Abstract
Many viruses, either naturally occurring or as a result of genetic manipulation, exhibit conditional replication in transformed cells. This principle is the basis for experimental therapeutic approaches exploiting the oncolytic potential of such agents without the danger of collateral damage to resistant normal tissues. One of the potential obstacles to these approaches is the possibility of genetic adaptation of oncolytic viruses upon replication in susceptible tumor tissues. Genetic variation can reverse genetic manipulations of parental viral genomes that determine attenuation of virulence, selective tumor cell tropism or other desirable traits. Alternatively, it may convey new properties not originally associated with parental strains, e.g. adaptation to a human host range. We examined genetic stability of an oncolytic non-pathogenic poliovirus recombinant considered for therapy of recurrent glioblastoma multiforme. This was done by serial passage experiments in glioma xenografts in vivo and investigation of phenotypic and genotypic markers of attenuation. Intratumoral inoculation of oncolytic poliovirus produced efficient tumor regress and elimination without altering temperature sensitive growth, selective cytotoxicity or genetic markers of attenuation of virus recovered from inoculated animals. Our studies demonstrate that active viral oncolysis of malignant glioma does not alter the conditional replication properties of oncolytic non-pathogenic poliovirus recombinants.
INTRODUCTION
A variety of viruses are under consideration for therapeutic applications against cancer. These include strategies using viruses for the delivery of exogenous genetic material or for direct cytolysis of cancerous cells. Certain veterinary pathogens (e.g. Newcastle disease virus,1 vesicular stomatitis virus),2 or naturally weak human pathogens (e.g. reovirus)3 have been investigated for therapeutic purposes without genetic manipulation. Generally, inherent conditional replication of these viruses in cancerous cells is due to defects of innate immune pathways in malignancy, which thwart viral growth in normal tissues.2 In contrast, a variety of human pathogenic viruses have been genetically engineered to confer desirable properties, most importantly attenuated virulence, tumor-selective propagation or cancer tropism. Genetic engineering strategies include deletion of critical virulence genes dispensable for replication in rapidly dividing tumor cells (e.g. the herpes simplex virus γ34.5 gene)4, insertion of tumor-specific promoter elements,5 or designs to re-direct virus tropism by manipulating structural features of virus particles interacting with the host cell exterior.6
All viruses have the capacity for genetic adaptation upon replication in permissive hosts. Since important safety features of most oncolytic viruses rely on cognate host range restrictions or genetic manipulation of their genomes, this adaptability is of grave concern for clinical application. This concern is more pronounced for RNA viruses, due to the inherent high error rates of RNA-dependent RNA polymerases and resulting genetic variability.7 Although mutations in viral RNA polymerases associated with increased fidelity have been reported,8 there are few strategic options to circumvent the fundamental principle of genetic variability associated with RNA viruses. Conversely, genetic variability of RNA viruses has been implicated in efficient viral propagation and invasion in vivo9 and, hence, may contribute to anti-cancer activity of oncolytic viruses. Given the possibility of genetic adaptation and emergence of undesirable pathogenic properties, it is important to test the potential for such events upon oncolytic virus replication in target tumor tissues.
We report here studies of the genetic stability of PVS-RIPO, a non-pathogenic oncolytic poliovirus recombinant under consideration for therapeutic application against glioblastoma multiforme (GBM). Poliovirus exhibits natural target tropism for GBM, because these tumors, as well as a range of other cancers, exhibit widespread ectopic expression of the poliovirus receptor CD155.10,11 PVS-RIPO consists of the genome of the live attenuated serotype 1 (Sabin) vaccine strain of poliovirus containing an internal ribosomal entry site (IRES) of human rhinovirus type 2 (HRV2). PVS-RIPO thus combines attenuation of neurovirulence mediated by the heterologous HRV2 IRES10,12 with the known attenuation determinants mapping to the coding regions for the type 1 (Sabin) capsid13 and RNA-dependent RNA polymerase.14,15 We demonstrated that the HRV2 IRES interacts with a heterodimer composed of the dsRNA binding protein 76 (DRBP76) and the nuclear factor (NF) 45 in neuronal cells, but not GBM cells.16,17 The DRBP76:NF45 heterodimer is preferentially localized in cytoplasm and associates with the translation machinery specifically in neurons.16,17 Binding of the DRBP76:NF45 heterodimer intercepts translation initiation at the HRV2 IRES, thus preventing efficient viral translation in neuronal cells.17
We used a serial in vivo passage approach to investigate whether PVS-RIPO replication in GBM cells selects for variants with altered pathogenic properties. Our findings indicate that serial in vivo passage in GBM xenografts does not alter phenotypic markers of the attenuated neurovirulence phenotype of PVS-RIPO. Accordingly, sequence analyses revealed that the genetic determinants for attenuated neurovirulence do not change upon serial in vivo passage in GBM.
RESULTS
Study design
The objectives of this study were to evaluate the evolution of PVS-RIPO genotypes upon serial passage in GBM xenografts. The study comprised two separate identical in vivo passages of PVS-RIPO in HTB-15 glioma xenografts, each followed by several amplification steps to generate sufficient material for subsequent assays. The target cell line, HTB-15 is derived of an anaplastic astrocytoma and representative of the intended clinical target,18 the route of administration was the intended (intratumoral) clinical route and the dose administered was the anticipated maximal clinical dose. The study was designed to favor a high number of viral replication events and, thus, genetic adaptation in an environment recapitulating GBM in patients. The first passage was initiated by inoculating PVS-RIPO from a good manufacturing practice (GMP) toxicology lot (L0603006) into established HTB-15 xenografts. The second passage involved virus recovered from xenografts 10 days after virus inoculation and subsequent amplification in HTB-15 cells in vitro. Finally, virus re-isolated from animals 10 days after PVS-RIPO injection in the second passage was amplified in HTB-15 cells in vitro and subjected to pheno- and genotypic analysis. All study results reported below describe the second in vivo passage and analyses of material recovered thereof.
