SUMMARY
Interactions between CD40 and its ligand CD154 are involved in the progression of both cell mediated and innate immunity. These interactions are brought about by the transient expression of CD154 on activated CD4+ T cells, which is regulated, in part, at the level of mRNA turnover. Here we have focused on analyzing the pattern of post-transcriptional regulation in mouse CD4+ T cells in response to activation. Initial experiments identify a region of the murine CD154 mRNA that binds a polypyrimidine-tract binding protein (PTB)-containing complex (mComplex I), which is activation-dependent and binds to a single CU-rich site within the 3’ untranslated region (UTR). Subsequent findings demonstrate that in vivo polyclonal activation of T cells leads to a pattern of differential CD154 mRNA stability that is directly dependent on extent of activation. Furthermore, in vitro activation of antigen-primed T cells shows that the CD154 mRNA half-life (t1/2) increases relative to that of unprimed cells. Importantly, this is the first report demonstrating that the regulation of CD154 in vivo is connected to an activation-induced program of mRNA decay and thus provides strong evidence for posttranscriptional mechanisms having a physiological role in regulating CD154 expression during an ongoing immune response.
Keywords: CD154, posttranscriptional regulation, polypyrimidine tract-binding protein, mRNA stability
INTRODUCTION
CD40 ligand (CD40L, CD154) expression on activated CD4+ T cells has far reaching consequences for the development of both innate and adaptive immunity through the interaction of CD40 expressed on antigen presenting cells (APCs), activated endothelial cells, microglia and astrocytes [1–4]. Initial kinetic studies of CD154 expression indicated a rapid, but transient response following T cell activation [5–7]. However, additional studies suggested that activated T cells following an initial peak, continued to express a lowered level of CD154 for an extended period of time (up to 4 d) [8–10]. Overall expression could be separated into an early phase and an extended phase (after 24 h) that were reciprocally regulated by IL-4 and IL-12 [11]. These studies revealed that CD154 on CD4+ T cells is modulated throughout an immune response however the molecular events that control differential expression remain only partially defined. Expression is controlled in part at the transcriptional level through the binding of NF-AT and NF-κB transcription factors [12–18] and at the posttranscriptional level through a process of activation-induced differential mRNA decay [19–22]. In human CD4+ T cells stimulated ex vivo for up to 8 hr with anti-CD3 mAb the CD154 transcript was shown to decay with a rapid half-life (t1/2) of less than 40 min whereas at extended times of T cell activation (i. e. 24 and 48 h) the transcript became significantly more stable (t1/2 ~ 2.2 h) [23]. Increased stabilization was shown to be concomitant with the binding of polypyrimidine-tract binding (PTB) complexes (Complex I and II) to three cis-acting elements within the 3’UTR [24–26]. Notably both early and late pathways functioned independently of A+U-rich element (ARE) binding [25–27], a pathway critical for regulating the steady-state levels of multiple cytokines and growth factors including TNFα, GM-CSF, IL-2, and IL-10 [28].
Because the majority of studies that analyzed CD154 mRNA turnover utilized human CD4+ T cells stimulated ex vivo, findings from these studies precluded an understanding of how regulated decay interfaced with the controlled expression of CD154 during an immune response. Therefore, our present work has utilized mouse T cells to investigate the role antigen challenge plays in shaping the posttranscriptional regulation of CD154. Our findings demonstrate that murine CD154 mRNA decays through an activation-induced program of posttranscriptional regulation and following secondary antigen challenge, the stability of this transcript is markedly increased suggesting a mechanism for sustained CD154 expression at extended times of activation.
RESULTS
The murine CD154 transcript degrades via an activation-induced pattern of mRNA decay
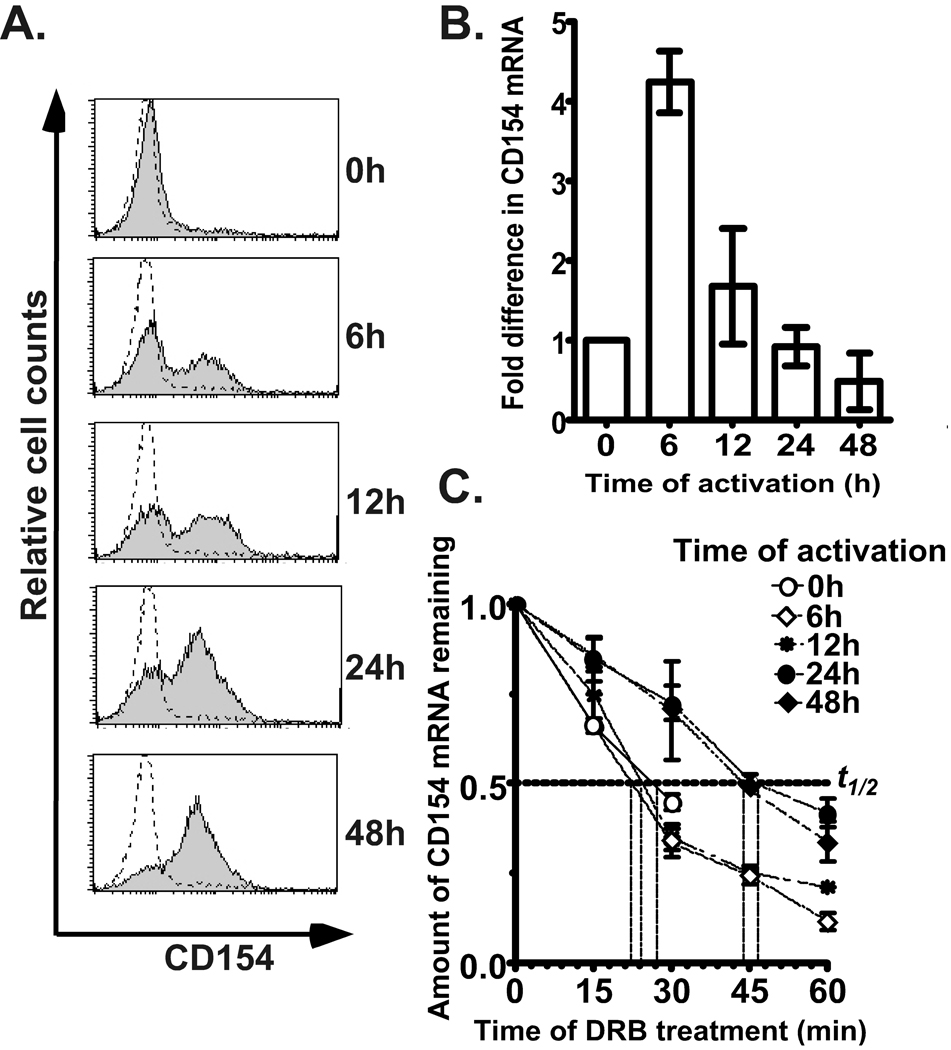
To understand how CD154 expression is regulated in the mouse, CD4+ T cells were isolated from spleens and incubated in vitro with anti-CD3 and anti-CD28 mAbs over a 48 h time course. Surface CD154 expression increased progressively so that a significant majority of cells were CD154 positive at 24 h and beyond (Fig. 1A). Analysis of steady-state levels of CD154 mRNA levels over the same time course revealed that levels were highest at 6 h followed by a decline to near basal levels between 24 and 48 h of activation (Fig. 1B).
