Abstract
Purpose
Detection of micrometastases in sentinel lymph nodes (SLNs) is important for accurate staging and prognosis in melanoma patients. However, a significant number of patients with histopathology-negative SLNs subsequently develop recurrent disease. We hypothesized that a quantitative realtime reverse transcriptase polymerase chain reaction (qRT) assay using multiple specific mRNA markers could detect occult metastasis in paraffin-embedded (PE) SLNs to upstage and predict disease outcome.
Patients and Methods
qRT was performed on retrospectively collected PE SLNs from 215 clinically node-negative patients who underwent lymphatic mapping and sentinel lymphadenectomy for melanoma and were followed up for at least 8 years. PE SLNs (n = 308) from these patients were sectioned and assessed by qRT for mRNA of four melanoma-associated genes: MART-1 (antigen recognized by T cells-1), MAGE-A3 (melanoma antigen gene-A3 family), GalNAc-T (β1→4-N-acetylgalactosaminyl-transferase), and Pax3 (paired-box homeotic gene transcription factor 3).
Results
Fifty-three (25%) patients had histopathology-positive SLNs by hemotoxylin and eosin and/or immunohistochemistry. Of the 162 patients with histopathology-negative SLNs, 48 (30%) had nodes that expressed at least one of the four qRT markers, and these 48 patients also had a significantly increased risk of disease recurrence by a Cox proportional hazards model analysis (P < .0001; risk ratio, 7.48; 95% CI, 3.70 to 15.15). The presence of ≥ one marker in histopathology-negative SLNs was also a significant independent prognostic factor by multivariate analysis for overall survival (P = .0002; risk ratio, 11.42; 95% CI, 3.17 to 41.1).
Conclusion
Molecular upstaging of PE histopathology-negative SLNs by multiple-marker qRT assay is a significant independent prognostic factor for long-term disease recurrence and overall survival of patients with early-stage melanoma.
INTRODUCTION
With an incidence that is one of the highest among cancers diagnosed in the United States, malignant melanoma continues to be a major health problem.1,2 The 5-year survival rate approaches 90% for American Joint Committee on Cancer (AJCC) stage I malignant melanoma and 70% for AJCC stage II melanoma, but decreases significantly to 25% to 50% for AJCC stage III melanoma, depending on the number of nodes involved.3 Because identification of regional lymph node metastases is one of the major prognostic factors for tumor recurrence and survival,4 accurate staging is highly important for optimal management of early-stage disease.
Historically, complete dissection of regional lymph nodes was the standard treatment for patients diagnosed with primary melanoma.5,6 However, most patients with early-stage disease have lymph nodes that are tumor-free by routine histopathology. Therefore, we developed sentinel lymphadenectomy (SLND), a less-invasive method to assess a tumor-draining lymph node basin.7–9 Because the sentinel lymph node (SLN) represents the first lymph node in the regional lymphatic basin to receive drainage from the primary tumor it is the initial site of early nodal metastases. We have shown that identification of a tumor-negative SLN accurately predicts the absence of tumor metastases in other regional lymph nodes (non-SLNs) of the lymphatic basin that drains a primary tumor.8,9 Lymphatic mapping with SLND has dramatically changed the surgical approach to early-stage melanoma.7 This procedure allows a more focused and efficient pathologic analysis of micrometastatic disease. The addition of serial sectioning and immunohistochemical (IHC) analysis of lymph nodes with HMB-45 and anti-S-100 antibodies has further improved occult tumor cell detection as compared to hematoxylin and eosin (H&E) staining alone.7,10,11 Nevertheless, 20% to 30% of patients with tumor-negative lymph nodes will develop recurrent disease within 10 years.
To date, we and others have utilized several defined melanoma-associated markers in multiple-marker (MM) reverse transcriptase polymerase chain reaction (RT-PCR) assays that were developed to detect occult metastatic melanoma cells in blood and lymphoid tissues.11–17 Multiple specific markers can compensate for a tumor’s heterogeneous gene expression and variable mRNA levels, thereby improving assay sensitivity.11–15 The rationale of using multiple-markers also addresses problems of tumor heterogeneity among patients has been reviewed previously.11,17 Although the clinical significance of nodal metastases detected by RT-PCR cannot be established without long-term follow-up, multiple-marker RT-PCR analysis can be more sensitive and predictive than conventional pathology staining for the detection of cancer cells.11,13 The use of single-marker RT-PCR assays for detection of metastatic disease has significant problems in detection. In a pilot study of SLN frozen sections, we demonstrated that the tumor status of histopathology-negative SLNs could be significantly upstaged by a gel-based RT-PCR assay based on multiple mRNA markers.11
Sampling approach of lymph nodes for molecular staging is an important issue. The employment of arbitrary node dissection for RT-PCR and pathology analysis has its limitation for detection of occult disease.18 Bivalving and use of serially sectioned frozen SLNs for RT-PCR assessment improves the procedure; however, this is time-consuming and logistically not practical for routine histopathology analysis. Moreover, electrophoresis-based RT-PCR assays are generally subjective, with limited quantification and sensitivity.11 The development of a quantitative realtime RT-PCR (qRT) assay for detection of occult tumor cells in paraffin-embedded (PE) SLNs would be a major logistic improvement.19–21 We developed an MM qRT assay for analysis of PE SLNs. In this study, we assessed its clinical utility by examining PE specimens from melanoma patients with long-term follow-up.
