Summary
Mobile group I introns are RNA splicing elements that have been invaded by endonuclease genes. These endonucleases facilitate intron mobility by a unidirectional, duplicative gene-conversion process known as homing [1]. Survival of the invading endonuclease depends upon its ability to promote intron mobility. Therefore, the endonuclease must either quickly change its cleavage specificity to match the site of intron insertion, or it must already be pre-adapted to cleave this sequence. Here we show that the group I intron in the DNA polymerase gene of T7-like bacteriophage ΦI is mobile, dependent upon its intronic HNH homing endonuclease gene, I-TslI. We also show that gene 5.3 of phage T3, located adjacent to its intronless DNA polymerase gene, is a homologous homing endonuclease gene whose protein product initiates efficient spread of gene 5.3 into empty sites in related phages. Both of these endonucleases cleave intronless DNA polymerase genes at identical positions. This shared feature between an intronic and free-standing endonuclease is unprecedented. Based on this evidence we propose that introns and their homing endonucleases evolve separately to target the same highly conserved sequences, uniting afterwards to create a composite mobile element.
Results
The gene 5.3 locus is polymorphic
Gene 5.3 is a small open reading frame (ORF) of unknown function located adjacent to the DNA polymerase gene (gene 5) in both T7 and the related phage T3. Despite a lack of similarity to each other, each gene shows similarity to homing endonucleases from different families. T7 gp5.3 shows slight similarity to the cyanobacterial intron endonuclease I-Ssp6803I [2–5], while T3 gp5.3 (and homologous ORFs in Yersinia phage ΦYeO3-12 [6] and Pseudomonas phage PA11 [7]) contains the HNH homing endonuclease motif [8, 9].
The T7-like phages ΦI, W31 and K1F lack gene 5.3, having just a 19-bp separation between genes 5 and 5.5 that is AT rich and contains the ribosome binding site for gene 5.5. However, these phages have a 601-bp group I intron inserted 156 bp from the end of gene 5 [10] [11]. These introns encode a 131 codon ORF belonging to the HNH endonuclease gene family, inserted into stem P6a [10]. Yersinia pestis phage A1122 has neither a gene 5.3 nor a group I intron [12]. PCR analysis of the gene 5.3 locus from five additional phages in this family (ΦIIP, ΦIIW, H, ViIII and C21R) did not detect any additional insertions (data not shown). Phage ΦIIP, which lacks any insertion in this region, was selected for further study. Schematic representations of this region in the phages used in this study are presented in Figure 1.
Figure 1. Schematic representation of the gene 5.3 locus of T7-like phages.
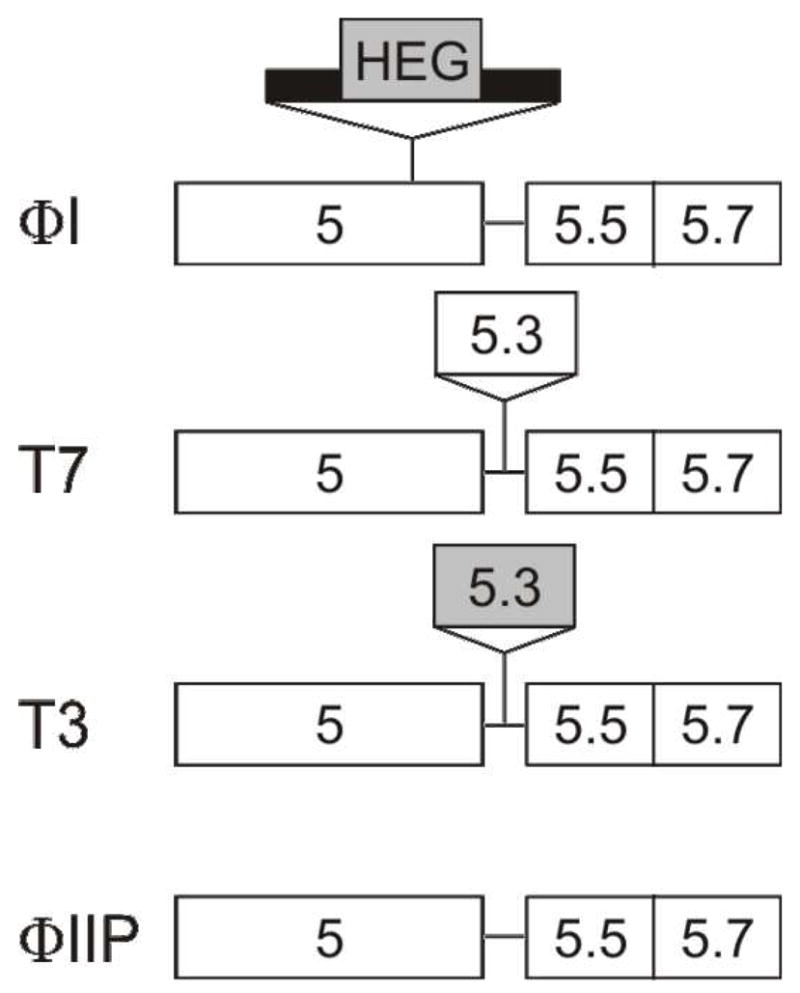
Names of representative phages are on the left. Gene names are indicated in the boxes. Thick line indicates intron sequences. Shaded boxes indicate ORFs with HNH motifs.
