Abstract
Studies were conducted to examine the population genetic structure of Anopheles arabiensis (Patton) in Mwea Rice Irrigation Scheme and surrounding areas in Central Kenya, under different agricultural systems. This study was motivated by observed differences in malaria transmission indices of An. arabiensis within the scheme compared with adjacent nonirrigated areas. Agricultural practices can modify local microclimate and influence the number and diversity of larval habitats and in so doing may occasion subpopulation differentiation. Thirty samples from each of the three study sites were genotyped at eight microsatellite loci. Seven microsatellite loci showed high polymorphism but revealed no genetic differentiation (FST = 0.006, P = 0.312) and high gene flow (Nm = 29–101) among the three populations. Genetic bottleneck analysis showed no indication of excess heterozygosity in any of the populations. There was high frequency of rare alleles, suggesting that An. arabiensis in the study area has a high potential of responding to selective pressures from environmental changes and vector control efforts. These findings imply that An. arabiensis in the study area occurs as a single, continuous panmictic population with great ability to adapt to human-imposed selective pressures.
Keywords: Microsatellites, An. arabiensis, gene flow, agricultural practices, Kenya
Malaria in sub-Saharan Africa is transmitted mainly by members of the Anopheles gambiae (Giles) and An. funestus Giles complexes. Within the An. gambiae complex, An. gambiae and An. arabiensis (Patton) are the most widespread and important vectors of the disease. Both species coexist throughout the continent, but An. arabiensis is more adapted to arid environments. In rice growing areas of East Africa, An. arabiensis is the only sibling species of the An. gambiae complex present (Ijumba et al. 2002, Mutero et al. 2004a). Notable exception to this occurs in Ahero Rice Scheme in western Kenya where both An. gambiae and An. arabiensis coexist (Githeko et al. 1996). An. arabiensis is a versatile species with ability to feed on multiple host species both indoors and outdoors and to adapt to a wide range of larval habitats (Muriu et al. 2008, Mwangangi et al. 2008).
Currently, indoor residual spraying (IRS) and insecticide treated bednets (ITNs) are the most widely used vector control measures. Although these methods have proved useful in reducing malaria burden (Lengeler 2004, Fegan et al. 2007), their effectiveness has been threatened by increasing prevalence of insecticide resistance by the major malaria vectors as well as the phenotypic flexibility of An. arabiensis. The need for effective, sustainable alternatives to current vector control strategies is therefore urgent and obvious. One such strategy that has dominated current literature is the prospect of manipulating the vectors by introduction of genes conferring refractoriness to the parasite (Collins 1994, Collins and Besansky 1994). This approach requires adequate knowledge of genetic structure of the target mosquito populations and the extent of gene flow among populations. Gene flow indicates the direction and rates of dispersal among populations and may help in development and evaluation of mosquito control strategies, as well as the management of insecticide resistance (Slatkin 1987, Boete and Koella 2003). It could also explain the role of vectors in spatial and temporal heterogeneity in malaria transmission (Donnelly and Townson 2000).
Microsatellites, short tracts of tandemly repeated DNA sequences of up to 6 bp (Schlotterer 2000) are widely used genetic markers because they are codominant, widely distributed in the genome, highly polymorphic, and easy to score. At least 150 polymorphic microsatellite loci have been characterized in An. gambiae s.l. (Zheng et al. 1993, Zheng et al. 1996) and have been used to elucidate the population structure and gene flow within and between members of the An. gambiae complex (Kamau et al. 1999, Kamau et al. 2007, Moreno et al. 2007). Most of these studies have mainly been conducted on An. gambiae and less frequently on An. arabiensis. Existing literature on An. arabiensis has revealed the absence of subpopulation differentiation in relation to larval habitat exploitation (Kamau et al. 2007) as well as the lack of annual bottlenecks in response to changes in weather conditions (Simard et al. 2000, Kent et al. 2007). However, there is evidence in support of (Simard et al. 1999, Donnelly and Townson 2000) and against (Kamau et al. 1999, Kent et al. 2007) restricted gene flow among widely separated populations. Where genetic differentiation has been reported, geographic distance and habitat change have been suggested as the main contributors of this differentiation. Conversely, large effective population size and/or recent range expansion as opposed to mass migration (Donnelly and Townson 2000, Simard et al. 2000) has been attributed to extensive gene flow observed in other studies given the short flight range reported for this species (Thomson et al. 1995). In general, past studies have indicated that An. arabiensis has variable deme sizes ranging from as little as 25 km (Donnelly and Townson 2000) to a few thousand kilometers (Kent et al. 2007).
In Mwea rice scheme of Central Kenya, An. arabiensis mosquito densities decrease as you move away from the scheme (Mutero et al. 2004b, Muturi et al. 2006). Conversely, the human blood index (Muturi et al. 2008) and malaria transmission (Mutero et al. 2004b) by this species is significantly lower within than outside the rice scheme. Agricultural practices can modify local microclimate (Karaca et al. 2008) as well as influence the number and diversity of larval habitats, all of which affect mosquito reproductive fitness, survivorship, and fecundity (Afrane et al. 2006). Such changes may alter malaria transmission indices (Ijumba and Lindsay 2001) and can lead to subpopulation differentiation (Toure et al. 1994, 1998). However, the lack of obvious geographical barriers that might restrict gene flow between mosquitoes in the surrounding areas, favors existence of a single panmictic population. Some studies have suggested that the presence of mosquitoes in neighboring non-irrigated areas during the dry season is maintained through migration of a few individuals from irrigated areas (Dolo et al. 2004).
