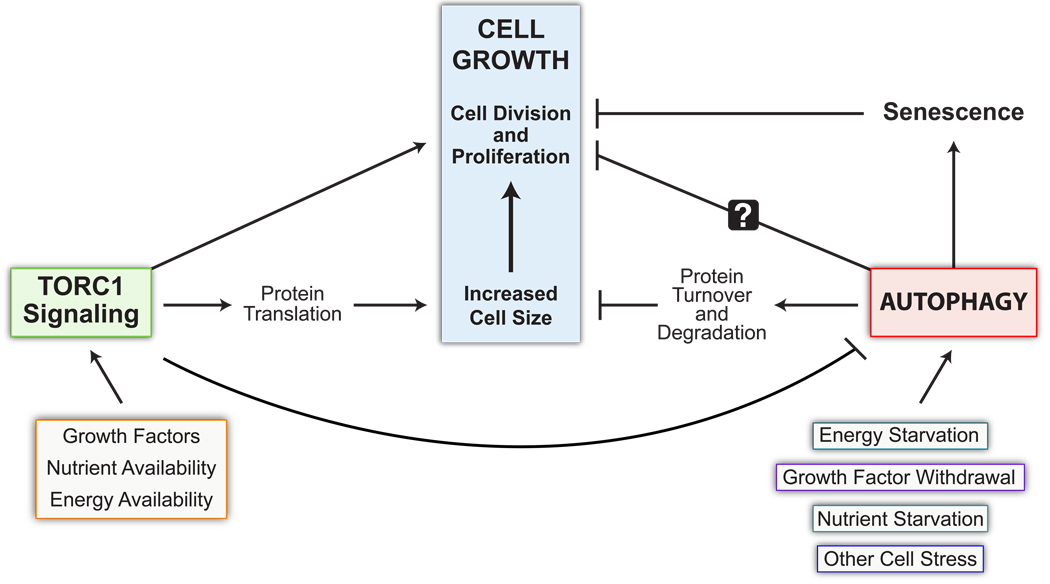
FIGURE 1. Interrelations between TOR complex signaling, autophagy, and cell growth control.
TORC1 signaling promotes cell growth through its effects both on increasing cell proliferation and increasing cell size and potentially through its inhibitory effects on autophagy. TORC1 increases protein synthesis and cell size through intermediates such as 4E-BP and S6K; it promotes cell proliferation at least in part through its effects on cyclin and cyclin-dependent kinase (CKD) inhibitors. In contrast, autophagy is believed to inhibit cell growth at least in part by promoting protein and/or organelle turnover. In addition, autophagy may have indirect effects on inhibiting cell proliferation through promoting senescence. Additional, not yet defined, mechanisms may also contribute to the inhibitory effects of autophagy on cell proliferation. Note that the presence of growth factors, nutrients, and sufficient energy are all required for the full activation of TORC1 signaling, whereas the absence of any of these factors or other types of cellular stressors are sufficient to induce autophagy.
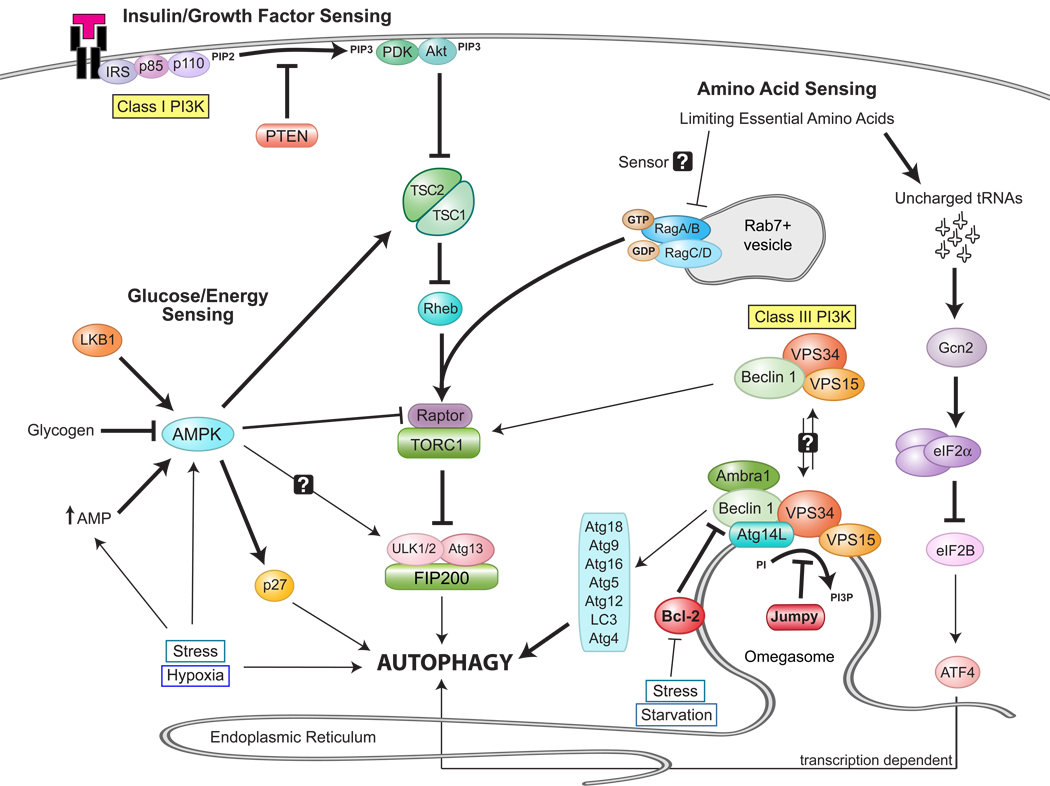
FIGURE 2. Coordinated regulation of TORC1 signaling and autophagy.
