Abstract
Foxo transcription factors have a conserved role in the adaptation of cells and organisms to nutrient and growth factor availability. Here we show that Foxo1 has a crucial, nonredundant role in T cells. In naive T cells, Foxo1 controlled the expression of the adhesion molecule L-selectin, the chemokine receptor CCR7 and the transcription factor Klf2, and its deletion was sufficient to alter lymphocyte trafficking. Furthermore, Foxo1 deficiency resulted in a severe defect in interleukin 7 receptor α-chain (IL-7Rα) expression associated with its ability to bind an Il7r enhancer. Finally, growth factor withdrawal induced a Foxo1-dependent increase in Sell, Klf2 and Il7r expression. These data suggest that Foxo1 regulates the homeostasis and life span of naive T cells by sensing growth factor availability and regulating homing and survival signals.
Throughout adult life, the number and diversity of peripheral T cells depends on de novo cell development and cell division, balanced against programmed cell death. A growing number of studies show that this ‘homeostasis’ of T cells is controlled by cytokines, such as interleukin 7 (IL-7), as well as by interactions between T cell antigen receptor (TCR) and major histocompatibility complex (MHC)1,2. However, the cell-intrinsic factors responsible for the integration of environmental signals and the manner in which they manifest changes in cell populations remain poorly defined.
The Foxo subfamily of transcription factors has a highly conserved role in the regulation of life span, cell cycle progression, apoptosis, glucose metabolism and stress resistance by integrating information pertaining to the abundance of nutrients, growth factors and stress signals3. In mammals, the Foxo subfamily consists of four members: Foxo1 (A000944), Foxo3, Foxo4 and Foxo6. These factors often act as direct transcriptional activators or as coregulatory molecules through interactions with factors such as β-catenin, STAT3, Runx3, Smad3 or Smad4 (ref. 4). In response to growth factors such as insulin or cytokines, kinases downstream of phosphatidylinositol-3-OH kinase (PI(3)K), including Akt and/or SGK, phosphorylate Foxo proteins, resulting in their translocation to the cytoplasm and subsequent proteasomal degradation. Conversely, cell starvation and oxidative stress trigger the relocalization of Foxo members from the cytoplasm to the nucleus5.
Foxo1 and Foxo3 have been detected in Tand B cells6. After antigen or cytokine stimulation, they are rapidly phosphorylated and deactivated in a PI(3)K-dependent manner7–11, whereas cytokine withdrawal causes their dephosphorylation and activation8,12,13. In T and B cell lines, overexpression of Foxo3 induces arrest in the G1 phase of the cell cycle and apoptosis, associated with induction of the cell cycle inhibitor p27Kip1 and proapoptotic molecules FasL and Bim8,10,13,14. Moreover, Foxo1 and Foxo3 overexpression in the pro-B cell line Ba/F3 synergizes with the transcription factor EF1-δ to activate the transcription of Ccng2 (encoding cyclin G2) and Rbl2 (encoding retinoblastoma p130), two genes implicated in Foxo-dependent quiescence of fibroblasts15,16.
Although these studies suggest that Foxo transcription factors promote apoptosis induced by quiescence or growth factor withdrawal in lymphocytes, the function of Foxo1 and Foxo3 in T cells remains poorly understood. Mice harboring a targeted deletion of Foxo3 by retroviral insertion develop a mild lymphoproliferative syndrome associated with inflammatory lesions in multiple organs, CD4+ T cell autoreactivity and increased cytokine production by T cells after in vitro restimulation17. However, phenotypic and functional analysis of T cells from two different strains of Foxo3-deficient mice did not reveal any spontaneous or autoimmune-driven T cell activation (A.S.D., D.R.B., Y.M.K., A. Babour, K. Arden et al., unpublished observations; and refs. 18,19). In addition, studies involving acute deletion of Foxo1, Foxo3 and Foxo4 revealed a level of redundancy suggesting possible compensation between Foxo1 and Foxo3 in T cells20,21. Finally, the recent implications of Foxo1 and Foxo3 in the regulation of Rag1 and Rag2 expression and other aspects of B cell development revealed that these transcription factors could have unanticipated functions22–24.
Here we report that, consistent with its preferential expression in lymphoid cells, conditional deletion of Foxo1 substantially affected T cell homeostasis in vivo. Unexpectedly, whereas Foxo1 deletion did not result in spontaneous T cell activation, Foxo1 was required to maintain naive T cell homeostasis through the regulation of several genes crucially involved in T cell trafficking and survival. Finally, we provide evidence that Foxo1 is key to negative feedback circuits that dynamically balance growth factor signaling with homing and survival of naive T cells.
RESULTS
Foxo1 is preferentially expressed in lymphoid cells
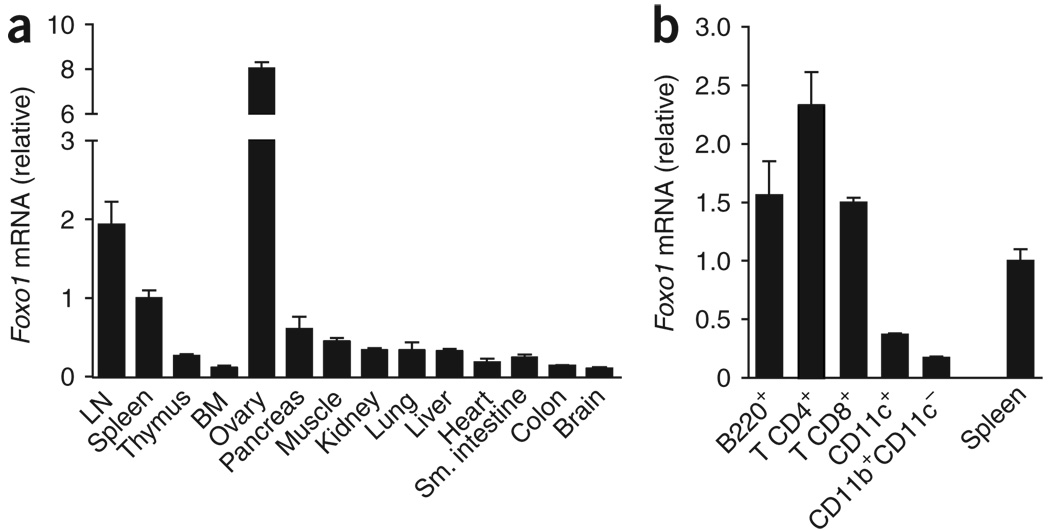
We compared the expression patterns of Foxo1 and Foxo3 in a variety of mouse tissues to explore their relative functional importance. Foxo3 showed a ubiquitous expression pattern, whereas the highest expression of Foxo1 mRNA was detected in the ovary, peripheral lymph nodes and spleen (Fig. 1a and data not shown). Quantitative PCR (QPCR) and immunoblot analyses of purified subsets revealed that Foxo1 was highly expressed in CD4+ T cells, CD8+ T cells and B cells compared with dendritic cells (CD11c+) and macrophages (CD11b+ CD11c−; Fig. 1b, Supplementary Fig. 1a online and data not shown). Consistent with these results, the expression of Foxo1 and Foxo3 mRNA reported in mouse and human expression databases25 further showed that this pattern is conserved across species (Supplementary Fig. 1b). These observations support the idea of an essential role for Foxo1 in T and B cells.
Figure 1.
Foxo1 is preferentially expressed in lymphoid cells. (a) QPCR analysis of Foxo1 mRNA expression in tissues from C57BL/6 mice. LN, lymph node; BM, bone marrow. (b) QPCR analysis of Foxo1 mRNA expression in purified cell subsets from C57BL/6 mice lymph nodes and spleen. The abundance of Foxo1 mRNA in each sample was normalized to that of Hprt1 mRNA and then normalized to the amount obtained for the spleen (set to 1). Data in a,b are mean ± s.d. of duplicate samples. Results are representative of two independent experiments.