The current study investigated the population structure of An. arabiensis from three sites with different agricultural practices in Central Kenya. We believe that inundated rice fields are important sources of vector mosquitoes to the neighboring non-irrigated areas. Therefore, we tested the hypothesis that An. arabiensis in the study area is a single panmictic population that is adapted to a wide range of environmental gradients.
Materials and Methods
Study Area
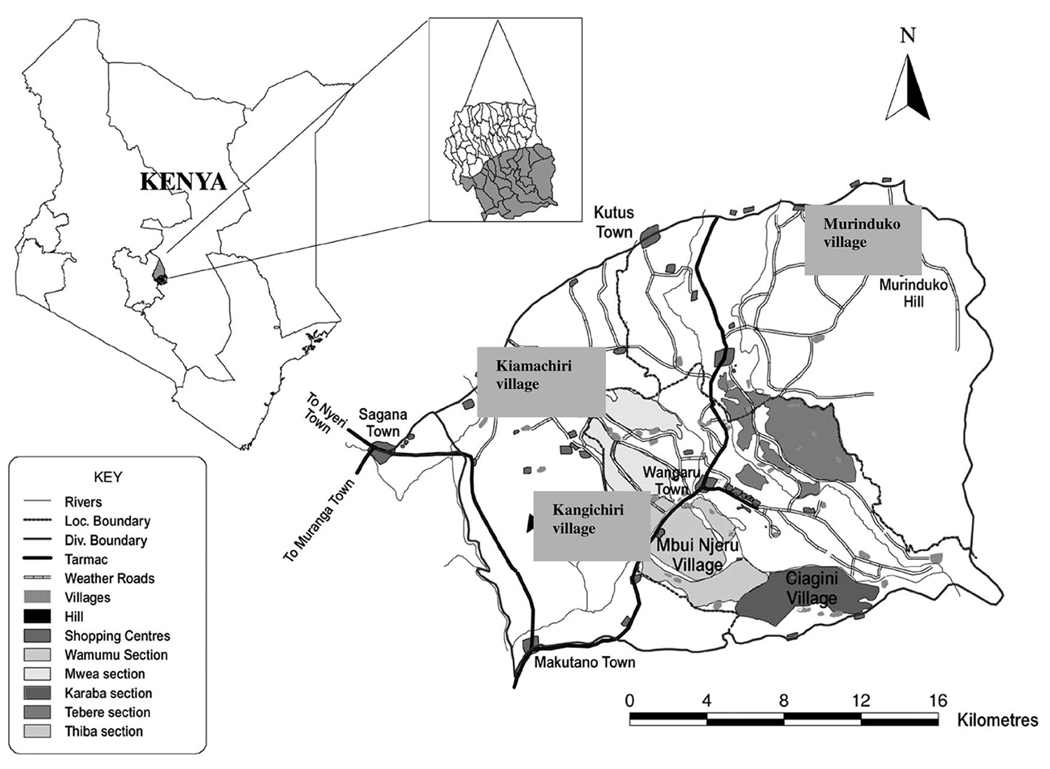
The study was conducted in three sites, Kangichiri, Kiamachiri, and Murinduko villages located in Mwea division, Kirinyaga District, 100 km North East of Nairobi, Kenya (Fig. 1). The study area has been the focus of numerous entomological, parasitological and epidemiological studies and has been described in details elsewhere (Muturi et al. 2007, Muturi et al. 2008). Kangichiri is within the Mwea Rice Irrigation Scheme and ≈75% of the land is under rice cultivation. Farmers in this village follow a defined rice cropping cycle as determined by the National Irrigation Board (planned rice cultivation). Kiamachiri is ≈5 km away from the rice scheme and ≈20% of the village land is used for rice cultivation. Individual farmers decide their own cropping cycle depending on water availability. Murinduko is ≈15–20 km from the scheme and is generally a nonrice growing village, mainly because of its hilly terrain that renders much (≈90%) of the area unsuitable for rice cultivation. Limited rice growing activity (<5% of the total area) is done along one major river valley that runs along the edge of the village.
Fig. 1.
A map of Kenya showing the sampling sites.
Mosquito Collection
Mosquitoes were collected in 10 randomly selected houses by pyrethrum spray catch (World Health Organization 1975) as described previously (Muturi et al. 2006, Muturi et al. 2008). The collections were done twice per month between 0700 and 1100 hours during August and September 2006.
Genomic DNA Extraction
DNA from individual An. gambiae s.l. mosquitoes was extracted using DNeasy Blood and Tissue Kit (QIAGEN, Valencia, CA). Each mosquito was homogenized with the aid of a microtube pestle (USA Scientific, Enfield, CT) in a 1.5 ml tube containing 180 µl phosphate buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4) and subjected to DNA extraction according to manufacturer’s protocol. Isolated DNA was reconstituted in 100 µl AE buffer (Qiagen, 10mM Tris-Cl, 0.5 mM EDTA, pH 9.0) and stored at −20°C for subsequent analysis by polymerase chain reaction (PCR).