Schematic diagram depicting the different inputs that contribute to the regulation of TORC signaling and autophagy and, ultimately, cell growth. Bold lines represent an activation or inhibition for which there is molecular evidence for a direct interaction. Thin lines represent activation steps that are likely indirect. Some activation steps which have not yet been demonstrated in mammlian systems cells are marked with a question mark.
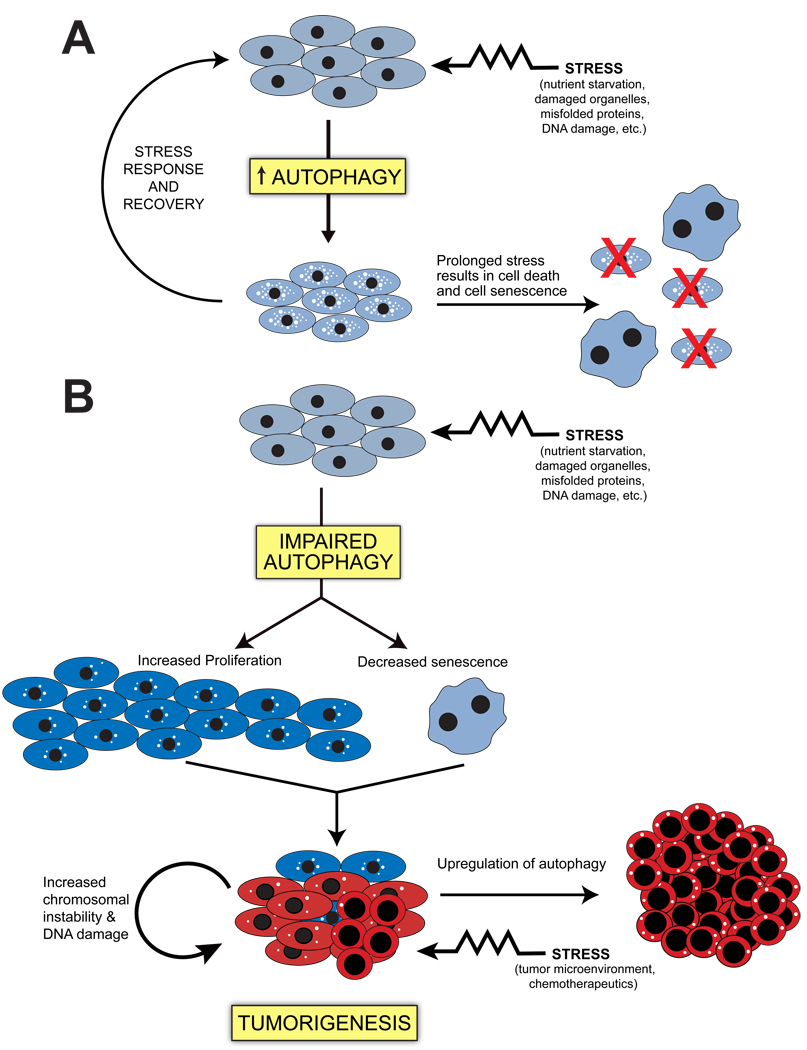
FIGURE 3. Possible mechanisms by which impaired autophagy promotes tumorigenesis.