The intron-encoded ORFs are most similar to T3 gene 5.3
Database searches with BLASTP [13] using the translated intron-encoded ORF from phage ΦI reveal the next highest similarities to intron-encoded ORFs from W31 and K1F followed by ORFs at the gene 5.3 locus of phages T3, ΦYE03-12 and PA11. These relationships were confirmed by phylogenetic reconstruction using PHYML (not shown). The 101-amino acid T3 gp5.3 aligns over its entire length with the intron-encoded ORFs, with especially strong conservation in the HNH region (Fig. S1). This similarity between the intron-encoded ORFs and T3 gp5.3 prompted us to determine the function of these proteins.
The ΦI and W31 introns encode endonucleases that nick double-stranded DNA
The in vitro synthesized ΦI and W31 intron-encoded proteins were assayed for endonuclease activity on synthetic DNA substrates corresponding to intronless ΦI and W31 DNA polymerase sequences. Both proteins nick the bottom strand of each target DNA, but are not active on the top strands (Fig. 2A). This endonuclease has been named I-TslI (Intron-encoded endonuclease from T-seven-like phages, I).
Figure 2. Endonuclease activity of I-TslI and F-TslI.
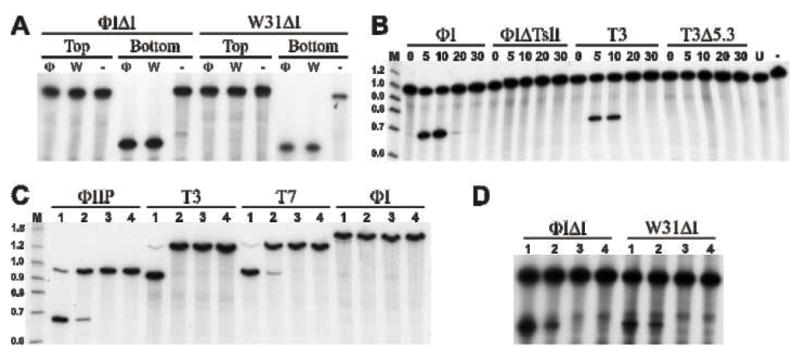
A. In vitro synthesized I-TslI from ΦI (Φ) and W31 (W), or mock translation products (−) were incubated with 60-mer target DNAs representing the fused exons of the ΦIΔI or W31ΔI DNA polymerase genes. Substrate DNAs (indicated at the top of the figure) are 5′ end-labeled on the coding (top) or template (bottom) strand, as indicated. Reaction products were separated by electrophoresis on a 6% denaturing polyacrylamide gel. B. Endonuclease activity in phage infected cells. Extracts from phage-infected cells (noted above the line) were prepared at various times after infection (noted above each lane). Bottom strand labeled DNA was amplified from ΦIIP. C. and D. Substrate specificity of I-TslI and F-TslI. Extracts prepared 10 min post-infection from phage ΦI (lane 1), T3 (lane 2), ΦIΔTsl (lane 3) and T3Δ5.3 (lane 4) were incubated with substrate DNAs (indicated above the line) generated by (C) PCR from phages or (D) oligonucleotides representing the fused exons from ΦI and W31 (indicated above the line). Only the bottom strand is 5′-end labeled.
Endonuclease activity from phage-infected cell lysates
Free-standing (non-intron or -intein encoded homing endonucleases) ORFs of bacteriophages, eubacteria and eukaryotes have also been identified that contain homing endonuclease signature motifs [14]. Notably, the genome of bacteriophage T4 is infested with ORFs bearing similarity to homing endonucleases [15]. In addition to the endonuclease genes in its three self-splicing group I introns, T4 has at least 12 homing endonuclease-like ORFs inserted between genes that are conserved in other T-even phages. Several of these encode endonucleases [15–21] that are themselves mobile, invading vacant sites in other phages by a gene conversion mechanism analogous to intron homing [16–18, 21, 22], a process called ‘intronless homing’ [16].
In vitro protein synthesis of T3 or T7 gene 5.3 gave poor expression yields, and endonuclease activity could not be demonstrated (data not shown). Therefore, we assayed phage-infected cell extracts for endonuclease activity using PCR-amplified ΦIIP substrate DNA. No activity was observed from ΦIIP or T7 extracts (data not shown). However, both ΦI and T3 extracts demonstrated cleavage activity, with optimal activity appearing 10 min post-infection (Fig. 2B). Cleavage of top-strands was not detected (data not shown). Deletions that remove most of the coding sequences of I-TslI and gp5.3 eliminate activity (Fig. 2B). These results indicate that I-TslI and T3 gp5.3 are transcribed and translated during phage infection and that the endonuclease activity in T3 extracts is due, at least in part, to gp5.3. However, since we were unable to synthesize active protein in vitro it remains to be determined if gp5.3 is sufficient for the activity. Following the convention suggested for free-standing homing endonucleases, the activity of gp5.3 is designated F-TslI [23].