Species Identification
Although An. arabiensis is the only known member of the An. gambiae complex occurring in the study area, species identification was confirmed using rDNA PCR method (Scott et al. 1993). Confirmed An. arabiensis samples were used for microsatellite analysis.
Microsatellite Genotyping
Nine microsatellite loci originally identified in An. gambiae and previously used for analysis of An. arabiensis population structure (Simard et al. 1999, Simard et al. 2000) were evaluated and eight were optimized for use in this study (Table 1). Thirty samples were genotyped for each study site using previously published primers (Zheng et al. 1996). PCR amplification was done using the Veriti Thermo Cycler (Applied Biosystems Inc., Foster City, CA) in a 25 µl reaction volume containing 12.5 µl of 2X TaqMan Universal PCR Master Mix (containing AmpliTaq Gold DNA Polymerase, AmpErase UNG, dNTPs with dUTP, and optimized buffer components; Applied Biosystems), 0.5 µl each of 10 µM forward and reverse primer, and 2.0 µl of template DNA. Either forward or reverse primer of each pair was labeled with a fluorescent dye (6FAM, HEX, NED, or PET). Preliminary amplification of An. arabiensis DNA from individual samples was conducted at denaturation temperature of 96°C for 5 min, followed by 40 cycles of denaturation at 96°C for 30 s, five different annealing temperatures (55, 57, 59, 61, and 63°C) for 30 s, extension at 72°C for 1 min and a final extension step at 72°C for 10 min. The aim of this initial amplification was to determine the optimum annealing temperature for each set of primers. Optimal PCR conditions were an initial denaturation at 96°C for 5 min, followed by 40 cycles of denaturation at 96°C for 30 s, allele-specific annealing temperature (Table 1) for 30 s, extension at 72°C for 1 min and a final extension step at 72°C for 10 min. Each PCR amplicon (2 µl aliquot diluted 30 times in nuclease free water) was mixed with 11.5 µl of formamide followed by addition of 0.5 µl of GeneScan-600 LIZ size standard (Applied Biosystems) for reproducible sizing of the fragments, and analyzed by 3130 Genetic Analyzer (Applied Biosystems). Compatible primer sets were multiplexed to increase the overall assay throughput. A positive and a negative control were included in every genotyping analysis and approximately one-sixth of the specimens for each population were genotyped in duplicate to ensure consistency of allele amplification. Data were analyzed using GeneMapper software version 4.0 (Applied Biosystems) to derive microsatellite allele sizes and genotypes. To ensure correct genotype scoring, manual visual inspection was carried out.
Table 1.
An. arabiensis microsatellite loci: cytological location, repeat motif, primer sequences, and locus-specific annealing temp
| Locus | Cytological location |
Primer sequences (5′–3′) | Repeat motif |
Allele size range |
Annealing temp (°C) |
|---|---|---|---|---|---|
| 24D | 2 La: 24D | F: GGCGAGCAGTTCATTCAAGT R: CGTCTGGAAGTTTCGTTGAG* |
CT | 103–121 | 59 |
| 141 | 2 Rd | F: CGGAGCAAATCTGAACCGTG R: CCTTGGCCACAACAACATCG* |
GT | 97–107 | 63 |
| 26 | 2 Rb: 12 | F: GGTTCCTGTTACTTCCTGCC R: CCGGCAACACAAACAATCGG* |
GT | 75–105 | 61 |
| 7 | X: 1C | F: CACGATGGTTTTCGGTGTGG* R: ATTTGAGCTCTCCCGGGTG |
GT | 81–93 | 63 |
| 49 | X: 1D | F: CAGCGCCTCCATATAGAACG R: GATCATTCAGCTGAACCTGC* |
GT | 79 | 57 |
| 803 | 2 R: 7B | F: CTCGATAAATCCCGTCGGTG R: GTCGGTTTGAGGTTGTAAAGC* |
TG | 117–123 | 63 |
| 45C | 3 liters: 45C | F: AAAAGTGGTGACCGAGTGAC* R: ATCTTCAACACTTCAGCACG |
GA | 142–158 | 59 |
| 147 | 2 R:19 | F: CTGCTGTTGCTGCCAAAATG R: AGCTTCACGGAAAGCAAAGG* |
GT | 155–171 | 57 |
| 29C | 2 R:19 | F: CTGCTGTGCTGCCAAAATG* R: AGCTTCACGGAAAGCAAAGG |
GT | – | Nd |
F, forward;
R, reverse;
fluorescence-labeled primer;
–, primer did not amplify;
Nd, value was not determined.
Statistical Analysis
Genetic diversity of mosquito populations was assessed by the number of observed (Na) and effective (Ne) alleles per locus, heterozygosity (He), and unbiased genetic diversity (HE). Intraspecific genetic diversity was also assessed by Shannon’s index (I) (Shannon 1948), a robust measure that is relatively insensitive to the skewing effects caused by failure to detect heterozygotes (Dawson et al. 1995). Nei’s genetic distances (GD) among population pairs were also determined. GenAlEX6.1 (Peakall and Smouse 2006) was used for these analyses. Hierarchical analysis of molecular variance (AMOVA) was performed to determine the genetic variation portioned at two levels: among individuals within populations and among populations. In addition, estimates of gene flow (Nm) and Wright’s FST among population pairs were carried out, using ARLEQUIN 3.1.1 (Schneider et al. 1997). Linkage disequilibrium (LD) can give important clues about population structure and history. To test the null hypothesis that genotypes for one locus are independent of genotypes for other loci, pairwise LD between markers was examined using GENEPOP 3.4 (Raymond and Rousset 1995), with 10,000 iterations. GENEPOP creates contingency tables for all pairs of loci in each sample, and then performs a probability test (Fisher exact test) for each table using a Markov-Chain-Method.