(A) In normal tissues, autophagy functions to promote cell survival in response to a variety of cellular insults including viral infection, DNA damage, nutrient starvation, or misfolded proteins. If the stressor is irreparable, the cell has a number of mechanisms to prevent the proliferation of damaged cells including apoptotic cell death, non-apoptotic cell death, and senescence. (B) In autophagy-deficient tissues, a cell is unable to respond to stressors, and several mechanisms have been proposed to contribute to tumorigenesis. (1) Autophagy has a role in the induction of senescence (Young et al., 2009) and non-apoptotic cell death (Yu et al., 2004), so its loss may result in a decreased ability to execute these potential tumor suppressor mechanisms; (2) Decreased autophagy has also been shown to increase cell proliferation through as-of-yet unknown mechanisms (Fimia et al., 2007; Qu et al., 2003); (3) Decreased autophagy results in increased chromosomal instability and mutagenesis possibly through the accumulation of damaged organelles and increased ROS (Karantza-Wadsworth et al., 2007; Mathew et al., 2009; Mathew et al., 2007). In contrast, the upregulation of autophagy in established tumors may promote the survival of tumor cells in response to metabolic stress in the tumor microenvironment. In this figure, normal cells are depicted as light blue; cells with decreased autophagy that are hyperproliferative are depicted as dark blue; and transformed cells are depicted as red. In general, there is an inverse correlation between levels of autophagy and cell size (Hosokawa et al., 2006; Lum et al., 2005a) which is depicted in this figure; however, multiple factors other than levels of autophagy contribute to the increased nuclear/cytoplasmic ratio of tumor cells. Senescent cells display a large and flattened morphology.
Table 1.
Summary of Roles of Autophagy Genes in Cellular Growth Control
| Organism | Relevant Gene(s) Mutation* |
Phenotype | Reference |
|---|---|---|---|
| C. elegans |
unc-51 (Atg1/ULK); bec-1 (Atg6/beclin 1) |
Decreased worm length** |
(Aladzsity et al., 2007) |
| daf-2 (Igf-1) | Increased cell size rescued by unc-51 or bec-1 mutants |
(Aladzsity et al., 2007); (McCulloch and Gems, 2003) |
|
| Drosophila |
Atg1 (ULK) overexpression |
Decreased cell size | (Scott et al., 2007) |
| Atg1−/− | Normal cell size in well- fed animals; increased size in rapamycin-treated animals |
(Scott et al., 2007) | |
| Atg1−/−, dTOR |
Atg1−/− partially rescues the decreased cell size of dTOR mutants |
(Lee et al., 2007) | |
| Mammalian | Atg5−/− (mouse cell lines) | Increased cell size of starved fibroblasts; no change in cell cycle profile; no impairment of entry into quiescence |
(Hosokawa et al., 2006); (Valentin and Yang, 2008) |
| beclin 1+/− (mice) | Increased proliferation in mammary epithelial cells and splenic germinal center lymphocytes |
(Qu et al., 2003) | |
| Ambra1−/− (mice) | Increased cell proliferation in fetal brain |
(Fimia et al., 2007) | |
|
Atg7 shRNA, Atg5 shRNA (human cell lines) |
Delayed onset of senescence |
(Young et al., 2009) |
Gene names in other organisms listed in parentheses
Decreased cell size in autophagy-deficient C. elegans is proposed to occur through impaired ability to utilize cytosolic materials for cell remodeling and elongated cell shape.