In this report, four melanoma tumor mRNA markers from non-overlapping biologic pathways were utilized for detection of tumor cells. MART-1 (antigen recognized by T cells-1) is a major melanocyte differentiation antigen and MAGE-A3 (melanoma antigen gene-A3 family) is a tumor antigen; both are frequently expressed in melanomas.14,16,20,22,23 MAGE-A3 is not found in normal tissues except male germline cells and placenta.24 GalNAc-T (β1→4-N-acetylgalactosaminyl-transferase) is a key glycosyltransferase enzyme in the biosynthetic pathway for the cell-surface tumor-associated gangliosides GM2 and GD2, which are found at elevated levels during melanoma progression.25–27 GM2 and GD2 are not found in melanocytes or nevi. We have previously described the high incidence of GalNAc-T mRNA expression in melanoma cells and its potential as a molecular marker for melanoma progression.27 Pax3 (paired-box homeotic gene transcription factor 3) is involved in the regulation of melanin synthesis and cellular physiologic functions, such as migration and antiapoptosis.28,29 Pax3 is well-expressed in early-stage melanomas and advanced metastatic melanomas; it is not detected in normal skin melanocytes or benign nevi by RT-PCR and in situ hybridization.28
This report represents the largest study with the longest follow-up to date for molecular staging of PE histopathology-negative SLNs from melanoma patients. It shows that molecular upstaging of PE SLN is a significant independent prognostic factor for disease-free survival (DFS) and overall survival (OS) in patients with early-stage melanoma.
PATIENTS AND METHODS
Patients and Tumors
PE SLN tissues from surgeries at John Wayne Cancer Institute (JWCI; Santa Monica, CA) from 1992 to 1996 were obtained in consultation with surgeons and pathologists at the JWCI. All eligible patients (n = 281) who underwent SLND for primary cutaneous melanoma between 1992 and 1996 were initially identified and then sequentially selected based on long-term follow-up, available PE SLN blocks, informed consent, and IHC analysis. Informed human subjects Saint John’s Health Center/JWCI institutional review board–approved consent was obtained for all patient specimens. All patients were diagnosed with early-stage clinically node-negative malignant melanoma and underwent preoperative lymphoscintigraphy to identify the tumor-draining lymph node basin(s). SLND was performed after intraoperative lymphatic mapping of the SLNs with a combination of isosulfan blue dye (Lymphazurin; Hirsch Industries Inc, Richmond, VA) and radioisotope (99mtechnetium sulfur colloid).7–9 A final total of 226 melanoma patients were selected by the melanoma database management personnel, independently of investigators and biostatisticians. These patients used for the study were selected based on the above defined criteria by database management.
All SLNs were stained with H&E, and IHC was performed using HMB-45 and anti-S-100 antibodies.9,11 Eleven patients were excluded from the study because PE SLNs RNA was undetectable (poor quality) for qRT analysis. Patients with tumor-positive SLNs by H&E and/or IHC underwent complete lymph node dissection of the involved lymphatic basin. All patients were followed up with standard clinical and diagnostic examinations. Patients who had melanoma recurrence after SLND received elective surgery, immunotherapy, and/or biochemotherapy. The JWCI melanoma computer database with patients’ follow-up and history was independently provided to the biostatisticians (R.E. and D.E.) for analysis.
Histopathologic Examination and RNA Isolation
Personnel performing the MM qRT assay or pathology analysis did not know the clinicopathology parameters of the patients. All SLNs were prepared for histopathologic examination with H&E and IHC using antibodies to HMB-45 and S-100 as previously described.9,11 If melanoma cells were identified in the frozen section, a complete lymph node dissection was then performed.11 Six immediately adjacent frozen sections were cut on the cryostat to a thickness of 12 μm each and stored at −80°C until processed at a later date.11 The remainder of the bisected node was placed in 10% formalin and embedded in paraffin. Paraffin sections 4 μm-thick sections were cut and examined with H&E staining; adjacent 4 μm sections were evaluated by IHC. This PE section evaluation was performed at two different levels separated by approximately 40 μm.
For the MM qRT assay, 10 additional 10 μm thick sections were cut from each PE SLNs with a sterile microtome blade for each tissue block, deparaffinized, and digested with proteinase K before RNA extraction using a modified protocol of the Paraffin Block RNA Isolation Kit (Ambion, Austin, TX) as previously described.13 Pellet Paint (Novagen, Madison, WI) was also used in the RNA precipitation procedure. The RNA was quantified by ultraviolet spectrophotometry and the RiboGreen detection assay (Molecular Probes, Eugene, OR). Total cellular RNA from specimens was extracted, isolated, and purified as previously described.11,14,20 If the quality of the RNA was poor, specimens were not used in the study. Established melanoma cell lines and PE melanoma tumors were used as positive controls for the MM qRT assay. To verify the sensitivity of detecting occult metastases, we assessed total RNA from ~1,000 melanoma cells microdissected from PE metastatic melanomas using the PixCell II Laser Capture Microdissection System (LCM; Arcturus Engineering, Mountain View, CA). Total RNA isolated from 1,000 melanoma cells was serially diluted and qRT was performed for each mRNA marker.
A receiver operating characteristic (ROC) curve30 for each mRNA marker was established using an independent set of 32 PE H&E-verified metastatic melanomas and 39 PE histopathology cancer-negative lymphoid tissues obtained from breast cancer patients and patients who underwent surgery for benign disease (ie, tonsillitis).
Primers and Probes
Primer and probe sequences were designed for qRT assay as previously described.20 Fluorescence resonance energy transfer probe sequences were designed to enhance the specificity of the assay. The probes used were: MART-1, 5′-FAM-CAGAACAGTCACCACCACCTTATT-BHQ-1-3′; MAGE-A3, 5′-FAM-AGCTCCTGCCCACACTCCCGCCTGT-BHQ-1-3′; GalNAc-T, 5′-CALRED-ATGAGGCTGCTTTCACTATCCGCA-BHQ-2-3′; Pax3, 5′-FAM-CAGACTGATTACGCGCTCTCCC-BHQ-1-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-FAM-CAGCAATGCCTCCTGCACCACCAA-BHQ-1-3′. PE melanomas, histopathology-verified tumor negative PE lymph nodes, and melanoma lines were used as controls to optimize the assay. Expression of housekeeping-gene GAPDH served as an internal reference for mRNA integrity.