The samples from the three sites were also examined for evidence of any recent severe decrease in population size (genetic bottleneck). Recently bottle-necked populations exhibit heterozygosity excess at majority of loci. Wilcoxon Signed-Rank Test for heterozygosity excess was employed to detect bottlenecks under three models: the infinite alleles model (IAM), stepwise mutation model (SMM), and two-phase model (TPM), using BOTTLENECK 1.2.02 (Piry et al. 1999). Evidence for allele frequency distortion that occurs at neutral loci during bottlenecks was also assessed using the “mode-shift” indicator (Luikart et al. 1998).
Results
Genetic Diversity Among Populations
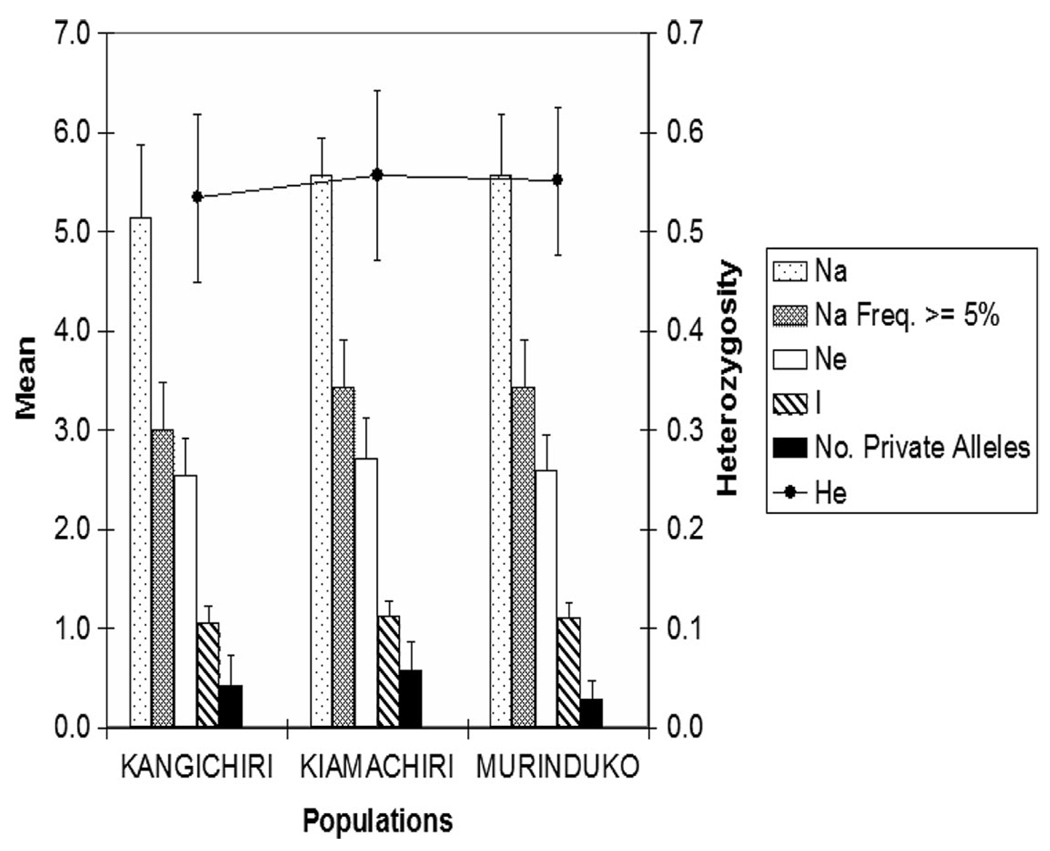
Nine microsatellite loci were initially screened across the three populations (Table 1). Two markers, 29C and 49 were excluded from further analysis because the former did not amplify well and the later was monomorphic (79 bp) across all populations. The number of alleles per locus among populations ranged from three (locus 141) to 10 (locus 26); mean 5.29 ± 0.87–5.71 ± 0.47. The pooled population number of alleles was however higher, being five (loci AR7, AR141, AR803), six (AR24D), seven (AR45, AR147), and 12 (AR26); overall pooled mean Na per locus = 6.7 ± 0.94. There was no significant difference in the mean number of effective alleles (Ne), Information index (I), and heterozygosity (Ho, He, HE) (Fig. 1) among the three samples. However, the mean number of private alleles per locus was lower at Murinduko (0.286 ± 0.184) compared with Kangichiri (0.429 ± 0. 297) or Kiamachiri (0.571 ± 0. 297) (Fig. 2). In general, Ne was much lower than Na, reflecting a high frequency of rare alleles.
Fig. 2.