Acknowledgements
The work in the authors’ own laboratory was supported by NIH grants RO1 CA84254 and ROI CA109618 to B.L. The authors thank Angela Diehl for expert medical illustration. We apologize to those authors whose work could not be cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, Yoo NJ, Lee SH. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115:1344–1349. doi: 10.1111/j.1600-0463.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- Aladzsity I, Toth ML, Sigmond T, Szabo E, Bicsak B, Barna J, Regos A, Orosz L, Kovacs AL, Vellai T. Autophagy genes unc-51 and bec-1 are required for normal cell size in Caenorhabditis elegans. Genetics. 2007;177:655–660. doi: 10.1534/genetics.107.075762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- Christie GR, Hajduch E, Hundal HS, Proud CG, Taylor PM. Intracellular sensing of amino acids in Xenopus laevis oocytes stimulates p70 S6 kinase in a target of rapamycin-dependent manner. J Biol Chem. 2002;277:9952–9957. doi: 10.1074/jbc.M107694200. [DOI] [PubMed] [Google Scholar]
- Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2009 doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenan RM, Liu X, Bertram PG, Zheng XF. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J Biol Chem. 2004;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL, Prescott AR, Cooper J, Brachmann SM, Wang L, Tang X, Backer JM, Lucocq JM. Inhibition of autophagy in mitotic animal cells. Traffic. 2002;3:878–893. doi: 10.1034/j.1600-0854.2002.31204.x. [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans. Autophagy. 2007;3:569–580. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23:3898–3904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu Rev Genet. 2009;43:389–410. doi: 10.1146/annurev-genet-102108-134815. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell. 2002;1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hu P, Han Z, Couvillon AD, Kaufman RJ, Exton JH. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol Cell Biol. 2006;26:3071–3084. doi: 10.1128/MCB.26.8.3071-3084.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23:639–645. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106:14564–14569. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Vogt PK. Constitutively active Rheb induces oncogenic transformation. Oncogene. 2008;27:5729–5740. doi: 10.1038/onc.2008.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Koneri K, Goi T, Hirono Y, Katayama K, Yamaguchi A. Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res. 2007;27:1453–1457. [PubMed] [Google Scholar]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kim S, Lee J, Park J, Lee G, Kim Y, Kim JM, Chung J. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8:360–365. doi: 10.1038/sj.embor.7400917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Pineda V, Pan Y, Chen H, Kilberg MS. Induction of p21 and p27 expression by amino acid deprivation of HepG2 human hepatoma cells involves mRNA stabilization. Biochem J. 2004;379:79–88. doi: 10.1042/BJ20031383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10:637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Li DD, Wang LL, Deng R, Tang J, Shen Y, Guo JF, Wang Y, Xia LP, Feng GK, Liu QQ, et al. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene. 2009;28:886–898. doi: 10.1038/onc.2008.441. [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005a;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005b;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- McCulloch D, Gems D. Body size, insulin/IGF signaling and aging in the nematode Caenorhabditis elegans. Exp Gerontol. 2003;38:129–136. doi: 10.1016/s0531-5565(02)00147-x. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, Harris AL. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, Mannucci S, De Nisi MC, Toscano M, Malagnino V, et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30:429–436. [PubMed] [Google Scholar]
- Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, Vicencio JM, Soussi T, Kroemer G. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier-Denis E, Pattingre S, El Benna J, Codogno P. Erk1/2-dependent phosphorylation of Galpha-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J Biol Chem. 2000;275:39090–39095. doi: 10.1074/jbc.M006198200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2009;284:2719–2728. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- Pimkina J, Humbey O, Zilfou JT, Jarnik M, Murphy ME. ARF induces autophagy by virtue of interaction with Bcl-xl. J Biol Chem. 2009;284:2803–2810. doi: 10.1074/jbc.M804705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Samari HR, Seglen PO. Inhibition of hepatocytic autophagy by adenosine, aminoimidazole-4-carboxamide riboside, and N6-mercaptopurine riboside. Evidence for involvement of amp-activated protein kinase. J Biol Chem. 1998;273:23758–23763. doi: 10.1074/jbc.273.37.23758. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Shen Y, Li DD, Wang LL, Deng R, Zhu XF. Decreased expression of autophagy-related proteins in malignant epithelial ovarian cancer. Autophagy. 2008;4:1067–1068. doi: 10.4161/auto.6827. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Tajeddine N, Vitale I, Criollo A, Vicencio JM, Hickman JA, Geneste O, Kroemer G. Cell cycle-dependent induction of autophagy, mitophagy and reticulophagy. Cell Cycle. 2007;6:2263–2267. doi: 10.4161/cc.6.18.4681. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Valentin M, Yang E. Autophagy is activated, but is not required for the G0 function of BCL-2 or BCL-xL. Cell Cycle. 2008;7:2762–2768. doi: 10.4161/cc.7.17.6595. [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase jumpy. EMBO J. 2009;28:3780. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Proud CG. Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol. 2009;19:260–267. doi: 10.1016/j.tcb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001;21:5742–5752. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008a;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won KY, Kim GY, Kim YW, Song JY, Lim SJ. Clinicopathologic correlation of beclin-1 and bcl-2 expression in human breast cancer. Hum Pathol. 2010;41:107–112. doi: 10.1016/j.humpath.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu J, Li CR, Momen B, Kohanski RA, Pick L. Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proc Natl Acad Sci U S A. 2009;106:19617–19622. doi: 10.1073/pnas.0905083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–6688. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]