Foxo1 deficiency impairs peripheral T cell homeostasis
Foxo1-null mice die at embryonic day 10.5 from defects in vasculogenesis18,26. To study the role of Foxo1 in T cell physiology in vivo, we crossed mice in which exon 2 of Foxo1 is flanked by loxP sites (Foxo1f/f mice) to mice carrying the Tg(Cd4-cre)1Cwi transgene (Cd4Cre mice) to induce a T cell–specific recombination. Efficient and specific deletion of Foxo1, but not Foxo3, was evidenced by PCR of genomic DNA and immunoblot analysis, thereby allowing us to specifically study the effect of Foxo1 deficiency (Supplementary Fig. 2 online).
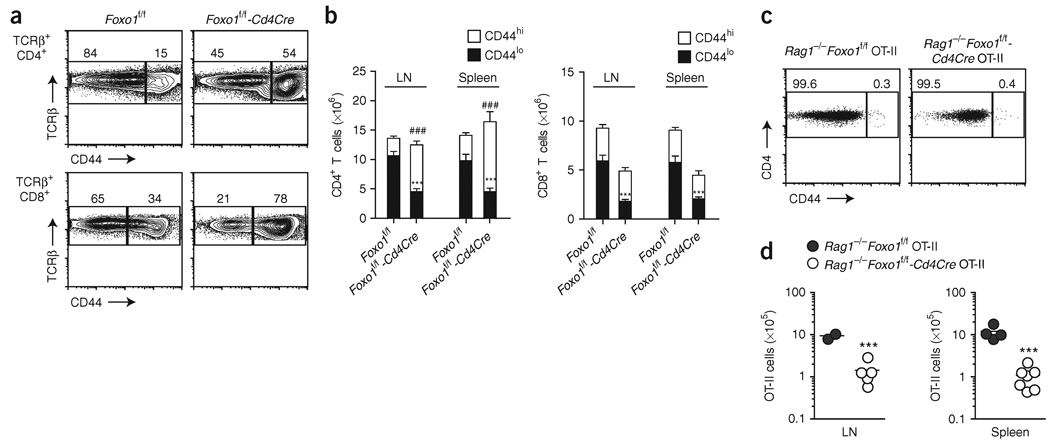
Phenotypic analysis of various T cell subpopulations in 8- to 12-week-old Foxo1f/f-Cd4Cre mice revealed a substantially higher proportion of activated-memory phenotype (CD44hi) CD4+ and CD8+ T cells compared to littermate controls (Fig. 2a). Accordingly, we noted a considerable increase in the proportion of cytokine-secreting cells of the T helper type 1, 2 and 17 subsets and in CD8+ T cells secreting both interferon-γ and tumor necrosis factor after restimulation ex vivo (Supplementary Fig. 3a online). Further analysis of the numbers of naive and activated-memory T cells indicated that this phenotype was the consequence of a reduced number of CD44loCD4+ T cells, balanced by an expansion of the CD44hi population; for CD8+ T cells, the deletion of Foxo1 seemed to specifically affect the number of CD44lo cells (Fig. 2b). Finally, most Foxo1-deficient CD4+ T cells showed characteristics typical of acutely activated T cells, as indicated by the concomitant increase in the proportion of cells expressing the early activation marker CD69 and the reduced expression of L-selectin (A001417) on CD44hi cells (Supplementary Fig. 3b). In contrast, no exaggerated increase in CD69-expressing cells was noted among CD8+ T cells, and the majority of the CD44hi cells were L-selectinhi, indicating that these cells were phenotypically related to central memory T cells (Supplementary Fig. 3b).
Figure 2.
Foxo1 is required for maintenance of T cell homeostasis. (a,b) CD44 expression by lymph node TCRβ+ CD4+ and CD8+ gated cells (a) and corresponding cell counts (b; mean ± s.e.m.) of 8- to 12-week-old mice. LN, lymph node. ### and ***, P < 0.0001 wild-type versus knockout for CD44hi and CD44lo populations, respectively). Data represent n = 16 littermate controls and n = 13 Cd4Cre mice analyzed in four independent experiments. (c) CD44 expression by lymph node Vβ5+ cells and (d) total counts of Vβ5+CD4+ cells of 8-week-old mice analyzed in two (spleen) or one (lymph node) experiments. Each circle indicates one mouse (***, P < 0.001).
Consistent with a role in T cell quiescence, we reasoned that the increase in activated CD4+ T cells in Foxo1f/f-Cd4Cre mice could be the result of spontaneous activation. To analyze this, we generated Foxo1f/f-Cd4Cre mice carrying the Tg(TcraTcrb425)Cbn transgene (called ‘OT-II’ here), which encodes a TCR specific for ovalbumin residues 323–339 in association with H-2Ab (ref. 27), and Rag1−/− alleles such that the resulting T cells did not recognize environmental or self-encoded antigens. Vβ5+CD4+ cells from mice Rag1−/− Foxo1f/f-Cd4Cre OT-II showed a typical naive CD44loCD69− phenotype, with no spontaneous proliferation, as measured by incorporation of bromodeoxyuridine (Fig. 2c and data not shown). However, reminiscent of the lower number of naive CD44lo T cells in Foxo1f/f-Cd4Cre mice, Foxo1 deletion in Rag1−/−Foxo1f/f-Cd4Cre OT-II mice resulted in a reduction of T cell numbers in secondary lymphoid organs to 10% of that in littermate controls (Fig. 2d). We concluded that Foxo1 has an essential and nonredundant role in T cell homeostasis independent of its potential role in quiescence. In addition, we hypothesized that the phenotype observed in Foxo1f/f-Cd4Cre mice is caused in part by a specific reduction in the number of naive CD8+ and CD4+ T cells.
Foxo1 is dispensable for T cell development
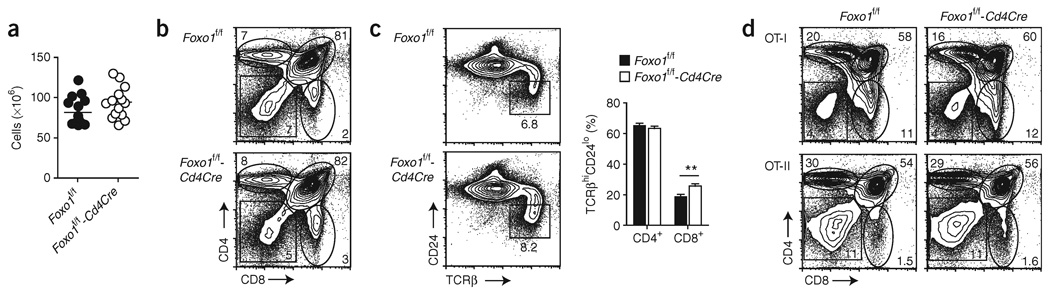
Foxo1 has been implicated in B and T cell development22–24,28. We therefore examined whether the observed peripheral phenotype could arise from a defect in thymic differentiation. Analysis of the numbers and proportions of double-negative, double-positive and single-positive thymic subsets did not reveal any significant differences between Foxo1f/f and Foxo1f/f-Cd4Cre mice (Fig. 3a,b), but we did note a trend in mice of the latter genotype toward a larger proportion of mature thymocytes (TCRβhiCD24lo) and an increased frequency of CD8+ cells (Fig. 3c). In addition, when Foxo1 deficiency was associated with transgenic expression of TCRs specific for MHC class I (Tg(TcraTcrb)1100Mjb transgene, or OT-I) or MHC class II (OT-II), there were again no significant differences in thymic cell subsets (Fig. 3d). Finally, we found that even early deletion of Foxo1, using the proximal Lck promoter to turn on Cre expression at the double-negative stage 3 of development (Tg(Lck-cre)1Cwi; called ‘LckCre’ here), had no effect on the principal thymic cell populations. The phenotype of these mice was similar to that of the Foxo1f/f-Cd4Cre mice, with a trend toward a greater number of mature CD8+ T cells compared with Foxo1+/+-Lck Cre mice (Supplementary Fig. 4 online). Consistent with the low expression of Foxo1 in the thymus (Fig. 1a) and reports showing that Foxo1 is expressed in only the most mature thymocytes (ref. 29 and http://www.immgen.org/index_content.html), these results indicate that the reduced number of peripheral, naive T cells in Foxo1f/f-Cd4Cre mice does not stem from a lack of progression through thymic development.