Microsatellite diversity patterns across populations. Whiskers represent standard errors. Na = Number of different alleles; Ne = Number of effective alleles =1/(Sum pi2̂); I = Shannon’s Information Index = − 1* Sum (pi * Ln (pi)); He = Expected Heterozygosity = 1 - Sum pi2̂; Where pi is the frequency of the ith allele for the population and Sum pi2̂ is the sum of the squared population allele frequencies.
Hardy-Weinberg Equilibrium (HWE) and LD
Deviations from HWE were detected in three (Kangichiri and Kiamachiri) and four (Murinduko) out of the 21 HWE tests. However, after Bonferroni correction, only one locus for Kangichiri (AR26), two loci for Murinduko (AR26 and AR147), and three loci for Kiamachiri (AR26, AR141, and AR147) remained outside HWE expectations. Exact tests found no locus pair showing LD in any population after Bonferroni correction, suggesting independent segregation of the typed loci.
Gene Flow and Population Structure
High levels of gene flow (Nm range = 29–101) were detected between all population pairs (Table 2). The highest gene flow was between Kangichiri and Kiamachiri, which have the shortest geographic distance (5 km) apart. Nei genetic distances were miniscule (<0.05), but followed the trend of gene flow. Wright’s FST estimates showed no significant differentiation among populations. Hierarchical AMOVA corroborated FST estimates in showing no significant difference among populations, with only 0.6% of the variation among populations (P = 0.3), while a huge amount (99.4%) of the variation was partitioned within populations (Table 3).
Table 2.
Genetic distance, gene flow, and FST among An. arabiensis pop pairs
| Populations | Geographic Distance (km) |
Nei GD |
Nm | FST | P* |
|---|---|---|---|---|---|
| Kangichiri vs Kiamachiri | 5 | 0.026 | 101 | 0.005 | 0.386 |
| Kangichiri vs Murinduko | 20 | 0.033 | 29 | 0.017 | 0.068 |
| Kiamachiri vs Murinduko | 15 | 0.027 | 95 | 0.005 | 0.352 |
P value for FST, estimated over 10100 permutations using Arlequin. Overall FST = 0.006, P = 0.312.
Table 3.
AMOVA among An. arabiensis populations in three areas of Kenya with different agricultural practices
| Source of variation | Degrees of freedom |
Sum of squares |
Variance components |
% variation | P |
|---|---|---|---|---|---|
| Among populations | 2 | 5.2 | 0.011 | 0.6 | 0.312 |
| Within populations | 177 | 334.1 | 1.95 | 99.4 | <0.001 |
| Total | 179 | 339.3 | 1.96 | 100.0 |
Bottleneck Analysis
Analyses for evidence of bottlenecks under all known microsatellite mutation models gave insignificant results (P > 0.05), with alleles showing a normal, L-shaped distribution across all populations (Table 4).
Table 4.
Bottleneck analysis of An. arabiensis populations from three areas with differing agricultural systems in Kenya
| Modela | IAM | SMM | TPM | Mode-shift |
|---|---|---|---|---|
| Area | ||||
| Kangichiri (n = 30) | 0.6875 | 0.05469 | 0.6875 | Normal |
| Kiamachiri (n = 30) | 0.9375 | 0.07813 | 0.8125 | Normal |
| Murinduko (n = 30) | 0.8125 | 0.07813 | 0.375 | Normal |
The Wilcoxon test was used to test for heterozygosity excess under infinite allele (IAM), stepwise mutation (SMM), and two-phase (TPM) models. P values are indicated under each model type. Parameters for TPM were: variance = 30; proportion of SMM = 70%. Estimates based on 1000 replications.
n, no. samples tested.
Discussion
Of the eight microsatellite loci analyzed in the current study, seven were found to be polymorphic, with pooled population allele numbers per locus ranging from 5 to 12 (3–10 among individual populations). This level of allelic polymorphism provided powerful measures to identify population subdivision. The mean heterozygosities observed in the current study are comparable to those observed using the same loci in West Africa (0.558–0.608) and Madagascar, but higher than those observed in the outer, eastern Africa Islands of Réunion and Mauritius (Simard et al. 1999, Simard et al. 2000). Results of Hardy-Weinberg tests, linkage disequilibrium and AMOVA indicated that samples from the three study sites belonged to a single, continuous population in panmixia. These findings were further supported by the low genetic divergence (as measured by Wright’s FST) that resulted in large estimations of average migration index (Nm > 1 in all cases). Results across the African continent have shown that An. arabiensis has large deme sizes with little or no differentiation among mosquitoes collected at sites <200–250 km apart (Kamau et al. 1999, Donnelly and Townson 2000, Simard et al. 2000). In fact, recent studies in Zambia found An. arabiensis samples collected within a 2,000 km2 to be panmictic (Kent et al. 2007). The authors of this study concluded that this species was unlikely to be differentiated at shorter distances except in the presence of a strong topographic barrier. It has been suggested that the high gene flow observed between widely separated mosquito samples is mostly because of large effective population sizes and/or recent range expansion rather than extensive gene flow or mass migration (Donnelly and Townson 2000, Simard et al. 2000, Kent et al. 2007). While the observed panmixia signature may as well represent an ongoing influence of An. arabiensis recent demographic history (i.e., its recent population expansion), our study cannot rule out the possibility of extensive gene flow or mass migration.