Response to virus inoculation in HTB-15 xenografts
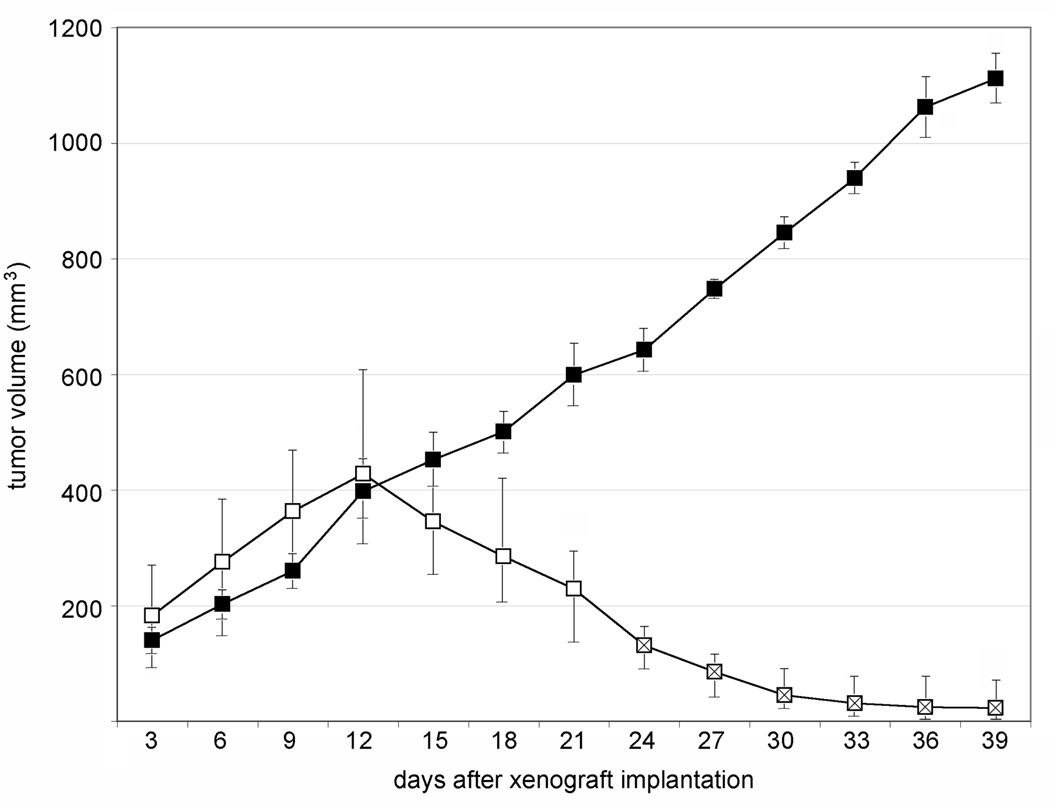
Groups of 12 athymic Balb/c mice, ~8 weeks of age were enrolled in each phase of the experiment. To simultaneously monitor tumor regress and enable virus recovery from the same animal, bilateral HTB-15 xenografts were implanted in the flanks of mice. The inoculum consisted of 6 × 106 cells on each flank and caliper measurements of tumors were initiated 3 days after implantation. Tumor measurements for the second phase of the study are shown (Figure 1). Ten animals were inoculated bilaterally with PVS-RIPO at a tissue culture infectious dose (TCID50) of 1 × 108 administered to the tumor; two animals received bilateral inoculations of vehicle. We observed steady tumor progress in all animals until the day of virus/vehicle inoculation (day 12 after xenograft implantation; Figure 1). Ten days after PVS-RIPO inoculation the median xenograft size had shrunk by ~45%, but tumors retained an average size slightly larger than 3 days after xenograft implantation (Figure 1). At this interval, six virus-treated animals were removed from the study, euthanized, and both tumors were dissected for histopathological analysis (Figure 2) and virus recovery (Figure 3), respectively.
Figure 1.
HTB-15 xenograft growth after vehicle- or PVS-RIPO treatment. All tumor sizes were determined by caliper measurement. Open squares represent average sizes of 20 xenografts (bilateral tumors in 10 animals) and crossed open squares are the median sizes of 8 xenografts (bilateral tumors in 4 animals observed for 40 days post xenograft implantation). Closed squares are the median sizes of 4 bilateral xenografts in 2 vehicle-treated animals. The size range of all xenografts at any given interval is indicated. Virus/vehicle inoculation occurred on day 12, after measuring of the tumors.
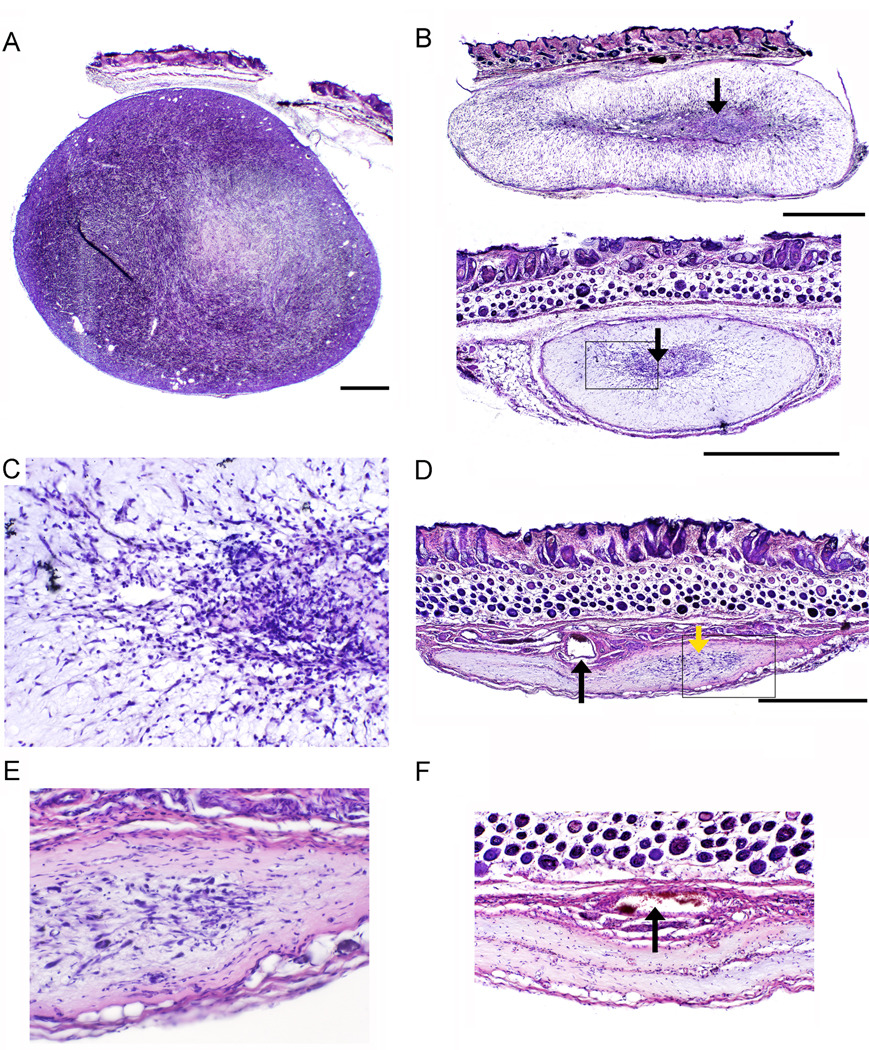
Figure 2.