Figure 1. Expression pattern of murine CD154 during a time course of T cell activation ex vivo.
A). Shown is representative of 3 flow cytometric analyses of CD154 surface expression using a PE-conjugated mAb with purified spleenic CD4+ T cells activated for 0, 6, 12, 24 and 48 h with anti-CD3 and anti-CD28 mAbs. Dotted line indicates isotype-PE control. B). Steady-state expression of CD154 mRNA was determined in samples after stimulation with anti-CD3 and anti-CD28 mAbs for indicated times. Fold differences of CD154 mRNA molecules relative to resting cells were normalized using β-actin mRNA and results are the average +/− SEM of 3 independent experiments. C). CD4+ T cells were stimulated for 0, 6, 12, 24 and 48 h with anti-CD3 and anti-CD28 mAbs followed by treatment with DRB for 15, 30, 45 and 60 min. RNA was isolated and CD154 expression analyzed by real-time PCR to determine the t1/2 of the CD154 transcript. Results were normalized using β-actin transcripts and plotted as the fraction of CD154 mRNA remaining relative to time 0 for each DRB time point. These results represent the average +/− SEM of 3 independent experiments. In resting (0 h) no CD154 mRNA was detectable beyond 30 min of DRB treatment.
The contribution of RNA decay to the steady state level of CD154 transcript was assessed by treating resting and differentially activated CD4+ T cells with the transcriptional inhibitor DRB for 1 h and evaluating the percent of mRNA remaining at 15 min intervals by real-time PCR. We found that a very low, but detectable level of transcript was expressed in resting T cells and this species rapidly degraded with a t1/2 of approximately 27 min. Further analysis across a 48 h time course revealed a regulated pattern of decay where at early time points of activation the t1/2 of the CD154 message was between 18 and 24 min whereas at late time points there was a an approximate two-fold increase in the t1/2 to between 43 and 46 min (Fig. 1C). These findings demonstrated that the mouse transcript was regulated at the posttranscriptional level by T cell activation and that overall the message was much less stable than the human transcript across the activation time course.
mComplex I is regulated in T cells upon activation
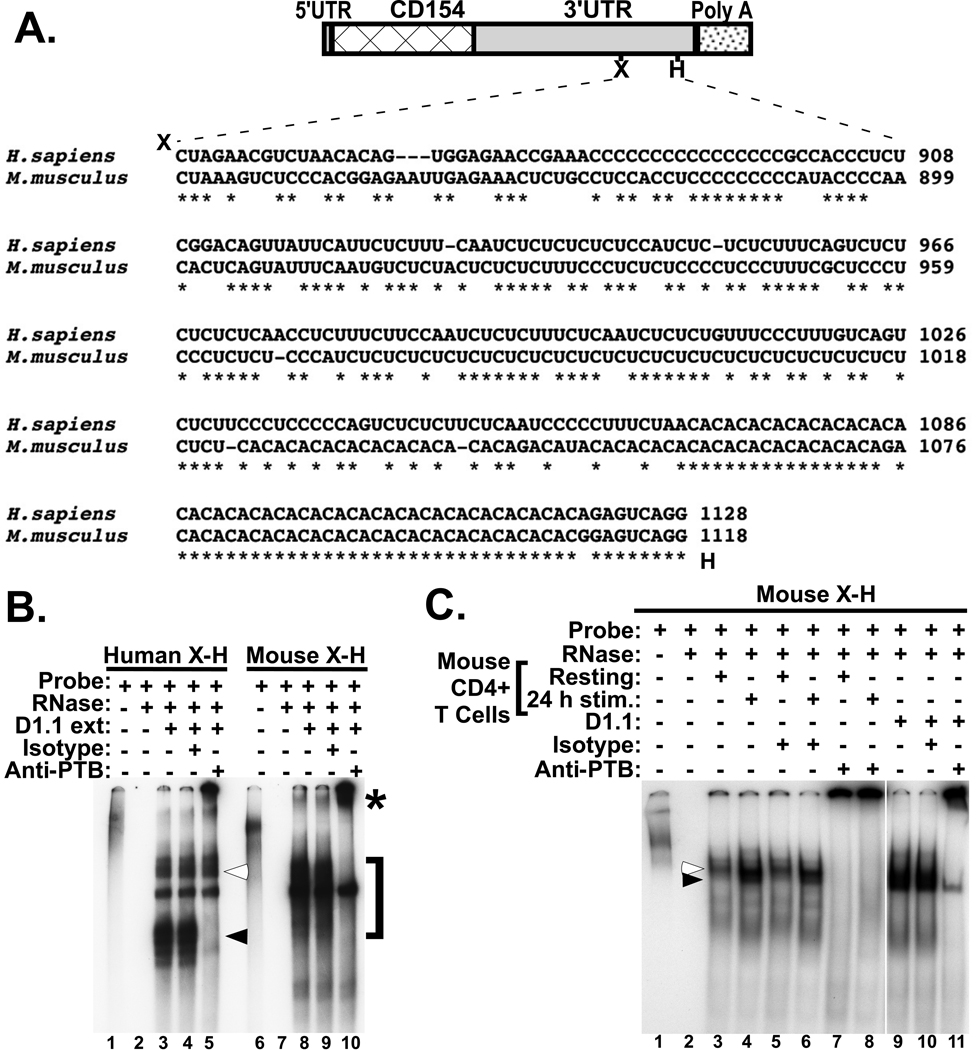
The critical element for human CD154 mRNA stability has been mapped to a sequence extending 350 nucleotides between the Xba I and Hae III sites (X-H region) in the 3’UTR and encodes three binding sites for PTB-containing complexes [26]. Sequence alignment between the human and mouse CD154 3’UTR revealed a high level of homology (76% using ClustaW [29]) and similarity of many features of the human X-H region including a highly pyrimidine-rich region adjacent to an upstream poly-(C) stretch and a (CA)-rich region lying immediately downstream (Fig. 2A). Binding studies with cytoplasmic extracts from Jurkat/D1.1 cells, which recapitulate activated human CD4+ T cells in terms of CD154 mRNA posttranscriptional regulation [25], and either the human or mouse X-H probe demonstrated that PTB complexes bound to both X-H probes (Fig. 2B, lanes 3–10). With the human X-H probe the predicted PTB-containing Complex I and slower migrating Complex II (lanes 3–5) were observed whereas the mouse X-H probe detected a broad complex that ran with a higher mobility than Complex I (brackets, lane 8) and was partially super-shifted with anti-PTB mAb (asterisk, lane 10). Using resting and activated mouse splenic CD4+ T cell extracts and anti-PTB antibodies we found that PTB-containing complexes were formed with both extracts although the migration patterns were different (see empty and filled arrows, lanes 3 – 6, Fig. 2C). This finding demonstrated that while PTB was a common component of the two complexes, the overall difference in composition appeared to be directly linked to T cell activation. Based on the similarity to its human counterpart we refer to the activation-induced complex as murine Complex I, or mComplex I (filled arrow).