I-TslI and F-TslI have similar but distinct substrate specificities
The DNA polymerase genes from the T7-like phages described in this study provide a sampling of natural target site variants for I-TslI and F-TslI. We tested extracts from cells infected with phages ΦI, ΦIΔTsl (lacking 73 of 131 residues of I-TslI, including the HNH catalytic domain), T3, and T3Δ5.3 (lacking 94 of 101 residues of F-TslI) on DNA substrates amplified from phage ΦIIP, T3, T7, and ΦI, and also on double-stranded oligonucleotides representing fused exon sequences from both ΦI and W31. I-TslI and F-TslI cleave the ΦIIP, T7, and intronless ΦI and W31 substrates, but are inactive on a substrate containing the ΦI intron (Fig. 2C and D). Interestingly, the T3 DNA polymerase sequence is sensitive to cleavage by I-TslI, but resistant to cleavage by T3-encoded F-TslI. A summary of the cleavage results is shown in Fig. 3B.
Figure 3. I-TslI and F-TslI cleave at the identical location.
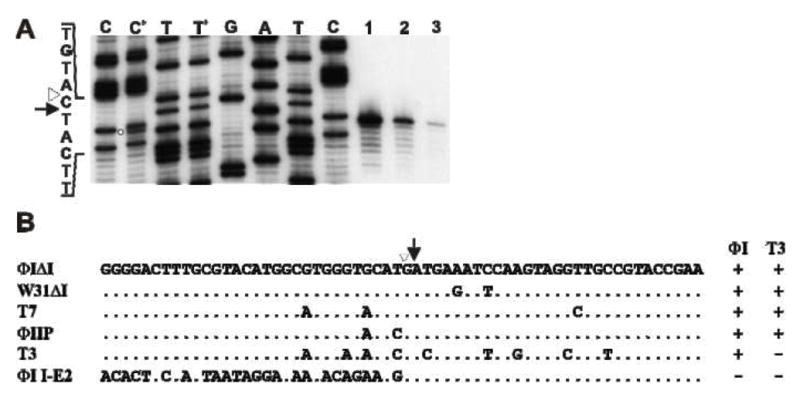
A. Cleavage site mapping. DNA containing the IIS was amplified by PCR directly from pT7Δ5.3 (using primers ΦI5.0 and 5′-end labeled ΦI5.0rev) and incubated with in vitro synthesized ΦI I-TslI (lane 1), ΦI-infected cell extract (lane 2) and T3-infected cell extract (lane 3) for 15 min. Reaction products were separated by denaturing polyacrylamide gel electrophoresis, next to sequencing reactions generated with the same labeled primer and template DNA used in the PCR. Lanes C+ and T+ contain a mixture of the contents of lane 2 with the C and T sequencing reactions, respectively. The open circle indicates the ΦI cleavage product in the C+ lane. The site of intron insertion is indicated by an open triangle and the position of cleavage is indicated by an arrow. B. Summary of cleavage specificity. The sequences (top/coding strand) of target DNAs from T7-like phages, surrounding the site of intron insertion, are aligned with the ΦI and W31 fused exon sequences. Results of cleavage by the ΦI and T3 extracts are indicated on the right. Identical positions are indicated by dots; only sequence differences are shown. The site of intron insertion is indicated by an open triangle and the position of cleavage on the bottom strand is indicated by an arrow.
I-TslI and F-TslI cleave at the same position near the intron insertion site (IIS)
The exact positions of cleavage by I-TslI and F-TslI were determined on the T7 target sequence. In vitro synthesized I-TslI and extracts from ΦI- and T3-infected cells cut the template strand of the T7 DNA polymerase gene at the same position, one nucleotide 5′ of the IIS (Fig. 3A). A portion of the I-TslI cleavage reaction (lane 1) was mixed with the C and T sequencing reactions to eliminate any discrepancy in migration (Fig. 3A, lanes C+ and T+, respectively). An extra band corresponding to the cleavage product is apparent in the C+ lane, while no extra band can be seen in the T+ lane, indicating that the cleavage product co-migrates with the T band.
I-TslI and F-TslI promote homing
To investigate homing of the ΦI intron, we constructed an isogenic derivative, ΦIΔI, with an exact deletion of the intron in its DNA polymerase gene. After co-infection of ΦIΔI and ΦI, the fraction of intron-containing individuals increased from an average of 54% of the input phages to ~92% of the progeny (Table 1). Preferential inheritance of the intron is dependent on integrity of I-TslI, since it did not occur in co-infection between ΦIΔI and ΦIΔTsl.
Table 1.