Histopathological analysis of PVS-RIPO induced oncolysis of HTB-15 xenografts in athymic mice. The bar shown with low-magnification images represents 1 mm. (a) Cross-section of a vehicle-injected tumor in an animal sacrificed 40 days after tumor implantation. The xenograft displays typical dense, hyper-cellular morphology. (b) Representative xenografts 10 days post PVS-RIPO inoculation. Only a minor, central portion of the xenograft retains the characteristic appearance of tumor (arrows). The boxed region is shown at higher magnification in (c). (c) Detail of (b) (bottom panel). Dense infiltrates surrounding and invading the central part of the xenograft can be distinguished at higher magnification. (d) Overview of a HTB-15 xenograft 28 days after PVS-RIPO inoculation. A scar had replaced the tumor, but a minor focus of remaining active tissue re-arrangement is visible (yellow arrow). A large patent vessel (black arrow) and surrounding collapsed minor vessels may represent the former tumor’s vascular supply. The boxed region is shown at higher magnification in (e). (e) Detail of (d) showing the area of active tissue re-modeling at higher magnification. (f) Two out of 4 tumors had entirely been eliminated 28 days post PVS-RIPO and replaced by a scar. The remnants of several large vessels are visible at the center of the scar’s base (arrow).
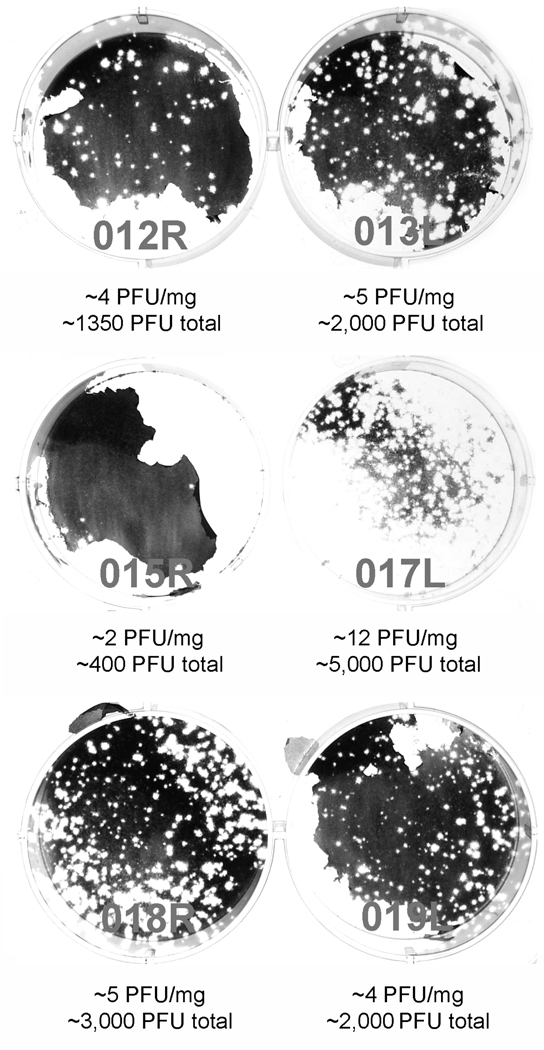
Figure 3.
Recovery of PVS-RIPO from HTB-15 xenografts 10 days post virus inoculation. Numbering refers to the animal and location of the xenograft the virus originated from. Estimates of PFU/mg xenograft tissue and total PFU/xenograft recovered are shown below for each tumor. The plaque assay of tumor 015R shows an artifact due to detachment of the cell layer during staining. The numbers shown were calculated from a repeat assay.
The remaining 4 virus-treated animals left on the study were observed for another 18 days. In these animals, tumors continued to regress (Figure 1). By the end of the study the xenograft had almost obliterated in all animals. The implantation sites, however, were clearly distinguishable macroscopically. As with animals 10 days after PVS-RIPO inoculation, the remaining tissue was excised and processed for histopathology and virus recovery. In vehicle-treated animals, xenografts continued to progress throughout the entire observation interval (Figure 1). A monolateral growth was removed from euthanized vehicle-treated animals for histopathological analysis only.
Histopathology of HTB-15 xenografts
In addition to gross measurements of tumor size in vivo, histopathological analysis of xenografts was performed on tissues from each animal enrolled in the study. The growth contralateral to the tumor removed for virus recovery was analyzed from each mouse euthanized 10 or 28 days post PVS-RIPO inoculation. To enable comprehensive analysis of tumor/host response to PVS-RIPO, the xenografts were sectioned in their entirety and processed for histopathological analysis.
Histopathology of xenografts 10 days after PVS-RIPO inoculation revealed essentially identical responses in all 6 animals analyzed. Representative examples from two animals are shown (Figure 2b). Our analyses revealed substantially more advanced tumor lysis than in vivo measurements suggested. A small central area of the xenograft retained the dense, hypercellular architecture of vehicle-treated tumors (compare Figure 2a, 2b). However, the bulk of the tumor no longer had the appearance of proliferating tumor. Instead, most of the tumor was replaced by tissue with dramatically altered appearance that no longer contained cells with the morphology typical of the HTB-15 xenograft. The majority of the tumor mass had vastly reduced cell content and was diffusely invaded by infiltrates (Figure 2c). Based on analyses of xenografts 28 days post PVS-RIPO inoculation (see below), we assume that the tissue rearrangement represents the host’s reaction to tumor destruction and a transition towards scar formation (Figure 2b, c). Both the central area of the xenograft, presumably containing residual tumor cells (Figure 2c) and the periphery were characterized by abundant and diffuse infiltrates. In addition to the characteristic central area (Figure 2b), xenografts in all animals contained multiple foci of what appeared to be remaining active tumor. These were in various stages of tissue rearrangement and invariably surrounded by intense infiltrates (data not shown).