An. arabiensis occurs more abundantly in irrigated than in nonirrigated areas because inundated rice fields provide permanent and extensive aquatic habitats for larval development. During the dry season when most larval habitats are dry, inundated rice fields not only continue to support large numbers of mosquitoes, but also elevate relative humidity that aids mosquito survival (Ijumba and Lindsay 2001). There has been a speculation that migration of mosquitoes from irrigated to nonirrigated areas ensures continuous presence of adult mosquitoes in the latter, especially during the dry periods (Dolo et al. 2004). Considering that the three study sites were <25 km apart, it is possible that dispersal of individuals across the three areas partly accounts for the high amounts of gene flow observed in the current study. While no data from direct estimates of dispersal by mark-release-recapture (MRR) experiments is available for An. arabiensis, MRR data from its sibling species, An. gambiae suggest that this mosquito species does not disperse beyond 1 km (Costantini et al. 1996, Midega et al. 2007). However, some MRR studies have reported collecting mosquitoes as far as 6 km from the release point yet most studies restrict their recapture efforts to distances <2 km(Costantini et al. 1996, Toure et al. 1998). Given its relatively high versatility, it is possible that An. arabiensis may disperse distances greater than the present MRR estimates for An. gambiae. The lack of bottleneck effect on any of our study populations supports the idea of mass dispersal within the deme (Simard et al. 2000).
An. arabiensis possess high adaptive potential in the choice of larval habitats, blood meal hosts, feeding and resting sites. In the current study, we found high number of rare alleles in An. arabiensis suggesting that this species has a high potential of responding to environmental changes and pressures imposed by control measures. This may explain why this species thrives in harsher environments compared with its sister species An. gambiae.
The lack of conformance to HWE at some loci was associated with heterozygote deficiency. Such deficiency could be caused by Wahlund effect, inbreeding or presence of null alleles. We attributed the large positive values associated with some loci to the presence of null alleles rather than Wahlund effect or inbreeding for two reasons. First, there was no evidence of linkage disequilibrium that should be detected in the case of pooling separate gene pools or inbreeding (Simard et al. 2000). Second, some individuals failed to amplify at some of the affected loci, but provided PCR product for other loci. Microsatellite null alleles are caused by mutations in the primer-binding sites, leading to PCR-amplification failures. They have been reported widely in An. gambiae and An. arabiensis population studies (Lehmann et al. 1996, Kamau et al. 1998, Simard et al. 1999, Simard et al. 2000) and complicate interpretation of microsatellite data by causing heterozygote deficits.
An important limitation of the current study is that the three agro-ecosystems were each represented by only one study site, with a few (7) microsatellite markers to evaluate the genetic structure of An. arabiensis populations. Future studies should consider using multiple study sites for each agro-ecosystem and a larger number of samples and genetic markers.
In summary, this study records high levels of gene flow, with no evidence of population differentiation or genetic bottlenecks among three An. arabiensis populations in areas under different agricultural practices. The significance of these results can be argued in two ways. On the positive side, if the genetically modified mosquitoes maintain the same behavior and adaptive abilities as the wild types, extensive gene flow would ensure that essential genes such as those conferring refractoriness to disease transmission are easily disseminated into the mosquito population. However, under such extensive gene flow, insecticide resistant genes can spread rapidly within this geographic region. This is worrisome, especially in our study area, given the present widespread usage of pesticides in rice cultivation as well as ITNs for malaria control.
Acknowledgments
We are grateful to Josephat Shililu for his support in facilitating the day-to-day activities of the field work. We acknowledge the technical support provided by ICIPE field staff in Mwea especially Simon Muriu, James Wauna, Peter Barasa, and Enock Mpanga. We also thank the two anonymous reviewers whose comments helped improve the manuscript. This research was supported by NIH/NIAID Grant #U01A1054889 (Robert Novak).