MM qRT Assay
Moloney murine leukemia virus reverse-transcriptase was used for first strand cDNA synthesis with both oligo-dT and random hexamers.15,17 qRT assays were performed as previous described;20 each PCR reaction mixture consisted of cDNA from 250 ng of total RNA. Samples were amplified with a precycling hold at 95°C for 10 minutes, followed by 45 cycles of denaturation at 95°C for 1 minute, annealing at 55°C for 1 minute for GAPDH, at 58°C for MAGE-A3 (40 cycles), at 62°C for MART-1, at 62°C for GalNAc-T (42 cycles), and at 65°C for Pax3, and extension at 72°C for 1 minute using the iCycler iQ RealTime PCR Detection System (Bio-Rad Laboratories, Hercules, CA). For ROC curve analysis, all reactions were performed under the same conditions at 45 cycles. The specificity of PCR conditions and annealing temperatures for each marker were optimized and assessed by ROC curve analysis.
All assays were incorporated with standard curves, positive controls, and negative controls. Positive controls (melanoma lines, PE metastatic melanomas), negative controls (normal or tumor-free lymph nodes), and reagent controls (reagents without RNA or cDNA) for qRT were included in each assay. Each assay was performed in triplicate for verification, and the mean copy number was used for analysis. In patients who had two or more SLNs, the highest copy number was used. The standard curve for quantifying mRNA copy number was constructed using templates with known numbers of the cloned MAA cDNA template.20 Standard curves for each marker were generated by using the threshold cycle of dilutions of known number templates. Mock cDNA of individual genes were initially used to optimize the assay and determine potential contamination. The mRNA copy number was calculated using RealTime Detection System Software. Pre- and post-PCR were carried out in separate rooms with designated instruments for the study. Stringent standard operation procedures were used to maintain contamination-free reactions.11 Any reaction that was contaminated or suspected was repeated once or twice. Random repeat analysis were employed with individual markers and SLNs to verify results.
Statistical Analysis
To assess the association between mRNA marker(s) and clinicopathologic parameters, t test was used for continuous variables and χ2 test was used for categoric variables. Kappa analysis and Spearman correlation coefficient analysis were used to compare the copy numbers of each mRNA marker. The cumulative DFS and OS rates for patient groups were calculated using Kaplan-Meier methods and compared with the log-rank test. Cox proportional hazards (Cox-PH) models were used for multivariate analysis of the following standard prognostic variables: Breslow thickness, Clark level, primary tumor site, sex, and age. Backwards step-wise model selection procedures were employed to find the optimal set of predictors for DFS and OS. All P values were assessed as two-sided and were significant at ≤.05.
RESULTS
MM qRT Sensitivity and Specificity
MART-1, MAGE-A3, GalNAc-T, and Pax3 mRNA expression was measured by qRT assay in 10 melanoma cell lines, and in frozen and PE melanoma tumors for optimization. All four markers were expressed in every melanoma cell line. ROC curve analysis was performed to define the potential accuracy of the MM qRT assay for the detection of metastatic melanoma in PE specimens. Marker mRNA copy levels were measured in 32 PE histopathology-verified metastatic melanomas and 39 PE histopathology-defined cancer-negative lymph nodes. The ROC value (W is area under the ROC curve ± SE, sensitivity and specificity at 95% CI) of each marker is as follows: MART-1, 0.906 ± 0.039, 0.813, 1.0; MAGE-A3, 0.903 ± 0.039, 0.781, 1.0; GalNAc-T, 0.813 ± 0.053, 0.594, 1.0; and Pax3, 0.968 ± 0.023, 0.969, 0.923, respectively (Fig 1). Markers were not detected under the assay’s optimal conditions in the 39 normal PE lymph nodes. ROC analysis demonstrated no false-positives for all markers, verifying their specificity. MAGE-A3, GalNAc-T, and Pax3 mRNAs were not detected in nevi.
Fig 1.
Receiver operator characteristic (ROC) curve analysis of each mRNA marker. The area under the ROC curve (W) of each marker is as follows: MART-1, 0.906; MAGE-A3, 0.903; GalNAc-T, 0.813; and Pax3, 0.968.
To validate the sensitivity of the qRT assay, marker mRNA copy levels in PE specimens were compared to the copy levels in parallel frozen sections from 10 histopathology-positive SLNs and 21 histopathology-negative SLNs (Table 1). Positivity of each marker in PE SLN sections coincided with positivity in corresponding frozen SLN sections. The Spearman correlation coefficient analysis was significant (P < .0001) for all markers, and κ analysis revealed significant correlation (P < .003) between each marker’s mRNA copy levels in parallel PE and frozen SLN sections. These studies demonstrated the relative assay efficiency in assessing PE sections as compared to the gold standard of paired frozen sections. The sensitivity of mRNA marker detection by the qRT assay was verified by serial dilution analysis of total RNA from 1,000 melanoma cells microdissected by LCM from PE metastatic melanoma tumors. In multiple samples, MART-1, MAGE-A3, GalNAc-T, and Pax3 mRNA copy levels were detectable in 1/100 diluted total RNA, which is approximately equivalent to total RNA from less than 10 melanoma cells.
Table 1.
Consistency of Marker mRNA Expression Between PE and Frozen SLNs
| Consistency of Results |
κ Analysis |
Spearman Correlation Coefficient |
|||||
|---|---|---|---|---|---|---|---|
| Marker | No. of Patients | % | Coefficient | 95% CI | P | Coefficient | P |
| MART-1 | 30/31 | 97 | 0.912 | 0.744 to 1.081 | <.0001 | 0.963 | <.0001 |
| MAGE-A3 | 27/31 | 87 | 0.597 | 0.260 to 0.934 | .0022 | 0.868 | <.0001 |
| GalNAc-T | 27/31 | 87 | 0.688 | 0.409 to 0.968 | .0003 | 0.835 | <.0001 |
| Pax3 | 30/31 | 97 | 0.912 | 0.744 to 1.081 | <.0001 | 0.970 | <.0001 |
Abbreviations: PE, paraffin-embedded; SLN, sentinel lymph node.