Figure 2. Distinct PTB-containing complexes from both resting and activated mouse extracts bind to the mouse stability element.
A). Diagram showing sequence alignment and relative location of the X-H region in the 3’UTR of human and murine CD154 mRNA. B). Human (lanes 1–5) and murine (lanes 6–10) X-H region probes labeled with alpha-32P-UTP were used in binding studies with cytoplasmic extracts from human Jurkat/D1.1 cells. Anti-PTB (lanes 5 and 10) or isotype control (lanes 4 and 9) antibodies were included to identify PTB-containing complexes (denoted by asterisk). Lanes 1 and 6 show probes alone and reactions in lanes 2 and 7 contained probe plus RNase without extract. Closed and open arrows designate human Complex I and II, respectively, whereas the square bracket indicate the broad complex binding to the mouse X-H region. C). Cytoplasmic extracts were prepared from splenic CD4+ T cells (lanes 3–8) that were either resting (lanes 3, 5 and 7) or activated with anti-CD3 and anti-CD28 mAbs for 24 h (lanes 4, 6 and 8). RNA-EMSA was carried out using the mouse X-H region probe labeled with alpha-32P-UTP. Comparison reactions were established using Jurkat/D1.1 cytoplasmic extracts (lanes 9–11). Anti-PTB (lanes 7, 8 and 11) and control isotype (lanes 5, 6 and 10) antibodies were added to specific reactions to establish the presence of PTB. The open and closed arrows indicate the PTB-containing complexes in resting and activated CD4+ T cells, respectively.
Characterization of the mComplex I minimal binding region
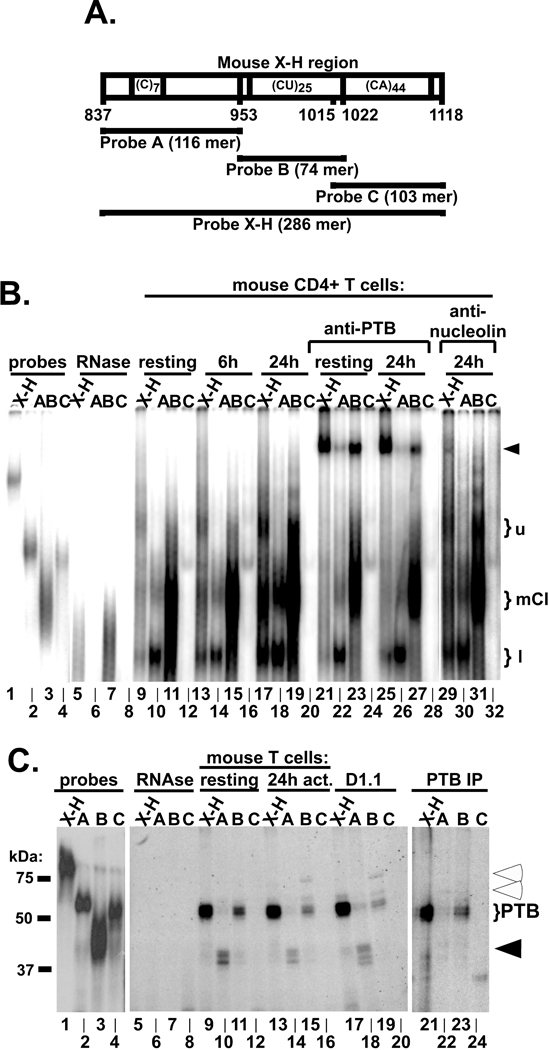
Additional analysis of complex formation was carried out using resting, 6 h and 24 h activated mouse splenic CD4+ T cell extracts with probes that subdivided the X-H region into three parts loosely corresponding to sites A, B and C of the human CD154 X-H region [26] (Fig. 3A). In this experiment, gels were run longer to separate the different complexes observed in Fig. 2B and probes were labeled with alpha-32P-CTP to identify any poly(C)- or CA-specific complexes. Notably, the mouse X-H region was found to bind three distinct PTB-containing complexes in 24 h activated extracts (Fig. 3B, brackets, lanes 11 and 25) which were also weakly visible in extracts from resting (lane 9) and 6 h activated (lane 13) extracts. One main complex in 24 h activated extracts that ran with intermediate mobility and showed the highest intensity corresponded to mComplex I (see Fig. 2C). Probe A interacted weakly with this complex and also bound a faster migrating complex under all conditions tested (Fig 3B, lanes 10, 14 and 18). This lower complex was only supershifted when bound to the X-H probe (lanes 21 and 25) and not probe A (lanes 22 and 26). Surprisingly, site B was found to constitutively bind both PTB and non-PTB containing complexes (lanes 11, 15, 19 and 27). Only faint complexes were observed with probe C at 0 and 6 h of activation (lanes 12 and 16) and these complexes did not supershift with anti-PTB mAb (lanes 24 and 28). Since nucleolin is known to be part of human Complex I [27] we tested whether anti-nucleolin mAb could also supershift mComplex I. We observed that upon addition of these antibodies ribonucleoprotein complexing on the X-H probe was strongly weakened and lightly supershifted (lane 29), was weakened on probe A (lane 30) but was not affected on probe B (lane 31). Together these findings demonstrated that the X-H and B regions contained the primary sites for binding of PTB complexes that were present in both resting, 6 h and 24 h activated cells. Furthermore, while PTB is a common component of mComplex I in differentially activated cells, the overall composition and binding activity involves other proteins including nucleolin, whose binding activity is directly linked to T cell activation.
Figure 3. Characterization of the X-H region of mouse CD154 mRNA.
A). Diagram showing the relative location of sequence features within the mouse X-H region and the sequence location of probes A, B and C. B). Cytoplasmic extracts were prepared from splenic CD4+ T cells (lanes 9–28) that were either resting (lanes 9–12 and 21–24) or activated with anti-CD3 and anti-CD28 mAbs for 6 h (lanes 13–16) or 24 h (lanes 17–20 and 25–28). RNA-EMSA was carried out using the mouse X-H probe (lanes 9, 13, 17, 21 and 25) and probes A (lanes 10, 14, 18, 22 and 26), B (lanes 11, 15, 19, 23 and 27) and C (lanes 12, 16, 20, 24 and 28) labeled with alpha-32P-CTP. Anti-PTB (lanes 21–28) and anti-nucleolin (lanes 29–32) mAbs were added to reactions to super-shift complexes (filled arrow). mComplex I is indicated by middle bracket (mCI) and an upper (u) and lower (l) minor complexes are indicated. C). Alpha-32P-UTP-labeled RNA probes corresponding to the mouse X-H, A, B and C regions were incubated with cytoplasmic extracts from resting (lanes 9–12) or 24 h activated (lanes 13–16) mouse CD4+ T cells or human Jurkat/D1.1 cells (lanes 17–20) followed by RNAse treatment. The migration of the PTB-mRNA complex is shown with a bracket while filled and empty arrows point to slow and fast migrating ribonucleoproteins, respectively. The same sets of probes were incubated with cytoplasmic extracts from 24 h activated mouse CD4+ T cells and the mRNA-PTB complex immunoprecipitated (PTB-IP) using PTB mAb (lanes 21 to 24). The same reaction was repeated using isotype control, which did not yield any band (data not shown). Intact probes are shown in lanes 1–4, while RNase treated probes in the absence of extract are shown in lanes 5–8.