References Cited
- Afrane YA, Zhou G, Lawson BW, Githeko AK, Yan G. Effects of microclimatic changes caused by deforestation on the survivorship and reproductive fitness of Anopheles gambiae in western Kenya highlands. Am. J. Trop. Med. Hyg. 2006;74:772–778. [PubMed] [Google Scholar]
- Boete C, Koella JC. Evolutionary ideas about genetically manipulated mosquitoes and malaria control. Trends Parasitol. 2003;19 doi: 10.1016/s1471-4922(02)00003-x. 32-28. [DOI] [PubMed] [Google Scholar]
- Collins FH. Prospects for malaria control through the genetic manipulation of its vectors. Parasitol. Today. 1994;10:370–371. doi: 10.1016/0169-4758(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Collins FH, Besansky NJ. Vector biology and the control of malaria in Africa. Science. 1994;264:1874–1875. doi: 10.1126/science.8009215. [DOI] [PubMed] [Google Scholar]
- Costantini C, Li SG, Della Torre A, Sagnon N, Coluzzi M, Taylor CE. Density, survival and dispersal of Anopheles gambiae complex mosquitoes in a west African Sudan savanna village. Med. Vet. Entomol. 1996;10:203–219. doi: 10.1111/j.1365-2915.1996.tb00733.x. [DOI] [PubMed] [Google Scholar]
- Dawson IK, Simons AJ, Waugh R, Powell W. Diversity and genetic differentiation among subpopulations of Gliricidia sepium revealed by PCR-based assays. Heredity. 1995;74(Pt 1):10–18. doi: 10.1038/hdy.1995.2. [DOI] [PubMed] [Google Scholar]
- Dolo G, Briet OJT, Dao A, Traore SF, Bouare M, Sogoba N, Niare O, Bagayogo M, Sangare D, Teuscher T, Toure YT. Malaria transmission in relation to rice cultivation in the irrigated Sahel of Mali. Acta Trop. 2004;89:147–159. doi: 10.1016/j.actatropica.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Donnelly MJ, Townson H. Evidence for extensive genetic differentiation among populations of the malaria vector Anopheles arabiensis in Eastern Africa. Insect Mol. Biol. 2000;9:357–367. doi: 10.1046/j.1365-2583.2000.00197.x. [DOI] [PubMed] [Google Scholar]
- Fegan GW, Noor AM, Akhwale WS, Cousens S, Snow RW. Effect of expanded insecticide-treated bednet coverage on child survival in rural Kenya: a longitudinal study. Lancet. 2007;370:1035–1039. doi: 10.1016/S0140-6736(07)61477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Githeko A, Service M, Mbogo C, Atieli F. Resting behaviour, ecology and genetics of malaria vectors in large scale agricultural areas of Western Kenya. Parassitologia. 1996;38:481–489. [PubMed] [Google Scholar]
- Ijumba J, Lindsay S. Impact of irrigation on malaria in Africa: paddies paradox. Med. Vet. Entomol. 2001;15:1–11. doi: 10.1046/j.1365-2915.2001.00279.x. [DOI] [PubMed] [Google Scholar]
- Ijumba J, Mosha F, Lindsay S. Malaria transmission risk variations derived from different agricultural practices in an irrigated area of northern Tanzania. Med. Vet. Entomol. 2002;16:28–38. doi: 10.1046/j.0269-283x.2002.00337.x. [DOI] [PubMed] [Google Scholar]
- Kamau L, Hawley WA, Lehmann T, Orago AS, Cornel A, Ke Z, Collins FH. Use of short tandem repeats for the analysis of genetic variability in sympatric populations of Anopheles gambiae and Anopheles arabiensis. Hered. 1998;80:675–682. doi: 10.1046/j.1365-2540.1998.00327.x. [DOI] [PubMed] [Google Scholar]
- Kamau L, Mukabana WR, Hawley WA, Lehmann T, Irungu LW, Orago AA, Collins FH. Analysis of genetic variability in Anopheles arabiensis and Anopheles gambiae using microsatellite loci. Insect. Mol. Biol. 1999;8:287–297. doi: 10.1046/j.1365-2583.1999.820287.x. [DOI] [PubMed] [Google Scholar]
- Kamau L, Munyekenye GO, Vulule JM, Lehmann T. Evaluating genetic differentiation of Anopheles arabiensis in relation to larval habitats in Kenya. Infect. Genet. Evol. 2007;7:293–297. doi: 10.1016/j.meegid.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Karaca M, Ekici A, Sen O, Yesilnacar I, Kindap T. Irrigation induced climate change in Southeastern Turkey. Geophysical Research Abstracts. 2008;10 EGU2008-A-08390. [Google Scholar]
- Kent RJ, Mharakurwa S, Norris DE. Spatial and temporal genetic structure of Anopheles arabiensis in Southern Zambia over consecutive wet and drought years. Am. J. Trop. Med. Hyg. 2007;77:316–323. [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Hawley WA, Kamau L, Fontenille D, Simard F, Collins FH. Genetic differentiation of Anopheles gambiae populations from East and west Africa: comparison of microsatellite and allozyme loci. Heredity. 1996;77:192–200. doi: 10.1038/hdy.1996.124. [DOI] [PubMed] [Google Scholar]
- Lengeler C. Insecticide-treated bed nets and curtains for preventing malaria. Cochrane Db. Syst. Rev. 2004;2004 doi: 10.1002/14651858.CD000363.pub2. CD000363. [DOI] [PubMed] [Google Scholar]
- Luikart G, Allendorf FW, Cornuet JM, Sherwin WB. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 1998;89:238–247. doi: 10.1093/jhered/89.3.238. [DOI] [PubMed] [Google Scholar]