Xenografts 28 days post PVS-RIPO injection had essentially been replaced by a scar (Figure 2d, f). In two out of four tumors analyzed, we detected minor foci of active tissue remodeling, characterized by the presence of cells with diverse morphology surrounded by infiltrates (Figure 2e). All scars contained visible remnants of the previous tumor. The base was composed of collapsed tissue containing profuse infiltrates. A large central artery and minor vessels situated at the center of the scar base presumably formed the vascular supply of the former xenograft (Figure 2d, f). Our histopathological analyses indicate that active PVS-RIPO replication in xenografts and a vigorous host response to the receding infected tumor induced complete tumor elimination resulting in scar formation.
Re-isolation of virus from HTB-15 xenografts
Tumors from xenograft-bearing mice were isolated at day 10 or 28 post PVS-RIPO administration and processed for virus recovery. Tumor lysates obtained as described in Materials & Methods were first tested by plaque assay to confirm the presence of virus. All xenografts obtained from 6 animals 10 days after PVS-RIPO inoculation contained infectious material (Figure 3). We estimate the amounts of PVS-RIPO contained in the xenografts to be very low, considering that at the height of infection, spinal cord tissue of poliovirus-infected CD155-transgenic mice may contain up to 105 plaque forming units (PFU)/mg.10 The titers associated with HTB-15 xenografts 10 days after virus inoculation ranged from 2–12 PFU/mg (Figure 3). Based on our histopathological analyses of xenografts 10 days post PVS-RIPO inoculation (Figure 2b), we speculate that viral titers decline with the demise of HTB-15 cells in which PVS-RIPO can replicate. These analyses also suggest that PVS-RIPO may exclusively reside in the central area morphologically resembling viable tumor in the receding xenograft (Figure 2b). Thus, local concentrations of virus in remaining viable tumor may be much higher.
We were unable to recover infectious material from tumors 28 days after PVS-RIPO inoculation (data not shown). This is not surprising given the low amount of xenograft remaining at that interval (Figure 2d, f), its histopathological appearance (complete or almost complete replacement by a scar; Figure 2d, f) and the very low viral titers in xenografts at 10 days post PVS-RIPO inoculation (Figure 3). Our observations indicate that PVS-RIPO is unable to persist in tumor, once the supply of viable tumor cells has disappeared.
The amounts of PVS-RIPO recovered from HTB-15 xenografts 10 days after PVS-RIPO inoculation were insufficient for direct analysis. Therefore, the tissue lysates from all six xenografts tested were pooled and the resulting combined homogenate was amplified in cultured HTB-15 cells. The resulting ‘xenograft passaged’ stock (lot 022208) was purified according to standard methods12 and used for further analyses.
Testing serially passaged PVS-RIPO in selective cytotoxicity assays
A series of analytic procedures were carried out in tissue culture to examine if xenograft passaged PVS-RIPO retains the hallmarks of the attenuated neurovirulence phenotype. First, we conducted a replicative capacity at temperature (rct) 40°C assay. This assay determines the sample titer of infectious PVS-RIPO at 36°C and 40°C by plaque formation on Vero indicator cells enabling calculation of the fold-reduction in infectious virus at the two temperatures to determine thermoresistance. This assay essentially follows the WHO-established standard.19 The rationale for this assay is the fact that PVS-RIPO exhibits a temperature-sensitive (ts) growth phenotype characterized by severely impaired viral propagation at 40°C.20 A ts phenotype has been documented for all three Sabin vaccine strains and it is considered an important correlate of neurovirulence attenuation.21,22 The rct assay of xenograft passaged PVS-RIPO suggests that the virus retains the ts phenotype characteristic of the type 1 (Sabin) backbone and parental virus from the toxicology lot (Table 1).
Table 1.
Sensitivity to growth at 40°C of xenograft passaged PVS-RIPO.
| Controls | Test Sample | ||
|---|---|---|---|
| Resistant Control RCT 40+ |
Sensitive Control RCT 40− |
PVS-RIPO (xenograft passage) |
|
| 36°C | 6.03 logs | 8.54 logs | 8.29 logs |
| 40°C | 6.01 logs | ≤1.22 logs | ≤1.22 logs |
| 36°C:40°C differential | 0.02 logs | ≥7.32 logs | ≥7.08 logs |
Second, we tested neuron-specific replication deficits of PVS-RIPO in a selective cytotoxicity assay. PVS-RIPO is characterized by a severe, specific replication deficit in cells of neuronal origin.23 This phenotype can be recapitulated in tissue cultured cells of neuronal origin, either neuroblastoma cells12,23,24 or HEK-293 cells.25 We prefer the latter, because most neuroblastoma cell lines are notorious for spontaneously interconverting between neuroblast-and fibroblast-like phenotypes.26,27 HEK-293 cells, whose neuronal origin has been documented by their originator,28 closely and faithfully recapitulate attenuation phenotypes observed in animal models for poliomyelitis, e.g. with poliovirus type 1 (Sabin) or PVS-RIPO.25 Based on these considerations, we developed a selective cytotoxicity assay testing viral replication in HEK-293 vs. HTB-14 malignant glioma cells. Oncolytic efficacy of PVS-RIPO or its progenitor based on the genome of wild type serotype 1 (Mahoney) has been documented in HTB-14 and HTB-15 glioma cells alike.10
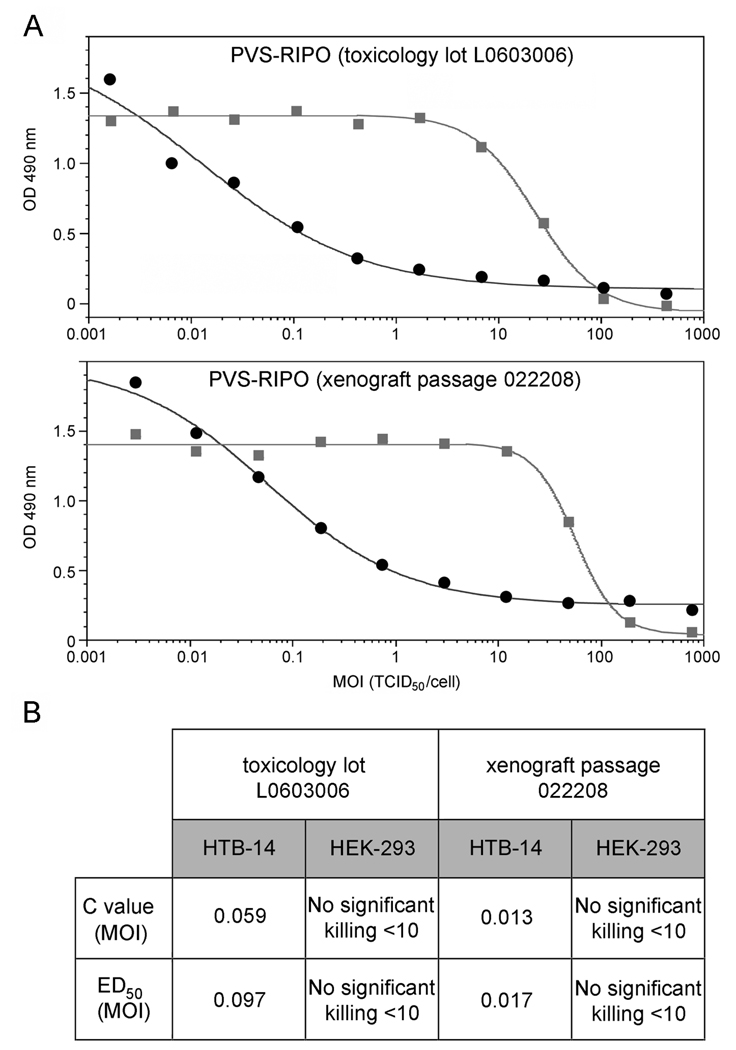
Toxicology lot L0603006 and xenograft passaged PVS-RIPO lot 022208 were tested in the selective cytotoxicity assay using HTB-14 and HEK293 cells (Figure 4). The assay measures cell viability after PVS-RIPO infection over a wide range of multiplicities of infection (MOI). In HTB-14 malignant glioma cells, the original toxicology lot (L0603006) and xenograft passaged PVS-RIPO (022208) showed similar dose-dependent cell killing (Figure 4). More importantly, we did not observe altered cytotoxicity in HEK-293 cells in relation to HTB-14 cells with xenograft passaged PVS-RIPO. These results demonstrate that in vivo serial passage of PVS-RIPO in malignant glioma does not change the selective cytotoxicity profile of the original toxicology lot (Figure 4).