Endonuclease Dependent Homing
| Input | Progeny | |||||
|---|---|---|---|---|---|---|
| Cross | Exp | a MOI | b CSR | c gene5.3 | b CSR | c gene5.3 |
| T3/CSR × T3Δ5.3/CSS | 1 | 16 | 0.57 | 0.57 | 0.91 | 0.89 |
| 2 | 29 | 0.61 | 0.61 | 0.95 | 0.94 | |
| 3 | 35 | 0.57 | 0.57 | 0.92 | 0.92 | |
| T3Δ5.3 × T3Δ5.3/CSS | 1 | 17 | 0.42 | d NA | 0.49 | d NA |
| 2 | 33 | 0.47 | d NA | 0.48 | d NA | |
| 3 | 35 | 0.47 | d NA | 0.45 | d NA | |
|
e I+ |
e I+ |
|||||
| ΦI × ΦIΔI | 1 | 9 | 0.65 | 0.92 | ||
| 2 | 30 | 0.44 | 0.94 | |||
| 3 | 25 | 0.52 | 0.88 | |||
| ΦIΔTslI × ΦIΔI | 1 | 7 | 0.64 | 0.47 | ||
| 2 | 18 | 0.46 | 0.41 | |||
| 3 | 18 | 0.48 | 0.39 | |||
Multiplicity of Infection
Oligonucleotides T3IIS and T3mut3 were used to identify the cleavage resistant wild-type ( CSR ) and cleavage sensitive mutant (CSS) T3 sequences, respectively.
Oligonucleotides T35.3 and T3del3 were used to identify intact and deleted T3 gene 5.3.
Not Applicable
Oligonucleotide ΦId2 wasused to identify the presence of the intron.
Phage T3 is not compatible with most other T7-like phages in mixed infections [24]. To investigate homing directed by F-TslI, we constructed a T3 variant whose sequence at the cleavage site in gene 5 is sensitive to its own endonuclease. The fused exon sequence of ΦIΔI, which contains nine base pair differences from the T3 sequence near the cleavage site, is cleaved by F-TslI (Fig. 2D & 3B). We incorporated all nine of these substitutions in T3Δ5.3to create a sensitive cleavage site (CSS) in a T3 genetic background. In mixed infections, both the wild-type cleavage site (CSR) and the intact gene 5.3 were preferentially inherited in an F-TslI dependent manner (Table 1). Interestingly, a small fraction of the progeny contains the CSR allele from the donor and the Δ5.3 allele from the recipient, illustrating that recombination is readily detected between these closely spaced loci. The reciprocal recombinant, uniting CSS and intact gene 5.3, should be lethal and was not detected.
Discussion
The self-splicing group I intron in Φ1, W31 and K1F interrupts a conserved, functionally important region of the phage-encoded DNA polymerase gene [10], being inserted immediately adjacent to the aspartate residue that coordinates Mg2+ in the enzyme catalytic center (Fig. S2). We have shown that this intron encodes I-TslI, an HNH-like homing endonuclease (Figs. 2 and 3). It is intriguing that the closest relatives to I-TslI are the three genes 5.3 from phages PA11, Ye03-12 and T3, which also encode an endonuclease, F-TslI (Fig. 2; Fig. S1). In addition to sequence similarity, I-TslI and F-TslI share functional characteristics. Both enzymes generate a single-strand nick at precisely the same position of the template DNA strand, one nucleotide 5′ of the IIS (Fig. 3A) and promote homing reactions in vivo (Table I).
Prevention of self-cleavage is a hallmark of homing, whether it is mediated by an intron/intein-encoded, or a free-standing endonuclease. For intron and intein homing, protection from endonuclease cleavage is generally a consequence of the DNA binding site and/or the cleavage site being split by insertion of the respective intervening sequence [25]. However, in intronless homing the target site is not afforded this protection. Instead, the endonuclease distinguishes self from non-self by using sequence polymorphisms in the DNA target [16, 18]. In this respect I-TslI and F-TslI act as typical homing endonucleases as neither enzyme cleaves its own DNA under the same conditions where the target sequence of related phages is cleaved extensively (Fig. 2). T3 gene 5 has unique sequence polymorphisms in the highly conserved DNA target region (Fig. 3B), one or more of which may enable F-TslI to discriminate self from non-self in DNA binding and/or cleavage reactions. Interestingly, F-TslI is also unable to cleave the intron-containing target (Fig. 2C) implying that the intron lies within the cleavage/recognition sites for both I-TslI and F-TslI.
Selfish DNA meets catalytic RNA
It has been proposed that homing endonuclease genes are selfish DNAs that have invaded splicing elements. This association is thought to be mutually advantageous, offering a phenotypically silent home for the endonuclease gene while providing mobility to the splicing element [25]. Multiple lines of evidence support this hypothesis [26–28]. Although most homing endonucleases apparently confer no selective advantage on their host genomes and could be quickly eliminated by mutation, homing allows their propagation and maintenance in the population [29]. While free-standing endonuclease genes are usually restricted to intergenic, non-essential locations, endonuclease invasion of an intron (or intein) greatly expands their potential insertion sites to include essential coding sequences.
The most interesting feature shared by endonucleases I-TslI and F-TslI is their ability to cleave DNA at the identical position within the DNA polymerase gene (Fig. 3A). This feature, along with the similarity of their sequences, strongly suggests that they have a recent common ancestor. In addition, the close proximity of insertion sites of the genes encoding these endonucleases, only 156 bp apart in the genomes of closely related phages, is highly provocative. Either I-TslI escaped from its intron home into the adjacent intercistronic space, or F-TslI jumped from its intercistronic position into a nearby intron. As we will explain below, we find the latter to be the more likely sequence of events. This scenario could help us to understand the origin of group I intron homing.