- Midega JT, Mbogo CM, Mwnambi H, Wilson MD, Ojwang G, Mwangangi JM, Nzovu JG, Githure JI, Yan G, Beier JC. Estimating dispersal and survival of Anopheles gambiae and Anopheles funestus along the Kenyan coast by using mark-release-recapture methods. J. Med. Entomol. 2007;44:923–929. doi: 10.1603/0022-2585(2007)44[923:edasoa]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno M, Salgueiro P, Vicente JL, Cano J, Berzosa PJ, de Lucio A, Simard F, Caccone A, Do Rosario VE, Pinto J, Benito A. Genetic population structure of Anopheles gambiae in Equatorial Guinea. Malar. J. 2007;6:137. doi: 10.1186/1475-2875-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriu SM, Muturi EJ, Shililu JI, Mbogo CM, Mwangangi JM, Jacob BG, Irungu LW, Mukabana RW, Githure JI, Novak RJ. Host choice and multiple blood feeding behaviour of malaria vectors and other anophelines in Mwea rice scheme, Kenya. Malar. J. 2008;7:43. doi: 10.1186/1475-2875-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutero C, Ng‘ang’a P, Wekoyela P, Githure J, Konradsen F. Ammonium sulphate fertilizer increases larval populations of Anopheles arabiensis and culicine mosquitoes in rice fields. Acta Trop. 2004a;89:187–192. doi: 10.1016/j.actatropica.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Mutero C, Kabutha C, Kimani V, Kabuage L, Gitau G, Ssennyonga J, Githure J, Muthami L, Kaida A, Musyoka L, Kiarie E, Oganda M. A transdisciplinary perspective on the links between malaria and agroecosystems in Kenya. Acta Trop. 2004b;89:171–186. doi: 10.1016/j.actatropica.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Muturi J, Shililu J, Jacob B, Githure J, Gu W, Novak R. Mosquito species diversity and abundance in relation to land use in a riceland agroecosystem in Mwea, Kenya. J. Vector Ecol. 2006;31:129–137. doi: 10.3376/1081-1710(2006)31[129:msdaai]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Muturi J, Muriu S, Shililu J, Mwangangi J, Jacob B, Mbogo C, Githure J, Novak R. Effect of rice cultivation on malaria transmission in central Kenya. Am. J. Trop. Med. Hyg. 2008;78:270–275. [PubMed] [Google Scholar]
- Mwangangi J, Muturi E, Shililu J, Muriu S, Jacob B, Kabiru E, Mbogo C, Githure JI, Novak R. Contribution of different aquatic habitats to adult Anopheles arabiensis and Culex quinquefasciatus (Diptera: Culicidae) production in a rice agroecosystem in Mwea, Kenya. J. Vector Ecol. 2008;33:129–138. doi: 10.3376/1081-1710(2008)33[129:codaht]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Mol. Ecol. Notes. 2006;6:288–295. [Google Scholar]
- Piry S, Luikart G, Cornuet JM. BOTTLENECK: a Computer software program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999;90:502–503. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J. Heredity. 1995;86:248–249. [Google Scholar]
- Schlotterer C. Evolutionary dynamics of microsatellite DNA. Chromosoma. 2000;109:365–371. doi: 10.1007/s004120000089. [DOI] [PubMed] [Google Scholar]
- Schneider S, Kueffer JM, Roessli D, Excoffier L. Genetics and biometry laboratory, Department of Antrhopology. Geneva, Switzerland: University of Geneva; 1997. ARLEQUIN, a software Package for population genetics. [Google Scholar]
- Scott J, Brogdon W, Collins F. Identification of single species specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- Shannon CE. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. 623–656. [Google Scholar]
- Simard F, Fontenille D, Lehmann T, Girod R, Brutus L, Gopaul R, Dournon C, Collins FH. High amounts of genetic differentiation between populations of the malaria vector Anopheles arabiensis from West Africa and eastern outer islands. Am. J. Trop. Med. Hyg. 1999;60:1000–1009. doi: 10.4269/ajtmh.1999.60.1000. [DOI] [PubMed] [Google Scholar]
- Simard F, Lehmann T, Lemasson JJ, Diatta M, Fontenille D. Persistence of Anopheles arabiensis during the severe dry season conditions in Senegal: an indirect approach using microsatellite loci. Insect Mol. Biol. 2000;9:467–479. doi: 10.1046/j.1365-2583.2000.00210.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Thomson MC, Connor SJ, Quinones ML, Jawara M, Todd J, Greenwood BM. Movement of Anopheles gambiae s.l. malaria vectors between villages in The Gambia. Med. Vet. Entomol. 1995;9:413–419. doi: 10.1111/j.1365-2915.1995.tb00015.x. [DOI] [PubMed] [Google Scholar]
- Toure YT, Dolo G, Petrarca V, Traore SF, Bouare M, Dao A, Carnahan J, Taylor CE. Mark-release-recapture experiments with Anopheles gambiae s.l. in Banambani Village, Mali, to determine population size and structure. Med. Vet. Entomol. 1998;12:74–83. doi: 10.1046/j.1365-2915.1998.00071.x. [DOI] [PubMed] [Google Scholar]
- Toure YT, Petrarca V, Traore SF, Coulibaly A, Maiga HM, Sankare O, Sow M, Di Deco MA, Coluzzi M. Ecological genetic studies in the chromosomal form Mopti of Anopheles gambiae s.str. in Mali, west Africa. Genetica. 1994;94:213–223. doi: 10.1007/BF01443435. [DOI] [PubMed] [Google Scholar]
- [WHO] World Health Organization. Manual on practical entomology in malaria, part II. Geneva, Switzerland: World Health Organization; Methods and techniques. 1975
- Zheng L, Benedict MQ, Cornel AJ, Collins FH, Kafatos FC. An integrated genetic map of the African human malaria vector mosquito, Anopheles gam-biae. Genetics. 1996;143:941–952. doi: 10.1093/genetics/143.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Collins FH, Kumar V, Kafatos FC. A detailed genetic map for the X chromosome of the malaria vector, Anopheles gambiae. Science. 1993;261:605–608. doi: 10.1126/science.8342025. [DOI] [PubMed] [Google Scholar]