Figure 3.
Foxo1 is dispensable for T cell development. (a) Total numbers of thymocytes. Each dot indicates one mouse. (b) Expression of CD4 and CD8 on thymocytes of the indicated genotypes. (c) Left, expression of TCRβ and CD24 on thymocytes of the indicated genotypes. Right, percentages of CD4+ and CD8+ single-positive cells within the population of mature TCRβhiCD24lo thymocytes (mean ± s.e.m.) from 8-week-old mice. Data in a–c represent n = 17 littermate controls and n = 14 Cd4Cre mice, analyzed in five independent experiments (**, P < 0.01). (d) CD4 and CD8 expression on thymocytes from OT-II and OT-I transgenic mice. Data represent three to eight mice per genotype analyzed in two or three independent experiments.
Foxo1 controls naive T cell homing
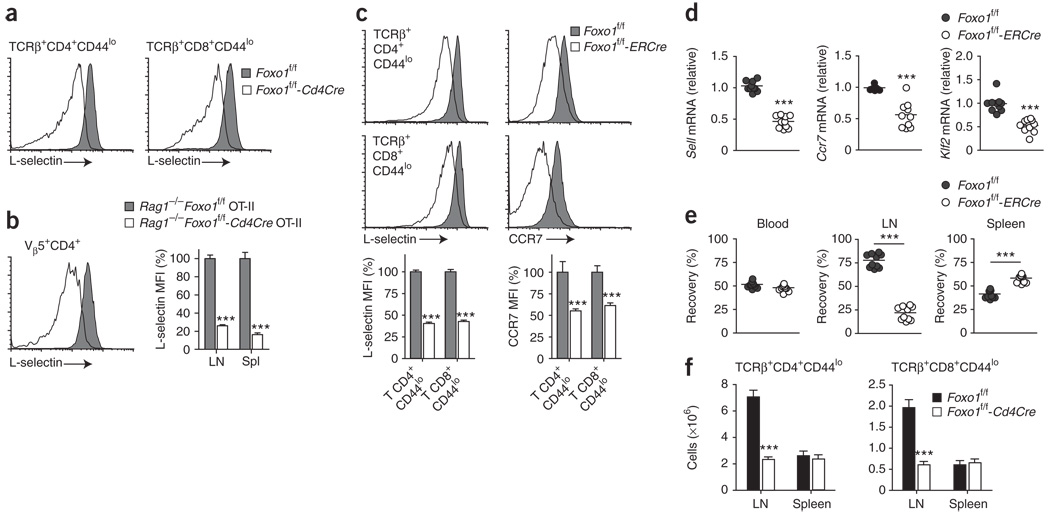
Initial phenotypic analysis of Foxo1-deficient T cells revealed a consistent reduction of L-selectin expression on naive CD4+ and CD8+ CD44lo T cells compared with wild-type cells, whereas CD11a (LFA1α, αL integrin), another receptor involved in lymph node migration, was unaffected (Fig. 4a and data not shown). In addition, although CD44 expression was unaffected, mature (TCRβhi) single-positive thymocytes from Foxo1f/f-Cd4Cre mice showed reduced surface expression of L-selectin, as did T cells from Rag1−/−Foxo1f/f-Cd4Cre OT-II mice (Fig. 4b and Supplementary Fig. 5 online). These results suggested that Foxo1 deficiency could alter naive T cell homing.
Figure 4.
Foxo1 regulates L-selectin, CCR7 and Klf2 expression and T cell homing in vivo. (a) L-selectin expression on CD44loTCRβ+ CD4+ and CD8+ cells of 8- to 12-week-old mice. Results are representative of n = 14 littermate controls and n = 11 Cd4Cre mice analyzed in four independent experiments. (b) Quantification of L-selectin expression on Vβ5+CD4+ cells of 8-week-old mice (mean ± s.e.m.). LN, lymph node. Data represent n = 4 littermate controls and n = 7 Cd4Cre mice analyzed in two independent experiments (***, P < 0.0001). MFI, mean fluorescence intensity. (c–e) Foxo1f/f-ERCre mice and littermate controls (CD45.2+) were treated for 5 d with tamoxifen and rested for 5 d. (c) Quantification of L-selectin and CCR7 expression on lymph node CD44loTCRβ+ CD4+ and CD8+ cells (mean ± s.e.m.). Data represent n ≥ 4 mice per genotype, analyzed in two to three independent experiments (***, P < 0.0001). (d) QPCR analysis of Sell, Ccr7 and Klf2 mRNA expression, normalized to Hprt1 mRNA, in purified lymph node T cells. Each circle indicates one mouse (***, P < 0.0001). (e) lymph node T cells were purified, and one of two populations was labeled with CFSE. The two populations were mixed at a 1:1 ratio and injected into C57BL/6 CD45.1+ mice (10 × 106 cells per mouse). Donor cell recovery was analyzed 18 h later in peripheral blood, lymph nodes and spleen (CD45.2+-gated CFSE+ versus CFSE− cells). Each circle indicates one host mouse. Results are from two independent experiments (***, P < 0.0001). (f) Number of naive T cells (mean ± s.e.m.) in 3-week-old mice. Data represent n = 6 littermate controls and n = 9 Cd4Cre mice analyzed in two independent experiments (***, P < 0.0001).
To further analyze this effect, we used mice expressing a chimeric Esr1-cre gene recombined into the ubiquitously expressed Rosa26 locus30 (Gt(ROSA)26Sor; called ‘ERCre’ here). After treatment with tamoxifen, the estrogen receptor (ER)-Cre fusion protein, normally sequestrated in the cytoplasm, translocates to the nucleus, allowing Cre-mediated deletion of loxP-flanked alleles. We treated Foxo1f/f and Foxo1f/f-ERCre mice for 5 d with tamoxifen and then rested them for 5 d. QPCR and immunoblot analysis of purified T cells showed that acute activation of the ER-Cre fusion protein resulted in efficient reduction of Foxo1 mRNA and protein, whereas Foxo3 mRNA expression remained unaltered (Supplementary Fig. 6a,b online). Notably, this short-term deficiency did not significantly affect the proportion of naive and activated-memory T cell populations (Supplementary Fig. 6c), indicating that the phenotype observed in adult Foxo1f/f-Cd4Cre mice requires prolonged insufficiency of Foxo1. In addition, this experimental system allowed us to exclude the contribution of potential secondary effects induced by excessive T cell activation, a lymphopenic environment or abnormal thymic T cell maturation. Similar to the effect of Cd4Cre-mediated Foxo1 deletion, acute tamoxifen-mediated deletion of Foxo1 induced a 60% reduction of L-selectin protein expression on CD44lo CD4+ and CD8+ T cells, associated with a 50% reduction of Sell mRNA (L-selectin) in purified lymph node T cells (Fig. 4c,d).
Further analysis of homing receptor expression on CD44lo T cells in Foxo1f/f-ERCre and Foxo1f/f-Cd4Cre mice revealed that Foxo1 deficiency also affects both protein and mRNA expression of CCR7 (A000630; Fig. 4c,d, Supplementary Fig. 5 and data not shown). Because both L-selectin and CCR7 expression are dependent on the transcription factor Klf2 (ref. 31), we examined Klf2 mRNA expression. The results showed that tamoxifen treatment caused a 50% decrease in Klf2 expression in T cells from Foxo1f/f-ERCre mice relative to that of T cells from Foxo1f/f mice (Fig. 4d).
Finally, we investigated the functional effects of Foxo1 deletion on T cell trafficking in vivo. We purified lymph node T cells from tamoxifen-treated Foxo1f/f and Foxo1f/f-ERCre mice, labeled them with the cytosolic dye CFSE, mixed them at a ratio of 1:1 and transferred them into wild-type CD45.1 recipients. The number of Foxo1-deficient T cells in lymph nodes was severely impaired. After 18 h, although we recovered an equal proportion of transferred T cells from blood, we found that the ability of Foxo1-deficient T cells to migrate into the lymph nodes was considerably impaired relative to that of Foxo1-sufficient cells (Fig. 4e). Consistent with the altered migratory properties of L-selectin– and CCR7-deficient T cells, as well as pertussis toxin–treated T cells32–34, Foxo1-deficient T cells accumulated in the spleen (Fig. 4e). Together, these data show that Foxo1 regulates the expression of L-selectin, CCR7 and Klf2 and controls homing of naive T cells in vivo.