In order for an endonuclease to be maintained in its new environment after being inserted into an intron, it must be able to promote the spread of the intron by recognizing and cleaving intronless alleles. Two alternative scenarios, both of which require rare events, were considered by Loizos et al. [30] as explanations of how this may have occurred. One proposal was suggested by the observation that the IIS of the sunY/nrdB intron resembles short sequences flanking its homing endonuclease gene, I-TevII. Moreover, when these flanking sequences were fused, the resultant sequence could be cleaved by the enzyme. They suggested that the endonuclease gene had originally evolved to cut a closely related sequence, and was inserted into the intron as a consequence of repair of a cleavage event at this ectopic site. Since there would not necessarily be specificity for the IIS at this new location, survival of the endonuclease would depend on transposition of the intron to a new genomic location; presumably the site for which the enzyme was already adapted. However, this pathway will produce functional gene products only if there is sufficient sequence similarity between the donor and recipient DNA to initiate replicative repair, if the intron is capable of splicing in its new environment, and if the changes brought about by co-conversion of flanking DNA are compatible with the function of the final gene product. These requirements are especially unlikely to be met at locations where many group I introns reside: coding sequences of highly conserved enzyme active centers.
In a second scenario, considered by Loizos et al. to be less likely [30], a rare, non-homologous recombination event could have inserted an endonuclease, which already had specificity for the IIS, into the intron. We suggest that the intron in phages ΦI, W31 and K1F acquired its endonuclease gene by this route, and that this may be a general pathway for the origin of these composite mobile elements.
Group I introns and homing endonucleases favor highly conserved target sequences
Group I introns and inteins tend to be inserted into highly conserved sequences encoding functionally significant regions of essential genes [31–37]. The only way a gene can rid itself of an intron in such a position is by exact deletion. Inexact deletion would leave a lethal insertion/deletion or frame shift [33, 36]. We assume that prior to acquiring homing endonuclease genes, group I introns were inserted randomly but survived only where they could not be easily eliminated. Conversely, successful elimination of introns from nonessential coding regions may be responsible for some of the variability in contemporary genomes.
On the other hand, for free-standing endonuclease genes to survive in their intercistronic locations they must be propagated by intronless homing, using target sequences in an adjacent gene [16, 18]. Functionally important regions of essential genes provide optimal targets, since these will be most frequently encountered and not easily changed. Since optimal IISs and endonuclease cleavage sites share the same properties, introns and endonuclease genes are likely to converge independently on the same set of almost universally conserved sequences.
By residing within an intron, an endonuclease can transpose to new sites within coding sequences without necessarily inactivating them and, having acquired an endonuclease that is pre-adapted for the IIS, the intron is immediately able to increase its frequency in a population by homing. Thus, when an intron and an endonuclease happen to converge on the same conserved DNA target, their combination into a single genetic element should provide substantial selective advantage to both of them, assuring recovery of rare recombination events.
Figure 4 illustrates how an intron and an endonuclease gene could have come together in the DNA polymerase gene of T7-like phages. An ancestral phage (A), such as ΦIIP, acquires an intron, perhaps by reverse splicing into mRNA followed by reverse transcription [38, 39], to create phage B. The intron is stably inserted in this site, but cannot spread through the population due to lack of a homing mechanism. Other ancestral phages independently acquire endonuclease F-TslI by insertion into the intercistronic region between genes 5 and 5.5. Presumably cleavage activity is initially weak, and this phage (C) is under selection to alter the target sequence in its gene 5, mostly by third position changes, creating a phage similar to phage T3. Simultaneously, to facilitate its spread through the population by homing, the endonuclease is under selection for more efficient cleavage of the unaltered target sequence. Recombination in T7-like phages is very efficient [40], so the eventual encounter of phages B and C in a mixed infection will lead to creation of the recombinant phage (D). This combination is stable and should spread through the population by homing to intronless sites, with very efficient co-conversion of the intron and nearby endonuclease gene. Since it requires the participation of two independent genetic units, we have named this process collaborative homing to distinguish it from conventional intron homing (QZ, RPB and DAS, Curr. Biol. in press).
Figure 4. Proposed pathway for establishment of intron homing in the DNA polymerase gene of T7-like phages.