To further identify PTB binding sites within the X-H region UV-cross-linking experiments were performed with extracts prepared from resting and 24 h activated CD4+ T cells and Jurkat/D1.1 cells (Fig. 3C). The mouse X-H probe bound two proteins of 50 and 55 kDa in resting, activated and Jurkat extracts that were consistent with the mw of the PTB-1 and PTB-4 isoforms (Fig 3C, lanes 9, 13 and 17). These species were also detected with Probe B but at reduced intensity (lanes 11 15 and 18). Two slower migrating proteins also bound probe B in extracts from activated and Jurkat cells but not resting cells (open arrows, lane 15 and 19). Probe A cross-linked to three lower molecular weight species (filled arrows, lanes 10, 14 and 18), which were not visible with the X-H probe and no bands were detected with Probe C (lanes 12 and 16). Immunoprecipitation with anti-PTB mAb following UV-cross-linking, pulled down the two most prominent 50 and 55 kDa proteins indicating that the major RNA binding proteins correspond to PTB1 and PTB4, respectively (lanes 21–24). These findings demonstrated that unlike the human X-H sequence, which has been shown to contain three distinct binding regions for PTB-containing complexes, only a single discrete PTB binding region was identified in the equivalent mouse region. However, based on the enhanced binding to the X-H probe we cannot exclude that additional binding sites may span the individual regions.
CD154 RNA is stabilized in vivo by extended TCR stimulation
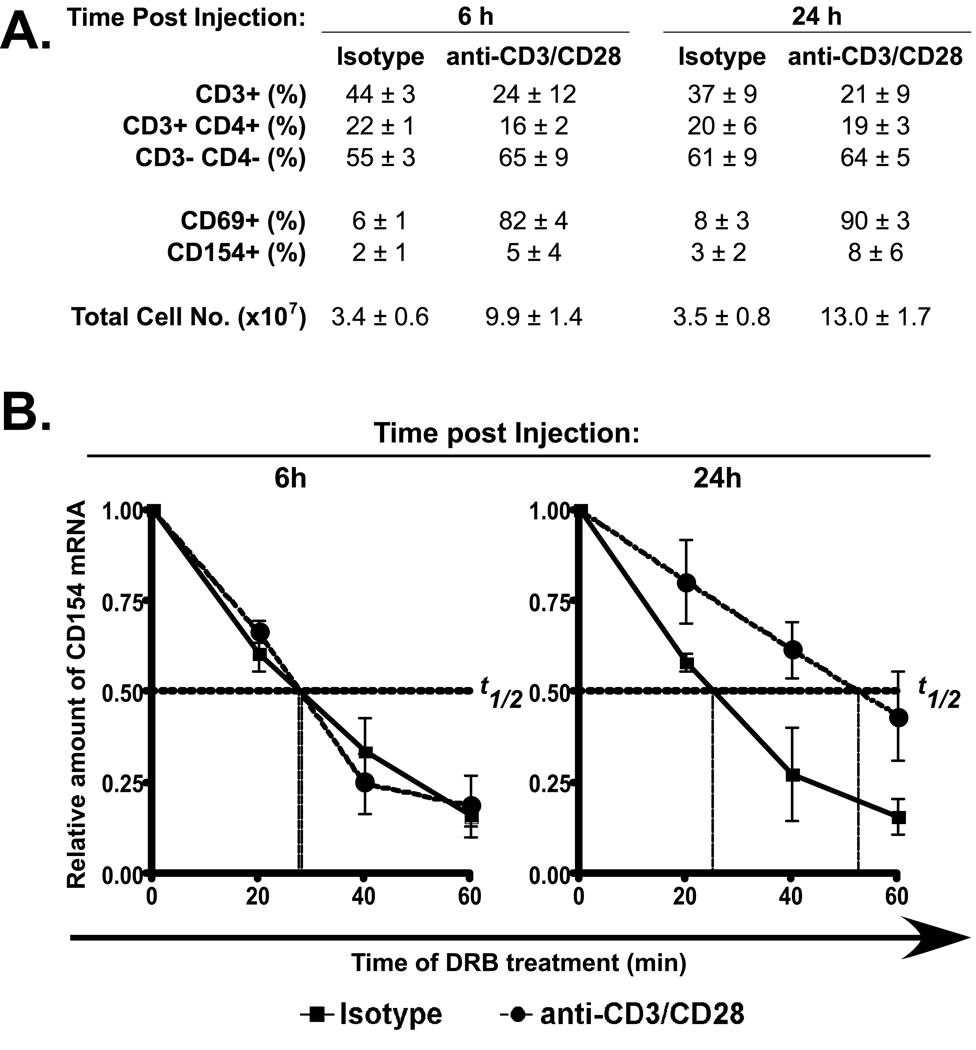
To establish whether a pattern of regulated CD154 mRNA decay occurs during an in vivo time course of T cell activation, healthy mice were injected with a cocktail of anti-CD28 and anti-CD3 mAbs and changes in CD154 mRNA turnover were measured in activated cells. It has been previously shown that injection of anti-CD28 antibodies results in severe splenomegaly [30]. In our experiments, we also observed splenomegaly after injection (see Total Cell No., Fig. 4A). CD69 expression was assayed at discrete time points following injection of mAbs to assess the extent of activation (Fig. 4A). The expression level of CD154 on the activated population was only slightly above that found on resting cells, however this is consistent with down-regulation of CD154 via a pathway of receptor-mediated endocytosis [31]. As shown in Fig. 4B, after 6 h of treatment the t1/2 of CD154 mRNA was approximately 30 min, which is comparable to the isotype control group and demonstrated that the CD154 mRNA in resting and early-activated T cells is rapidly turned over (left graph). Analysis of cells at 24 h of activation revealed that the t1/2 increased approximately 2-fold to greater than 1 h (center and right graphs). Under these same conditions, injection of isotype mAb failed to affect message stability supporting our hypothesis that CD154 mRNA stabilization requires T cell activation. Although these experiments were carried out with injections of both anti-CD3 and anti-CD28 mAbs similar results were obtained with anti-CD3 mAb alone (data not shown) indicating a direct relationship between TCR stimulation and the pathways that regulate CD154 message decay.
Figure 4. CD154 mRNA is stabilized in vivo in response to activation with anti-CD3 and anti-CD28 mAbs.
A). Changes in number, ratio and phenotype of splenocytes detected by FACS analysis are recorded over time (6 and 24 h) following injection of anti-CD3 and anti-CD28 mAbs compared to injection of isotype control antibodies. This data reflects the average and SEM of 3 independent experiments. B). The t1/2 of CD154 mRNA in splenocytes isolated from mice injected with anti-CD3 and anti-CD28 mAbs or isotype control antibodies for 6 and 24 h was measured by real-time PCR following incubation with the transcription inhibitor DRB for periods of 0, 20, 40 and 60 min. This data represent the average and +/− SEM (error bars) of 3 independent experiments.