MM qRT Assay of PE SLNs
MART-1, GalNAc-T, Pax3, and MAGE-A3 mRNA copy levels were assessed using the MM qRT assay on 308 SLNs from 215 patients (Table 2). The patients ranged in age from 18.7 to 91.3 years (mean age, 50.9 ± 15.9 standard deviation); there were 127 males and 88 females. Of the 53 patients with SLN histopathology-proven metastatic melanoma cells (46 detected by H&E and IHC; seven by IHC alone), MART-1, GalNAc-T, Pax3, and MAGE-A3 mRNA markers were detected in 42 (79%), 37 (70%), 39 (74%), and 25 (47%) patients, respectively (Table 2). Among the histopathology-positive SLNs, 51 (96%) of 53 had one or more markers, and 20 (38%) demonstrated all four markers. MART-1, GalNAc-T, Pax3, and MAGE-A3 markers were detected in 162 patients with histopathology-negative (H&E/IHC) SLNs as follows: 10 patients (6%), 27 patients (17%), 28 patients (17%), and eight patients (5%), respectively. Forty-eight (30%) of 162 histopathology-negative SLNs had one or more markers, and 19 (12%) had two or more markers (Table 2). Detection of individual markers was significantly (P < .0001) higher in histopathology-positive SLNs than in histopathology-negative SLNs.
Table 2.
Correlation of Risk Factors With SLN Histopathology
| H&E/IHC-Positive SLN (n = 53) |
H&E/IHC-Negative SLN (n = 162) |
||||||
|---|---|---|---|---|---|---|---|
| Risk Factors | No. of Patients | % | No. of Patients | % | P | ||
| Age, years | |||||||
| Mean | 46.5 | 52.3 | .02 | ||||
| SD | 14.9 | 16.0 | |||||
| Sex | |||||||
| Male (n = 127) | 30 | 57 | 97 | 60 | |||
| Female (n = 88) | 23 | 43 | 65 | 40 | .80 | ||
| Primary site | |||||||
| Head/neck | 5 | 9 | 26 | 16 | |||
| Trunk | 20 | 38 | 66 | 41 | |||
| Extremity | 28 | 53 | 70 | 43 | .35 | ||
| Breslow thickness, mm | |||||||
| ≤ 1.00 | 6 | 11 | 58 | 36 | .001 | ||
| 1.01–2.00 | 20 | 38 | 65 | 40 | |||
| 2.01–4.00 | 19 | 36 | 26 | 16 | |||
| > 4.00 | 7 | 13 | 13 | 8 | |||
| Unknown | 1 | 2 | 0 | 0 | |||
| Mean | 2.9 | 1.9 | .003 | ||||
| SD | 2.6 | 1.8 | |||||
| Clark level | |||||||
| I | 0 | 0 | 2 | 1 | .03 | ||
| II | 4 | 8 | 11 | 7 | |||
| III | 10 | 19 | 65 | 40 | |||
| IV | 35 | 66 | 72 | 44 | |||
| V | 3 | 6 | 8 | 5 | |||
| Unknown | 1 | 2 | 4 | 2 | |||
| MART-1 mRNA | |||||||
| Positive | 42 | 79 | 10 | 6 | |||
| Negative | 11 | 21 | 152 | 94 | < .0001 | ||
| GalNAc-T mRNA | |||||||
| Positive | 37 | 70 | 27 | 17 | |||
| Negative | 16 | 30 | 135 | 83 | < .0001 | ||
| Pax3 mRNA | |||||||
| Positive | 39 | 74 | 28 | 17 | |||
| Negative | 14 | 26 | 134 | 83 | < .0001 | ||
| MAGE-A3 mRNA | |||||||
| Positive | 25 | 47 | 8 | 5 | |||
| Negative | 28 | 53 | 154 | 95 | < .0001 | ||
| mRNA markers | |||||||
| 0 | 2 | 4 | 114 | 70 | |||
| 1 | 9 | 17 | 29 | 18 | |||
| 2 | 12 | 23 | 13 | 8 | |||
| 3 | 10 | 19 | 6 | 4 | |||
| 4 | 20 | 38 | 0 | 0 | < .0001 | ||
Abbreviations: SLN, sentinel lymph node; H&E, hematoxylin and eosin; IHC, immunohistochemistry; SD, standard deviation.
Individual marker mRNA copy number per 250 ng of total RNA in histopathology-positive SLNs varied as expected with size of the metastasis. Mean mRNA copy number for each marker in histopathology-positive SLNs (n = 53) was as follows: MART-1, 24,040 copies; GalNAc-T, 332 copies; Pax3, 701 copies; and MAGE-A3, 114 copies. Mean mRNA copy number for each marker was significantly higher in histopathology-positive SLNs than in histopathology-negative SLNs overall (t test). All SLN specimens were positive for GAPDH, showing high integrity of viable mRNA extracted from the tumor-positive or tumor-negative PE SLNs. Patient age, tumor Breslow thickness, and Clark level were also significantly different between histopathology-positive and -negative SLNs (Table 2).
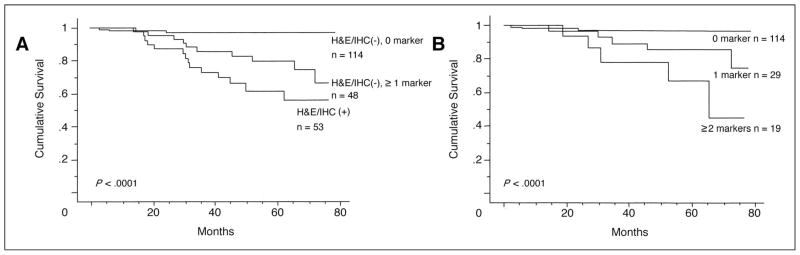
During an overall median follow-up period of 60.4 months with more than 8 years for survivors, 32 (60%) of 53 patients with histopathology-positive SLNs and 39 (24%) of 162 patients with histopathology-negative SLNs developed recurrence. The median DFS was significantly lower for 53 patients with histopathology-positive SLNs than for 162 patients with histopathology-negative SLNs (29 months v not reached; log-rank test P < .0001; Fig 2A). Among 32 recurrent patients with histopathology-positive SLNs, 10 (31%) patients developed locoregional recurrence (two patients with regional lymph node metastasis and eight patients with in-transit skin metastasis), and 22 (69%) patients developed distant metastasis.