Foxo1 is required for naive T cell survival
L-selectin and CCR7 deficiencies are associated with decreased T cell numbers in the lymph nodes and normal or increased T cell numbers in the blood and spleen32,33; in contrast, Klf2 deficiency induces a general decrease in the number of peripheral T cells owing to retention of mature T cells in the thymus and abnormal homing31,35. Consistent with an important role for Foxo1 in naive T cell homing and with the acute deletion experiments, 3-week-old Foxo1f/f-Cd4Cre mice showed a specific reduction in the number of naive lymph node T cells (Fig. 4f). Our results also showed that Foxo1 deletion does not significantly affect thymic cell numbers, but leads to a similarly lower number of naive CD44lo T cells in the lymph nodes and spleen of adult Foxo1f/f-Cd4Cre mice, Foxo1f/f-LckCre mice and Foxo1f/f-ERCre mice 5 weeks after tamoxifen treatment (Figs. 2 and 3 and data not shown). Moreover, compared with Foxo1f/f mice, adult Foxo1f/f-Cd4Cre mice showed a substantial decrease in the proportion and relative number of circulating blood T cells, relative to wild-type, and those present were mostly CD44hi (Fig. 5a and data not shown). We therefore considered whether, in addition to the role of Foxo1 in the regulation of T cell trafficking, prolonged loss of Foxo1 might also affect T cell survival.
Figure 5.
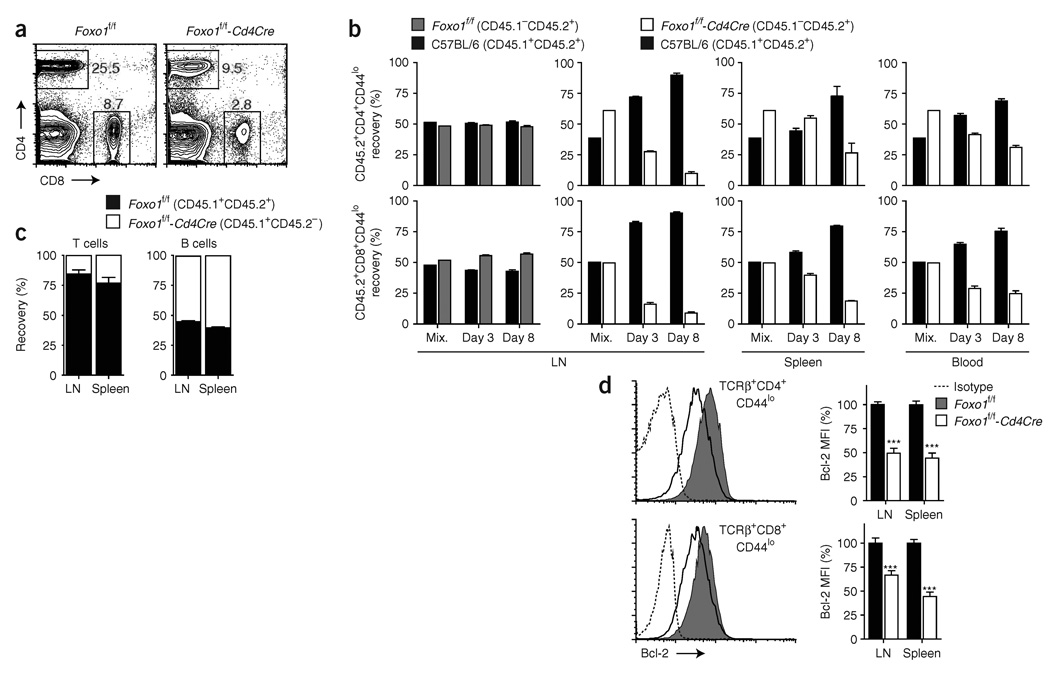
Foxo1 is required for naive T cell survival. (a) CD4+ and CD8+ cells in peripheral blood of 8- to 12-week-old mice, as assessed by flow cytometry. Results are representative of n = 5 littermate controls and n = 9 Cd4Cre mice analyzed in two independent experiments. (b) Purified lymph node T cells were mixed at a 1:1 ratio based on the number of CD8+CD44lo cells and transferred into C57BL/6 CD45.1+ hosts (2 × 106 total CD8+CD44lo cells). Donor cell recovery was analyzed before injection (Mix.) and at the indicated time points (CD45.2+-gated CD45.1+ versus CD45.1− cells). LN, lymph node. Representative results from two independent experiments with n ≥ 3 mice per group and per time point (initial difference in CD4+CD44lo proportion comes from a modification of the CD4:CD8 ratio in Foxo1f/f-Cd4Cre mice). (c) T and B cell recovery in mixed bone chimeras 8 weeks after reconstitution (mean ± s.e.m. of n = 3 mice per group). (d) Quantification of Bcl-2 expression in CD44lo T cells from 8- to 12-week-old mice (mean ± s.e.m.). Data represent n = 9 littermate controls and n = 12 Cd4Cre mice analyzed in two independent experiments (***, P < 0.0001).
To test this hypothesis, we adoptively transferred wild-type T cells with Foxo1f/f or Foxo1f/f-Cd4Cre T cells from adult mice into wild-type hosts. As opposed to naive Foxo1-sufficient T cells, which were normally maintained, the proportion of naive CD44lo CD4+ and CD8+ T cells from Foxo1f/f-Cd4Cre mice rapidly decreased in all organs tested (Fig. 5b and data not shown). Notably, we consistently recorded a diminished T cell recovery in lymph nodes compared to spleen and blood, suggestive of altered T cell homing. The impaired maintenance of Foxo1-deficient cells was not caused by Cre-induced toxicity or rejection, as Foxo1f/+-Cd4Cre T cells were normally maintained after transfer (data not shown). Additionally, generation of mixed bone marrow chimeras showed that Foxo1f/f-Cd4Cre bone marrow cells were unable to reconstitute a normal T cell compartment (Fig. 5c).
T cell survival is dependent on balanced expression of the proapoptotic protein Bim and prosurvival proteins including Bcl-2 and Bcl-xL1,36. Although previous studies have shown a role for Foxo3 as a transcriptional regulator of Bim expression in T cells, we did not detect changes in Bim mRNA expression after acute deletion of Foxo1 (Supplementary Fig. 6d). However, we did find decreased expression of Bcl-2 in CD44lo CD4+ and CD8+ T cells (Fig. 5d). These results collectively indicate that Foxo1 is required to maintain the expression of Bcl-2 and the survival of naive T cells in vivo.
Foxo1 controls IL-7Rα expression in naive T cells
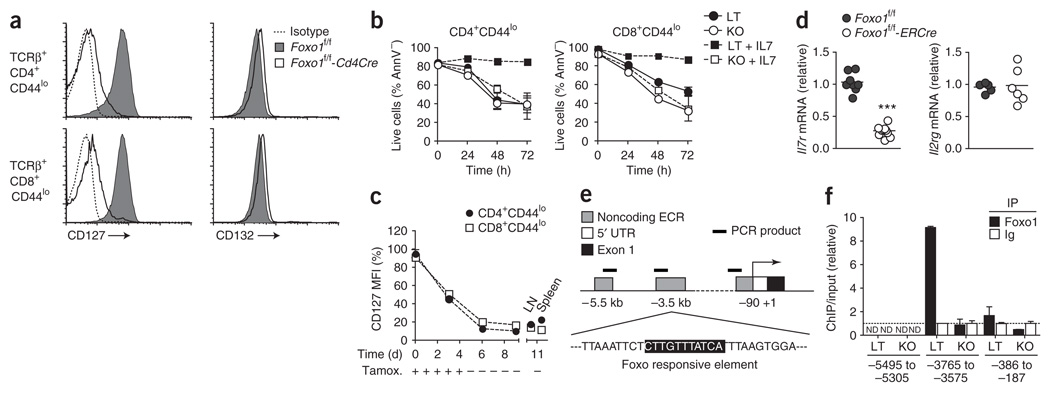
IL-7 is required for the survival of naive T cells in vivo, and one consequence of IL-7R (A001267) signaling is Bcl-2 induction1,2,37,38. As Foxo transcription factors are inactivated after cytokine stimulation and thus are unlikely to be directly responsible for this effect, we considered that Foxo1 deletion could alter the expression of IL-7R. Wild-type, naive CD44lo CD4+ and CD8+ T cells expressed high amounts of both the IL-7Rα chain (CD127) and the common cytokine receptor γ-chain (CD132) constituting the IL-7R. Notably, IL-7Rα expression was severely impaired on CD44lo CD4+ and CD8+ T cells from Foxo1f/f-Cd4Cre mice, whereas expression of γ-chain was unaffected (Fig. 6a). Consistent with this phenotype, addition of IL-7 did not rescue naive Foxo1-deficient T cells from death induced by ex vivo growth factor withdrawal, and STAT5 phosphorylation induced by IL-7 was markedly reduced (Fig. 6b and data not shown).