Figure 4.
Differential cell killing assay of PVS-RIPO. (a) Assays measuring differential cell killing of the control lot (L0603006; top panel) and xenograft passaged lot (022208; bottom panel) in HTB-14 cells (●) and HEK-293 cells ( ) yielded similar results. (b) C values refer to the virus titer at half maximal cell killing; ED50 values represent virus titers at 50% cell killing. C and ED50 values were unchanged upon PVS-RIPO xenograft passage.
) yielded similar results. (b) C values refer to the virus titer at half maximal cell killing; ED50 values represent virus titers at 50% cell killing. C and ED50 values were unchanged upon PVS-RIPO xenograft passage.
Genotyping serially passaged PVS-RIPO
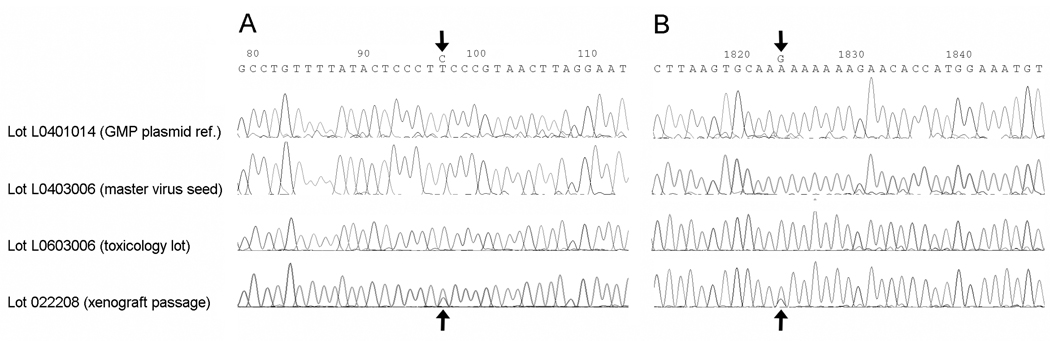
The pooled virus isolates from 6 xenografts 10 days after PVS-RIPO inoculation were subjected to sequencing analysis. This was accomplished by RT-PCR of viral RNA isolated from pooled virus amplified in HTB-15 glioma cell culture as described in Materials & Methods. The genome sequence of the xenograft passaged virus (lot 022208) was compared to the GMP plasmid reference, the master virus seed and the toxicology lot L0603006 (Figure 5).
Figure 5.
Comparison of representative sequence chromatograms of the polymorphic base positions 97 (a) and 1824 (b) identified through PVS-RIPO sequencing. Sequencing results for xenograft passaged PVS-RIPO (lot 022208) were compared with those for the GMP reference plasmid, the master virus seed and the toxicology lot L0603006 The nucleotide numbers refer to the genome of PVS-RIPO, the polymorphic bases are arrowed.
The nucleotide sequence for PVS-RIPO lot 022208 was found to be 100% homologous to all 7303 base positions sequenced when compared to the cloned viral cDNA contained in the PVS-RIPO plasmid reference (lot L0401014), the master viral seed sequence (lot L0403006), and to the toxicology lot L0603006 (data not shown). Two base positions in the xenograft passaged pool (lot 022208) were polymorphic with the predominant base matching the PVS-RIPO plasmid and viral reference sequences (Figure 5). These polymorphic sites are located at nucleotide positions 97 (mapping to the 5’UTR) and 1824 (mapping to the coding region for the VP3 capsid protein) in the PVS-RIPO genome (Figure 5). Base position 97 is located within the cloverleaf, a highly conserved structure involved in genome replication, comprising the 5’ terminal ~100 nucleotides of all enterovirus 5’UTRs.15 The polymorphism at position 1824 results in an Lys➔ Arg substitution in VP3.
In order to eliminate the possibility of base mis-incorporation during RT-PCR, two independent RT-PCR assays were performed for each polymorphic site. Both polymorphisms were observed regardless of the RT-PCR amplicons utilized for sequencing (data not shown). Since the sequencing method employed can only detect polymorphisms greater than ~15–20% of the total base population, and since all reads across the two sites identified the same polymorphic bases, the ‘strain variants’ observed are considered to be at least 15% of the total virus population in the sample. The degree to which the polymorphic sites are linked is unknown. None of the polymorphic sites map to the engineered IRES (base position 97 is upstream of the proximal insertion site) or to genetic determinants of attenuated neurovirulence of poliovirus type 1 (Sabin) [base position 1824 is not one of the capsid mutations distinguishing type 1 (Sabin) from its neurovirulent wild-type progenitor].29
A BLASTn analysis of the two locations indicated that both sites are known genetic variation ‘hot-spots’ in PV type 1 and 2 strains. The site at position 97 is known to be polymorphic for cytidine and thymidine and the surrounding bases constitute a hot-spot in the viral genome (see poliovirus serotype 1 isolate sequences in GenBank# AY928383, AY928384, and AY928386). The site at position 1824 is within a polyadenosine region in the VP3 coding region and also has been recognized as a polymorphic site with much greater sequence variability than position 97 (see poliovirus serotype 1 isolate sequences in GenBank# DQ264351–DQ264353, DQ264355, and DQ264358). The observed guanosine polymorphism at 1824 has been previously reported in GenBank (AF538841 and DQ264354) but more typically cytidine is observed at this locus, although all four bases have been reported as substitutions within this polyadenosine region of the viral genome.