Fig 2.
(A) Kaplan-Meier curve analysis of disease-free survival (DFS) for 162 sentinel lymph node (SLN) histopathology-negative patients versus 53 SLN histopathology-positive patients; (B) Kaplan-Meier curve analysis of DFS according to multiple marker quantitative realtime reverse transcriptase polymerase chain reaction status (zero markers v one or more markers) for 162 SLN histopathology-negative patients; (C) Kaplan and Meier curve analysis of DFS according to number of molecular markers in 162 SLN histopathology-negative patients.
Among the 114 patients who had neither histopathologic nor molecular marker evidence of SLN metastasis, only 12 (11%) developed recurrent disease (Table 3). In contrast, disease recurrence was diagnosed in 27 (56%) of 48 patients whose histopathology-negative SLNs expressed at least one marker. Fourteen of the 27 patients (52%) whose histopathology-negative SLNs expressed one or more markers had local recurrence (eight patients with regional lymph nodes metastasis and six patients with intransit skin metastasis) and 13 patients (48%) had distant metastasis as an initial recurrence site. Median DFS was significantly shorter when histopathology-negative SLNs expressed one or more markers (54 months v not reached; P < .0001; Fig 2B). An increasing number of markers detected in histopathology-negative SLN was associated with a progressive decrease in DFS (Fig 2C); the number of recurrences was 12 (41%) of 29 patients whose SLNs expressed one marker, nine (69%) of 13 patients with two markers, and six (100%) of six patients with three markers.
Table 3.
Correlation of Risk Factors With Disease Recurrence in Patients With H&E/IHC-Negative SLN
| No Recurrence (n = 123) |
Recurrence (n = 39) |
||||||
|---|---|---|---|---|---|---|---|
| Risk Factors | No. of Patients | % | No. of Patients | % | P | ||
| Age, years | .58 | ||||||
| Mean | 51.9 | 53.5 | |||||
| SD | 16.3 | 15.0 | |||||
| Sex | .53 | ||||||
| Male (n = 97) | 72 | 59 | 25 | 64 | |||
| Female (n = 65) | 51 | 41 | 14 | 36 | |||
| Primary site | .011 | ||||||
| Head/neck | 15 | 12 | 11 | 28 | |||
| Trunk | 57 | 46 | 9 | 33 | |||
| Extremity | 51 | 41 | 19 | 49 | |||
| Breslow thickness, mm | .025 | ||||||
| ≤ 1.00 | 48 | 39 | 10 | 26 | |||
| 1.01–2.00 | 52 | 42 | 13 | 33 | |||
| 2.01–4.00 | 14 | 11 | 12 | 31 | |||
| > 4.00 | 9 | 7 | 4 | 10 | |||
| Mean | 1.7 | 2.4 | .034 | ||||
| SD | 1.6 | 2.0 | |||||
| Clark level | .41 | ||||||
| I | 2 | 2 | 0 | 0 | |||
| II | 7 | 6 | 4 | 10 | |||
| III | 54 | 44 | 11 | 28 | |||
| IV | 52 | 42 | 20 | 51 | |||
| V | 6 | 5 | 2 | 5 | |||
| Unknown | 2 | 2 | 2 | 5 | |||
| MART-1 mRNA | .0005 | ||||||
| Positive | 3 | 2 | 7 | 18 | |||
| Negative | 120 | 98 | 32 | 82 | |||
| GalNAc-T mRNA | < .0001 | ||||||
| Positive | 8 | 7 | 19 | 49 | |||
| Negative | 115 | 93 | 20 | 51 | |||
| Pax3 mRNA | .00006 | ||||||
| Positive | 13 | 11 | 15 | 38 | |||
| Negative | 110 | 89 | 24 | 62 | |||
| MAGE-A3 mRNA | .00002 | ||||||
| Positive | 1 | 1 | 7 | 18 | |||
| Negative | 122 | 99 | 32 | 82 | |||
| MRNA markers | < .0001 | ||||||
| 0 | 102 | 83 | 12 | 31 | |||
| 1 | 17 | 14 | 12 | 31 | |||
| 2 | 4 | 3 | 9 | 23 | |||
| 3 | 0 | 0 | 6 | 15 | |||
| 4 | 0 | 0 | 0 | 0 | |||
Abbreviations: H&E, hematoxylin and eosin; IHC, immunohistochemistry; SLN, sentinel lymph node; SD, standard deviation.
A Cox-PH model was used to evaluate the correlation of standard prognostic factors, molecular markers, and rate of recurrence for histopathology-negative SLN patients. Backwards step-wise model selection removed all factors from the prognostic models except for Breslow thickness and the molecular markers. Analysis of each of the four markers gave a relative risk (RR) of 2.09 to 5.81. An increasing number of markers expressed was associated with a decreased DFS (RR, 3.26; 95% CI, 2.40 to 4.43; P < .0001; Table 4). A comparison of zero versus one or more positive markers yielded a RR of 7.48 (95% CI, 3.70 to 15.15; P < .0001; Table 4).
Table 4.