Figure 6.
Foxo1 is required for IL-7Rα expression in naive T cells and binds to an Il7r enhancer. (a) CD127 and CD132 expression on CD44lo T cells from 8- to 12-week-old mice. Data represent n = 9 littermate controls (LT) and n = 10 Cd4Cre mice (KO) analyzed in three independent experiments. (b) Lymph node (LN) cells were cultured in medium supplemented or not with IL-7 for 3 d, and the proportion of live (annexin V–negative, AnnV−) CD44lo CD4+ and CD8+ T cells was measured by flow cytometry at the indicated time points (mean ± s.d. of triplicate cultures). Results are representative of three independent experiments. (c) Quantification of CD127 expression on CD44lo CD4+ and CD8+ T cells after tamoxifen treatment in Foxo1f/f-ERCre mice (mean ± s.e.m.). MFI, mean fluorescence intensity. Results are representative of three independent experiments with n = 7–11 mice per time point and per genotype. (d) QPCR analysis of Il7r and Il2rg mRNA expression in purified lymph node T cells on day 11 after the beginning of tamoxifen treatment. Each dot indicates one mouse (***, P < 0.0001). (e) Il7r locus. (f) Chromatin immunoprecipitation analysis of Foxo1 binding to the Il7r locus in purified lymph node T cells from littermate controls and Cd4Cre mice. Results are relative to the value obtained for the control immunoprecipitation (Ig), with Foxo1-sufficient T cells set as 1 (mean ± s.d. of duplicate samples). ND, not detected. Results are representative of four independent experiments.
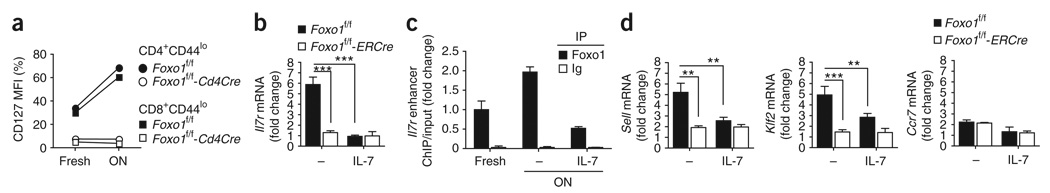
The expression of IL-7Rα was also impaired on thymic, mature T cells from Foxo1f/f-Cd4Cre mice (Supplementary Fig. 5). We thus wished to determine whether the impaired expression of IL-7Rα on peripheral, naive T cells arose from a blockade of its expression during T cell maturation or whether Foxo1 was required for continuous expression. We treated Foxo1f/f and Foxo1f/f-ERCre mice with tamoxifen and followed the expression of IL-7Rα on peripheral CD44lo T cells over time. Acute deletion of Foxo1 induced a rapid and profound downregulation of IL-7Rα expression associated with a significant reduction of Il7r mRNA, but not Il2rg mRNA, in purified lymph node T cells (Fig. 6c,d).
Among the genes we found to be affected by Foxo1 deletion, Il7r showed the most pronounced regulation. We therefore sought to determine whether Foxo1 directly regulates Il7r expression. Genomic alignment of the Il7r locus across several mammalian species indicated the presence of three evolutionarily conserved noncoding regions (ECRs) upstream of the transcription initiation site, including one corresponding to the defined promoter region (Supplementary Fig. 7 online and ref. 39). Detailed bioinformatic analysis of transcription factor binding sites in each of these regions revealed the presence of a highly conserved Foxo binding sequence in ECR2 located 3.5 kb upstream of the transcription initiation site (Fig. 6e and Supplementary Fig. 7). To determine whether Foxo1 can directly bind within the Il7r locus, we conducted chromatin immunoprecipitation experiments using primer sets designed to amplify regions located in each of these ECRs. In purified lymph node T cells, Foxo1 bound to the Il7r locus specifically within the region containing this putative binding site (Fig. 6f). Collectively, these data strongly suggest that Foxo1 regulates IL-7Rα expression by binding directly to this ECR in the Il7r locus.
Dynamic regulation of Foxo1 activity
Growth factor withdrawal leads to Foxo dephosphorylation and increased activity, whereas TCR or cytokine (including IL-7) stimulation induces Foxo phosphorylation and decreased transcription of target genes. This finding is consistent with the observation that T cells cultured in the absence of growth factors show increased IL-7Rα expression, whereas stimulation with IL-7, IL-2, IL-4, IL-6 or IL-15 decreases IL-7Rα expression40,41. Accordingly, overnight culture of Foxo1-sufficient T cells in culture medium without added growth factors resulted in a strong increase in both IL-7Rα surface expression and Il7r mRNA, an effect that was completely inhibited by the addition of IL-7 (Fig. 7a,b and data not shown). A deficiency in Foxo1 completely prevented this increase, and the addition of IL-7 had no effect (Fig. 7a,b). Moreover, overnight growth factor starvation of lymph node T cells was associated with recruitment of Foxo1 to the Il7r locus, an effect inhibited by the addition of IL-7 (Fig. 7c).
Figure 7.
Foxo1-mediated control of Il-7Rα and trafficking receptors after cell starvation. (a) CD127 expression on lymph node T cells freshly isolated and rested overnight (ON) in medium (mean ± s.d. of triplicate cultures). (b–d) Foxo1f/f-ERCre mice and littermate controls were treated with tamoxifen for 5 d and rested for 3 d. Lymph node T cells were then purified and cultured overnight in medium supplemented with IL-7 (10 ng/ml) as indicated. (b) QPCR analysis of Il7r mRNA, normalized to Hprt mRNA, after overnight culture. c, Chromatin immunoprecipitation analysis of Foxo1 binding to IL7r ECR2. (d) QPCR analysis of Sell, Klf2 and Ccr7 mRNA. Results are presented as fold change (mean ± s.d. of triplicate cultures) relative to the value obtained for freshly isolated T cells set to 1. Results are representative of two (a,c) or three (b,d) independent experiments (**, P < 0.01; ***, P < 0.0001).
Previous studies have shown that culturing T cells in absence of any stimulation also induces an increase in the surface expression of L-selectin on both CD4+ and CD8+ T cells42, although the expression of CCR7 does not seem to be affected43. Considering our previous results, we analyzed the role of Foxo1 in these effects and observed that growth factor withdrawal induces a similar Foxo1-dependent increase in the expression of Sell and Klf2 mRNA in naive T cells (Fig. 7d). This increase was again prevented by the addition of IL-7. Notably, Ccr7 mRNA expression was unaffected by any of these culture conditions, independent of Foxo1 expression. These results also revealed that the expression of L-selectin, CCR7 and Klf2 does not require IL-7R signaling, thus showing that the defective expression of these molecules in Foxo1-deficient T cells is not secondary to the defective IL-7Rα expression. Furthermore, L-selectin and CCR7 were not decreased on CD44lo T cells from Il7r−/− mice (Supplementary Fig. 8 online), in agreement with the phenotype of TCR-transgenic Il2rg−/− T cells44.