DISCUSSION
Our work shows that PVS-RIPO causes sustained tumor regress and elimination in a glioma xenograft model without acquiring phenotypic or genetic alterations that might alter its beneficial safety profile. This confirms previous observations from prolonged passage in cancer cell tissue culture12 or from in vivo tests of non-GMP PV1-RIPO [identical to PVS-RIPO, except the genetic backbone is wild-type PV type 1 (Mahoney)].10 Parallel studies of tumor regress and virus recovery suggest that virus titers sharply decline in step with receding tumor and indicate the inability of PVS-RIPO to persist in remnant scars, once viable tumor tissue has been eliminated. We observed a vigorous host response to viral tumor cell lysis, evident by extensive infiltrative lesions within and surrounding the xenograft, perivascular cuffing, active tissue remodeling and, eventually, scarring. Similar observations implicating host reactions against infected tumor were previously obtained in a syngeneic mouse model for neuroblastoma with a different type of genetically engineered oncolytic poliovirus.30 It is conceivable that host inflammatory reactions contribute to virus removal, once tumor obliteration eliminates the site of active viral replication.
PVS-RIPO is unique among oncolytic viruses, because its safety and tumor-specific replication partly rely on an abnormal environment for translation initiation in malignant glioma cells.16,17 It is conceivable that the foreign IRES element in PVS-RIPO could mediate general viral translation and propagation deficits that would contribute to the attenuation phenotype by globally reducing viral replication. This scenario would be of concern regarding genetic stability, since a genotype associated with a global growth deficit might revert upon replication in patients’ tumor tissue. The present studies, previous observations in tissue culture25 and tests in conjunction with large scale, GMP-grade production of PVS-RIPO argue against this possibility. It is difficult to precisely determine the combined number of passages in HTB-15 cells in this study, due to multi-step amplification of very low titers recovered from xenografts 10 days post PVS-RIPO inoculation and the difficulty to project virus dynamics in xenografts in vivo. Based on the available information, we estimate the combined passage number from both phases of the study to be at least 15. The absence of genetic adaptation upon serial passage in glioma cells in vivo suggests that the heterologous HRV2 IRES functions optimally in a PVS-RIPO context in susceptible, non-neuronal cells, e.g. GBM.
Serial passage of PVS-RIPO in HTB-15 xenografts in vivo did not generate adapted variants with changes in the genetic signatures known to mediate attenuated neurovirulence. However, we did identify polymorphisms emerging in two distinct regions of the viral genome mapping to known ‘hot-spots’ for genetic variability in PV. Neither polymorphism was present in the PVS-RIPO plasmid, master viral seed, nor the toxicology lot. Therefore, the observed polymorphisms resulting from serial passage in GBM xenografts are most likely the result of random genetic drift leading to a potential founders' effect, or the result of weak selection pressure in vivo. The polymorphisms were identified in virus recovered and pooled from several xenografts; we did not attempt to locate the origin of the polymorphisms to individual tumors in specific animals.
Our studies presented here are an essential part of thorough investigations to prepare PVS-RIPO for toxicology evaluations in Cynomolgus macaques and to support consideration of this agent for clinical trials against recurrent GBM. The results obtained suggest that intratumoral propagation of PVS-RIPO does not select for genetic variants with altered pathogenic properties.
MATERIALS & METHODS
PVS-RIPO
PVS-RIPO is poliovirus type 1 (Sabin) (GenBank #V01150) containing a heterologous IRES of HRV2. Construction of the recombinant and insertion of the HRV2 IRES, and re-derivation of virus from plasmid cDNA was carried out as described previously.23 For this work, a GMP-produced PVS-RIPO lot (L0603006) generated in Vero cells was used for the initial phase of in vivo serial passaging. The viral titer of the lot was confirmed by TCID50 assay according to World Health Organization (WHO)-standardized practices.19,31 The subsequent in vivo phase was conducted with virus re-isolated from HTB-15 xenografts and amplified in HTB-15 cell culture according to standard procedures described elsewhere.10
HTB-15 cell culture, xenograft implantation, intratumoral PVS-RIPO inoculation
HTB-15 (also referred to as U-118), derived from a human malignant glioma case, were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cells were grown as instructed by ATCC. All in vivo experiments were conducted with cells implanted into animals at their 3rd passage. To harvest cells for xenograft implantation, 16 sub-confluent 750mL tissue culture flasks were trypsinyzed and the resulting dispersed cells resuspended in 50mL of sterile phosphate-buffered saline (PBS) and centrifuged twice. Cell viability and numbers were determined by microhemocytometer. Female, ~8 week-old athymic Balb/c mice (bred in-house) were used in this study. Mice were anesthetized with isofluorane, the surgical area in the flank was sterilized with betadine, and a subcutaneous inoculation with a 27G1/2 insulin syringe (Becton & Dickinson, Franklin Lakes, NJ) was performed to deposit 6 × 106 HTB-15 cells in a volume of ~40µL PBS. Intratumoral vehicle/virus inoculations were performed in isofluorane-anesthetized animals as follows. Vehicle (sterile PBS) or PVS-RIPO suspension in PBS (1 × 108 TCID50) was injected with a 28G1/2 insulin syringe (Becton & Dickinson, Franklin Lakes, NJ) bilaterally. The volume of the inoculum was 20µL. Special emphasis was given to ensuring deposit of the inoculum within the firm xenograft. This was accomplished by very slow infusion of the inoculum while slowly withdrawing the syringe from within the tumor. The injection technique was optimized to prevent leakage of the inoculum out of the injection site or formation of subcutaneous vesicles in the tumor vicinity. Tumors were measured by Vernier caliper in 3-day intervals and their size was calculated as follows: v=(length) × (width)2 × (π/6). At the time of sacrifice, animals were euthanized by CO2 and bilateral thoracotomy for dissection of xenograft tissues. All vertebrate animal procedures were carried out with approval of the institutional animal care and use committee.