Cox Proportional Hazard Models for Disease-Free Survival in Patients With H&E/IHC-Negative SLN
| Coefficient | Risk Ratio | 95% CI | P | |
|---|---|---|---|---|
| Comarison of (+) markers (0/1/2/3) | ||||
| Molecular positivity | 1.183 | 3.26 | 2.40 to 4.43 | < .0001 |
| Breslow thickness | 0.188 | 1.21 | 1.05 to 1.39 | .0076 |
| Comparison of zero v one or more (+) markers | ||||
| Molecular positivity | 2.013 | 7.48 | 3.70 to 15.15 | < .0001 |
| Breslow thickness | 0.137 | 1.15 | 1.00 to 1.32 | .053 |
Abbreviations: H&E, hematoxylin & eosin; IHC, immunohistochemistry; SLN, sentinel lymph node.
When MM qRT results were correlated with OS using a Cox-PH model, the OS was significantly (P <.0001) lower for patients with histopathology-positive SLNs than for patients with histopathology-negative SLNs (Fig 3A). Among the 162 patients with histopathology-negative SLNs, the survival rate was significantly lower for patients with one or more markers than for patients with no markers (P <.0001; Fig 3A). Furthermore, an increasing number of markers expressed was associated with a decreased OS on Cox multivariate analysis (RR, 3.61; 95% CI, 2.21 to 5.90; P < .0001; Table 5). Comparing zero versus one or more positive markers yielded a RR of 11.42 (95% CI, 3.17 to 41.1; P = .0002; Table 5). No other factors were retained in the Cox-PH models except Breslow thickness. Kaplan-Meier curves for OS according to number of positive markers are shown in Figure 3B.
Fig 3.
(A) Kaplan-Meier curve analysis of overall survival (OS) according to multiple marker quantitative realtime reverse transcriptase polymerase chain reaction and histopathology status in 215 patients; (B) Kaplan-Meier curve analysis of OS according to number of molecular markers in 162 sentinel lymph node histopathology-negative patients. H&E, hemotoxylin and eosin; IHC, immunohistochemistry.
Table 5.
Cox Proportional Hazard Models for Overall Survival in Patients With H&E/IHC-Negative SLN
| Coefficient | Risk Ratio | 95% CI | P | |
|---|---|---|---|---|
| Comparison of (+) markers (0/1/2/3) | ||||
| Molecular positivity | 1.284 | 3.61 | 2.21 to 5.90 | < .0001 |
| Breslow thickness | 0.201 | 1.22 | 0.99 to 1.51 | .062 |
| Comparison of zero v one or more (+) markers | ||||
| Molecular positivity | 2.435 | 11.42 | 3.17 to 41.1 | .0002 |
| Breslow thickness | 0.131 | 1.14 | 0.92 to 1.41 | .23 |
Abbreviations: H&E, hematoxylin & eosin; IHC, immunohistochemistry; SLN, sentinel lymph node.
DISCUSSION
In this study, we demonstrated that molecular upstaging of PE histopathology-negative SLNs using MM qRT assay has diagnostic and prognostic utility. Our previous studies reported clinicopathologic utility using frozen sections of SLN and PE SLN from a limited number of melanoma patients.11,13 This is the largest study with the longest follow-up demonstrating the clinical significance of molecular upstaging of PE SLN. Our findings show that the combination of lymphatic mapping, SLND, and molecular pathology provides a highly significant advance in staging melanoma.
Lymph node status remains the single most important prognostic factor in patients with early-stage melanoma.3,4 Although recent studies have focused on the evaluation of primary tumor signatures for disease outcome,31,32 primary melanoma tumor gene expression has not been shown to be predictive of SLN metastasis. Diagnosis of early regional lymph node metastasis is an important factor for planning elective adjuvant therapy. We have shown that molecular analysis of the SLN allows earlier detection of regional node micrometastasis and predicts disease outcome.
Previous studies have demonstrated that tumor markers in melanoma primaries and metastases are heterogeneous in presence and in mRNA copy levels.14,20 An optimal panel of molecular markers was developed based on sensitivity and specificity in PE lymph nodes. These mRNA markers were selected based on their respective proteins from independent biochemical pathways with no direct interaction in their functions. Our qRT assessment of PE SLN was more sensitive and specific than our previous assessment of SLNs study using a different combination of markers in an RT-PCR assay.13 In the previous study, we used markers tyrosinase, MART-1, TRP-1, and TRP-2 related to the melanogenesis pathway. The markers can be expressed in low levels in normal melanocytes and nevi, and therefore have a potential risk of false-positives. In the present study, we selected markers that had functional relation to tumor progression and of independent biochemical pathways from each other. We did not use tyrosinase because of the problematic nature of this marker. Nevi cells can be found in lymph nodes and may be detected by tyrosinase, and therefore can be a source of false-positive results.
There were no false-positive qRT results when normal or inflamed PE lymph nodes were assessed. There were two (4%) of 53 histopathology-positive SLNs that were not detected by any of the markers. One of the possible explanations may be that the total RNA obtained from each of these SLNs was low, and SLNs were diagnosed as H&E-negative/IHC-positive with tumor cell clusters. Alternatively, false-negative qRT results may be due to the lack of markers in the occult metastatic tumor cells or the loss of tumor cells through removal of sections used in histopathology analysis (H&E and IHC). The latter can be a problem in molecular analysis, since a portion of the SLNs must always be used for conventional histopathology analysis. There were histopathology-negative SLNs that expressed just one of the markers alone, indicating the significance of individual markers and heterogeneity of marker expression in metastasis. GalNAc-T and Pax3 are promising molecular markers for detecting occult melanoma cells in combination with MAA markers such as MART-1 and/or MAGE-A3 in lymph nodes. The latter two MAAs are known to be immunogenic in melanoma patients and may potentially influence disease outcome.20 In addition, expression of GalNAc-T and Pax3 can indicate more aggressive metastatic clones.25–29,33 GalNAc-T mRNA is a surrogate marker for GM2 and GD2, which are characteristic of a metastatic melanoma phenotype.25–27 Pax3 is an embryonic transcription factor activated in neuroectoderm cells; it is involved in regulation of many cell functions such as cell cycle, apoptosis, and migration.28,29,33 Both GalNAc-T and Pax3 are activated during fetal tissue development, such as migration of neuroectoderm cells. Reactivation of these genes in melanomas may be critical in the establishment of a successful melanoma metastasis.