As the expression of Il7r and Sell increased in wild-type T cells but not Foxo1-deficient T cells ex vivo, this suggested that basal inhibition of Foxo1 in vivo limits their expression. Indeed, acute deletion of the PI(3)K-dependent pathway inhibitor PTEN was sufficient to decrease the expression of IL-7Rα and L-selectin in naive T cells in vivo (Supplementary Fig. 9a,b online). Moreover, Pten deletion prevented IL-7Rα upregulation after cytokine withdrawal ex vivo and enhanced its downregulation after IL-7 stimulation (Supplementary Fig. 9c), thus showing that unrestrained activation of the PI(3)K pathway inhibits IL-7Rα expression. Collectively, these results indicate that pathways controlling homing and survival of naive T cells are coordinately and dynamically regulated by growth factor availability through the transcription factor Foxo1.
DISCUSSION
Maintenance of lymphocyte homeostasis is crucial to prevent immunopathology while promoting the generation of protective immunity. Studies over the past 30 years have shown that the T cell population is regulated by homeostasis, albeit with a high degree of plasticity45,46. More recent studies have established the importance of TCR-MHC interactions in maintaining T cell viability and of IL-7 as the essential T cell survival cytokine47–50. However, the cell-intrinsic factors acting as the ‘control center’ to integrate these environmental signals and determine the appropriate response remain elusive. To achieve this function, T cells may use mechanisms universally used in eukaryotes. In particular, the Foxo family of transcription factors has a highly conserved role in the regulation of cellular and organismal metabolism depending on nutrient or growth factor availability.
To date, most studies on Foxo transcription factors have shown their involvement in quiescence of naive T cells and apoptosis induced by growth factor withdrawal7,9,51,52, but these hypotheses have not been exhaustively tested in vivo. Our experiments show that Foxo1 deletion alone does not result in spontaneous T cell proliferation, increased numbers of cells, modification of Bim expression or increased resistance to apoptosis induced by growth factor withdrawal. Rather, we provide compelling evidence that Foxo1 plays a crucial role in naive T cell homeostasis by promoting homing to lymph nodes and survival of naive T cells. However, as Foxo1-deficient T cells still express Foxo3, these results do not rule out the likely possibility of redundancy in the regulation of target genes, which may include those involved in quiescence and apoptosis.
Deletion of Foxo1 resulted in a specific reduction of naive T cells in the lymph nodes of young Foxo1f/f-Cd4Cre mice, associated with decreased cell surface expression of L-selectin and CCR7 and diminished expression of Sell, Ccr7 and Klf2 mRNAs. Consistent with these data, naive Foxo1-deficient T cells showed defective homing after transfer in vivo. Naive T cells were not completely absent from peripheral lymph nodes, and thymus egress seemed fairly unperturbed. The expression of L-selectin, CCR7 and Klf2 was diminished in Foxo1-deficient naive T cells but not entirely abrogated, indicating that other mechanisms are involved in the regulation of these genes. Furthermore, these results suggest that the remaining expression may be sufficient to allow thymic egress and homing of naive T cells into lymph nodes, albeit at a lower rate. Notably, a previous study showed that the inhibition of Klf2, L-selectin and CCR7 expression is controlled by PI(3)K signaling during T cell activation53. We thus consider the possibility of a redundancy between Foxo1 and Foxo3. In support of this hypothesis, preliminary results indicate that loss of both Foxo1 and Foxo3 affects T cell egress from the thymus (Y.M.K. and S.M.H., unpublished observations). However, the transcriptional regulation of Klf2, L-selectin and CCR7 is, at least partially, rapamycin sensitive, suggesting the existence of alternative PI(3)K-dependent regulatory mechanisms that coexist with Foxo1 to control expression53.
Klf2 deficiency is associated with defective L-selectin expression, and Klf2 can transactivate the Sell promoter in reporter assays in vitro31,54. The decreased L-selectin expression in Foxo1-deficient T cells can therefore originate from the Foxo1-mediated control of Klf2 transcription. Indeed, one study reports that overexpression of a constitutively active form of human FOXO1 induces the expression of L-selectin and KLF2, and FOXO1 binds to the KLF2 promoter in human T cells55. Consistent with our results, this study also showed that overexpression of active FOXO1 in a Jurkat T cell line leads to the induction of CCR7 mRNA expression. As KlF2 deficiency affects the cell surface expression, but not the transcription, of Ccr7 (refs. 31,35), these data indicate that Foxo1 may also act through other regulatory mechanisms to regulate CCR7 expression and thus naive T cell homing. Supporting this view and further indicating the uncoupled regulation of L-selectin and CCR7, previous work and our results show that T cell starvation induces a parallel Foxo1-dependent increase of both Sell and Klf2 transcription, whereas Ccr7 expression is unchanged.
The results presented here show that Foxo1 further links the regulation of homing with the regulation of cell viability by controlling IL-7Rα expression in naive T cells. Foxo1f/f-Cd4Cre mice represent the first model with such a profound defect in IL-7Rα expression on peripheral naive T cells, without the complications associated with T cell development in mice deficient in IL-7 or IL-7Rα. Despite the well-known role of IL-7 signaling in naive T cell survival, we detected a substantial number of naive T cells in secondary lymphoid organs of Foxo1f/f-Cd4Cre mice. Consistent with our results, anti-IL7Rα treatment in vivo induced a comparable decrease in T cell numbers29. In addition, as observed in Il7r−/− mice49, the expression of Bcl-2 is not completely abrogated. One conclusion is that other pathways contribute to Bcl-2 expression and naive T cell survival in vivo. Of note, we observed that expression of the OT-II TCR on Foxo1-deficient T cells accentuates the decrease in naive T cell numbers, consistent with a role for TCR-MHC interactions.
Despite the loss of IL-7Rα expression and paucity of naive T cells, we observed normal or higher numbers of activated-memory phenotype cells in Foxo1f/f-Cd4cre mice. These results therefore indicate that other growth factors, such as IL-15, could substantially contribute to the survival of these cells; however, the TCR-dependent expansion and increased proportion of CD4+ T cells expressing the early-activation marker CD69 suggest that Foxo1 has other important functions in the regulation of T cell homeostasis.
The expression of IL-7Rα is regulated by growth factor–induced negative feedback41. Growth factor withdrawal resulted in Foxo1 recruitment to a region of the Il7r locus previously characterized for glucocorticoid receptor–dependent enhancer activity56 and was associated with Foxo1-dependent IL-7Rα expression. At least for CD8+ T cells, this regulation may be further modulated by another factor, GFI41, but these data establish an essential role for Foxo1 in the regulation of IL-7Rα by negative feedback.
One theory regarding the logic underlying the role of Foxo1 in the common control of homing receptors and IL-7Rα is that the amount of available IL-7 is fixed and limiting for T cell survival; and this IL-7 concentration determines, in part, the size of the T cell population37. For example, at least some IL-7 transgenic mice have greatly expanded numbers of T cells57,58. Also, naive T cells compete for limited self-peptide–MHC complexes to maintain their survival47,59, and both IL-7Rα and TCR signaling activate PI(3)K and Akt, resulting in the inactivation of Foxo factors9,60,61. Furthermore, as we show here, such signaling inhibits IL-7Rα, L-selectin and CCR7 expression, so those T cells receiving the most stimulation will subsequently be disadvantaged in two ways. First, they will home to secondary lymphoid organs at a reduced rate—and this in itself is a requirement for survival43,62; second, they will compete less effectively for limited IL-7. This negative feedback is predicted to cause an oscillation of survival and homing signals and prevent the most avaricious T cells from dominating the population. Thus, we predict that this feedback circuit is crucial to prevent narrowing of the naive repertoire in the periphery once generated and selected in the thymus. Together, the results presented here reveal an unanticipated connection between homing and survival of naive T cells through the transcription factor Foxo1 and emphasize its importance in the regulation of naive T cell homeostasis.