CD154 mRNA is stabilized in response to antigen (Ag) recognition
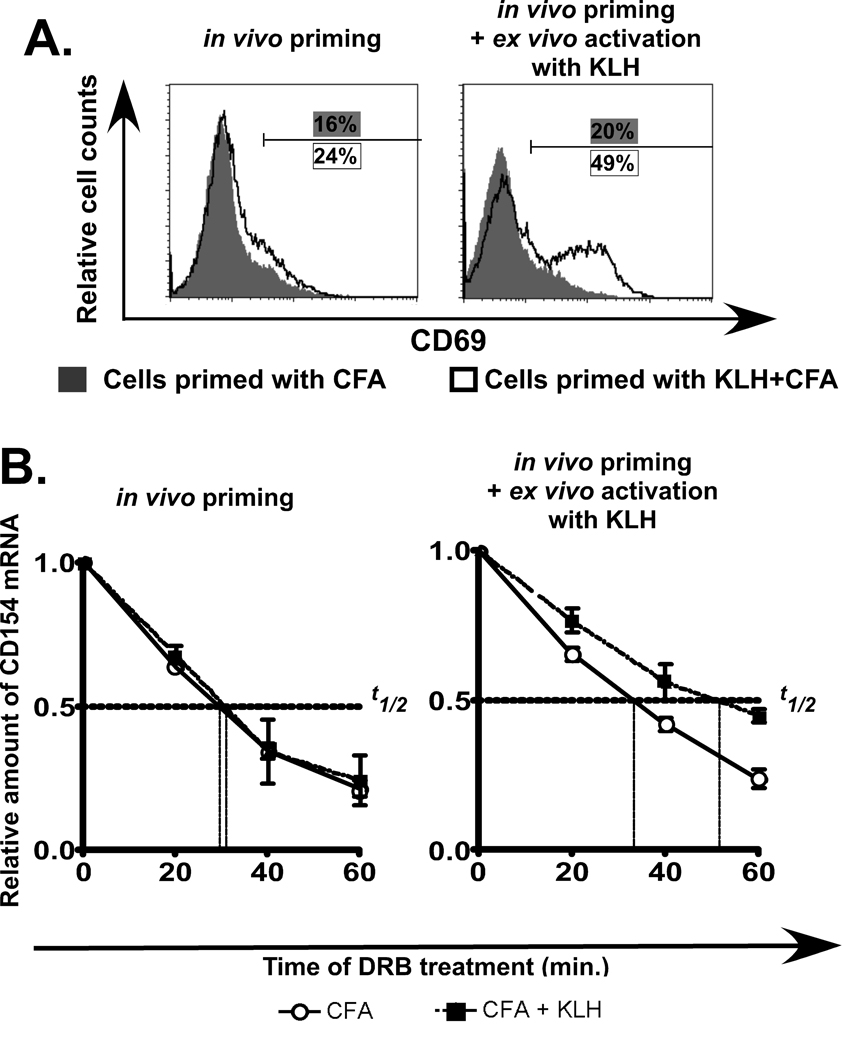
We next extended our study to analyze CD154 mRNA decay in T cells following Ag stimulation. Ag-specific T cells were generated by priming animals with 50.0 µg KLH in Complete Freund’s Adjuvant (CFA) or CFA alone. After 9 days, total lymphocytes from both sets of mice were isolated from draining lymph nodes and half of each population was re-stimulated with KLH. Draining lymph nodes were collected at day 9 since primed, but resting cells were needed. Prior to ex vivo stimulation, all cells were found to be resting based on their low CD69 expression (Fig. 5A, left panel). Upon secondary stimulation with KLH, lymphocytes from animals primed with CFA alone showed a low level of activation (grey, filled peak in right panel). In contrast, lymphocytes from mice primed with KLH and CFA together were more responsive to re-exposure to KLH as revealed by the increased expression of CD69 (black, open peak in right panel). This difference in response between the two populations confirmed that the primed lymphocytes retained KLH recognition upon re-exposure to Ag.
Figure 5. Antigen specific T cell stimulation coincides with an increase in CD154 mRNA stability.
A). CD69 expression in cells suspensions of draining lymph nodes of mice primed with either CFA alone (left graph grey, filled peak) or KLH + CFA (left graph, black, open peak). The same analysis was carried out following ex vivo incubations of primed cells with KLH for 24 h (right graph). This data is representative of three independent experiments. B). Analysis of CD154 mRNA decay in cells isolated from mice that were primed 9 days earlier with either CFA alone (open circle) or KLH + CFA (closed square) (left graph). The same samples were incubated ex vivo with KLH for 24 h (right graph) and real-time PCR was carried out on both samples following incubation with DRB for periods of 0, 20, 40 and 60 min. This data represents the average and +/− SEM of 3 independent experiments.
Analysis of RNA t1/2 profiles of the unprimed and primed lymphocyte populations demonstrated that 9 days following injection, CD154 mRNA decayed with a t1/2 of approximately 30 minutes in all two samples (Fig. 5B, left graph). This confirmed that although two groups of mice were exposed to Ag at day 0 (injected with KLH+CFA or CFA alone), 9 days later the resulting Ag-specific lymphocytes had engaged a program of fast transcript decay. When the two differentially primed populations were exposed to KLH ex vivo the t1/2 of CD154 mRNA in the population primed with KLH and CFA together, increased two-fold with a change from ~30 to ~50 min (compare KLH+CFA in left and right graphs). Notably, transcript stability in lymphocytes primed with CFA alone remained the same before and after ex vivo incubation with KLH. These findings reveal for the first time that Ag-specific stimulation of CD4+ T cells results in an increase in CD154 transcript stability.
DISCUSSION
Our present findings demonstrate several novel aspects of CD154 posttranscriptional regulation in murine CD4+ T cells. First, characterization of the mouse stability element identified PTB binding regions and revealed similarities and differences with the human CD154 transcript. A major difference in the murine element was that it bound cytoplasmic PTB- complexes in both resting and activated cells. Although the intensity of complex formation is markedly different in resting versus activated mouse T cell extracts, we predict that the ubiquitously bound mouse PTB could be modified to recruit nucleolin and other factors in response to activation. This is highlighted by the fact that subdividing the X-H region into discrete regions resulted in the binding of complexes to probes A and B that were highly distinct from mComplex I at each stage of cell activation; suggesting that the whole X-H region is needed to bring about the activation dependent mComplex I binding. Also by the fact that anti-nucleolin antibody decreased ribonucloprotein complexing to the X-H probe using extracts from 24 h activated cells. Based on our gel shift data other factors are recruited into mComplex I through protein-protein interactions when the whole X-H sequence is present and not protein-RNA interactions as is the case with the fragmented region. This type of mechanism is highly reminiscent of the posttranscriptional regulation of murine inducible nitric-oxide synthetase (iNOS) where PTB binds to the iNOS 3’UTR in both resting and septic-shock treated murine hepatocytes and recruits hnRNP-L into a complex to increase the stability of iNOS mRNA [32]. The fact that increased CD154 mRNA stability and mComplex I activation were detected at a time when transcript levels are reproducibly low and protein expression is at a maximum suggests this complex may play a role in regulating CD154 translation. This possibility is supported by the finding that PTB increases the translational rate of the Hypoxia-Inducible Factor I (HIF-I) transcript under hypoxia conditions [33] and that a sequences within the CD154 stability element may affect both poly(A) tail length and translational efficiency [34].