MM qRT was positive in 30% of the histopathology-negative SLNs. The number of markers expressed significantly correlated with a worse DFS and OS, demonstrating the prognostic significance associated with MM analysis. Interestingly, the mRNA copy number of each marker in histopathology-negative SLNs was significantly higher for patients who recurred; only 12 patients with no histopathologic or molecular evidence of metastasis developed disease recurrence. In these patients, metastasis may have occurred exclusively via a hematogenous route15,17; or systemic and regional metastasis may have occurred, while metastatic cells in the SLN were eliminated by host factors such as immune responses.20,34 Nevertheless, our study established that patients whose histopathology-negative SLNs express MMs are at higher risk for recurrence. Higher number of metastatic tumor cells and/or specific gene expression profile in aggressive or established metastatic tumor cells may explain why patients with a greater number of markers have an increased risk for recurrence. The frequency of local recurrence was higher in patients whose histopathology-negative SLNs expressed one or more markers than in patients with histopathology-positive SLNs. At our institute, patients with histopathology-negative SLN receive no further surgical treatment after SLND, whereas patients with histopathology-positive SLNs undergo complete lymph node dissection (CLND). These results suggest that CLND after SLND may prevent local recurrences in a subset of patients with histopathology-negative but PCR-positive SLNs. The therapeutic value of SLND and CLND is currently under phase III evaluation in a randomized international multicenter selective lymphadenectomy trial, which has completed patient accrual.9
In summary, the MM qRT assay can detect occult metastatic melanoma cells in PE SLN, and thereby upstage early malignant melanoma. The approach we have shown is rapid, reproducible, and quantitatively significant for improved assessment of patients diagnosed with primary cutaneous melanoma. This study provides a baseline for developing a regional node pathology classification for RT-PCR tumor-positive lymph nodes, which could be included in AJCC staging. Uniform use of RT-PCR to stage patients’ lymph nodes will initially require utilization of PE sections for RT-PCR with matched H&E/IHC stained sections. This will help ensure quality analysis as well as address initial logistic sampling problems. Additionally, this approach allows participation of multi-institutions, as well as community hospitals in SLN staging studies.
The application of RT-PCR analysis of SLN in other cancers has been investigated but has not been developed as successfully. In breast cancer SLN analysis using RT-PCR, there are problems in the markers specificity and sensitivity.35 As a result of these technical problems, clinical pathologic utility has not been demonstrated. On the other hand, melanoma markers are better defined and more specific, and the significance of RT-PCR positive SLNs in melanoma patients has demonstrated potential clinical pathologic utility. Our future investigations will prospectively validate the assay in PE SLN obtained from an ongoing international, multicenter, randomized SLND trial. In our study, we have demonstrated that molecular detection of occult metastasis in SLN is an independent predictor of disease outcome and provides a potential stratification factor for managing patients diagnosed with early-stage melanoma.
Acknowledgments
Supported in part by: National Institutes of Health, National Cancer Institute, PO1 CA 20925 Project II; National Institutes of Health, National Cancer Institute, PO1 CA 12528 Project II; and Roy E. Coates Foundation.
Footnotes
Authors’ Disclosures of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Rigel DS, Carucci JA. Malignant melanoma: Prevention, early detection, and treatment in the 21st century. CA Cancer J Clin. 2000;50:215–236. doi: 10.3322/canjclin.50.4.215. [DOI] [PubMed] [Google Scholar]
- 2.Jemal AJ, Murray T, Samuels A, et al. Cancer statistics. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Balch CM, Buzaid AC, Soong SJ, et al. Final version of the American Joint Committee on cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–3648. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 4.Balch CM, Soong SJ, Gershenwald JE, et al. Prognostic factors analysis of 17,600 melanoma patients: Validation of the American Joint Committee on Cancer Melanoma Staging System. J Clin Oncol. 2001;16:3622–3634. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- 5.Morton DL, Wanek L, Nizze JA, et al. Improved long-term survival after lymphadenectomy of melanoma metastatic to regional nodes. Analysis of prognostic factors in 1134 patients from the John Wayne Cancer Clinic. Ann Surg. 1991;214:491–499. doi: 10.1097/00000658-199110000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascinelli N, Morabito A, Santinami M, et al. Immediate or delayed dissection of regional nodes in patients with melanoma of the trunk: A randomized trial. WHO Melanoma Programme. Lancet. 1998;351:793–796. doi: 10.1016/s0140-6736(97)08260-3. [DOI] [PubMed] [Google Scholar]
- 7.Morton DL, Wen DR, Wong JH, et al. Technical details of intraoperative lymphatic mapping for early stage melanoma. Arch Surg. 1992;127:392–399. doi: 10.1001/archsurg.1992.01420040034005. [DOI] [PubMed] [Google Scholar]
- 8.Bostick P, Essner R, Glass E, et al. Comparison of blue dye and probe-assisted intraoperative lymphatic mapping in melanoma to identify sentinel nodes in 100 lymphatic basins. Arch Surg. 1999;134:43–49. doi: 10.1001/archsurg.134.1.43. [DOI] [PubMed] [Google Scholar]