Virus recovery from xenograft tissues and RT-PCR sequencing
The area of the xenograft was generously excised and the tumor itself was dissected from its surroundings. The tissue was weighed, immersed in 750µL DMEM and snap frozen in dry ice. After thawing the samples, the tissue was homogenized thoroughly in a Potter Elvehjem micro tissue grinder (Wheaton, Milville, NJ). Dedicated grinders were used for each sample to avoid cross-contamination. The resulting lysate was freeze-thawed twice, centrifugated at 2,000 rpm for 5 min to remove large debris and analyzed by plaque assay as described previously.9 Virus recovered from xenograft was amplified in HTB-15 cultures as described before10 and subjected to RT-PCR sequencing. Genomic sequencing essentially followed procedures reported previously.12,32 Briefly, viral RNA was purified from the pooled cell culture isolates using the Qiagen QIAamp viral RNA extraction procedure with AVL buffer (Qiagen, Valencia, CA). Recovered RNA was quantified and reverse transcribed using SuperScript III (Invitrogen, Carlsbad, CA) and PCR amplified in 8 (~1200 bp) overlapping sections. Two additional RT-PCR assays were subsequently performed with the purified viral RNA extract to investigate the origin of two polymorphisms observed in initial sequencing reactions. RT-PCR products were purified using Qiagen QIAquick PCR Cleanup columns and analyzed by agarose gel electrophoresis prior to sequencing. Four-fold coverage of the entire PVS-RIPO viral genome (2-fold in each direction) was achieved using 59 independently primed overlapping sequence reactions with an average span of ~250 bases between primer annealing sites. Fluorescent dye-terminator sequencing reactions were performed using ABI BigDye® Terminator v1.1 Cycle Sequencing (Applied Biosystems, Foster City, CA) reagents in an MJ Research PTC-225 Peltier thermal cycler and subsequently purified using Centriflex Gel Filtration cartridges (Edge Biosystems, Gaithersburg, MD). Automated DNA sequence analysis was performed using an ABI 3130×l DNA sequencer and the raw sequence reads were scored, trimmed, and aligned to generate a complete viral contig with Sequencher software v4.7 (GeneCodes, Ann Arbor, MI).
Histopathology
Xenografts were excised with the overlying dermis intact for orientation and fixed in fresh 4% neutral buffered paraformaldehyde for 12 hrs at 4°C. After rinsing in PBS for 2 hrs, the samples were dehydrated in serial dilutions of ethanol, cleared in toluene and paraffin-embedded for histological sectioning as described previously.33 The entire paraffin blocks were sectioned at 20µm thickness and all sections recovered were mounted for further analysis. The sections on slides were de-paraffinized in Citrus Clearing Solvent (Richard Allan Scientific, Kalamazoo, MI), re-hydrated in serial dilutions of ethanol, stained with hematoxylin/eosin, dehydrated, cleared and sealed as described before.33
RCT40 assay
This assay follows WHO-established guidelines.19,31 Briefly, serial dilutions of each virus sample were inoculated onto replicate ~90% confluent Vero cells in 60mm culture dishes and incubated at 36°C or 40°C. Virus adsorption was allowed to proceed for 60 min at 36°C or 40°C with periodic rocking of the inoculated dishes. Afterward, the virus suspension was removed and cells were overlaid with 4 mL of 2× EMEM/agarose (1:1). Cells were incubated at 36°C or 40°C until plaques were fully formed. Cultures were stained with neutral red and plaques enumerated. After calculation of titers, the log reductions were determined. As per WHO: a 40°C rct− reference standard, fulfilling the WHO definition of >5.0 log 36°C:40°C titer differential, and a 40°C rct+ reference standard fulfilling the WHO definition of <5.0 log 36°C:40°C titer differential, derived from the WHO Sabin type 1 standard, were included.
Selective cytotoxicity assay
HTB-14, or U87 astrocytoma cells, obtained from J. Sampson (Duke University, Durham, NC) and HEK-293 cells (Magenta Corp., Rockville, MD) were cultured as described previously.20,25 HTB-14 or HEK-293 cells were plated in a 96-well plate at 4 × 104 cells/well (4 × 105 cells/mL, 0.1 mL/well). Test articles were diluted to an initial titer of 2 × 108 TCID50/ml (based on the titer of PVS-RIPO determined on Vero cells) followed by serial 3–4 fold dilutions. The diluted virus samples (100 µL/well) were transferred into the plates containing the cells within 2 hrs. The final MOI of test articles ranged from ~500 to 0.002 TCID50/ml at the time of seeding or infection. The plates were incubated at 37°C in a 5% CO2 and 80% humidity for 48 hrs. After 48 hrs incubation, CellTiter96® AQueous One Solution was added (20 µL/well) and incubated for another 4 hrs at 37°C in 5% CO2 and 80% humidity. SDS (25 µL/well of 10% solution) was added to the wells and the plates were read within 5 min at 490 nm on a plate reader. The background readings in the wells with medium (no cells) were subtracted from the sample well readings. The data were analyzed using a non-linear 4-parameter curve fit (SoftMax Pro; Molecular Devices, Sunnyvale, CA) or alternate programs. ED50 refers to the virus titer that causes 50% cell killing. The C value of the four-parameter curve fit is denoted as the half maximal cell killing titer.
ACKNOWLEDGMENTS
We gratefully acknowledge A. Nomoto (Univ. of Tokyo, Japan) for providing the genomic cDNA clone of poliovirus type 1 (Sabin) used to generate PVS-RIPO. This work was supported by PHS grants NS20023 (MG), CA124756 (MG) and Alex’s Lemonade Stand Foundation. This project has been supported in whole or in part through the NCI-RAID Program of the Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: Disclaimer: the content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or any organization imply endorsement by the U.S. Government.