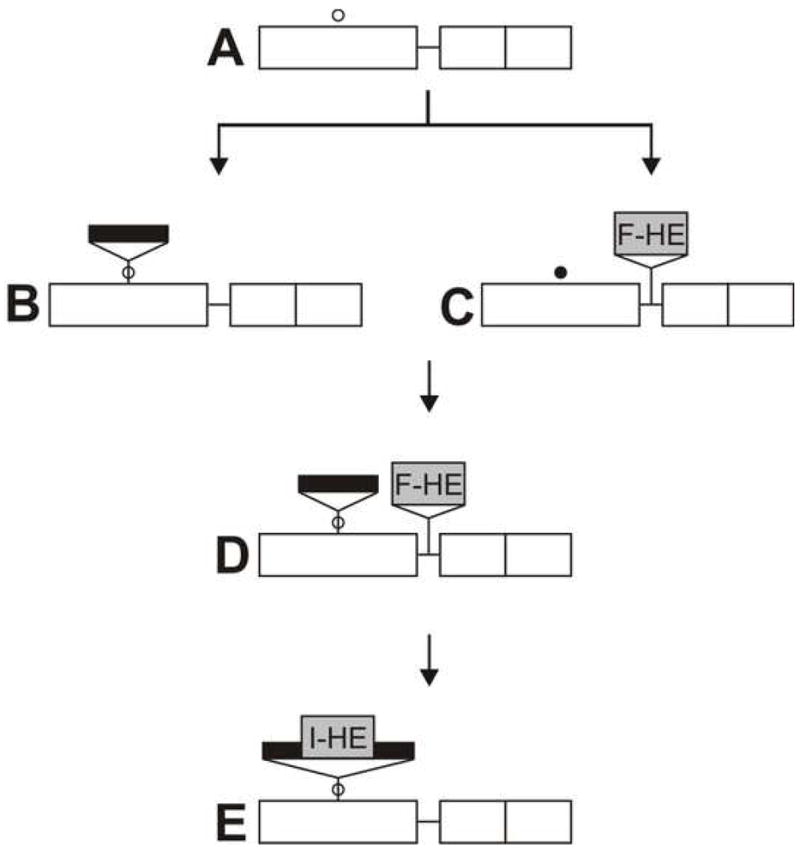
A. Ancestral gene arrangement prior to insertions, exemplified by phage ΦII, with location of a highly conserved, functionally importantsequence indicated by an open circle (○). B. Insertion of a self splicing group I intron (black bar) into the highly conserved sequence in gene 5. C. A free-standing endonuclease gene (F-TslI) is inserted between genes 5 and 5.5, as in phage T3. The altered target sequence, resistant to F-TslI, is indicated by a closed circle (●). The endonuclease gene is propagated by intronless homing. D. Phage containing both the intron and the endonuclease gene, generated by homologous recombination during mixed infection between phages B and C. The intron and the endonuclease gene are propagated together by collaborative homing. E. Insertion of the endonuclease gene into the intron, creating a mobile intron.
However, a significant proportion of recombinational events will separate the intron and endonuclease gene, similarly to what has been shown here for F-TslI and its cleavage resistant site (Table 1), and elsewhere for the T4-encoded free-standing endonuclease SegG [18]. A rare DNA rearrangement can result in insertion of the endonuclease gene into the intron (E). This coupling is the most stable, optimal arrangement for both entities. It ensures that the mobility element (the endonuclease) and the protective element (the intron) can never be separated. The chimeric mobile element can spread to intronless sites via homing and, occasionally, transpose to non-allelic coding sequences. Finally, even if eliminated by exact deletion, this recreates an IIS that is subject to re-invasion in a subsequent homing event.
Two examples of the arrangement proposed in Figure 4D, where the intron and endonuclease have come together in the same chromosome but where the endonuclease gene has not yet been transferred into the intron, have recently been identified. Aeromonas salmonicida phage 25, which is related to E. coli phage T4, has a group I intron inserted into the gene for a DNA polymerase subunit [41]. Adjacent to the DNA polymerase coding sequence is a gene encoding an endonuclease of the GIY-YIG family that cleaves the homologous intronless sequence of phage T4 (V. Petrov and J.D. Karam, personal communication). Similarly, we have shown that a free-standing homing endonuclease gene is adjacent to the intron-containing psbA gene in cyanobacterial phage S-PM2. The endonuclease (F-CphI) cannot cut intron-containing DNA, but it does cut both the in vitro synthesized IIS of S-PM2 and intronless psbA genes of related phages (QZ, RPB, and DAS, Curr. Biol. in press).
Conclusion
We propose a model whereby group I introns and homing endonuclease genes individually target the same set of highly conserved DNA sequences for insertion and cleavage, respectively, ultimately uniting to form a chimeric mobile element. The same opportunities for the confluence of homing endonuclease genes and group I introns by genetic recombination that we propose here also apply to other genomes (eubacteria, fungal mitochondria, chloroplasts, nuclei of unicellular eukaryotes) where mobile group I introns are frequently found. Since inteins and group I introns have been found as insertions in the same highly conserved DNA targets [33, 34], this process may also apply to the origin of mobile inteins.
Experimental Procedures
Strains
T7-like phages [24] ΦI, ΦIIP, ΦIIW, H, W31, T7 and T3L were from W. F. Studier and A1122, C21R and ViIII were from I. Molineux. DNA from phages A1122 and ViIII, which infect Y. pestis and Citrobacter spp. Ci23, respectively, was obtained directly from the donated samples. All other phages were propagated on E. coli BE.
Synthetic Oligonucleotides
Sequences are shown in Supplemental Methods.
Preparation of DNA substrates
Synthetic ΦI and W31 intronless DNAs were made with the complementary oligonucleotide pairs, ΦItop and ΦIbot or W31top and W31bot, respectively. Six pmol of [32P] 5′-end labeled oligonucleotide was annealed to 60 pmol unlabeled complementary oligonucleotide in 50 mM Tris-HCl (pH 7.5) and 50 mM NaCl by heating to 100°C for 5 min and slow cooling to room temperature. PCR amplified substrates were prepared from phage DNA templates with primers ΦI5.0 and 5′-end labeled T75.7a, so only the bottom strand of the DNA is labeled, using 6 pmol each of 5′-end labeled and unlabeled primers.