- 9.Morton DL, Thompson JF, Essner R, et al. Validation of the accuracy of intraoperative lymphatic mapping and sentinel lymphadenectomy for early-stage melanoma: Multicenter trial. Multicenter Selective Lymphadenectomy Trial Group. Ann Surg. 1999;230:453–463. doi: 10.1097/00000658-199910000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cochran AJ, Wen DR, Herschman HR. Occult melanoma in lymph nodes detected by antiserum to S-100 protein. Int J Cancer. 1984;34:159–163. doi: 10.1002/ijc.2910340204. [DOI] [PubMed] [Google Scholar]
- 11.Bostick PJ, Morton DL, Turner RR, et al. Prognostic significance of occult metastases detected by sentinel lymphadenectomy and reverse transcriptase-polymerase chain reaction in early-stage melanoma patients. J Clin Oncol. 1999;17:3238–3244. doi: 10.1200/JCO.1999.17.10.3238. [DOI] [PubMed] [Google Scholar]
- 12.Shivers SC, Wang X, Li W, et al. Molecular staging of malignant melanoma: Correlation with clinical outcome. JAMA. 1998;280:1410–1415. doi: 10.1001/jama.280.16.1410. [DOI] [PubMed] [Google Scholar]
- 13.Kuo CT, Hoon DSB, Takeuchi H, et al. Prediction of disease outcome in melanoma patients by molecular analysis of paraffin-embedded sentinel lymph nodes. J Clin Oncol. 2003;21:3566–3572. doi: 10.1200/JCO.2003.01.063. [DOI] [PubMed] [Google Scholar]
- 14.Sarantou T, Chi DDJ, Garrison DA, et al. Melanoma-associated antigens as messenger RNA detection markers for melanoma. Cancer Res. 1997;57:1371–1376. [PubMed] [Google Scholar]
- 15.Hoon DS, Bostick P, Kuo C, et al. Molecular markers in blood as surrogate prognostic indicators of melanoma recurrence. Cancer Res. 2000;60:2253–2257. [PubMed] [Google Scholar]
- 16.Palmieri G, Ascierto PA, Cossu A, et al. Detection of occult melanoma cells in paraffin-embedded histologically negative sentinel lymph nodes using a reverse transcriptase polymerase chain reaction assay. J Clin Oncol. 2001;19:1437–1443. doi: 10.1200/JCO.2001.19.5.1437. [DOI] [PubMed] [Google Scholar]
- 17.Hoon DS, Wang Y, Dale PS, et al. Detection of occult melanoma cells in blood with a multiple-marker polymerase chain reaction assay. J Clin Oncol. 1995;13:2109–2116. doi: 10.1200/JCO.1995.13.8.2109. [DOI] [PubMed] [Google Scholar]
- 18.Cochran AJ, Balba BR, Starz H, et al. The Augsburg Consensus. Techniques of lymphatic mapping, sentinel lymphadenectomy, and completion lymphadenectomy in cutaneous malignancies. Cancer. 2000;89:236–241. [PubMed] [Google Scholar]
- 19.Palmieri G, Strazzullo M, Ascierto PA, et al. Polymerase chain reaction-based detection of circulating melanoma cells as an effective marker of tumor progression. Melanoma Cooperative Group. J Clin Oncol. 1999;17:304–311. doi: 10.1200/JCO.1999.17.1.304. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi H, Kuo C, Morton DL, et al. Expression of differentiation melanoma-associated antigen genes is associated with favorable disease outcome in advanced-stage melanomas. Cancer Res. 2003;63:441–448. [PubMed] [Google Scholar]
- 21.Takeuchi H, Bilchik A, Saha S, et al. C-Met expression level in primary colon cancer: A predictor of tumor invasion and lymph node metastases. Clin Cancer Res. 2003;9:1480–1488. [PubMed] [Google Scholar]
- 22.Gaugler B, van den Eynde B, van der Bruggen P, et al. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawakami Y, Eliyahu S, Delgado CH, et al. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci U S A. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyashiro I, Kuo C, Huynh K, et al. Molecular strategy for detecting metastatic cancers with use of multiple tumor-specific MAGE-A genes. Clin Chem. 2001;47:505–512. [PubMed] [Google Scholar]
- 25.Tsuchida T, Saxton RE, Morton DL, et al. Gangliosides of human melanoma. J Natl Cancer Inst. 1987;78:45–54. doi: 10.1093/jnci/78.1.45. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchida T, Saxton RE, Irie RF. Gangliosides of human melanoma: GM2 and tumorigenicity. J Natl Cancer Inst. 1987;78:55–60. doi: 10.1093/jnci/78.1.55. [DOI] [PubMed] [Google Scholar]
- 27.Kuo CT, Bostick PJ, Irie RF, et al. Assessment of messenger RNA of β 1→4-N-acetylgalactosaminyl-transferase as a molecular marker for metastatic melanoma. Clin Cancer Res. 1998;4:411–418. [PubMed] [Google Scholar]
- 28.Scholl FA, Kamarashev J, Murmann OV, et al. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001;61:823–826. [PubMed] [Google Scholar]
- 29.Maschhoff KL, Baldwin HS. Molecular determinants of neural crest migration. Am J Med Genet. 2000;97:280–288. doi: 10.1002/1096-8628(200024)97:4<280::aid-ajmg1278>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 30.Henderson AR. Assessing test accuracy and its clinical consequences: A primer for receiver operating characteristic curve analysis. Ann Clin Biochem. 1993;30:521–529. doi: 10.1177/000456329303000601. [DOI] [PubMed] [Google Scholar]
- 31.Van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 32.Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 33.Kamaraju AK, Bertolotto C, Chebath J, et al. Pax3 down-regulation and shut-off of melanogenesis in melanoma B16/F10.9 by interleukin-6 receptor signaling. J Biol Chem. 2002;277:15132–15141. doi: 10.1074/jbc.M200004200. [DOI] [PubMed] [Google Scholar]
- 34.Marincola FM, Jaffee EM, Hicklin DJ, et al. Escape of human solid tumors from T-cell recognition: Molecular mechansims and functional significance. Adv Immunol. 2000;74:181–273. doi: 10.1016/s0065-2776(08)60911-6. [DOI] [PubMed] [Google Scholar]
- 35.Bostick PJ, Chatterjee S, Chi DD, et al. Limitations of specific reverse-transcriptase polymerase chain reaction markers in the detection of metastases in the lymph nodes and blood of breast cancer patients. J Clin Oncol. 1998;16:2632–2640. doi: 10.1200/JCO.1998.16.8.2632. [DOI] [PubMed] [Google Scholar]