A second major observation was that a program of regulated mRNA decay was functional during an on going immune response in vivo. This process would result in enhanced expression of CD154 following extended activation and supports the outcome of biological processes controlled by CD40 signaling in an established immune response. The requirement for extended activation suggests that before and during early times of activation CD154 expression is controlled primarily at the transcriptional level and that once this activation threshold is reached a more steady level of protein expression is driven by increased transcript stability. The requirement for this pattern of regulated decay may reflect the need for cognate interactions between APCs and Ag-specific T cells at discrete checkpoints and/or within specific differentiation niches such as germinal centers (GC) [35] or by impacting the course of B cell differentiation into short-lived APCs or memory B cells [36].
Regulated mRNA decay may also play a role in restricting CD154 expression in the absence of cellular activation. CD154 is minimally expressed by both CD4+CD8+ and CD4+CD8- thymocytes [37], however over-expression of CD154 causes severe disruption of thymic organization [38] and a decrease in cell viability [39]. The fact that resting naïve CD4+ T cells were found to have a pattern of fast degrading CD154 mRNA together with our observations that specific and extended TCR activation are required to induce stabilization of the transcript suggest that the level of TCR signaling required to induce mComplex I is only reached following APC contact in peripheral lymphoid tissues and not during maturation. Future work will focus on defining how posttranscriptional regulation of CD154 expression contributes to the temporal expansion of the humoral immune response under both normal and pathological conditions.
MATERIALS and METHODS
Mice, antibodies and cell lines
Female C57BL/6J (B6) mice were obtained from the National Cancer Institute and used between 8 and 9 weeks of age. Animal studies were approved by the Institutional Animal Care and Use Committee at Rutgers University. The anti-CD28 and anti-CD3 antibodies were purified as previously described [30]. Anti-PTB mAb were affinity purified from the supernatant of BB7 hybridoma culture (ATCC number: CRL-2501). Anti-nucleolin mAb from clone MS-3 were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). The Jurkat/D1.1 cell line was obtained from Dr. S. Lederman (Columbia University, NY) and cultured as previously described [25]. Flow cytometry mAbs were purchased from eBioscience (San Diego, CA).
Isolation of mouse CD4+ T cells
Freshly isolated spleens were minced to produce single cell suspensions and incubated in RBC lysis buffer (150mM NH4Cl, 1mM KHCO3, 100µM NaEDTA) for 7 min. The reaction was stopped by addition of 50 ml RPMI 1640. The cell suspension was filtered through a 40 µm cell strainer and CD4+ T cells isolated using negative selection with the MACS CD4+ T Cell Isolation kit (Miltenyi Biotec, Auburn, CA). A purity of above 95% CD4+CD3+ cells was repeatedly obtained using this method. The activation state of isolated cell populations was assessed by incubating with FITC-anti-CD69 and PE-anti-CD154 mAbs followed by flow cytometric analysis.
In vitro activation
Purified CD4+ T cells were resuspended at a concentration of 1×106 cells/ml in 10 ml of complete RPMI (RPMI 1640, 10% FBS, 50 µM 2-ME) supplemented with (1 µg/ml) anti-CD28 antibodies together with 50 U/ml recombinant hIL-2 (Peprotech, Rocky Hill, NJ) and plated on anti-CD3 mAb coated plates. At the time of collection, cell activation was assessed by flow cytometry to determine the percentage of CD69+CD154+ cells.
Flow Cytometry
A total of 2×105 cells were incubated in 50 µl flow cytometry buffer (3% FBS, 1% NaN3 in PBS) supplemented with 250 ng of each fluorophore-conjugated mAb and incubated for 30 min at 4°C. Fluorescence acquisition was performed on a FACS Calibur system (Becton Dickinson, San Jose, CA) and data analyzed using Cell Quest Software (Becton Dickinson).
Steady state expression and decay analyses of the CD154 transcript in mouse CD4+ T cells
To analyze the level of RNA stability cells were incubated with 50 µg/ml 5,6-Dichlorobenzimidizole 1-β-D-ribofuranoside (DRB) (Sigma Chemical Co., St. Louis, MO) and aliquots collected every 15 min for 1 h. Total RNA was isolated using the High Pure RNA Isolation Kit (Roche, Indianapolis, IN) and reverse transcription achieved with the Transcriptor First Strand cDNA Kit (Roche). Real-time PCR reactions were carried out on the MX4000 Multiplex Quantitative PCR System (Stratagene, Cedar Creek, TX) with the FastStart Universal Probe Master (Roche). For the quantification of CD154 and the normalizing β-actin transcripts, Universal Probe #89 and #106 (Roche) were used, respectively, together with primer sets: CD154-fwd 5’-ACGTTGTAAGCGAAGCCAAC-3’, CD154-rev 5’-TATCCTTTCTTGGCCCAC TG-3’, β-actin-fwd 5’-TGACAGGATGCAGAAGGAGA-3’ and β-actin-rev 5’-CGCTCAGGAGGAGCAATG-3’. Primers were used at a concentration of 300 nM and probes at a concentration of 200 nM. Amplification conditions were 95°C for 5min, followed by 40 cycles of 15 sec at 95°C and 30 sec at 60°C. The ΔCt analysis was used to compare the amount of CD154 transcript at each time point of DRB treatment relative to the untreated sample. For steady-state analysis the cross-threshold (Ct) values of samples collected at 0, 6, 12, 24 and 48 h of activation were compared to a standard curve generated with known amounts of the same PCR products. Results were normalized to 5×106 β-actin transcripts.
RNA Electromobility Shift Assay (RNA-EMSA) and UV cross-link Assay
Cytoplasmic extracts were prepared as previously described [40]. The mouse X-H sequence (nucleotide 837 to 1118) was cloned into pGEM-T Easy (Promega, Madison, WI) using primers X-H- fwd: 5’-CTAAAGTCTCCCACGGAGAA-3’ and X-H-rev: 5’-CCAACAATAGCCTGACTC-3’. This construct was used with the following primers to PCR amplify probe templates to generate fragments incorporating the target region flanked by the T7 promoter and poly-(dT)60 sequence. Probe X-H (nucleotide 837 to 1118): X-H-Fwd-5’-CGTAATACGACTCACTATAGGGCCAAGTTCTAAAGTCTCCC-3’ and X-H-rev-5’-(T)60CCAACAATAGCCTGACTCCGTGTG-3’; probe A (nucleotide 837 to 953) A-fwd 5’-CGTAATACGACTCACTATAGGGCCAAGTTCTAAAGTCTCCC-3 ’, A-rev 5’-(T)60CGAAAGGGAGGGGAGAGAGG-3’; probe B (nucleotide 953 to 1022) B-fwd 5’-CGTAATACGACTCACTATAGGGCTCCCTCCCTCTCTCCCATC-3’, B-rev 5’-(T)60GTGTGAGAGAGAGAGAGAG-3’; probe C (nucleotide 1015 to 1118) C-fwd 5’-CGTAATACGACTCACTATAGGGTCTCTCTCACACACACACAC-3’, C-rev 5’-(T)60CCAACAATAGCCTGACTCCGTGTG-3’. Synthesis of the human X-H templates, in vitro transcription of 32P labeled RNA probes, RNA-EMSA and UV cross-linking experiments were performed as previously described [41] using either alpha-32P-CTP or alpha-32P-UTP as indicated.