REFERENCES
- 1.Lorence RM, Reichard KW, Katubig BB, Reyes HM, Phuangsab A, Mitchell BR, et al. Complete regression of human neuroblastoma xenografts in athymic mice after local Newcastle disease virus therapy. J Natl Cancer Inst. 1994;86:1228–1233. doi: 10.1093/jnci/86.16.1228. [DOI] [PubMed] [Google Scholar]
- 2.Stojdl DF, Lichty B, Knowles S, Marius R, Atkins H, Sonenberg N, et al. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med. 2000;6:821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- 3.Coffey MC, Strong JE, Forsyth PA, Lee PW. Reovirus therapy of tumors with activated Ras pathway. Science. 1998;282:1332–1334. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- 4.Andreansky SS, He B, Gillespie GY, Soroceanu L, Markert J, Chou J, et al. The application of genetically engineered herpes simplex viruses to the treatment of experimental brain tumors. Proc Natl Acad Sci USA. 1996;93:11313–11318. doi: 10.1073/pnas.93.21.11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kambara H, Okano H, Chiocca EA, Saeki Y. An oncolytic HSV-1 mutant expressing ICP34.5 under control of a nestin promoter increases survival of animals even when symptomatic from a brain tumor. Cancer Res. 2005;65:2832–2839. doi: 10.1158/0008-5472.CAN-04-3227. [DOI] [PubMed] [Google Scholar]
- 6.Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8:573–587. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo E, Holland JJ. Complications of RNA heterogeneity for the engineering of virus vaccines and antiviral agents. Genet Eng. 1992;14:13–31. doi: 10.1007/978-1-4615-3424-2_2. [DOI] [PubMed] [Google Scholar]
- 8.Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gromeier M, Lachmann S, Rosenfeld MR, Gutin PH, Wimmer E. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc Natl Acad Sci USA. 2000;97:6803–6808. doi: 10.1073/pnas.97.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merrill MK, Bernhardt G, Sampson JH, Wikstrand CJ, Bigner DD, Gromeier M. Poliovirus receptor CD155-targeted oncolysis of glioma. Neuro-oncol. 2004;6:208–217. doi: 10.1215/S1152851703000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobrikova E, Florez P, Bradrick S, Gromeier M. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3' nontranslated region. Proc Natl Acad Sci USA. 2003;100:15125–15130. doi: 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minor PD, Macadam AJ, Stone DM, Almond JW. Genetic basis of attenuation of the Sabin oral poliovirus vaccines. Biologicals. 1993;21:357–363. doi: 10.1006/biol.1993.1096. [DOI] [PubMed] [Google Scholar]
- 14.Paul AV, Mugavero J, Yin J, Hobson S, Schultz S, van Boom JH, et al. Studies on the attenuation phenotype of polio vaccines: poliovirus RNA polymerase derived from Sabin type 1 sequence is temperature sensitive in the uridylylation of VPg. Virology. 2000;272:72–84. doi: 10.1006/viro.2000.0354. [DOI] [PubMed] [Google Scholar]
- 15.Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 16.Merrill MK, Dobrikova EY, Gromeier M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J Virol. 2006;80:3147–3156. doi: 10.1128/JVI.80.7.3147-3156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrill MK, Gromeier M. The Double-Stranded RNA Binding Protein 76:NF45 Heterodimer Inhibits Translation Initiation at the Rhinovirus Type 2 Internal Ribosome Entry Site. J Virol. 2006;80:6936–6942. doi: 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullard DE, Schold SC, Bigner SH, Bigner DD. Growth and chemotherapeutic response in athymic mice of tumors arising from human glioma-derived cell lines. J Neuropathol Exp Neurol. 1981;40:410–427. doi: 10.1097/00005072-198107000-00005. [DOI] [PubMed] [Google Scholar]
- 19.World Health Organization. Recommendations for the production and control of poliomyelitis vaccine (oral) Geneva: WHO; Technical Report Series, No. 904. 2002
- 20.Cello J, Toyoda H, Dejesus N, Dobrikova EY, Gromeier M, Wimmer E. Growth phenotypes and biosafety profiles in poliovirus-receptor transgenic mice of recombinant oncolytic polio/human rhinoviruses. J Med Virol. 2008;80:352–359. doi: 10.1002/jmv.21063. [DOI] [PubMed] [Google Scholar]
- 21.Macadam AJ, Ferguson G, Burlison J, Stone D, Skuce R, Almond JW, et al. Correlation of RNA secondary structure and attenuation of Sabin vaccine strains of poliovirus in tissue culture. Virology. 1992;189:415–422. doi: 10.1016/0042-6822(92)90565-7. [DOI] [PubMed] [Google Scholar]
- 22.Macadam AJ, Pollard SR, Ferguson G, Dunn G, Skuce R, Almond JW, et al. The 5' noncoding region of the type 2 poliovirus vaccine strain contains determinants of attenuation and temperature sensitivity. Virology. 1991;181:451–458. doi: 10.1016/0042-6822(91)90877-e. [DOI] [PubMed] [Google Scholar]
- 23.Gromeier M, Alexander L, Wimmer E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc Natl Acad Sci USA. 1996;93:2370–2375. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gromeier M, Bossert B, Arita M, Nomoto A, Wimmer E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J Virol. 1999;73:958–964. doi: 10.1128/jvi.73.2.958-964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell SA, Lin J, Dobrikova EY, Gromeier M. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J Virol. 2005;79:6281–6290. doi: 10.1128/JVI.79.10.6281-6290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rettig WJ, Spengler BA, Chesa PG, Old LJ, Biedler JL. Coordinate changes in neuronal phenotype and surface antigen expression in human neuroblastoma cell variants. Cancer Res. 1987;47:1383–1389. [PubMed] [Google Scholar]
- 27.Ross RA, Spengler BA, Biedler JL. Coordinate morphological and biochemical interconversion of human neuroblastoma cells. J Natl Cancer Inst. 1983;71:741–747. [PubMed] [Google Scholar]
- 28.Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. Faseb J. 2002;16:869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 29.Nomoto A, Omata T, Toyoda H, Kuge S, Horie H, Kataoka Y, et al. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci USA. 1982;79:5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toyoda H, Yin J, Mueller S, Wimmer E, Cello J. Oncolytic treatment and cure of neuroblastoma by a novel attenuated poliovirus in a novel poliovirus-susceptible animal model. Cancer Res. 2007;67:2857–2864. doi: 10.1158/0008-5472.CAN-06-3713. [DOI] [PubMed] [Google Scholar]
- 31.World Health Organization. Manual of laboratory methods for testing the potency of final vaccines used in the WHO expanded programme on immunization. Geneva: WHO; Technical Report Series, No. 800. 1990
- 32.Florez de Sessions P, Dobrikova E, Gromeier M. Genetic adaptation to untranslated region-mediated enterovirus growth deficits by mutations in the nonstructural proteins 3AB and 3CD. J Virol. 2007;81:8396–8405. doi: 10.1128/JVI.00321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dufresne AT, Gromeier M. A nonpolio enterovirus with respiratory tropism causes poliomyelitis in intercellular adhesion molecule 1 transgenic mice. Proc Natl Acad Sci USA. 2004;101:13636–13641. doi: 10.1073/pnas.0403998101. [DOI] [PMC free article] [PubMed] [Google Scholar]