In vitro synthesized protein
The ΦI and W31 intron-encoded ORFs were amplified from phage lysates by PCR with primers ΦI/W31u and ΦI/W31d and subsequently transcribed and translated in vitro using rabbit reticulocyte lysate [3].
Preparation of extracts from phage infected cells
E. coli BE was grown in LB medium at 37°C to an OD590 0.3 – 0.35. Cultures were shifted to 30°C and infected with the appropriate phage at an M.O.I of 4. At various times after infection, 15 mL aliquots were centrifuged at 4300 × g for 10 min at 4°C, washed with 2 ml 0.1M HSB (0.1M NaCl, 20mM Tris pH 8.0, 0.1mM EDTA, 10% glycerol), resuspended in 0.5 ml 0.1M HSB, frozen at −70°C and thawed. Each cell suspension was mixed with 1 ml lysis buffer (0.7M NaCl, 20mM Tris pH 8.0, 0.1mM EDTA, 55% glycerol), and disrupted by sonication with three bursts of 10 s. Cell debris was removed by centrifugation at 13,600 × g for 2 min at 4°C and the extracts were stored at −20°C.
Endonuclease assays
Proteins were assayed in a final volume of 20 μL with ~105 cpm 5′-end labeled DNA in (50 mM NaCl, 50 mM Tris pH 7.5, 10 mM MgCl2, 0.5 μg Poly dI-dC). In vitro synthesis reactions (2 μL) were incubated for 30 min, while infected cell extracts (1 μL) were incubated for 15 min at 30°C. Reaction products were phenol extracted and ethanol precipitated before electrophoretic separation on 6% denaturing polyacrylamide gels.
Supplementary Material
Acknowledgments
We thank Caren Stark and Deborah Hinton for critical reading of the manuscript. This work was supported by the Northeast Biodefense Center through NIH grant U54-AI057158, and by a research development award from the College of Arts and Sciences, University at Albany.
Abbreviations
- IIS
intron insertion site
- ORF
open reading frame
- PCR
polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Belfort M, Derbyshire V, Parker MM, Cousineau B, Lambowitz AM. Mobile Introns: Pathways and Proteins. In: Craig N, editor. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 2.Biniszkiewicz D, Cesnaviciene E, Shub DA. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. Embo J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonocora RP, Shub DA. A novel group I intron-encoded endonuclease specific for the anticodon region of tRNA(fMet) genes. Mol Microbiol. 2001;39:1299–1306. doi: 10.1111/j.1365-2958.2001.02318.x. [DOI] [PubMed] [Google Scholar]
- 4.Orlowski J, Boniecki M, Bujnicki JM. I-Ssp6803I: the first homing endonuclease from the PD-(D/E)XK superfamily exhibits an unusual mode of DNA recognition. Bioinformatics. 2007;23:527–530. doi: 10.1093/bioinformatics/btm007. [DOI] [PubMed] [Google Scholar]
- 5.Zhao L, Bonocora RP, Shub DA, Stoddard BL. The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif. EMBO J. 2007;26:2432–2442. doi: 10.1038/sj.emboj.7601672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pajunen MI, Kiljunen SJ, Soderholm ME, Skurnik M. Complete genomic sequence of the lytic bacteriophage phiYeO3-12 of Yersinia enterocolitica serotype O:3. J Bacteriol. 2001;183:1928–1937. doi: 10.1128/JB.183.6.1928-1937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwan T, Liu J, Dubow M, Gros P, Pelletier J. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J Bacteriol. 2006;188:1184–1187. doi: 10.1128/JB.188.3.1184-1187.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shub DA, Goodrich-Blair H, Eddy SR. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 9.Gorbalenya AE. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 1994;3:1117–1120. doi: 10.1002/pro.5560030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonocora RP, Shub DA. A self-splicing group I intron in DNA polymerase genes of T7-like bacteriophages. J Bacteriol. 2004;186:8153–8155. doi: 10.1128/JB.186.23.8153-8155.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholl D, Merril C. The genome of bacteriophage K1F, a T7-like phage that has acquired the ability to replicate on K1 strains of Escherichia coli. J Bacteriol. 2005;187:8499–8503. doi: 10.1128/JB.187.24.8499-8503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia E, Elliott JM, Ramanculov E, Chain PS, Chu MC, Molineux IJ. The genome sequence of Yersinia pestis bacteriophage phiA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J Bacteriol. 2003;185:5248–5262. doi: 10.1128/JB.185.17.5248-5262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambowitz AM, Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 15.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belle A, Landthaler M, Shub DA. Intronless homing: site-specific endonuclease SegF of bacteriophage T4 mediates localized marker exclusion analogous to homing endonucleases of group I introns. Genes Dev. 2002;16:351–362. doi: 10.1101/gad.960302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadyrov FA, Shlyapnikov MG, Kryukov VM. A phage T4 site-specific endonuclease, SegE, is responsible for a non-reciprocal genetic exchange between T-even-related phages. FEBS Lett. 1997;415:75–80. doi: 10.1016/s0014-5793(97)01098-3. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Belle A, Shub DA, Belfort M, Edgell DR. SegG Endonuclease Promotes Marker Exclusion and Mediates Co-conversion from a Distant Cleavage Site. J Mol Biol. 2003;334:13–23. doi: 10.1016/j.jmb.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 19.Sharma M, Ellis RL, Hinton DM. Identification of a family of bacteriophage T4 genes encoding proteins similar to those present in group I introns of fungi and phage. Proc Natl Acad Sci U S A. 1992;89:6658–6662. doi: 10.1073/pnas.89.14.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shcherbakov V, Granovsky I, Plugina L, Shcherbakova T, Sizova S, Pyatkov K, Shlyapnikov M, Shubina O. Focused genetic recombination of bacteriophage t4 initiated by double-strand breaks. Genetics. 2002;162:543–556. doi: 10.1093/genetics/162.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brok-Volchanskaya VS, Kadyrov FA, Sivogrivov DE, Kolosov PM, Sokolov AS, Shlyapnikov MG, Kryukov VM, Granovsky IE. Phage T4 SegB protein is a homing endonuclease required for the preferred inheritance of T4 tRNA gene region occurring in co-infection with a related phage. Nucleic Acids Res. 2008;36:2094–2105. doi: 10.1093/nar/gkn053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandegren L, Nord D, Sjoberg BM. SegH and Hef: two novel homing endonucleases whose genes replace the mobC and mobE genes in several T4-related phages. Nucleic Acids Res. 2005;33:6203–6213. doi: 10.1093/nar/gki932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hausmann R. T7 Group. In: Calendar R, editor. Bacteriophages. New York: Plenum Press; 1988. [Google Scholar]
- 25.Belfort M, Reaban ME, Coetzee T, Dalgaard JZ. Prokaryotic introns and inteins: a panoply of form and function. J Bacteriol. 1995;177:3897–3903. doi: 10.1128/jb.177.14.3897-3903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambowitz AM. Infectious introns. Cell. 1989;56:323–326. doi: 10.1016/0092-8674(89)90232-8. [DOI] [PubMed] [Google Scholar]
- 27.Belfort M. Bacteriophage introns: parasites within parasites? Trends Genet. 1989;5:209–213. doi: 10.1016/0168-9525(89)90083-8. [DOI] [PubMed] [Google Scholar]
- 28.Mota EM, Collins RA. Independent evolution of structural and coding regions in a Neurospora mitochondrial intron. Nature. 1988;332:654–656. doi: 10.1038/332654a0. [DOI] [PubMed] [Google Scholar]
- 29.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci U S A. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loizos N, Tillier ER, Belfort M. Evolution of mobile group I introns: recognition of intron sequences by an intron-encoded endonuclease. Proc Natl Acad Sci U S A. 1994;91:11983–11987. doi: 10.1073/pnas.91.25.11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgell DR, Shub DA. Related homing endonucleases I-BmoI and I-TevI use different strategies to cleave homologous recognition sites. Proc Natl Acad Sci U S A. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landthaler M, Begley U, Lau NC, Shub DA. Two self-splicing group I introns in the ribonucleotide reductase large subunit gene of Staphylococcus aureus phage Twort. Nucleic Acids Res. 2002;30:1935–1943. doi: 10.1093/nar/30.9.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgell DR, Belfort M, Shub DA. Barriers to intron promiscuity in bacteria. J Bacteriol. 2000;182:5281–5289. doi: 10.1128/jb.182.19.5281-5289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derbyshire V, Belfort M. Lightning strikes twice: intron-intein coincidence. Proc Natl Acad Sci U S A. 1998;95:1356–1357. doi: 10.1073/pnas.95.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pietrokovski S. Intein spread and extinction in evolution. Trends Genet. 2001;17:465–472. doi: 10.1016/s0168-9525(01)02365-4. [DOI] [PubMed] [Google Scholar]
- 36.Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. Inteins: structure, function, and evolution. Annu Rev Microbiol. 2002;56:263–287. doi: 10.1146/annurev.micro.56.012302.160741. [DOI] [PubMed] [Google Scholar]
- 37.Goodwin TJ, Butler MI, Poulter RT. Multiple, non-allelic, intein-coding sequences in eukaryotic RNA polymerase genes. BMC Biol. 2006;4:38. doi: 10.1186/1741-7007-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roman J, Woodson SA. Reverse splicing of the Tetrahymena IVS: evidence for multiple reaction sites in the 23S rRNA. Rna. 1995;1:478–490. [PMC free article] [PubMed] [Google Scholar]
- 39.Roman J, Woodson SA. Integration of the Tetrahymena group I intron into bacterial rRNA by reverse splicing in vivo. Proc Natl Acad Sci U S A. 1998;95:2134–2139. doi: 10.1073/pnas.95.5.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studier FW. The genetics and physiology of bacteriophage T7. Virology. 1969;39:562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- 41.Petrov VM, Nolan JM, Bertrand C, Levy D, Desplats C, Krisch HM, Karam JD. Plasticity of the gene functions for DNA replication in the T4-like phages. J Mol Biol. 2006;361:46–68. doi: 10.1016/j.jmb.2006.05.071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