In vivo activation
Mice were injected i.p. with 100 µl of PBS supplemented with either a combination of 100 µg purified anti-CD28 mAb and 10 µg anti-CD3 mAb or 110 µg of Purified American Hamster IgG (eBioscience). Single splenocyte suspensions were prepared by mechanical grinding at indicated times and RNA isolated as described above.
In vivo priming with antigen and ex vivo activation
Mice were injected in the upper-side of the left posterial foot-paw with 50 µl of solution of 1 µg/µl Keyhole Limpet Hemocyanin (KLH) in Complete Freud’s Adjuvant (CFA) or CFA alone. Control mice were injected with 50 µl of PBS. Nine days post injection a homogeneous cell suspension was prepared from the left popliteal draining lymph node. One half of each of these cell suspensions was incubated with DRB to allow assessment of CD154 mRNA decay rates by real-time PCR. The remaining cells were then re-suspended in 10 ml complete RPMI supplemented with 5 µg/ml KLH. After 24 h at 37 °C cells were treated with DRB and the t1/2 of the CD154 message measured as by real-time PCR as described above.
ACKNOWLDEGEMENTS
The authors would like to thank Dr. Satish Devadas and past and present members of the Covey lab for their valuable technical input and comments. This work was supported in part by United States Public Health Service grant PO1 AI-57596 to LRC and YS (Dr. Sidney Pestka, PI) and a Grant-in-Aid from the American Heart Association to LRC.
Abbreviations
- PTB
polypyrimidine-tract binding protein
- ARE
A+U-rich elements
- 3’UTR
3' untranslated region
- t1/2
half-life
- DRB
Dichlorobenzimidizole 1-β-D-ribofuranoside
- KLH
Keyhole Limpet Hemocyanin
- RNA EMSA
RNA Electromobility Shift Assay
Footnotes
The authors have no conflicting financial interests.
REFERENCES
- 1.van Kooten C. Immune regulation by CD40-CD40-l interactions - 2; Y2K update. Front Biosci. 2000;5 doi: 10.2741/kooten. D880-693. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi JP, Kooten Cv, Liu YJ, Rousset F, Saeland S. The CD40 antigen and its ligand. Ann Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 3.Grewal IS, Flavell RA. CD40 and CD154 in cell-mediated immunity. Ann Rev Immunol. 1998;16:111–135. doi: 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- 4.van Kooten C, Banchereau J. CD40-CD40 ligand. J Leukoc Biol. 2000;67:2–17. doi: 10.1002/jlb.67.1.2. [DOI] [PubMed] [Google Scholar]
- 5.Armitage RJ, Fanslow WC, Strockbine L, Sato TA, Clifford KN, Macduff BM, Anderson DM, Gimpel SD, Davis-Smith T, Maliszewski CR, Clark EA, Smith CA, Grabstein KH, Cosman D, Spriggs MK. Molecular and biological characterization of a murine ligand for CD40. Nature. 1992;357:80–82. doi: 10.1038/357080a0. [DOI] [PubMed] [Google Scholar]
- 6.Castle BE, Kishimoto K, Stearns C, Brown ML, Kehry MR. Regulation of expression of the ligand for CD40 on T helper lymphocytes. J Immunol. 1993;151:1777–1788. [PubMed] [Google Scholar]
- 7.Roy M, Waldschmidt T, Aruffo A, Ledbetter JA, Noelle RJ. The regulation of the expression of gp39, the CD40 ligand, on normal and cloned CD4+ T cells. J Immunol. 1993;151:2497–2510. [PubMed] [Google Scholar]
- 8.de Boer M, Dasran A, Kwekkeboom J, Walter H, Vandenberghe P, Ceupens JL. Ligation of B7 with CD28/CTLA-4 on T cells results in CD40 ligand expression, interkeukin-4 secretion and efficient help for antibody production by B cells. Eur J Immunol. 1993;23:3120. doi: 10.1002/eji.1830231212. [DOI] [PubMed] [Google Scholar]
- 9.Johnson-Leger C, Christenson JR, Holman M, Klaus GG. Evidence for a critical role for IL-2 in CD40-mediated activation of naive B cells by primary CD4 T cells. J Immunol. 1998;161:4618–4626. [PubMed] [Google Scholar]
- 10.Peng X, Remacle JE, Kasran A, Huylebroeck D, Ceuppens JL. IL-12 up-regulates CD40 ligand (CD154) expression on human T cells. J Immunol. 1998;160:1166–1172. [PubMed] [Google Scholar]
- 11.Lee BO, Haynes L, Eaton SM, Swain SL, Randall TD. The biological outcome of CD40 signaling is dependent on the duration of CD40 ligand expression: reciprocal regulation by interleukin (IL)-4 and IL-12. J Exp Med. 2002;196:693–704. doi: 10.1084/jem.20020845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindgren H, Axcrona K, Leanderson T. Regulation of transcriptional activity of the murine CD40 ligand promoter in response to signals through TCR and the costimulatory molecules CD28 and CD2. J Immunol. 2001;166:4578–4585. doi: 10.4049/jimmunol.166.7.4578. [DOI] [PubMed] [Google Scholar]
- 13.Parra E, Mustelin T, Dohlsten M, Mercola D. Identification of a CD28 response element in the CD40 ligand promoter. J Immunol. 2001;166:2437–2443. doi: 10.4049/jimmunol.166.4.2437. [DOI] [PubMed] [Google Scholar]
- 14.Schubert LA, Cron RQ, Cleary AM, Brunner M, Song A, Lu LS, Jullien P, Krensky AM, Lewis DB. A T cell-specific enhancer of the human CD40 ligand gene. J Biol Chem. 2002;277:7386–7395. doi: 10.1074/jbc.M110350200. [DOI] [PubMed] [Google Scholar]
- 15.Schubert LA, King G, Cron RQ, Lewis DB, Aruffo A, Hollenbaugh D. The human gp39 promoter. J Biol Chem. 1995;270:29624–29627. doi: 10.1074/jbc.270.50.29624. [DOI] [PubMed] [Google Scholar]
- 16.Srahna M, Remacle JE, Annamalai K, Pype S, Huylebroeck D, Boogaerts MA, Vandenberghe P. NF-kappaB is involved in the regulation of CD154 (CD40 ligand) expression in primary human T cells. Clin Exp Immunol. 2001;125:229–236. doi: 10.1046/j.1365-2249.2001.01601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsytsykova AV, Tsitsikov EN, Geha RS. The CD40L promoter contains nuclear factor of activated T cells-binding motifs which require AP-1 binding for activation of transcription. J Biol Chem. 1996;271:3763–3770. doi: 10.1074/jbc.271.7.3763. [DOI] [PubMed] [Google Scholar]