METHODS
Mice and tamoxifen treatment
C57BL/6, C57BL/6 CD45.1+, Cd4Cre, LckCre, Rag1−/−, OT-II and OT-I mice were maintained in pathogen-free conditions. ERCre mice were provided by T. Ludwig30, and Ptenf/f-ERCre mice were provided by C. Murre. Gene-trapped Foxo3-deficient (Foxo3Kca) mice were provided by K. Arden and backcrossed ten times to C57BL/6 mice. Foxo1f/f mice have been described20,21. H-2b homozygous F2 mice from Foxo1f/f (FVB/N, H-2q) mice crossed to Cd4Cre (C57BL/6, H-2b) or ERCre (C57BL/6, H-2b) mice were used to set up breeding pairs. Analysis of Foxo1f/+-Cd4Cre mice did not reveal any significant phenotypic differences from Foxo1f/f mice, ruling out a Cre-mediated effect or hemizygous gene dosage effect as a cause of the Foxo1f/f-Cd4Cre mice phenotype (data not shown). Mice used in short-term T cell transfer experiments were generated from Foxo1f/+ mice backcrossed six times to C57BL/6 mice. CD45.1+CD45.2+ C57BL/6 mice were produced by breeding C57BL/6 mice with CD45.1+ C57BL/6 mice. All procedures were approved by the Animal Care and Use Committee of the University of California, San Diego. ERCre-mediated deletion of floxed alleles was induced by intraperitoneal injection of 1 mg of tamoxifen (Sigma) emulsified in 200 µl of sunflower seed oil (Sigma) every day for 5 d.
Flow cytometry
Cell suspensions prepared from the indicated organs were incubated for 20 min at 4 °C in PBS containing 1% FCS, 2 mM EDTA, 0.01% NaN3 and the indicated fluorochrome-conjugated antibodies, in the presence of an optimal concentration of 2.4G2 hybridoma culture supernatant (antibody to mouse FcγRII/III). CCR7 staining was done at 37 °C for 30 min with phycoerythrin-conjugated antibody to mouse CCR7 (eBioscience). All intracellular staining was done with BD Cytofix/Cytoperm (BD Biosciences). Antibodies were purchased from eBioscience or BD PharMingen; clone identifiers are listed in Supplementary Table 1 online. Data were collected on a FACSCalibur (BD Biosciences) and analyzed with FlowJo software (Tree Star). Mean fluorescence intensity quantifications across experiments were assessed by normalizing mean fluorescence intensity values obtained for each mouse, with the mean of the values obtained for control mice set as 100% for each experiment or time point.
Immunoblot
Whole-cell extracts were resolved on 4–12% SDS-PAGE gels (Invitrogen) and transferred to a polyvinylidene fluoride membrane (Millipore) using a semidry transfer cell (Bio-Rad). Blots were blocked and incubated with the primary antibody at 4 °C overnight, followed by a 2-h incubation at 25 °C with the appropriate horseradish peroxidase–conjugated secondary antibody. Primary antibodies to the following molecules were used: Foxo1 (rabbit polyclonal; 9462; Cell Signaling Technology), phospholipase C-γ (Upstate Biotechnology) and β-tubulin (Upstate Biotechnology). Rabbit polyclonal antibody to Foxo3 was provided by A. Brunet. The specificity of this antibody was confirmed by the inclusion of cells from Foxo3-deficient mice.
Cell isolation and culture
Lymph node T cells were isolated by magnetic depletion of unwanted cells stained with a mix of biotinylated antibodies to B220, CD19, MHCII, DX5 and CD11b (all from eBioscience) and streptavidin-coupled microbeads (Miltenyi Biotec). B cells were isolated by magnetic positive selection from spleen cell suspensions stained with biotinylated antibody to CD19 and streptavidin-coupled microbeads. CD11c+ and CD11b+CD11c− cells were sorted on a FACSAria (BD Biosciences) from collagenase D–treated spleen (1 mg/ml for 30 min at 37 °C). Cell purity was routinely over 95%. For overnight culture, purified T cells were cultured at 5 × 106 cells/ml in complete RPMI medium (Gibco) supplemented with 5% FCS (Omega). For cell survival experiments, total lymph node cell suspensions were cleared from dead cells by density gradient centrifugation on Lympholyte-M (Cedarlane) and then cultured at 5 × 106 cells/ml in complete RPMI medium (Gibco) supplemented with 10% FCS (Omega). When indicated, cells were treated with 10 ng/ml recombinant mouse IL-7 (eBioscience).
Quantitative PCR
Total RNA was extracted from tissues or purified cell populations with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, treated with DNase using a DNA-free kit (Ambion) and subjected to reverse transcription with SuperScript III reverse transcriptase and random hexamers (both from Invitrogen). cDNA was analyzed in duplicate by QPCR amplification using Power SYBR Green PCR Master mix (Applied Biosystems) supplemented with 30 nM of reference dye (Stratagene) on an Mx3005P system (Stratagene). Data were analyzed by comparative quantification with MxPro software. Primer sequences and PCR conditions are shown in Supplementary Table 2 online.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were done using a chromatin immunoprecipitation kit (17–295; Upstate Biotechnology) according to the manufacturer’s instructions, with minor modifications. Briefly, 10 × 106 to 15 × 106 cells were fixed with 1% formaldehyde in PBS for 10 min at 25 °C with agitation. Fixed cells were immediately lysed with SDS lysis buffer for 10 min at 4 °C and sonicated with a digital Sonifier 250 (six 10-s pulses at 20% amplitude; Branson Ultrasonics). Lysates were diluted ten-fold, precleared for 2 h at 4 °C with salmon sperm DNA–protein A agarose, divided into two equal fractions and incubated overnight at 4 °C with 7.5 µg of either antibody to the transcription factor FKHR (H-128; Santa Cruz Biotechnology) or rabbit IgG. Protein-DNA immune complexes were then collected with protein A agarose beads, washed, eluted from the beads and incubated with NaCl (200 mM final concentration) for 4 h at 65 °C to reverse cross-links. After treatment with proteinase K, DNA was extracted with phenol-chloroform, precipitated with 100% ethanol for 2 h at –20 °C, washed and resuspended in Tris-EDTA buffer. Immunoprecipitates and input fraction were analyzed in duplicate by QPCR (Supplementary Table 3 online).
Mixed bone marrow chimeras
T cell–depleted bone marrow cells from CD45.1+CD45.2+Foxo1f/f-Cd4Cre and CD45.1+Foxo1f/f littermates were mixed at a 1:1 ratio and injected intravenously into lethally irradiated CD45.2+ C57BL/6 mice. Mice were killed and analyzed 8 weeks after reconstitution.
Statistics
Unpaired two-tailed Student t tests were used for statistical analysis, with GraphPad Prism software.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. D’Souza and M. McGargill for discussions and assistance with adoptive transfer experiments; T. Ludwig (Columbia University) for ERCre mice; C. Murre (University of California at San Diego) for PTENf/f-ERCre mice; H. Cheroutre (La Jolla Institute for Allergy and Immunology) for Il7r−/− mice; S. Kaech for identifying the evolutionarily conserved regions of Il7r; and A. Goldrath and P. Marrack for critical reading of the manuscript. Supported by funds made available by the University of California, San Diego Division of Biological Sciences.
Footnotes
Accession codes. UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org): A000944, A001417, A000630 and A001267.
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
Y.M.K. designed and conducted all of the experiments, in collaboration with D.R.B., R.T. and A.S.D. The breeding and initial characterization of the Foxo1; Cd4Cre mice were carried out by D.R.B. and R.T. Mice with a lox P-targeted Foxo1 locus were produced by D.H.C. and R.A.D. S.M.H. initiated the project with R.A.D. and supervised the experimentation. Y.M.K. and S.M.H. wrote the manuscript with contributions from the other authors.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Marrack P, Kappler J. Control of T cell viability. Annu. Rev. Immunol. 2004;22:765–787. doi: 10.1146/annurev.immunol.22.012703.104554. [DOI] [PubMed] [Google Scholar]
- 2.Almeida AR, Rocha B, Freitas AA, Tanchot C. Homeostasis of T cell numbers: from thymus production to peripheral compartmentalization and the indexation of regulatory T cells. Semin. Immunol. 2005;17:239–249. doi: 10.1016/j.smim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 4.van der Vos KE, Coffer PJ. FOXO-binding partners: it takes two to tango. Oncogene. 2008;27:2289–2299. doi: 10.1038/onc.2008.22. [DOI] [PubMed] [Google Scholar]
- 5.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 6.Peng SL. Foxo in the immune system. Oncogene. 2008;27:2337–2344. doi: 10.1038/onc.2008.26. [DOI] [PubMed] [Google Scholar]
- 7.Charvet C, et al. Vav1 promotes T cell cycle progression by linking TCR/CD28 costimulation to FOXO1 and p27kip1 expression. J. Immunol. 2006;177:5024–5031. doi: 10.4049/jimmunol.177.8.5024. [DOI] [PubMed] [Google Scholar]
- 8.Stahl M, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 9.Barata JT, et al. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J. Exp. Med. 2004;200:659–669. doi: 10.1084/jem.20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijkers PF, et al. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1) Mol. Cell. Biol. 2000;20:9138–9148. doi: 10.1128/mcb.20.24.9138-9148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabre S, et al. Stable activation of phosphatidylinositol 3-kinase in the T cell immunological synapse stimulates Akt signaling to FoxO1 nuclear exclusion and cell growth control. J. Immunol. 2005;174:4161–4171. doi: 10.4049/jimmunol.174.7.4161. [DOI] [PubMed] [Google Scholar]
- 12.You H, et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J. Exp. Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 14.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Gac L, Marques M, Garcia Z, Campanero MR, Carrera AC. Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol. Cell. Biol. 2004;24:2181–2189. doi: 10.1128/MCB.24.5.2181-2189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kops GJ, et al. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol. Cell. Biol. 2002;22:2025–2036. doi: 10.1128/MCB.22.7.2025-2036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin L, Hron JD, Peng SL. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Hosaka T, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. USA. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- 20.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herzog S, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat. Immunol. 2008;9:623–631. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 24.Dengler HS, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat. Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuyama T, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J. Biol. Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- 27.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 28.Leenders H, Whiffield S, Benoist C, Mathis D. Role of the forkhead transcription family member, FKHR, in thymocyte differentiation. Eur. J. Immunol. 2000;30:2980–2990. doi: 10.1002/1521-4141(200010)30:10<2980::AID-IMMU2980>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Vivien L, Benoist C, Mathis D. T lymphocytes need IL-7 but not IL-4 or IL-6 to survive in vivo. Int. Immunol. 2001;13:763–768. doi: 10.1093/intimm/13.6.763. [DOI] [PubMed] [Google Scholar]
- 30.Guo K, et al. Disruption of peripheral leptin signaling in mice results in hyperleptinemia without associated metabolic abnormalities. Endocrinology. 2007;148:3987–3997. doi: 10.1210/en.2007-0261. [DOI] [PubMed] [Google Scholar]
- 31.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 32.Forster R, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 33.Arbones ML, et al. Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin-deficient mice. Immunity. 1994;1:247–260. doi: 10.1016/1074-7613(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 34.Cyster JG, Goodnow CC. Pertussis toxin inhibits migration of B and T lymphocytes into splenic white pulp cords. J. Exp. Med. 1995;182:581–586. doi: 10.1084/jem.182.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat. Immunol. 2008;9:292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 36.Wojciechowski S, et al. Bim/Bcl-2 balance is critical for maintaining naive and memory T cell homeostasis. J. Exp. Med. 2007;204:1665–1675. doi: 10.1084/jem.20070618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jameson SC. T cell homeostasis: keeping useful T cells alive and live T cells useful. Semin. Immunol. 2005;17:231–237. doi: 10.1016/j.smim.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Lee SK, Surh CD. Role of interleukin-7 in bone and T-cell homeostasis. Immunol. Rev. 2005;208:169–180. doi: 10.1111/j.0105-2896.2005.00339.x. [DOI] [PubMed] [Google Scholar]
- 39.Xue HH, et al. GA binding protein regulates interleukin 7 receptor alpha-chain gene expression in T cells. Nat. Immunol. 2004;5:1036–1044. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- 40.Park JH, et al. ‘Coreceptor tuning’: cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat. Immunol. 2007;8:1049–1059. doi: 10.1038/ni1512. [DOI] [PubMed] [Google Scholar]
- 41.Park JH, et al. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity. 2004;21:289–302. doi: 10.1016/j.immuni.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 42.Chao CC, Jensen R, Dailey MO. Mechanisms of L-selectin regulation by activated T cells. J. Immunol. 1997;159:1686–1694. [PubMed] [Google Scholar]
- 43.Cinalli RM, et al. T cell homeostasis requires G protein-coupled receptor-mediated access to trophic signals that promote growth and inhibit chemotaxis. Eur. J. Immunol. 2005;35:786–795. doi: 10.1002/eji.200425729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lantz O, Grandjean I, Matzinger P, Di Santo JP. Gamma chain required for naive CD4+ T cell survival but not for antigen proliferation. Nat. Immunol. 2000;1:54–58. doi: 10.1038/76917. [DOI] [PubMed] [Google Scholar]
- 45.Wallis VJ, Leuchars E, Chaudhuri M, Davies AJ. Studies on hyperlymphoid mice. Immunology. 1979;38:163–171. [PMC free article] [PubMed] [Google Scholar]
- 46.Berzins SP, Boyd RL, Miller JF. The role of the thymus and recent thymic migrants in the maintenance of the adult peripheral lymphocyte pool. J. Exp. Med. 1998;187:1839–1848. doi: 10.1084/jem.187.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freitas AA, Rocha B. Peripheral T cell survival. Curr. Opin. Immunol. 1999;11:152–156. doi: 10.1016/s0952-7915(99)80026-0. [DOI] [PubMed] [Google Scholar]
- 48.Maeurer MJ, Lotze MT. Interleukin-7 (IL-7) knockout mice. Implications for lymphopoiesis and organ-specific immunity. Int. Rev. Immunol. 1998;16:309–322. doi: 10.3109/08830189809042999. [DOI] [PubMed] [Google Scholar]
- 49.Schluns KS, Kieper WC, Jameson SC, Lefrancois L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat. Immunol. 2000;1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- 50.Tan JT, et al. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc. Natl. Acad. Sci. USA. 2001;98:8732–8737. doi: 10.1073/pnas.161126098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birkenkamp KU, Coffer PJ. FOXO transcription factors as regulators of immune homeostasis: molecules to die for? J. Immunol. 2003;171:1623–1629. doi: 10.4049/jimmunol.171.4.1623. [DOI] [PubMed] [Google Scholar]
- 52.Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat. Rev. Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- 53.Sinclair LV, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat. Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J. Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 55.Fabre S, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J. Immunol. 2008;181:2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 56.Lee HC, Shibata H, Ogawa S, Maki K, Ikuta K. Transcriptional regulation of the mouse IL-7 receptor alpha promoter by glucocorticoid receptor. J. Immunol. 2005;174:7800–7806. doi: 10.4049/jimmunol.174.12.7800. [DOI] [PubMed] [Google Scholar]
- 57.Samaridis J, et al. Development of lymphocytes in interleukin 7-transgenic mice. Eur. J. Immunol. 1991;21:453–460. doi: 10.1002/eji.1830210230. [DOI] [PubMed] [Google Scholar]
- 58.Mertsching E, Burdet C, Ceredig R. IL-7 transgenic mice: analysis of the role of IL-7 in the differentiation of thymocytes in vivo and in vitro. Int. Immunol. 1995;7:401–414. doi: 10.1093/intimm/7.3.401. [DOI] [PubMed] [Google Scholar]
- 59.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 60.Pallard C, et al. Distinct roles of the phosphatidylinositol 3-kinase and STAT5 pathways in IL-7-mediated development of human thymocyte precursors. Immunity. 1999;10:525–535. doi: 10.1016/s1074-7613(00)80052-7. [DOI] [PubMed] [Google Scholar]
- 61.Riou C, et al. Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. J. Exp. Med. 2007;204:79–91. doi: 10.1084/jem.20061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Link A, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat. Immunol. 2007;8:1255–1265. doi: 10.1038/ni1513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.