- 18.Brunner M, Zhang M, Genin A, Ho IC, Cron RQ. A T-cell-specific CD154 transcriptional enhancer located just upstream of the promoter. Genes Immun. 2008;9:640–649. doi: 10.1038/gene.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suarez A, Mozo L, AbelGayo, Zamorano J, Gutierrez C. Requirement of a second signal via protein kinase C or protein kinase A for maximal expression of CD40 ligand. Involvement of transcriptional and posttranscriptional mechanisms. Eur J Immunol. 1997;27:2822–2829. doi: 10.1002/eji.1830271112. [DOI] [PubMed] [Google Scholar]
- 20.Ford GS, Yin CH, Barnhart B, Sztam K, Covey LR. CD40 ligand exerts differential effects on the expression of Ig transcripts in subclones of an IgM+ human B cell lymphoma line. J Immunol. 1998;160:595–605. [PubMed] [Google Scholar]
- 21.Rigby WF, Waugh MG, Hamilton BJ. Characterization of RNA binding proteins associated with CD40 ligand (CD154) mRNA turnover in human T lymphocytes. J Immunol. 1999;163:4199–4206. [In Process Citation] [PubMed] [Google Scholar]
- 22.Murakami K, Ma W, Fuleihan R, Pober JS. Human endothelial cells augment early CD40 ligand expression in activated CD4+ T cells through LFA-3-mediated stabilization of mRNA. J Immunol. 1999;163:2667–2673. [PubMed] [Google Scholar]
- 23.Ford GS, Barnhart B, Shone S, Covey LR. Regulation of CD154 (CD40 ligand) mRNA stability during T cell activation. J Immunol. 1999;162:4037–4044. [PubMed] [Google Scholar]
- 24.Hamilton BJ, Genin A, Cron RQ, Rigby WF. Delineation of a novel pathway that regulates CD154 (CD40 ligand) expression. Mol Cell Biol. 2003;23:510–525. doi: 10.1128/MCB.23.2.510-525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnhart B, Kosinski PA, Wang Z, Ford GS, Kiledjian M, Covey LR. Identification of a complex that binds to the CD154 3' untranslated region: implications for a role in message stability during T cell activation. J Immunol. 2000;165:4478–4486. doi: 10.4049/jimmunol.165.8.4478. [DOI] [PubMed] [Google Scholar]
- 26.Kosinski PA, Laughlin J, Singh K, Covey LR. A Complex Containing Polypyrimidine Tract-Binding Protein Is Involved in Regulating the Stability of CD40 Ligand (CD154) mRNA. J Immunol. 2003;170:979–988. doi: 10.4049/jimmunol.170.2.979. [DOI] [PubMed] [Google Scholar]
- 27.Singh K, Laughlin J, Kosinski PA, Covey LR. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J Immunol. 2004;173:976–985. doi: 10.4049/jimmunol.173.2.976. [DOI] [PubMed] [Google Scholar]
- 28.Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem Soc Trans. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- 29.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 30.Yin D, Zhang L, Wang R, Radvanyi L, Haudenschild C, Fang Q, Kehry MR, Shi Y. Ligation of CD28 in vivo induces CD40 ligand expression and promotes B cell survival. J Immunol. 1999;163:4328–4334. [PubMed] [Google Scholar]
- 31.Yellin MJ, Sippel K, Inghirami G, Covey LR, Lee JJ, Sinning J, Clark EA, Chess L, Lederman S. CD40 molecules induce down-modulation and endocytosis of T cell surface T cell-B cell activating molecule/CD40-L. J Immunol. 1994;152:598–608. [PubMed] [Google Scholar]
- 32.Soderberg M, Raffalli-Mathieu F, Lang MA. Inflammation modulates the interaction of heterogeneous nuclear ribonucleoprotein (hnRNP) I/polypyrimidine tract binding protein and hnRNP L with the 3'untranslated region of the murine inducible nitric-oxide synthase mRNA. Mol Pharmacol. 2002;62:423–431. doi: 10.1124/mol.62.2.423. [DOI] [PubMed] [Google Scholar]
- 33.Galban S, Kuwano Y, Pullmann R, Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, Holcik M, Gorospe M. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamilton BJ, Wang XW, Collins J, Bloch D, Bergeron A, Henry B, Terry BM, Zan M, Mouland AJ, Rigby WF. Separate cis-trans pathways posttranscriptionally regulate murine CD154 (CD40 ligand) expression: a novel function for CA repeats in the 3'-untranslated region. J Biol Chem. 2008;283:25606–25616. doi: 10.1074/jbc.M802492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzman-Rojas L, Sims-Mourtada JC, Rangel R, Martinez-Valdez H. Life and death within germinal centres: a double-edged sword. Immunology. 2002;107:167–175. doi: 10.1046/j.1365-2567.2002.01494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erickson LD, Durell BG, Vogel LA, O'Connor BP, Cascalho M, Yasui T, Kikutani H, Noelle RJ. Short-circuiting long-lived humoral immunity by the heightened engagement of CD40. J Clin Invest. 2002;109:613–620. doi: 10.1172/JCI14110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson AJ, Dunn RJ, Peach R, Aruffo A, Farr AG. The murine homolog of human Ep-CAM, a homotypic adhesion molecule, is expressed by thymocytes and thymic epithelial cells. Eur J Immunol. 1996;26:401–408. doi: 10.1002/eji.1830260220. [DOI] [PubMed] [Google Scholar]
- 38.Dunn RJ, Luedecker CJ, Haugen HS, Clegg CH, Farr AG. Thymic overexpression of CD40 ligand disrupts normal thymic epithelial organization. J Histochem Cytochem. 1997;45:129–141. doi: 10.1177/002215549704500116. [DOI] [PubMed] [Google Scholar]
- 39.Clegg CH, Rulffes JT, Haugen HS, Hoggatt IH, Aruffo A, Durham SK, Farr AG, Hollenbaugh D. Thymus dysfunction and chronic inflammatory disease in gp39 transgenic mice. Int Immunol. 1997;9:1111–1122. doi: 10.1093/intimm/9.8.1111. [DOI] [PubMed] [Google Scholar]
- 40.Porter JF, Vavassori S, Covey LR. A polypyrimidine tract-binding protein-dependent pathway of mRNA stability initiates with CpG activation of primary B cells. J Immunol. 2008;181:3336–3345. doi: 10.4049/jimmunol.181.5.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laughlin J, Oghlidos S, Porter JF, Matus-Nicodemos R, Sinquett FL, Marcelli V, Covey LR. Functional analysis of a tripartite stability element within the CD40 ligand 3' untranslated region. Immunology. 2008;124:368–379. doi: 10.1111/j.1365-2567.2007.02783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]